The Sox2 transcription factor must be robustly transcribed in embryonic stem (ES) cells to maintain pluripotency. Zhou et al. identify three novel enhancers that, through the formation of chromatin loops, form a chromatin complex with the Sox2 promoter in ES cells. The distal cluster containing SRR107 and SRR111, located >100 kb downstream from Sox2, is required for cis-regulation of Sox2 in ES cells.
Keywords: chromatin, transcription, enhancer elements, Sox2, embryonic stem cell, gene expression regulation
Abstract
The Sox2 transcription factor must be robustly transcribed in embryonic stem (ES) cells to maintain pluripotency. Two gene-proximal enhancers, Sox2 regulatory region 1 (SRR1) and SRR2, display activity in reporter assays, but deleting SRR1 has no effect on pluripotency. We identified and functionally validated the sequences required for Sox2 transcription based on a computational model that predicted transcriptional enhancer elements within 130 kb of Sox2. Our reporter assays revealed three novel enhancers—SRR18, SRR107, and SRR111—that, through the formation of chromatin loops, form a chromatin complex with the Sox2 promoter in ES cells. Using the CRISPR/Cas9 system and F1 ES cells (Mus musculus129 × Mus castaneus), we generated heterozygous deletions of each enhancer region, revealing that only the distal cluster containing SRR107 and SRR111, located >100 kb downstream from Sox2, is required for cis-regulation of Sox2 in ES cells. Furthermore, homozygous deletion of this distal Sox2 control region (SCR) caused significant reduction in Sox2 mRNA and protein levels, loss of ES cell colony morphology, genome-wide changes in gene expression, and impaired neuroectodermal formation upon spontaneous differentiation to embryoid bodies. Together, these data identify a distal control region essential for Sox2 transcription in ES cells.
SOX2 is part of the core regulatory network of transcription factors required for pluripotency maintenance and cellular reprogramming (Takahashi and Yamanaka 2006). SOX2 co-occupies many sites in the genome with OCT4 and NANOG, which together act as master regulators of pluripotency (Chen et al. 2008). Sox2 is expressed in both the inner cell mass and trophectoderm of the blastocyst (Avilion et al. 2003). Sox2-null embryos die shortly after implantation due to a failure to form a pluripotent epiblast, and blastocyst outgrowths form only trophectoderm-like cells (Avilion et al. 2003). Sox2-null embryonic stem (ES) cells down-regulate Oct4 and Nanog and differentiate to trophectoderm-like cells; however, pluripotency can be rescued in Sox2-null ES cells by forced expression of Oct4 (Masui et al. 2007). Conversely, Sox2 knockdown in two-cell embryos by RNAi, which depletes both maternal and embryonic Sox2, revealed a requirement for Sox2 in trophectoderm formation and development to the blastocyst stage (Keramari et al. 2010). Overexpression of Sox2 in ES cells induces differentiation toward the neuroectodermal lineage, and Sox2 expression is maintained in the developing neuroectoderm (Avilion et al. 2003; Kopp et al. 2008; Thomson et al. 2011).
Transcriptional regulation of Sox2 is complex, as the gene is expressed at high levels in ES cells and down-regulated upon differentiation to endoderm or mesoderm while being maintained in the neuroectodermal lineage (Loh and Lim 2011). In fact, deletion of Sox2 in the embryonic brains of mice leads to complete perinatal loss of hippocampal stem cells (Favaro et al. 2009). Two gene-proximal enhancers, Sox2 regulatory region 1 (SRR1) and SRR2, are able to drive transgene expression in ES cells as well as multipotent neural progenitor cells in the ventricular zone of embryonic brains (Zappone et al. 2000; Tomioka et al. 2002; Miyagi et al. 2004). However, ES cells containing a deletion of SRR1 were able to contribute to chimeras and establish a fertile SRR1-deleted line, indicating that SRR1 is not required for pluripotency (Ferri et al. 2004). These mice displayed cerebral malformations, indicating that SRR1 is involved in regulating Sox2 in the neuroectodermal lineage. While there has been significant focus on the regulatory role that the SOX2 protein plays in maintaining the pluripotent phenotype, the regulatory sequences required for Sox2 transcription in ES cells remain largely uncharacterized.
Intergenic regions play an important role in regulating gene expression (Tuan et al. 1989; Sagai et al. 2005; Lomvardas et al. 2006); however, characterizing their regulatory role is complicated by the observation that they do not always regulate the closest gene in the linear genome (Lettice et al. 2003; Sagai et al. 2005; Sanyal et al. 2012). There are many examples of distal regulatory elements that regulate genes from several kilobases or megabases on the same chromosome and even from different chromosomes (Tuan et al. 1989; Lettice et al. 2003; Lomvardas et al. 2006). For example, the murine β-globin genes are regulated by a cluster of distal regulatory elements—the locus control region (LCR)—located 50 kb upstream of the Hbb-b1 gene (Tuan et al. 1989). Another striking example is that of the Sonic hedgehog (Shh) gene, regulated by a limb enhancer located 1 Mb away in the intron of another gene (Lettice et al. 2003). Single nucleotide polymorphisms (SNPs) in this limb enhancer result in preaxial polydactyly in humans, while deletions in mice cause a shortening of the limbs (Lettice et al. 2003; Sagai et al. 2005). In these two examples as well as many others, distal regulatory regions function via the formation of chromatin loops, allowing the distal region to contact the gene-proximal region (Carter et al. 2002; Tolhuis et al. 2002; Ferrai and Pombo 2009). Genome-wide investigation of chromatin conformation in mouse and human cells has revealed conserved topological domains—megabase-sized regions of increased chromatin interaction (Dixon et al. 2012). The ENCODE project has created a compendium of putative regulatory elements in the human and mouse genomes; however, the majority of these elements still need to be functionally characterized (Bernstein et al. 2012; Stamatoyannopoulos et al. 2012). There is mounting evidence for the role of distal intergenic regions in disease susceptibility and phenotypic traits, as genome-wide association studies (GWASs) have identified SNPs linked with various diseases, and phenotypes often lie in the intergenic regions of the genome. Furthermore, these intergenic regions display open chromatin in the cell type associated with the disease, indicating that they are accessible to DNA-binding transcription factors and may act as transcriptional enhancers (Maurano et al. 2012).
The Sox2 gene is located in a gene desert, yet there is a diverse set of occupied transcription factor-binding sites in ES cells within a 130-kb region surrounding the Sox2 gene (Chen et al. 2012a). More than 100 kb downstream from the gene is a 30-kb region bound by 10 different ES cell-expressed transcription factors, including the pluripotency master regulators OCT4, SOX2, and NANOG. This region also recruits the histone acetyltransferase EP300 (p300) in ES cells (Chen et al. 2012a). EP300 is a transcriptional coactivator that is known to be bound at active tissue-specific enhancers (Visel et al. 2009). In addition, the insulator-binding protein CCCTC-binding factor (CTCF), a protein involved in anchoring chromatin–chromatin interactions, is bound within both the distal region and the promoter-proximal region (Phillips and Corces 2009; Shen et al. 2012). We previously identified 10 putative enhancers surrounding Sox2 by integrative modeling using four enhancer features: p300, NIPBL, and MED12 binding as well as monomethylation of histone H3 at Lys4 (Chen et al. 2012a). Two of these predicted enhancers overlapped with SRR1 and SRR2, the two previously validated enhancers within 4 kb of the Sox2 TSS (transcription start site). In this study, we investigated the regulatory role that each of these 10 regions plays in regulating Sox2 transcription in ES cells and identified three additional ES cell enhancers surrounding Sox2: SRR18, SRR107, and SRR111. The downstream distal enhancers SRR107 and SRR111 contact the more gene-proximal enhancers SRR1, SRR2, and SRR18 through the formation of a large chromatin loop in ES cells but not in fibroblasts. Finally, deletion analysis revealed that only the region containing SRR107 and SRR111, which we term the Sox2 control region (SCR), is required for Sox2 transcription in ES cells.
Results
Identification of transcriptional enhancers surrounding Sox2
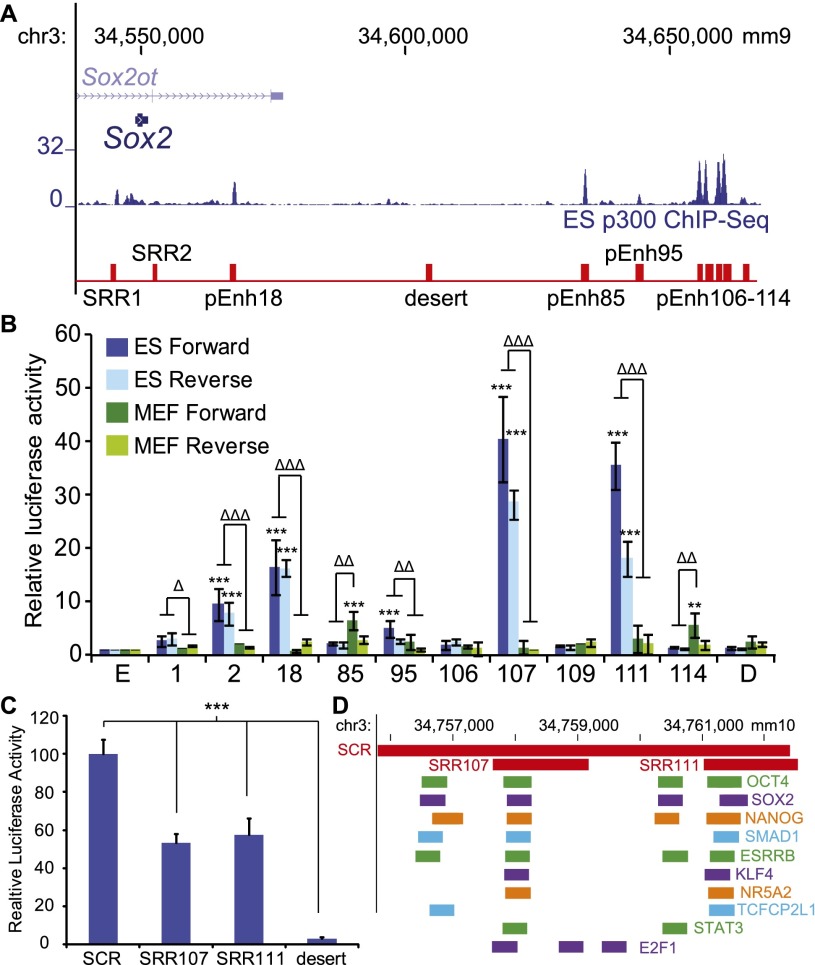
We previously predicted 10 enhancers (pEnh) surrounding Sox2, two of which overlapped SRR1 and SRR2 (Fig. 1A; Chen et al. 2012a). Here we used a luciferase reporter system to determine which of the additional eight predicted enhancers may regulate transcription of Sox2 in ES cells. The predicted enhancers were amplified and cloned downstream from the firefly luciferase gene, and their enhancer activity was assessed in ES cells and MEFs (mouse embryonic fibroblasts). We compared all regions displaying enhancer activity with SRR1 and SRR2, which are able to drive transgene expression in ES cells as well as multipotent neural progenitor cells in the ventricular zone of the embryonic brain (Zappone et al. 2000; Tomioka et al. 2002; Miyagi et al. 2004). We identified three novel functional enhancers that we termed SRR18, SRR107, and SRR111 based on their locations (SRR18, SRR107, and SRR111 are, respectively, 18, 107, and 111 kb downstream from the Sox2 TSS). Each of these regions was able to enhance transcription of the reporter when inserted in either the forward or reverse orientation in ES cells but not in MEFs (Fig. 1B). Interestingly, these distal enhancers were all more robust than the previously identified SRR1 and SRR2 enhancers. To determine whether SRR107 and SRR111 function synergistically in the distal enhancer cluster, we cloned the entire region from 105 to 112 kb downstream from Sox2 into the reporter vector and subsequently performed luciferase assays. This region displayed significantly increased activity that was additive compared with SRR107 or SRR111 alone, and we therefore termed this region the SCR (Fig. 1C). We examined available ES cell ChIP-seq (chromatin immunoprecipitation [ChIP] combined with deep sequencing) data compiled in CODEX, a next-generation sequencing experiment database, and determined that the SCR enhancers are bound by multiple transcription factors, including OCT4, SOX2, NANOG, SMAD1, ESRRB, KLF4, NR5A2, TCFCP2L1, STAT3, and E2F1, while the more gene-proximal enhancers (SRR1, SRR2, and SRR18) are not bound by KLF4, NR5A2, STAT3, or E2F1 (Fig. 1D; Supplemental Table S1; Sanchez-Castillo et al. 2014). We also found that the coactivator NCOA3, the cohesin-associated protein STAG2, and the transcription elongation factor SUPT5 were bound at the SCR enhancers but not the more gene-proximal enhancers (Supplemental Table S1). Based on the reporter assay, we narrowed our investigation to five regions with independent enhancer activity, two of which are located in the distal SCR and bound by multiple ES cell-expressed transcriptional activators.
Figure 1.
Distal enhancers downstream from Sox2 are active in ES cells. (A) The 130-kb transcription factor-bound region surrounding Sox2. ChIP-seq peaks for p300 in ES-Bruce4 cells from ENCODE/LICR are shown in blue. Predicted enhancer (pEnh) amplicons are shown in red. (B) Luciferase activity driven by predicted enhancers surrounding Sox2. Forward and reverse enhancer orientations were assayed in both ES cells and MEFs, the empty pGL4.23 vector (E), and the desert region (D); numbers correspond to enhancer notations in A. The ratios of firefly/Renilla luciferase activity were normalized to E. Values are an average of at least three biological replicate experiments, with each experiment performed in triplicate. Error bars represent SEM. A significantly higher activity over the empty vector is indicated by double asterisks (P < 0.01) or triple asterisks (P < 0.001). Significant differences between ES cells and MEFs for each enhancer are indicated by a single triangle (P < 0.05), double triangles (P < 0.01), or triple triangles (P < 0.001). (C) Forward enhancer orientations were assayed in ES cells for the SCR (105–112), SRR107, SRR111, and the desert region. The ratios of firefly/Renilla luciferase activity were normalized to the empty pGL4.23 vector. Values are represented as a percentage of the SCR value and are an average of at least three independent experiments, with each experiment performed in triplicate. Error bars represent SEM. The SCR drives significantly higher activity than SRR111 and SRR107 (P < 0.001) as well as the desert region. (D) Validated enhancers within the SCR are displayed in red on the University of California at Santa Cruz Genome Browser (mm10). Transcription factor-bound regions from ChIP-seq compiled in the CODEX database are shown below.
The SCR contacts the Sox2 gene in ES cells
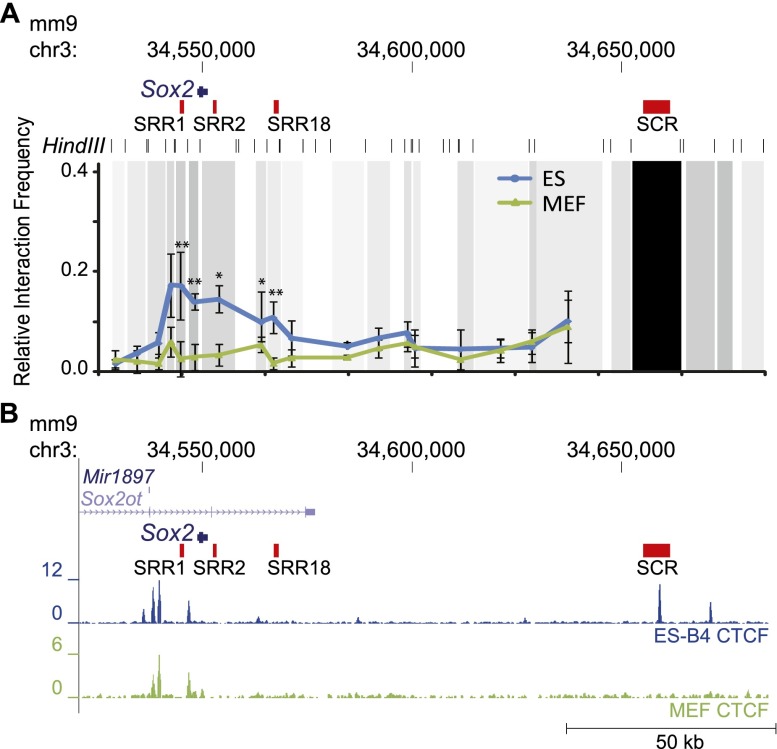
We next investigated the mechanisms through which the distal and proximal enhancers surrounding Sox2 may be involved in regulating Sox2 transcription in ES cells. Distal enhancers have been shown to regulate gene expression through physically contacting the proximal promoter of the regulated gene via the formation of chromatin loops (Carter et al. 2002; Tolhuis et al. 2002). Recently, interaction between the Sox2 promoter and SCR was recorded by 5C (chromosome conformation capture carbon copy) and ChIA-PET (chromatin interaction analysis with paired-end tag sequencing) in mouse ES cells (Kieffer-Kwon et al. 2013; Phillips-Cremins et al. 2013). To confirm and refine these interactions, we conducted 3C (chromosome conformation capture) analysis in ES cells and MEFs (Dekker et al. 2002). We found ES cell-specific chromatin loops between the SCR and the Sox2-proximal region (Fig. 2A; Supplemental Fig. S1). This looping brings the SCR distal enhancers SRR107 and SRR111 into contact with the gene-proximal enhancers SRR1, SRR2, and SRR18 in ES cells but not in MEFs, indicating that the SCR could be important in regulating Sox2 transcription in ES cells. CTCF binding has been shown to be associated with chromatin looping, and through inspection of ENCODE data (Bernstein et al. 2012; Stamatoyannopoulos et al. 2012), we noted the presence of ES cell-enriched CTCF-bound regions within the SCR overlapping pEnh109 as well as further downstream from the SCR (Fig. 2B; Supplemental Fig. S2). This cell type specificity of CTCF binding is unusual, as the majority of CTCF binding throughout the genome is not cell type-specific (Chen et al. 2012b), including additional CTCF-bound regions upstream of the Sox2 promoter that are present in many other cell types (Supplemental Fig. S2). In addition to the numerous transcription factors bound at the proximal and distal Sox2 enhancers, members of the mediator and cohesin complexes as well as the cohesin loading factor NIPBL are enriched in these regions, which may further support the formation and maintenance of the chromatin loop (Supplemental Fig. S1; Supplemental Table S1).
Figure 2.
The SCR contacts the Sox2 promoter and proximal enhancers in ES cells. (A) 3C was performed on mouse ES cells and MEFs. The frequency of interaction between the SCR (anchor fragment) and the surrounding regions was normalized to that between adjacent fragments at the α -aortic actin (α-Act) locus. The black bar represents the anchor fragment, and gray bars represent the interacting fragments. Values are an average of at least three biological replicate experiments, with each experiment performed in triplicate. Error bars represent standard deviation. A significant difference between ES cells and MEFs is indicated by a single asterisk (P < 0.05) or double asterisks (P < 0.01). (B) ChIP-seq peaks for CTCF are from ENCODE/LICR; ES-Bruce4 cells are shown in blue, and MEFs are shown in green.
The SCR is required for Sox2 transcription in ES cells
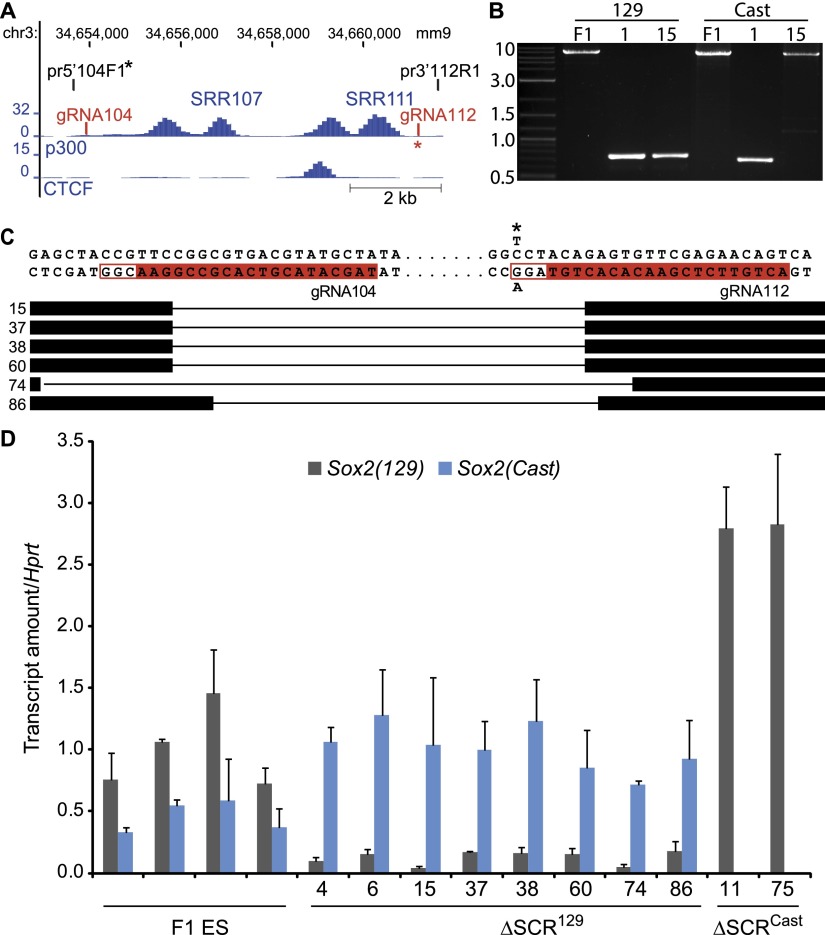
To determine whether the SCR is required for Sox2 transcription in ES cells, we used the CRISPR/Cas9 system (Ding et al. 2013; Mali et al. 2013) to delete the 7.3-kb region containing the SRR107 and SRR111 enhancers. We first carried out this deletion in mouse F1 (Mus musculus129 × Mus castaneus) ES cells, allowing allele-specific deletion screening and gene expression analysis (Mlynarczyk-Evans et al. 2006). We used two guide RNAs (gRNA104 and gRNA112) to generate the large deletion (Fig. 3). The presence of a SNP in the PAM sequence for gRNA112 on the M. castaneus (Cast) allele (C/G to T/A) caused preferential deletion of the SCR on the 129 allele (ΔSCR129) (Supplemental Fig. 3B/C). As expression from one Sox2 allele is sufficient to maintain pluripotency in ES cells (Avilion et al. 2003), we expected to obtain ES cell colonies even in the absence of Sox2 expression from the 129 allele. We monitored expression of Sox2 by allele-specific reverse transcription quantitative real-time PCR (RT-qPCR) due to the presence of SNPs within the Sox2 transcript (Fig. 3D; Supplemental Fig. S3A). In the ΔSCR129 clones, we observed a significant, eightfold reduction in the amount of 129 Sox2 mRNA (average of eight ΔSCR129 clones; P = 2.16 × 10−5), revealing that the SCR contributes to Sox2 transcription in ES cells (Fig. 3D). Unexpectedly, two of the clones screened contained a deletion within the SCR on the Cast allele (ΔSCRCast). These ΔSCRCast clones showed a dramatic reduction in the amount of Cast Sox2 mRNA (Fig. 3D). As expected, the levels of Oct4 mRNA were maintained in both the ΔSCR129 and ΔSCRCast clones (Supplemental Fig. S3), and these cells maintained ES cell colony morphology over several passages in culture. Interestingly, we noted a significant twofold increase (average of eight ΔSCR129 clones; P = 0.00036) in Cast Sox2 mRNA in ΔSCR129 clones and a similar increase in 129 Sox2 mRNA in ΔSCRCast clones, indicating that there is a feedback mechanism compensating for the loss of Sox2 expression from one allele that would mask the effects of a single deletion in ES cells derived from only one mouse strain.
Figure 3.
The SCR is required for Sox2 transcription in ES cells. (A) gRNA-binding regions 104 and 112 kb downstream from Sox2 are shown in red. Primers used to amplify the deleted region are shown in black. The locations of SNPs between 129 and Cast in primers or the PAM adjacent to gRNA112 are indicated by an asterisk. ES-Bruce4 ChIP-seq data for p300 and CTCF from ENCODE/LICR are shown in blue. (B) PCR amplification of the region targeted by gRNA104 and gRNA112 in F1 ES (F1), ΔSCR129/Cast clone 1 (1), and ΔSCR129 clone 15 (15). Primers specific to the 129 (5′104F1_129/3′112R1) or Cast (5′104F1_Cast/3′112R1) alleles were used to amplify each allele. (C) Sequences were obtained from selected 129 deleted clones; the clone number is indicated at the left, thick bars indicate that the sequence is present, and thin lines indicate missing sequences. Sequences of gRNAs 104 and 112 kb are highlighted in red, the locations of PAM sequences are shown by a red box, and the Cast SNP in the PAM adjacent to gRNA112 is indicated by an asterisk. (D) Deletion of the SCR dramatically reduces expression of the linked Sox2 allele. Allele-specific primers detected Sox2 129 mRNA or Sox2 Cast mRNA in RT-qPCR from F1 ES, ΔSCR129, and ΔSCRCast. Expression is shown relative to the levels in F1 ES cells. Error bars represent standard deviation.
The SCR is required for ES cell differentiation potential
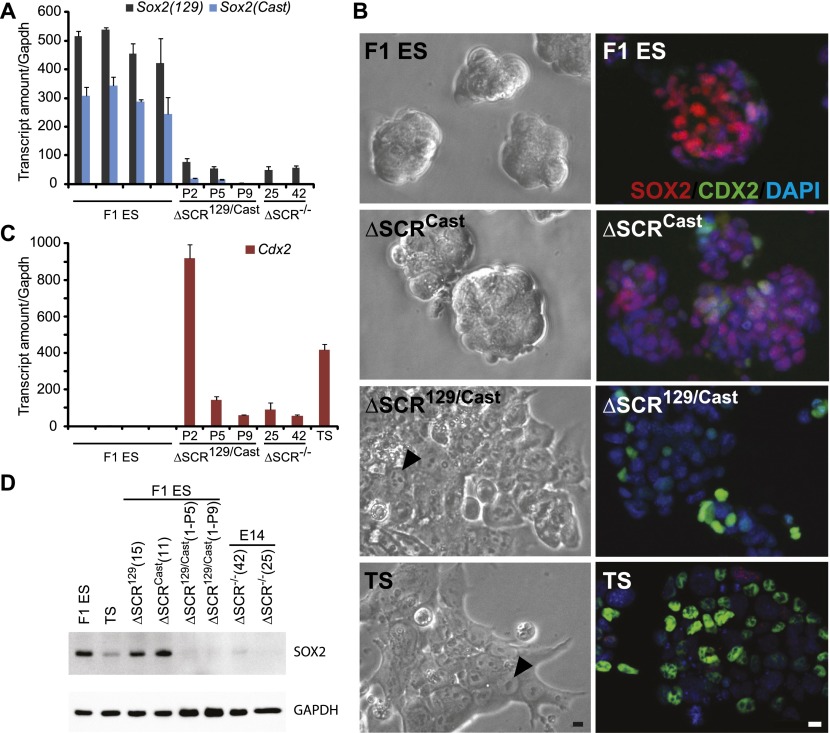
From the initial F1 ES cell targeting, we obtained one ES clone with a deletion of the SCR on both the 129 and Cast alleles (ΔSCR129/Cast). In this clone, Sox2 expression from both alleles was dramatically reduced by more than sixfold (Fig. 4A). Furthermore, although these cells were maintained in ES medium containing LIF and 2i, we observed a loss of ES colony features over time in culture and progression toward a trophectoderm-like phenotype (Fig. 4B). We profiled expression of the trophectoderm cell marker Cdx2 and found it to be up-regulated in the ΔSCR129/Cast clone (Fig. 4C; Supplemental Fig. S3). However, over time in culture (passages 2–9), expression of Cdx2 was reduced in ΔSCR129/Cast, indicating that these cells do not maintain stable trophoblast gene expression; this may be due in part to their maintenance in ES medium rather than trophoblast stem (TS) medium. In support of this possibility, we observed an increase in Cdx2 expression when the ΔSCR129/Cast clone was maintained in either ES medium without 2i inhibitors or TS medium (Supplemental Fig. S3). Immunofluorescence and immunoblot analysis confirmed a dramatic decrease in SOX2 protein levels in ΔSCR129/Cast, which is consistent with the requirement of the SCR for Sox2 transcription (Fig. 4D).
Figure 4.
The SCR is required for maintenance of the ES cell phenotype. (A) Homozygous deletion of the SCR disrupts Sox2 expression from both alleles. Allele-specific primers detect Sox2 129 mRNA or Sox2 Cast mRNA in RT-qPCR from F1 ES, ΔSCR129/Cast (clone 1 passages 2, 5, and 9), and ΔSCR−/− in E14 ES cells (clones 25 and 42). Error bars represent standard deviation. (B) The left panel displays bright-field images of F1 ES and ΔSCRCast (clone 11), which formed ES-like colonies over several passages in culture, compared with ΔSCR129/Cast (clone 1) cells, which showed features consistent with differentiation to a trophectoderm-like phenotype and F1 TS cells. Arrowheads indicate similar features in ΔSCR129/Cast and TS cells. The right panel displays SOX2 and CDX2 immunofluorescence in F1 ES, ΔSCRCast (clone 11), ΔSCR129/Cast (clone 1), and F1 TS cells. Bar, 10 μm. (C) Expression of differentiation marker Cdx2 is shown relative to the levels in F1 ES cells. Error bars represent standard deviation. (D) SOX2 protein levels in F1 ES cells, TS cells, ΔSCR129 (clone 15), ΔSCRCast (clone 11), ΔSCR129/Cast (clone 1) at passages 5 and 9 (P5 and P9), and ΔSCR−/− in E14 ES cells (clones 25 and 42). GAPDH levels reveal relatively equal protein loading in all samples.
To confirm the importance of the SCR in maintaining Sox2 expression in ES cells, we repeated the Cas9-mediated deletion in E14TG2a (E14) ES cells, where we expected both alleles to be equally targeted. We obtained two additional homozygous SCR-deleted clones (ΔSCR−/−) that showed a trophectoderm-like phenotype and an increase in Cdx2 expression (Fig. 4; Supplemental Fig. S4). This increase in Cdx2 expression was less than that observed in the F1 ΔSCR129/Cast clone at passage 2 and more similar to the levels of Cdx2 observed in the F1 ΔSCR129/Cast clone at later passages. As mentioned earlier, this may be due in part to the ES medium conditions (which are not designed for stable maintenance of Cdx2 expression) but could also indicate that SCR-deleted cells are not normal trophectoderm cells. In both E14 clones lacking the SCR, Sox2 mRNA and protein levels were dramatically reduced, similar to the ΔSCR129/Cast clone obtained from F1 ES cells (Fig. 4). Together, these data demonstrate that the SCR located 104–112 kb downstream from Sox2 is required to maintain Sox2 expression in ES cells and that the loss of this region alters the ES cell phenotype.
Interestingly, in clones containing a double deletion of the SCR, the mRNA levels of the pluripotency factor Oct4 was not affected by the dramatic reduction in SOX2 protein (Supplemental Fig. S3). This was unexpected, as other studies have shown that Sox2 knockdown or deletion leads to a decrease in Oct4 expression (Masui et al. 2007; Adachi et al. 2013). In contrast, we found that Oct4 mRNA levels are maintained despite the dramatic reduction in SOX2 protein levels. This difference did not appear to be due to the presence of the 2i inhibitors in our ES medium, as maintenance of ΔSCR129/Cast in ES medium without these inhibitors did not affect Oct4 transcript levels (data not shown). We next investigated the levels of OCT4 protein and found that, similar to mRNA levels, OCT4 protein levels were maintained in clones containing a homozygous deletion of the SCR (Supplemental Fig. S4).
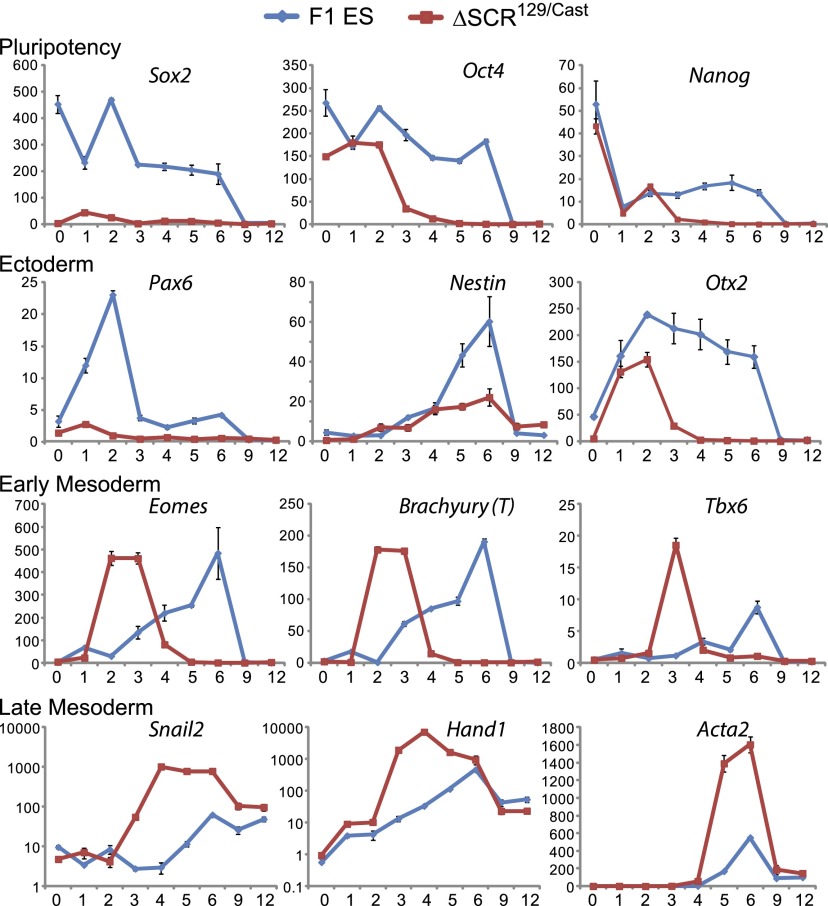
To assess whether SCR deletion affects the differentiation potential of F1 ES cells, we monitored the expression of pluripotency, ectoderm, mesoderm, and endoderm marker genes over 12 d of embryoid body (EB) formation (Fig. 5). In ΔSCR129/Cast EBs, we observed that the low levels of Sox2 persisted for 12 d, whereas in F1 EBs, the levels of Sox2 gradually decreased until day 9. Interestingly, although expression of Oct4 and Nanog were maintained in ΔSCR129/Cast cells cultured in LIF/2i, the expression of both decreased more rapidly during EB formation compared with F1 EBs. Sox2 overexpression in ES cells has been shown to promote differentiation to neuroectodermal cells (Kopp et al. 2008; Thomson et al. 2011). Consistent with this, we observed low expression of ectodermal and neuroectodermal markers Pax6, Nestin, Otx2, Foxd3, and Fgf5 in ΔSCR129/Cast EBs compared with F1 EBs. This indicates that SCR-deleted cells are impaired in their ability to form ectoderm (Fig. 5; Supplemental Fig. S5). Sox2 has been shown to repress differentiation to mesendoderm, whereas Oct4 promotes the formation of mesendoderm (Thomson et al. 2011; Wang et al. 2012; Radzisheuskaya et al. 2013). Consistent with maintained expression of Oct4 and the greatly reduced levels of Sox2 in the ΔSCR129/Cast ES cells, we observed that these cells are biased toward mesendoderm formation upon differentiation. Specifically, ΔSCR129/Cast EBs showed elevated expression of early mesodermal markers Eomes, Brachyury, Tbx6, and Bmp4 on day 2 compared with the F1 EBs in which these markers only increase expression after day 4 (Fig. 5; Supplemental Fig. S5). This led to a greater induction of late mesodermal markers, including Snail2, Hand1, Acta2, and Twist2 in the ΔSCR129/Cast EBs. Endodermal markers Gata4, Gata6, Sox17, and Sox7 showed a pattern similar to the early mesodermal markers, with an earlier increase in expression observed in ΔSCR129/Cast EBs compared with F1 EBs (Supplemental Fig. S5). Taken together, these data reveal that SCR-deleted cells have impaired ectoderm formation and a bias toward mesendoderm differentiation.
Figure 5.
EB formation reveals impaired differentiation to neuroectoderm and increased mesendoderm formation in SCR129/Cast cells. EB formation was induced by the hanging drop method, and changes in gene expression were monitored over 12 d by RT-qPCR. The Y-axis shows transcript levels relative to Gapdh. Snail2 and Hand1 are displayed on a log10 scale. The X-axis shows days of EB formation. Error bars represent standard deviation.
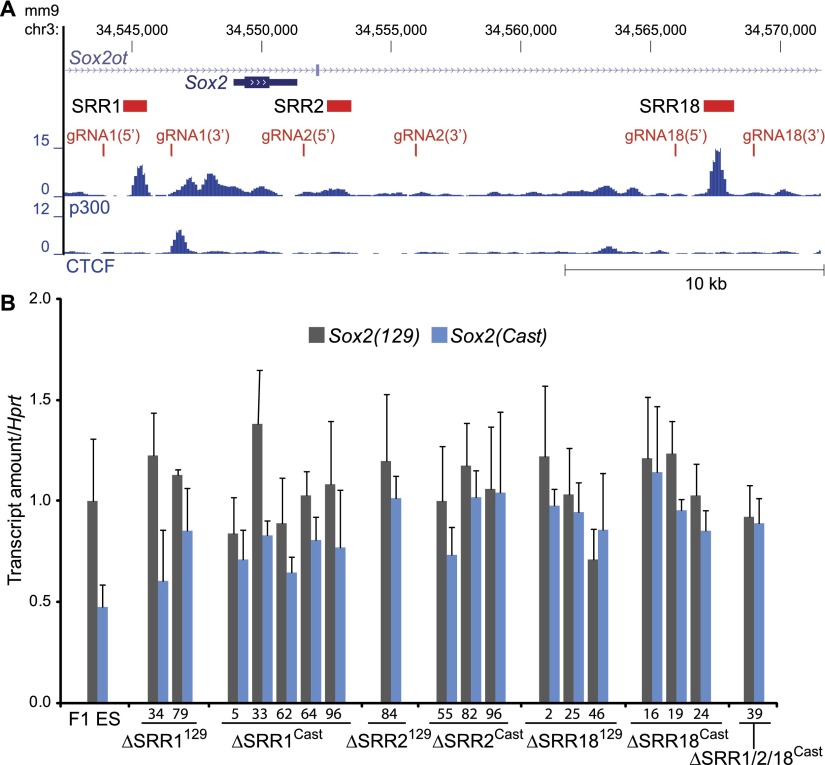
SRR1, SRR2, and SRR18 are not required for Sox2 transcription in ES cells
To determine whether any of the more proximal enhancers are required for Sox2 transcription in ES cells, we next deleted SRR1, SRR2, or SRR18 and monitored the effect on Sox2 transcription. For each deletion, we used two gRNAs to generate the large deletions (Fig. 6A). Earlier studies revealed that SRR1 is dispensable for Sox2 transcription in ES cells, although there is evidence that SRR1 is required in neural cells (Ferri et al. 2004). We found that deletion of SRR1 did not affect transcription of the linked Sox2 allele in ES cells (Fig. 6B). This is in agreement with the minimal amount of enhancer activity displayed in ES cells in the luciferase reporter assay (Fig. 1B). We also found that heterozygous and homozygous deletion of SRR2 or SRR18 did not impact transcription of the linked Sox2 allele (Fig. 6B; Supplemental Fig. S6). This was more surprising, as both of these regions displayed more robust enhancer activity than SRR1 in the reporter assay. To rule out the possibility that the gene-proximal enhancers have redundant functions, we deleted the entire region from 5′ of SRR2 to 3′ of SRR18 using gRNA2(5′) and gRNA18(3′) in a ΔSRR1Cast clone and generating a heterozygous enhancer triple-knockout clone (ΔSRR1/2/18Cast). In this clone, we still observed no significant effect on Sox2 transcription (Fig. 6B). Taken together, this deletion analysis reveals only a requirement for the enhancers of the SCR in maintaining robust Sox2 transcription in ES cells.
Figure 6.
Gene-proximal enhancers are not required for Sox2 transcription in ES cells. (A) SRR1, SRR2, and SRR18 were each deleted in F1 ES cells using two gRNAs. ΔSRR1/2/18Cast was generated using gRNA2(5′) with gRNA18(3′) in a ΔSRR1Cast clone. (B) Allele-specific RT-qPCR revealed that SRR1, SRR2, and SRR18 are all dispensable for Sox2 expression in ES cells. The Sox2 mRNA levels shown are relative to the levels in F1 ES cells. Error bars represent standard deviation between at least three technical replicates for each deleted clone.
RNA sequencing (RNA-seq) analysis reveals that the SCR is specific for Sox2 regulation
As enhancers can influence genes located at megabase distances away in the genome, we conducted RNA-seq to determine whether any additional genes are affected by deletion of the SCR. Initially, we investigated gene expression changes by RNA-seq in ΔSCR129/Cast. As expected, this clone displayed a dramatic reduction in total Sox2 transcript, consistent with the RT-qPCR results described above (Fig. 7A). We next investigated allele-specific effects in three heterozygous SCR-deleted clones (ΔSCR129 clones 15 and 37 and ΔSCRCast clone 11); Sox2 expression was affected only on the allele linked to the SCR deletion in these clones (Fig. 7A). As SOX2 protein levels are dramatically reduced in the ΔSCR129/Cast clone and as cells display a differentiated trophectoderm-like phenotype, we expected to observe genome-wide changes in gene expression in these cells. While Sox2 is the most dramatically down-regulated gene in the ΔSCR129/Cast clone, additional genes changed expression, consistent with the transition to the trophoblast lineage. For example, the TS cell-expressed genes Cdx2, Gata2, H19, Peg10, Krt18, Krt8, and Igf2 all displayed increased expression in ΔSCR129/Cast (Fig. 7B; Supplemental Table S2). Whereas there was a significant correlation between genes with increased (P < 1.032 × 10−39) or decreased (P < 2.704 × 10−211) expression in ΔSCR129/Cast and TS cells compared with those in ES cells, the transcriptome of ΔSCR129/Cast cells was more similar to that of ES cells than of TS cells, consistent with our earlier observation that the ΔSCR129/Cast clone does not stably maintain trophectoderm marker gene expression (Fig. 7B). Furthermore, we noted that of the major regulatory transcription factors expressed in ES cells (Oct4, Sox2, Sall4, Nanog, Klf4, Esrrb, and Tbx3), only Sox2 was significantly decreased in ΔSCR129/Cast. We did note in the RNA-seq data that Nanog transcripts were reduced 3.8-fold in ΔSCR129/Cast compared with F1 cells, and although this was not significant in the RNA-seq data, it may be biologically relevant. To investigate this possibility further, we examined NANOG protein levels in SCR-deleted clones and found that the levels of NANOG protein appeared reduced in clones containing a homozygous deletion of the SCR (Supplemental Fig. S4).
Figure 7.
RNA-seq analysis of SCR-deleted clones. (A) RNA-seq reads mapped to Sox2 in ΔSCR clones. Total reads are displayed in black, 129 reads are displayed in gray, and Cast reads are displayed in blue. Cast SNP identity is shown at the bottom. (B) Scatter plots reveal differences in transcript abundance between F1 ES cells, ΔSCR clones, and F1 TS cells. Transcript levels are log2 transformed. Sox2 is indicated in green, ES cell-expressed transcription factors are indicated in red, and TS cell-expressed genes are indicated in blue.
If the SCR is regulating additional genes on chromosome 3, we would expect to see an enrichment of genes with decreased expression in ΔSCR129/Cast on chromosome 3 (Supplemental Table S2). We did not observe an enrichment of genes on chromosome 3; however, we did note a striking enrichment of genes on the X chromosome in the set of genes with decreased expression in ΔSCR129/Cast. The F1 ES cells are female cells that may undergo X inactivation upon differentiation induced by SCR deletion. In support of this, we noted increased expression of the X-inactive-specific transcript (Xist) on both alleles in ΔSCR129/Cast at passage 5 (Supplemental Fig. S7). This increase in Xist expression was transient and not maintained to passage 9, whereas the decrease in gene expression on the X chromosome was most prominent at passage 9 and skewed toward inactivation of the 129 X chromosome. This observation was expected, as F1 ES cells display a bias toward inactivation of the 129 X chromosome over the Cast X chromosome upon differentiation (Ogawa and Lee 2003; Mlynarczyk-Evans et al. 2006).
We next investigated allele-specific effects in the three heterozygous SCR-deleted clones. As expected, these clones did not display the same changes in gene expression observed in the ΔSCR129/Cast clone, consistent with maintenance of the pluripotent phenotype in these cells (Fig. 7B). We hypothesized that if the SCR is responsible for regulating any additional genes on chromosome 3, we would observe a decrease in expression on the linked allele for those genes in all three heterozygous SCR-deleted clones. We found that the only gene in the genome that showed a significant (P < 0.05 with Benjamini and Hochberg multiple testing correction) decrease in expression on the linked allele in all three heterozygous clones was in fact Sox2. Furthermore, Sox2 is the only gene on chromosome 3 that decreases expression greater than twofold in all three heterozygous clones and the ΔSCR129/Cast clone, indicating that the SCR is specifically required in ES cells to regulate Sox2 transcription.
Discussion
Through the use of enhancer reporter assays, chromatin looping analysis, and Cas9-mediated deletion, we identified two independent enhancers (SRR107 and SRR111) clustered in a distal control region (the SCR) that regulate transcription of Sox2 in ES cells. Furthermore, the approach that we describe here revealed a functional requirement for the SCR in ES cell differentiation potential. Our targeted method of monoallelic Cas9-mediated enhancer deletion combined with allele-specific transcriptome analysis reveals a refined view of transcriptional regulation while avoiding the confounding effects that occur when critical transcriptional regulators are subjected to knockdown. Using this approach, we show that distal enhancer regions located >100 kb downstream from Sox2 are specifically required for Sox2 transcription, while the more gene-proximal regions with validated enhancer activity are dispensable for Sox2 transcription in naive ES cells.
The distal SCR functions through the formation of a large chromatin loop, allowing enhancer–promoter contact to occur in ES cells. In ES cells, knockdown of cohesin or mediator components has been shown to affect chromatin looping interactions at other loci and alter gene expression in ES cells, leading to differentiation (Kagey et al. 2010). In fact, an 8.7-kb region surrounding the Sox2 gene as well as a 27-kb region including the downstream SCR have been classified as superenhancers based on the density of mediator binding (Whyte et al. 2013). This raises the possibility that mediator and cohesin also support chromatin looping at the Sox2 locus. In support of this possibility, knockdown of SMC1A, MED12, or NIPBL led to a reduction in Sox2 transcript levels (Kagey et al. 2010). Based on these data, we propose a model in which Sox2 transcription in ES cells is regulated by the distal enhancers of the SCR through contact with the Sox2 promoter-proximal region. This contact is supported by the plethora of proteins bound at the distal SCR (transcription factors and the mediator and the cohesin complexes) and by CTCF bound at both the Sox2 promoter-proximal region and SCR.
Our data reveal that the distal SCR is required to maintain Sox2 expression and has more robust enhancer activity than the Sox2-proximal enhancers, which are dispensable, despite the observation that both regions appeared to be in contact through the formation of chromatin loops that extend across the two superenhancer domains. Although all three proximal enhancers were dispensable for Sox2 expression in ES cells, we suggest the possibility that SRR2 and SRR18, like SRR1, are important for Sox2 expression in other tissues. In addition to the role that Sox2 plays in ES cells, Sox2 is also required for osteoblast self-renewal and differentiation of mature neurons (Cavallaro et al. 2008; Favaro et al. 2009). Our deletion analysis in ES cells together with reporter assay data suggest that the robust additive activity of SRR107 and SRR111 in the distal SCR could override any more modest contributions to Sox2 transcription made by the proximal enhancers in ES cells. In neural progenitor cells, however, the SCR is not bound by chromatin proteins associated with enhancer activity (Phillips-Cremins et al. 2013), so the gene-proximal enhancers may be required for Sox2 transcription in this context, where the SCR is not active.
In the SCR itself, there are four predicted enhancers regions, each of which is bound by multiple transcription factors and p300. Only two of these regions (SRR107 and SRR111) have classical and independent enhancer activity in reporter assays. The two validated enhancers are each bound by OCT4, SOX2, NANOG, SMAD1, ESRRB, KLF4, and NR5A2 and recruit the coactivator NCOA3, whereas the two regions that failed to show enhancer activity and SRR1, SRR2, and SRR18 are not bound by all of these factors together, suggesting that these factors or a subset of these factors are required for enhancer activity of the SCR and transcription of Sox2 in ES cells. The master regulatory transcription factors in ES cells (SOX2, OCT4, NANOG, KLF4, and ESRRB) are thought to be controlled through a highly interconnected network of regulatory interactions (Loh and Lim 2011). All of these factors are bound within the SCR, and, taken together with the dramatic effect that SCR deletion has on Sox2 transcription and the lack of effect observed at other regions, our data suggest that the SCR is the region through which these master regulatory transcription factors exert their regulatory effect.
Transcriptome analysis of the ΔSCR129/Cast clone raises the question of what phenotype these cells acquire in the absence of Sox2 expression. Morphological inspection of these cells revealed a differentiated trophectoderm-like phenotype, consistent with earlier studies (Avilion et al. 2003; Masui et al. 2007); however, transcriptome analysis showed gene expression patterns that are more similar to the original F1 ES cells than TS cells. The similarity in the transcriptome to ES cells is likely due to the near-complete maintenance of the pluripotency transcription factor network, with the exception of Sox2. In contrast, the phenotypic appearance of these cells may be due to the increased expression of a few key transcriptional regulators expressed in trophoblast cells (namely, Cdx2 and Gata2) and the subsequent increased expression of additional trophoblast cell-expressed genes. Whereas the expression of other core pluripotency transcription factors is maintained in the ΔSCR129/Cast clone, we did observe a decrease in the expression of genes that enhance the final stages of reprogramming: Dppa2, Notum, Gtsf1l, Tuba3a, Klf2, Klf5, Fam25c, and Nanog (Supplemental Table S2; Golipour et al. 2012). Increased expression of endogenous Sox2 is one of the later events during the reprogramming process (Buganim et al. 2012; Golipour et al. 2012). We found that when we disrupted Sox2 transcription through SCR deletion, these genes involved in the final stages of reprogramming showed decreased expression, indicating that they are regulated by SOX2. Consistent with direct regulation by SOX2, we observed SOX2 binding within a 10-kb region upstream of Dppa2, Notum, Klf2, and Fam25c in ES cells.
The observation that Oct4 transcription is maintained in the almost complete absence of SOX2 protein is interesting, as knockdown or deletion of Sox2 in ES cells caused a reduction in Oct4 transcription in other studies (Masui et al. 2007; Adachi et al. 2013). We did observe increased expression of Sox3 in the ΔSCR129/Cast clone, which may partially compensate for the loss of Sox2 transcription and contribute to maintaining Oct4 transcription. However, we observed a deficiency in the ability of the ΔSCR129/Cast clone to induce the expression of ectodermal genes upon EB formation, indicating that the increase in Sox3 levels was not able to fully compensate for the loss of Sox2 expression. Whereas our data reveal a requirement for the SCR in maintaining Sox2 expression in ES cells and demonstrate that loss of Sox2 due to SCR deletion impairs the ability of ES cells to differentiate to all three germ layers, there are differences between Sox2-null ES cells generated by Cre-mediated deletion of Sox2 in ES cells (Masui et al. 2007) and our SCR-deleted ES cells. Our SCR-deleted ES cells express Oct4 at levels similar to those of the F1 cells and display increased expression of Cdx2, whereas Sox2-null ES cells do not maintain Oct4 expression or up-regulate Cdx2 (Masui et al. 2007). This difference could be due to residual low levels of SOX2 protein in our cells compared with the Sox2-null ES cells or differences in the way the experiment was conducted; Sox2-null ES cells were investigated for 4 d after Sox2 deletion, whereas our Cas9-mediated deletion required 10 d before gene expression analysis could be conducted in colonies expanded from single cells. This difference in time frame may result in differences in cellular phenotype in the two experiments.
High-throughput validation of enhancer activity in mouse tissues has previously been conducted mainly through the use of enhancer–reporter transgenes (Visel et al. 2007, 2009). Although such experiments reveal a regulatory potential for intergenic enhancers, they do not determine whether they are required for the expression of specific genes or, indeed, which genes they regulate. Due to the low efficiency of homologous recombination-based approaches in editing the genome of mammalian cells, few distal enhancers have been deleted from mammalian genomes to reveal their function, although the number is increasing with recent nuclease-mediated genome-editing techniques (Fiering et al. 1995; Sagai et al. 2005; Attanasio et al. 2013; Kieffer-Kwon et al. 2013). Using high-efficiency targeted deletion in F1 ES cells, we were able to profile cis-regulation by the SCR. Allele-specific transcriptome analysis in SCR-deleted clones revealed that Sox2 is the only gene significantly affected by deletion of the SCR in cis, indicating that the SCR specifically regulates Sox2 in ES cells. Other genes may be less likely to interact with the SCR simply due to genomic distance; Sox2 is the only annotated coding sequence within a 1.6-Mb gene-poor region on chromosome 3, with the next closest gene to the SCR >500 kb upstream of Sox2, although there are examples of cis-regulation at this distance. However, the topological domain surrounding Sox2 covers close to 2 Mb and contains four additional genes upstream of Sox2, which indicates interaction of these regions in the nucleus (Dixon et al. 2012). Despite this, we found that Sox2 is the only gene affected by SCR deletion in cis. It will be important to investigate what conveys this specificity in the regulation of Sox2 by the SCR. Possibilities include promoter-proximal sequences (although not the proximal enhancers themselves), chromatin domains created by CTCF that insulate other genes from interacting with the SCR, or other factors that influence chromatin loop formation and are lacking at other genes within the topological domain. From a thorough examination of predicted enhancer elements surrounding Sox2, we characterized the SCR, a distal transcriptional enhancer region required for Sox2 transcription and differentiation potential of ES cells. Furthermore, we highlighted the need for similar enhancer deletion studies to further our understanding of transcriptional regulation at other loci.
Materials and methods
Cell culture
E14 (obtained from the American Type Culture Collection) and F1 (M. musculus129 × M. castaneus obtained from Barbara Panning) mouse ES cells were cultured on 0.1% gelatin-coated plates in ES medium (DMEM containing 15% FBS, 0.1 mM MEM nonessential amino acids, 1 mM sodium pyruvate, 2 mM GlutaMAX, 0.1 mM 2-mercaptoethanol, 1000 U/mL LIF, 3 μM CHIR99021 [GSK3β inhibitor, Biovision], 1 μM PD0325901 [MEK inhibitor, Invivogen]) and maintained in a pluripotent state in the absence of a feeder layer (Mlynarczyk-Evans et al. 2006; Ying et al. 2008). MEFs were isolated from embryonic day 12.5 (E12.5) C57Bl/6 mouse embryos and cultured in DMEM containing 10% FBS and 2 mM GlutaMAX. F1 TS cells (M. musculusC57BL/6 × M. castaneus, obtained from Susannah Varmuza) were maintained as described in Tanaka (2006) (Miri et al. 2013). EBs were formed by the hanging drop method; to initiate differentiation, 1000 cells were suspended in 20-μL droplets of ES medium without LIF and 2i (Ohnuki and Kurosawa 2013). EBs were transferred to 0.1% gelatin on day 4 to facilitate further growth. EBs were collected at daily intervals up to day 12, and RNA was isolated for gene expression analysis by RT-qPCR. All animal experiments were approved by the University Animal Care Committee (UACC) at the University of Toronto and the Bioscience Local Animal Care Committee (LACC).
Enhancer validation and deletion
The activity of SRR1, SRR2, and eight predicted enhancer candidates was assayed using a dual-luciferase reporter assay (Promega). Briefly, PCR-amplified enhancer candidates were inserted downstream from the firefly luciferase gene in the pGL4.23 vector and cotransfected with a Renilla luciferase encoding plasmid (pGL4.75) into E14 ES cells on 96-well plates. Luciferase activity (firefly/Renilla) was measured on the Fluoroskan Ascent FL plate reader and normalized to the empty vector. To identify DNA looping interactions of putative enhancer candidates with the Sox2 gene, we adapted the 3C protocol from Dekker et al. (2002) (minor modifications are detailed in the Supplemental Material). To assay the role of putative enhancer candidates in Sox2 gene expression, we deleted these regions in mouse ES cells using the CRISPR/Cas-9 system. Pairs of plasmids encoding gRNAs (to direct Cas9 to regions flanking each candidate enhancer) were assembled using the gRNA empty vector (Addgene, ID no. 41824) as described by Mali et al. (2013) and cotransfected with pCas9_GFP (Addgene, ID no. 44719) into F1 ES cells (Ding et al. 2013; Mali et al. 2013). Targeted clones were screened by allele-specific qPCR to identify deletions, and deletions were confirmed by sequencing. Total RNA was purified and reverse-transcribed with random primers for gene expression analysis. Gene expression (normalized to Gapdh or Hprt) was quantified by qPCR using the standard curve method. Additional details are provided in the Supplemental Material.
Genome-wide sequencing data
RNA was isolated using Trizol (Life Technologies) from F1, ΔSCR129/Cast, ΔSCR129, and ΔSCRCast ES cells and DNase-treated prior to multiplexed massively parallel sequencing (paired-end 150 base pairs [bp]) using the Illumina platform. Sequence data were submitted to the Gene Expression Omnibus (GEO) repository (GSE58339). Paired-end reads were mapped to the mouse genome using TopHat2 running bowtie2 (Langmead et al. 2009; Trapnell et al. 2009; Langmead and Salzberg 2012; Kim et al. 2013). To prevent allelic bias, mapping was carried out to the M. musculus129 or M. castaneus genome generated from the NCBI37/mm9 reference by SNP substitution using variant files provided by the Sanger Mouse Genomes Project (Keane et al. 2011). Mapped reads were split into 129, Cast, or unknown using variant data to allow allele-specific quantification of transcripts; only variant bases sequenced with phred score >20 (on standard 33 offset) were considered for allele calling. Transcript quantification and scatter plot generation were done in SeqMonk (http://www.bioinformatics.babraham.ac.uk/projects/seqmonk). RNA-seq, RNA polymerase II phospo-S5 (RNAPII-Ser5), Mediator 12 (MED12), SMC1, and NIPBL ChIP-seq data for mouse ES cells were mapped using bowtie and displayed in the University of California at Santa Cruz Genome Browser (Langmead et al. 2009; Guttman et al. 2010; Kagey et al. 2010; Rahl et al. 2010). TS cell RNA-seq data (GSM967643) were from F1 (C57BL/6J × CAST/EiJ) TS cells (Calabrese et al. 2012).
Supplementary Material
Acknowledgments
We thank all of the members of the Mitchell laboratory for helpful discussions, and Janet Rossant and Susannah Varmuza for advice on ES and TS cell maintenance. We also thank the ENCODE Consortium for generating and releasing data to the scientific community. ENCODE data used in this publication were from the Ren laboratory (ENCODE/LICR). This work was supported by the Canadian Institutes of Health Research, the Canada Foundation for Innovation, and the Ontario Ministry of Research and Innovation (operating and infrastructure grants held by J.A.M.). Studentship funding was provided by the Natural Science and Engineering Research Council of Canada (CGSM held by H.Y.Z.) and Ontario Graduate Scholarships held by N.K.D. and H.Y.Z. J.A.M., H.Y.Z., and Y.K. conceived and designed the experiments. H.Y.Z. conducted the 3C analysis. H.Y.Z., F.C., and N.N.M. cloned and analyzed enhancer constructs in luciferase assays. Y.K. and J.A.M. conducted the CRISPR deletion analysis and conducted the RT-qPCR on deleted clones. M.S. conducted the EB formation assay and sequenced deleted clones. N.K.D. conducted the immunofluorescence and immunoblot analysis. S.D. analyzed ChIP-seq and RNA-seq data. H.Y.Z. and J.A.M. wrote the manuscript. All authors participated in critical review of the manuscript.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.248526.114.
References
- Adachi K, Nikaido I, Ohta H, Ohtsuka S, Ura H, Kadota M, Wakayama T, Ueda HR, Niwa H. 2013. Context-dependent wiring of Sox2 regulatory networks for self-renewal of embryonic and trophoblast stem cells. Mol Cell 52: 380–392. [DOI] [PubMed] [Google Scholar]
- Attanasio C, Nord AS, Zhu Y, Blow MJ, Li Z, Liberton DK, Morrison H, Plajzer-Frick I, Holt A, Hosseini R, et al. 2013. Fine tuning of craniofacial morphology by distant-acting enhancers. Science 342: 1241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. 2003. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev 17: 126–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. 2012. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell 150: 1209–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese JM, Sun W, Song L, Mugford JW, Williams L, Yee D, Starmer J, Mieczkowski P, Crawford GE, Magnuson T. 2012. Site-specific silencing of regulatory elements as a mechanism of X inactivation. Cell 151: 951–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. 2002. Long-range chromatin regulatory interactions in vivo. Nat Genet 32: 623–626. [DOI] [PubMed] [Google Scholar]
- Cavallaro M, Mariani J, Lancini C, Latorre E, Caccia R, Gullo F, Valotta M, DeBiasi S, Spinardi L, Ronchi A, et al. 2008. Impaired generation of mature neurons by neural stem cells from hypomorphic Sox2 mutants. Development 135: 541–557. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117. [DOI] [PubMed] [Google Scholar]
- Chen CY, Morris Q, Mitchell JA. 2012a. Enhancer identification in mouse embryonic stem cells using integrative modeling of chromatin and genomic features. BMC Genomics 13: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Tian Y, Shu W, Bo X, Wang S. 2012b. Comprehensive identification and annotation of cell type-specific and ubiquitous CTCF-binding sites in the human genome. PLoS ONE 7: e41374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. 2002. Capturing chromosome conformation. Science 295: 1306–1311. [DOI] [PubMed] [Google Scholar]
- Ding Q, Regan SN, Xia Y, Oostrom LA, Cowan CA, Musunuru K. 2013. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell 12: 393–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro R, Valotta M, Ferri AL, Latorre E, Mariani J, Giachino C, Lancini C, Tosetti V, Ottolenghi S, Taylor V, et al. 2009. Hippocampal development and neural stem cell maintenance require Sox2-dependent regulation of Shh. Nat Neurosci 12: 1248–1256. [DOI] [PubMed] [Google Scholar]
- Ferrai C, Pombo A. 2009. 3D chromatin regulation of Sonic hedgehog in the limb buds. Dev Cell 16: 9–11. [DOI] [PubMed] [Google Scholar]
- Ferri AL, Cavallaro M, Braida D, Di Cristofano A, Canta A, Vezzani A, Ottolenghi S, Pandolfi PP, Sala M, DeBiasi S, et al. 2004. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development 131: 3805–3819. [DOI] [PubMed] [Google Scholar]
- Fiering S, Epner E, Robinson K, Zhuang Y, Telling A, Hu M, Martin DI, Enver T, Ley TJ, Groudine M. 1995. Targeted deletion of 5′HS2 of the murine β-globin LCR reveals that it is not essential for proper regulation of the β-globin locus. Genes Dev 9: 2203–2213. [DOI] [PubMed] [Google Scholar]
- Golipour A, David L, Liu Y, Jayakumaran G, Hirsch CL, Trcka D, Wrana JL. 2012. A late transition in somatic cell reprogramming requires regulators distinct from the pluripotency network. Cell Stem Cell 11: 769–782. [DOI] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. 2010. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 28: 503–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, et al. 2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477: 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keramari M, Razavi J, Ingman KA, Patsch C, Edenhofer F, Ward CM, Kimber SJ. 2010. Sox2 is essential for formation of trophectoderm in the preimplantation embryo. PLoS ONE 5: e13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer-Kwon KR, Tang Z, Mathe E, Qian J, Sung MH, Li G, Resch W, Baek S, Pruett N, Grontved L, et al. 2013. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell 155: 1507–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp JL, Ormsbee BD, Desler M, Rizzino A. 2008. Small increases in the level of Sox2 trigger the differentiation of mouse embryonic stem cells. Stem Cells 26: 903–911. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice LA, Heaney SJ, Purdie LA, Li L, de Beer P, Oostra BA, Goode D, Elgar G, Hill RE, de Graaff E. 2003. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet 12: 1725–1735. [DOI] [PubMed] [Google Scholar]
- Loh KM, Lim B. 2011. A precarious balance: pluripotency factors as lineage specifiers. Cell Stem Cell 8: 363–369. [DOI] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. 2006. Interchromosomal interactions and olfactory receptor choice. Cell 126: 403–413. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, et al. 2007. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol 9: 625–635. [DOI] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et al. 2012. Systematic localization of common disease-associated variation in regulatory DNA. Science 337: 1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miri K, Latham K, Panning B, Zhong Z, Andersen A, Varmuza S. 2013. The imprinted polycomb group gene Sfmbt2 is required for trophoblast maintenance and placenta development. Development 140: 4480–4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi S, Saito T, Mizutani K, Masuyama N, Gotoh Y, Iwama A, Nakauchi H, Masui S, Niwa H, Nishimoto M, et al. 2004. The Sox-2 regulatory regions display their activities in two distinct types of multipotent stem cells. Mol Cell Biol 24: 4207–4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlynarczyk-Evans S, Royce-Tolland M, Alexander MK, Andersen AA, Kalantry S, Gribnau J, Panning B. 2006. X chromosomes alternate between two states prior to random X-inactivation. PLoS Biol 4: e159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y, Lee JT. 2003. Xite, X-inactivation intergenic transcription elements that regulate the probability of choice. Mol Cell 11: 731–743. [DOI] [PubMed] [Google Scholar]
- Ohnuki Y, Kurosawa H. 2013. Effects of hanging drop culture conditions on embryoid body formation and neuronal cell differentiation using mouse embryonic stem cells: optimization of culture conditions for the formation of well-controlled embryoid bodies. J Biosci Bioeng 115: 571–574. [DOI] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. 2009. CTCF: master weaver of the genome. Cell 137: 1194–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong CT, Hookway TA, Guo C, Sun Y, et al. 2013. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 153: 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radzisheuskaya A, Chia Gle B, dos Santos RL, Theunissen TW, Castro LF, Nichols J, Silva JC. 2013. A defined Oct4 level governs cell state transitions of pluripotency entry and differentiation into all embryonic lineages. Nat Cell Biol 15: 579–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. 2010. c-Myc regulates transcriptional pause release. Cell 141: 432–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagai T, Hosoya M, Mizushina Y, Tamura M, Shiroishi T. 2005. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development 132: 797–803. [DOI] [PubMed] [Google Scholar]
- Sanchez-Castillo M, Ruau D, Wilkinson AC, Ng FS, Hannah R, Diamanti E, Lombard P, Wilson NK, Gottgens B. 2014. CODEX: a next-generation sequencing experiment database for the haematopoietic and embryonic stem cell communities. Nucleic Acids Res doi: 10.1093/nar/gku895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J. 2012. The long-range interaction landscape of gene promoters. Nature 489: 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. 2012. A map of the cis-regulatory sequences in the mouse genome. Nature 488: 116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos JA, Snyder M, Hardison R, Ren B, Gingeras T, Gilbert DM, Groudine M, Bender M, Kaul R, Canfield T, et al. 2012. An encyclopedia of mouse DNA elements (mouse ENCODE). Genome Biol 13: 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676. [DOI] [PubMed] [Google Scholar]
- Tanaka S. 2006. Derivation and culture of mouse trophoblast stem cells in vitro. Methods Mol Biol 329: 35–44. [DOI] [PubMed] [Google Scholar]
- Thomson M, Liu SJ, Zou LN, Smith Z, Meissner A, Ramanathan S. 2011. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell 145: 875–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. 2002. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol Cell 10: 1453–1465. [DOI] [PubMed] [Google Scholar]
- Tomioka M, Nishimoto M, Miyagi S, Katayanagi T, Fukui N, Niwa H, Muramatsu M, Okuda A. 2002. Identification of Sox-2 regulatory region which is under the control of Oct-3/4-Sox-2 complex. Nucleic Acids Res 30: 3202–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuan DY, Solomon WB, London IM, Lee DP. 1989. An erythroid-specific, developmental-stage-independent enhancer far upstream of the human ‘β-like globin’ genes. Proc Natl Acad Sci 86: 2554–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. 2007. VISTA enhancer browser: a database of tissue-specific human enhancers. Nucleic Acids Res 35: D88–D92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457: 854–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Oron E, Nelson B, Razis S, Ivanova N. 2012. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell 10: 440–454. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. 2013. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153: 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. 2008. The ground state of embryonic stem cell self-renewal. Nature 453: 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappone MV, Galli R, Catena R, Meani N, De Biasi S, Mattei E, Tiveron C, Vescovi AL, Lovell-Badge R, Ottolenghi S, et al. 2000. Sox2 regulatory sequences direct expression of a β-geo transgene to telencephalic neural stem cells and precursors of the mouse embryo, revealing regionalization of gene expression in CNS stem cells. Development 127: 2367–2382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.