Histone chaperone HIRA deposits variant histone H3.3 and histone H4 into chromatin in a DNA replication-independent manner. Rai et al. show that nonproliferating senescent cells express and incorporate histone H3.3 and other canonical core histones into a dynamic chromatin landscape, and this depends on HIRA. In senescent cells, but not proliferating cells, promoters of active genes are exceptionally enriched in H4K16ac, and HIRA is required for retention of H4K16ac. HIRA is also required for retention of H4K16ac in vivo and suppression of oncogene-induced neoplasia.
Keywords: chromatin, senescence, HIRA, H4K16ac, dynamic, tumor suppression
Abstract
Cellular senescence is a stable proliferation arrest that suppresses tumorigenesis. Cellular senescence and associated tumor suppression depend on control of chromatin. Histone chaperone HIRA deposits variant histone H3.3 and histone H4 into chromatin in a DNA replication-independent manner. Appropriately for a DNA replication-independent chaperone, HIRA is involved in control of chromatin in nonproliferating senescent cells, although its role is poorly defined. Here, we show that nonproliferating senescent cells express and incorporate histone H3.3 and other canonical core histones into a dynamic chromatin landscape. Expression of canonical histones is linked to alternative mRNA splicing to eliminate signals that confer mRNA instability in nonproliferating cells. Deposition of newly synthesized histones H3.3 and H4 into chromatin of senescent cells depends on HIRA. HIRA and newly deposited H3.3 colocalize at promoters of expressed genes, partially redistributing between proliferating and senescent cells to parallel changes in expression. In senescent cells, but not proliferating cells, promoters of active genes are exceptionally enriched in H4K16ac, and HIRA is required for retention of H4K16ac. HIRA is also required for retention of H4K16ac in vivo and suppression of oncogene-induced neoplasia. These results show that HIRA controls a specialized, dynamic H4K16ac-decorated chromatin landscape in senescent cells and enforces tumor suppression.
Cellular senescence is a stable proliferation arrest associated with an altered proinflammatory secretory pathway (Salama et al. 2014). In response to acquisition of an activated oncogene, primary human cells enter this proliferation-arrested senescent state (oncogene-induced senescence [OIS]). By imposing proliferation arrest, OIS acts as a tumor suppressor mechanism (Braig et al. 2005; Chen et al. 2005; Collado et al. 2005; Michaloglou et al. 2005). Even in the absence of an activated oncogene, replicative senescence (RS) places an upper limit on the proliferative capacity of normal cells, also blocking tumor formation (Cosme-Blanco et al. 2007; Feldser and Greider 2007). The altered secretory program of senescent cells, comprised of proinflammatory cytokines, chemokines, and matrix proteases (the senescence-associated secretory phenotype [SASP]) (Krtolica et al. 2001; Acosta et al. 2008; Kuilman et al. 2008), can also contribute to tumor suppression by promoting clearance of some senescent cells by the immune system (Xue et al. 2007; Kang et al. 2011; Lujambio et al. 2013).
Extensive chromatin changes are apparent in senescent cells, most obviously in the form of domains of compacted heterochromatin called senescence-associated heterochromatin foci (SAHF) (Narita et al. 2003, 2006; Braig et al. 2005; Zhang et al. 2005; O’Sullivan et al. 2010; Chandra et al. 2012; Cruickshanks et al. 2013; De Cecco et al. 2013; Sadaie et al. 2013; Shah et al. 2013). These and other chromatin changes are thought to contribute to the onset and maintenance of viable senescence-associated proliferation arrest (Braig et al. 2005; Narita et al. 2006; Di Micco et al. 2011; Benhamed et al. 2012; Cruickshanks et al. 2013; Martin et al. 2013). Chromatin changes likely also contribute to activation of the SASP in senescent cells (Shah et al. 2013). Subunits of the SWI/SNF ATP-dependent chromatin remodeling complex are targets of mutation and inactivation in cancer (Wilson and Roberts 2011), and SWI/SNF is thought to contribute to tumor suppression in part through induction of senescence (Chai et al. 2005). In sum, several lines of evidence indicate that chromatin of senescent cells contributes to tumor suppression (Braig et al. 2005; Narita et al. 2006). However, the molecular basis of this is poorly understood.
Specifically, SAHF have been suggested to contribute to stable repression of proliferation genes and/or suppression of DNA damage signaling in senescent cells (Narita et al. 2003; Zhang et al. 2007; Di Micco et al. 2011). Either way, the apparent heterochromatinization of senescent cells intuitively suggests that their compacted, stable chromatin contributes to a senescence-mediated barrier to tumor progression. In line with this view of relatively static chromatin in senescent cells, compared with proliferating cells, senescent cells synthesize less total histone H3 and H4 (O’Sullivan et al. 2010). In fact, in mammalian cells, the canonical DNA replication-dependent histones—H2A, H2B, H3 (H3.1 and H3.2), and H4—are each coded for by 10–15 genes located mostly in clusters on chromosomes 1 and 6, and the corresponding mRNAs are typically unstable in nonproliferating cells (Marzluff et al. 2008). This is again consistent with the view that chromatin in nonproliferating senescent cells is a relatively fixed entity, intuitively suggestive of an immobile barrier to tumor progression. However, the extent of chromatin dynamics in senescent cells has not been properly investigated.
Histone variant H3.3 contributes to nucleosome destabilization (Jin and Felsenfeld 2007) and so is thought to facilitate nucleosome dynamics associated with transcription activation and ongoing transcription. Histone H3.3 is enriched at nucleosomes at transcription start sites (TSSs) of genes and at enhancers and gene bodies of actively transcribed genes (Ahmad and Henikoff 2002; Jin et al. 2009; Goldberg et al. 2010). H3.3 differs from canonical histones H3.1 and H3.2 by four or five amino acids and is expressed constitutively throughout the cell cycle and in proliferating and nonproliferating cells from two genes: H3F3A and H3F3B (Skene and Henikoff 2013). The HIRA chaperone complex, comprised of HIRA/UBN1/CABIN1, collaborates with histone-binding protein ASF1a to incorporate H3.3 into chromatin in a DNA replication-independent manner (Ray-Gallet et al. 2002; Tagami et al. 2004; Loppin et al. 2005). The HIRA protein is required for deposition of histone H3.3 at active and poised genes and enhancers (Goldberg et al. 2010; Ray-Gallet et al. 2011; Pchelintsev et al. 2013). Accordingly, HIRA is required for gene activation in some contexts (Placek et al. 2009; Dutta et al. 2010; Yang et al. 2011) as well as recruitment and function of polycomb complexes at gene promoters and dynamic restoration of chromatin after DNA damage repair (Adam et al. 2013; Banaszynski et al. 2013). On the other hand, the HIRA complex and its orthologs, together with histone H3.3, are also involved in chromatin silencing (Sherwood et al. 1993; van der Heijden et al. 2007). Importantly, several recent studies have shown histone H3.3 to be a target of recurrent missense mutations in human cancer, and transcription-coupled methylation of histone H3.3 is also thought to be tumor-suppressive (Schwartzentruber et al. 2012; Wu et al. 2012; Wen et al. 2014; Zhu et al. 2014). Appropriately for a DNA replication-independent chaperone, HIRA is also involved in control of chromatin in nonproliferating senescent cells (Zhang et al. 2005; Ye et al. 2007; Banumathy et al. 2009; Rai et al. 2011; Duarte et al. 2014). However, HIRA’s function in senescent cells is poorly defined, and there is currently no direct evidence for a role in tumor suppression.
Here we set out to better understand dynamic chromatin regulation in nonproliferating senescent cells and the role of HIRA in this process. We reveal chromatin in senescent cells to be a dynamic landscape linked to noncanonical regulation of histone mRNAs and exceptional marking by H4K16ac. The histone chaperone HIRA is required for regulation of this landscape and also for suppression of oncogene-induced neoplasia in a mouse model. This study points to shared roles of HIRA, histone H3.3, and H4K16ac in both cellular senescence and tumor suppression.
Results
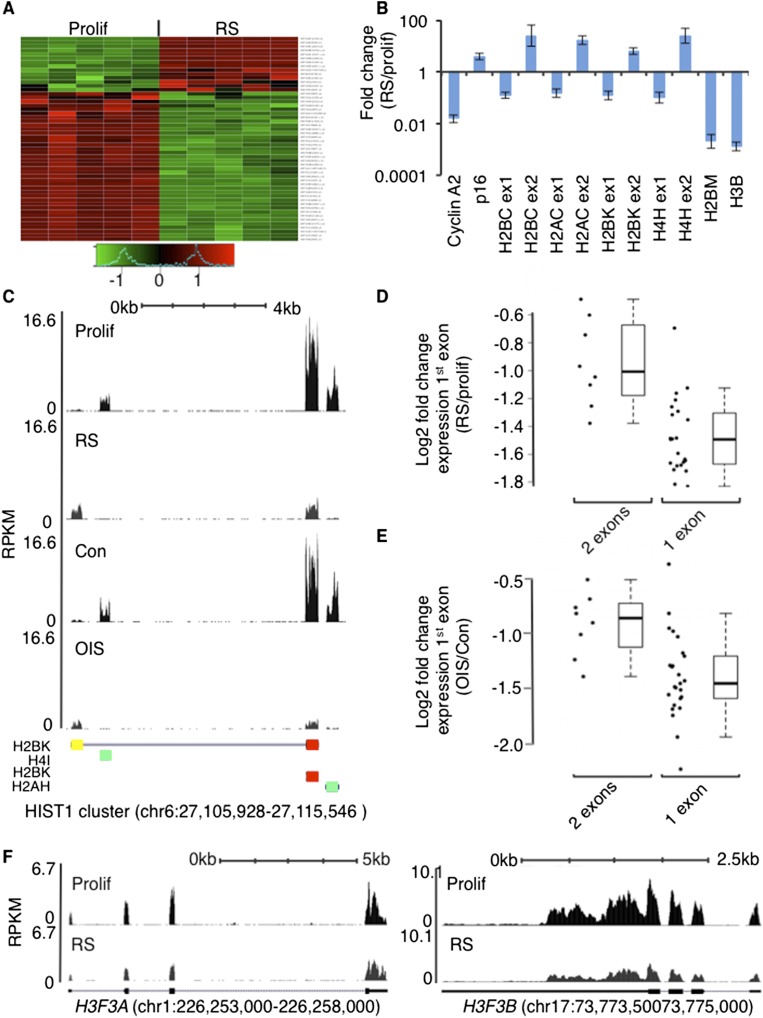
The mRNAs encoding the canonical DNA replication-dependent histones are generally regarded as unstable in nonproliferating cells (Marzluff et al. 2008). Moreover, compared with proliferating cells, senescent cells synthesize less total histone H3 and H4 (O’Sullivan et al. 2010). We were surprised, therefore, when gene expression microarray revealed that a subset of histone mRNAs was apparently up-regulated in RS cells (senescence confirmed by standard markers of senescence) (Fig. 1A; Supplemental Data Set 1; Supplemental Fig. 1A,B). Close examination showed that these are typically canonical DNA replication-dependent histones from the HIST1 cluster on chromosome 6 but are also annotated in Ensembl (hg19) to contain a noncoding second exon, and the Affymetrix probes target this second exon (Supplemental Data Set 2; Supplemental Fig. 1C). This second exon lacks the specialized stem–loop that terminates the canonical replication-dependent histone mRNAs to confer instability in nonproliferating cells (Marzluff et al. 2008) but instead directs mRNA polyadenylation. In both RS and OIS cells, we confirmed up-regulation of the second exon of several of these noncanonical histone mRNAs (and two others that lacked second exon probes on the array, HIST4H4 and HIST1H2BN) by quantitative RT–PCR (qRT–PCR) and RNA sequencing (RNA-seq) (Fig. 1B,C; Supplemental Data Sets 2, 3). Accumulation of these alternative spliced polyadenylated histone transcripts has been previously reported in some cancer cell lines after treatment with ionizing radiation and on differentiation of mesenchymal progenitor cells (Kari et al. 2013). Moreover, we observed up-regulation of the second exon in quiescent cells (Supplemental Fig. 1D), suggesting that accumulation of alternative spliced polyadenylated histone mRNAs occurs in other nonproliferating cells as well as senescent cells.
Figure 1.
Expression of histone mRNAs in senescent cells linked to alternative mRNA splicing. (A) Expression of multiple histone mRNAs assessed by Affymetrix microarray in proliferating (Prolif) and RS cells (five independent replicates of each). Gene expression is scaled for each gene individually, and resulting intensities are plotted between the green (low) and red (high) scale. (B) Fold change in abundance of first (coding) and second (noncoding, poly A) exons of selected HIST1 cluster mRNAs between proliferating and RS cells, determined by qRT–PCR. HIST1H2BM and HIST1H3B (coding for canonical histone H2B and H3.1, respectively) are expressed only as single-exon stem–loop mRNAs. Values are means ± standard deviation of three independent experiments normalized to GAPDH as a housekeeping control. (C) Exon read counts (reads per kilobase of exon model per million mapped reads [RPKM]) of single-exon (HIST1H4I and HIST1H2AH; green) and two-exon (HIST1H2BK; first coding exon in red and second noncoding exon in yellow) histone mRNAs in proliferating, RS, control (Con), and OIS cells. (D) Fold change in abundance of first coding exon between proliferating and RS cells, comparing single-exon, stem–loop, and no poly A mRNAs and those mRNAs with a poly A second exon whose expression increases in RS, as determined by RNA-seq. See Supplemental Data Set 3 for gene lists. P-value = 0.0002016. (E) As in D, but analysis performed in OIS. See Supplemental Data Set 3 for gene lists. P-value = 0.001837. (F) Expression of H3F3A and H3F3B genes assessed by RNA-seq in proliferating and RS cells.
RNA-seq also confirmed splicing of the first coding exon to the second exon in senescent cells, based on some reads spanning the two exons (Supplemental Fig. 1E). Significantly, qRT–PCR and RNA-seq data showed that the first exon of histone mRNAs spliced to a polyadenylated noncoding second exon was down-regulated less in senescence than canonical single coding exon stem–loop histone mRNAs, such as HIST1H2BM and HIST1H3B (Fig. 1B,D,E; Supplemental Data Set 3). Interestingly, the eight alternative-spliced histones include genes encoding the H2A, H2B, and H4 subtypes but no H3 gene. However, histone variant H3.3 is already known to be expressed in nonproliferating cells (Urban and Zweidler 1983; Grove and Zweidler 1984; Pantazis and Bonner 1984; Brown et al. 1985; Skene and Henikoff 2013). Indeed, substantial expression of genes encoding DNA replication-independent histone variant H3.3—H3F3A and H3F3B—was retained in RS and OIS cells (Fig. 1F; Supplemental Fig. 1F). This suggests that the alternative-spliced mRNAs coding for the canonical H2A, H2B, and H4 subtypes are counterparts of histone variant H3.3 (Supplemental Data Sets 2, 3). We conclude that senescent cells retain expression of a small subset of the >40 canonical histone genes whose expression is typically classed as replication-dependent, linked to alternative mRNA splicing to bypass a signal known to direct mRNA degradation in nonproliferating cells.
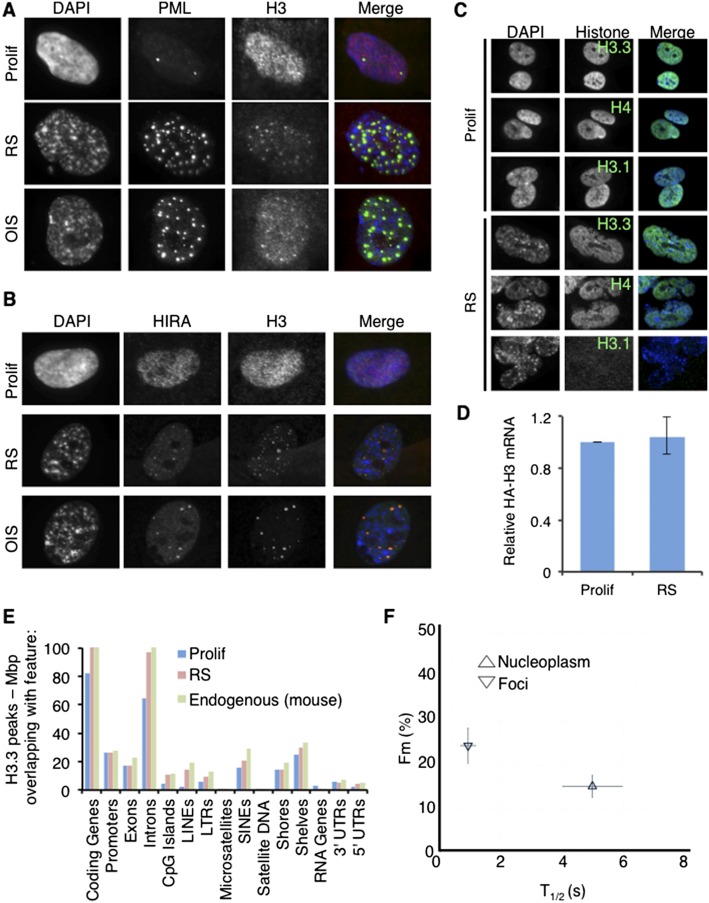
Next, we investigated whether newly synthesized histones are deposited into chromatin in senescent cells. Previous studies have indicated that recruitment of histone chaperones and histones to PML bodies is indicative of trafficking of newly synthesized histones into chromatin (Chang et al. 2013; Delbarre et al. 2013; Corpet et al. 2014). Consistent with ongoing histone deposition and nucleosome assembly in nonproliferating senescent cells, histone H3 was enriched in PML bodies of RS and OIS cells, where it colocalized with DNA replication-independent histone chaperone HIRA (Fig. 2A,B). While ectopically expressed epitope-tagged histone H3.3 was not detectably cell cycle inhibitory or toxic to proliferating or RS cells (Supplemental Fig. 2A), ectopically expressed H3.3 and H4 were incorporated into detergent-insoluble nuclear chromatin of senescent cells, directly demonstrating that newly synthesized histones can be incorporated into chromatin in senescent cells (Fig. 2C; Supplemental Fig. 2B). In contrast, DNA replication-dependent histone variant H3.1 was incorporated into chromatin of proliferating cells, but not senescent cells, despite comparable expression of ectopically expressed H3.1 mRNA between proliferating and senescent cells (Fig. 2C,D). Chromatin immunoprecipitation (ChIP) combined with sequencing (ChIP-seq) of ectopically expressed HA-tagged histone H3.3 in proliferating and RS cells confirmed that its distribution between different genome features was comparable with endogenous H3.3, as previously determined by Allis and coworkers (Goldberg et al. 2010) (Fig. 2E; Supplemental Table 1). Incorporation of ectopically expressed histones into chromatin implies a level of histone exchange in the chromatin of these senescent cells. Consistent with this idea, fluorescence recovery after photobleaching (FRAP) showed that a proportion of ectopically expressed nuclear GFP-H3.3 recovered from photobleaching within seconds (Fig. 2F). Together, these analyses of endogenous and ectopic histones suggest that histones H3.3 and H4 dynamically exchange at their physiological sites of incorporation in senescent cells.
Figure 2.
Dynamic incorporation of histones into chromatin of senescent cells. (A) Proliferating (Prolif), RS, and OIS cells fluorescently stained with antibodies to PML and histone H3. (B) Proliferating, RS, and OIS cells fluorescently stained with antibodies to HIRA and histone H3. (C) RS cells were infected with lentivirus encoding HA-histone H4, HA-histone H3.3, and HA-histone H3.1 and fluorescently stained with anti-HA. (D) Cells from C were analyzed for mRNA abundance of HA-histone H3.1 by qRT–PCR. The bar chart displays the mean HA-H3.1 mRNA abundance in senescent cells compared with proliferating cells, normalized to GAPDH as a housekeeping control. Standard error from three independent experiments. (E) Percentage of base pairs covered by H3.3 peaks that overlap with base pairs of specified genomic features for human proliferating and RS cells expressing ectopic HA-tagged H3.3 and mouse embryonic stem cells expressing endogenous HA-tagged H3.3. (F) The percentage of mobile fraction (Fm) and half-time of recovery in seconds (T1/2) for histones located in GFP-H3.3 foci (likely PML bodies, based on A,B) and nucleoplasm. Error bars represent ±SEM from three independent experiments.
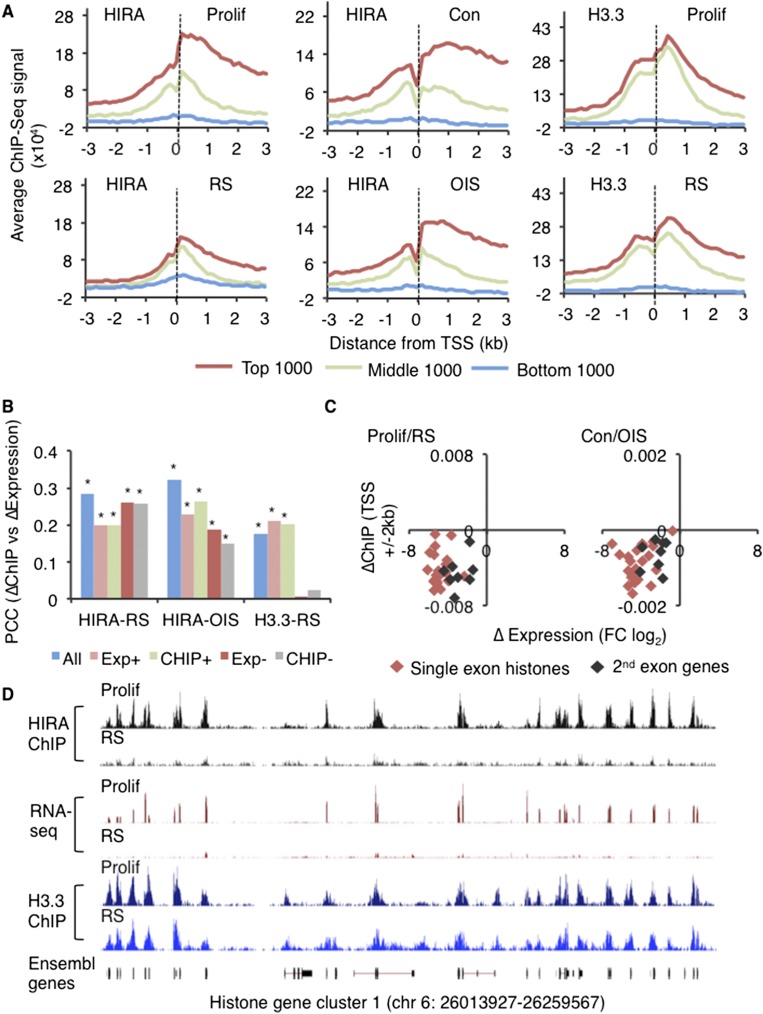
Given HIRA’s role in DNA replication-independent histone H3.3 deposition, we set out to compare the genomic distribution of HIRA and newly synthesized histone H3.3 in proliferating and senescent cells. HIRA ChIP-seq was performed on chromatin from proliferating, RS, and OIS (and control) cells. Comparison of the HIRA and histone H3.3 genome distribution from proliferating and RS cells confirmed a substantial and highly significant overlap of HIRA and H3.3 in both proliferating and senescent cells (Supplemental Tables 1, 2). Consequently, initial analyses were performed on the intersect of the HIRA and H3.3 data sets (Supplemental Table 3). Consistent with previous analyses, a large proportion of HIRA/H3.3 peaks localized at and near coding regions (Supplemental Fig. 3A; Ahmad and Henikoff 2002; Jin et al. 2009; Goldberg et al. 2010; Pchelintsev et al. 2013). In both proliferating and RS cells, HIRA/H3.3 peaks localized to ∼10,000 coding regions (Supplemental Fig. 3B). Specific enrichment was observed at CpG islands, promoters, exons, and 5′ untranslated regions (UTRs) (Supplemental Fig. 3C). At promoters, HIRA and H3.3 were both most enriched at the nucleosomes flanking the TSS, and for HIRA in proliferating, RS, control, and OIS cells and H3.3 in proliferating and RS cells, binding correlated strongly with gene expression, as determined by RNA-seq (Fig. 3A; Supplemental Fig. 3D; Supplemental Table 4).
Figure 3.
Genome distribution of HIRA and histone H3.3 between proliferating and senescent cells is regulated in parallel with gene expression. (A) HIRA and H3.3 enrichment profiles around (±3 kb) the TSSs of the top (red), middle (green), and bottom (blue) 1000 coding genes ordered by expression for proliferating (Prolif), RS, control (Con), and OIS cells. ChIP-seq signal was normalized to input. (B) At significantly changing coding genes (Benjamini and Hochberg false discover rate 5%), the Pearson correlation coefficients (PCCs) between the difference in ChIP-seq signal and the difference in expression. ChIP-seq signals were calculated for promoter regions (TSS ±2 kb) and normalized to input. Differences in expression were calculated as fold change (FC; log2). Differences were calculated from proliferating to senescent cells (RS or OIS, as indicated). The X-axis denotes the ChIP-seq data set used to generate the PCCs. Bar colors indicate the gene sets used for the comparison: all genes (All) and genes that gain expression (Exp+), lose expression (Exp−), gain ChIP-seq signal (ChIP+), and lose ChIP-seq signal (ChIP−). An asterisk indicates significant correlation at P < 0.05. (C) Scatter plot of single-exon, stem–loop, no poly A (red diamonds), and two-exon poly A (black diamonds) histone genes (Supplemental Data Set 3) showing the difference in input normalized ChIP-seq signal (TSS ± 2 kb) versus the difference in expression (fold change log2) for proliferating to RS cells and control to OIS cells. (D) Genome browser representation of HIRA, ribodepleted RNA, and H3.3 in proliferating and RS IMR90 cells at histone gene cluster one (HIST1).
Since proliferating and senescent cells exhibit distinct programs of gene expression, we asked whether changes in gene expression between proliferating and senescent cells correlated with change in HIRA and H3.3 binding. For HIRA, a change in gene expression, either up or down, directly correlated with a change in HIRA binding measured by ChIP (Fig. 3B) in both OIS and RS. Similarly, for histone H3.3, an increase in expression directly correlated with an increase in binding in RS (Fig. 3B; Supplemental Fig. 3E). Notably, 1919 genes acquired both HIRA and H3.3 in senescent cells, and these are enriched in genes related to TNF signaling, a component of the SASP (Supplemental Fig. 3B; Supplemental Data Set 4; Coppe et al. 2008). These correlations are also illustrated by a comparison of the 250 most up-regulated and down-regulated genes (Supplemental Fig. 3E). However, across all genes, a decrease in expression in senescence did not correlate with decreased H3.3 binding, suggesting that genes that are repressed on the transition from proliferating to RS cells continue to incorporate histone H3.3 (Fig. 3B; Supplemental Fig. 3E). These phenomena were most marked at histone genes. Decreased expression of the DNA replication-dependent histones in senescent cells was accompanied by a marked decrease in HIRA binding in both RS and OIS (Fig. 3C,D). However, H3.3 did not substantially decrease at these genes in senescent cells (Fig. 3D). In sum, HIRA and newly synthesized H3.3 are localized to promoters of expressed genes in proliferating and senescent cells and are partially relocalized between proliferating and senescent cells, in line with changes in gene expression—most notably in the case of HIRA at DNA replication-dependent histone genes.
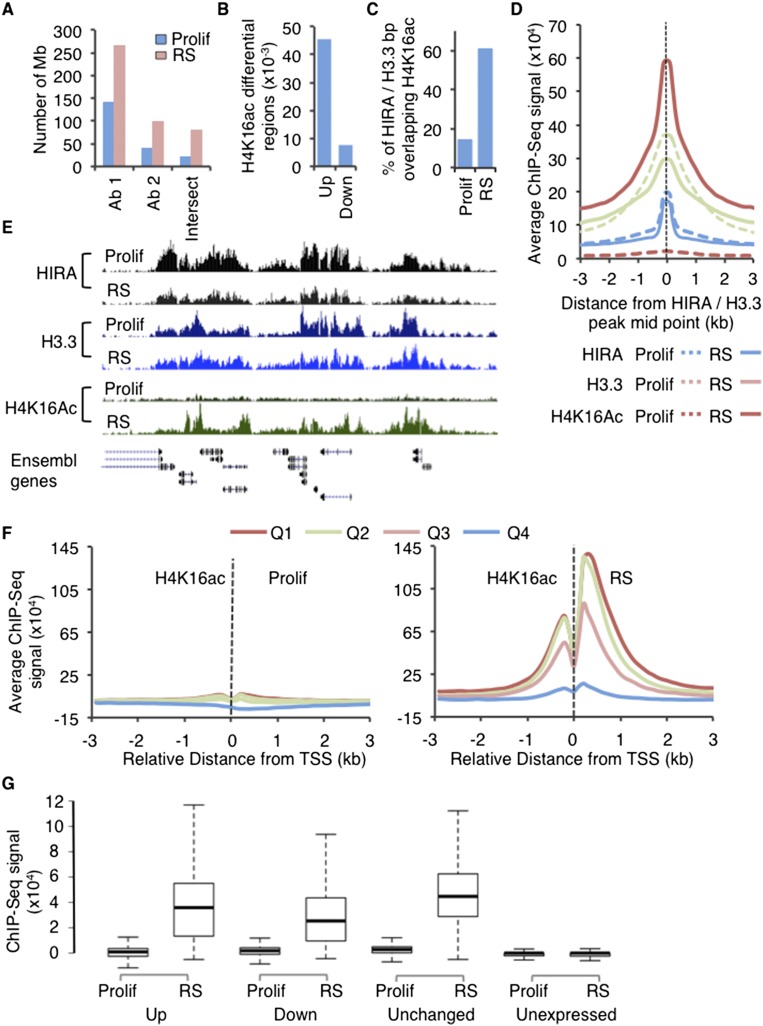
A recent study established a link between histone H3.3 and H4K16ac (Lin et al. 2013). Interestingly, H4K16ac also has candidate tumor suppressor activity (Fraga et al. 2005; Elsheikh et al. 2009). Therefore, to investigate this link between HIRA/H3.3 and H4K16ac in tumor-suppressive senescent cells, we performed ChIP-seq of H4K16ac in proliferating and RS cells using two different validated antibodies to H4K16ac. There was a substantial and highly significant overlap between the distribution of H4K16ac detected by the two antibodies, particularly in RS cells (Fig. 4A; Supplemental Tables 1, 5, 6). For downstream analyses, we evaluated the intersect of the two H4K16ac data sets. Qualitatively, the overall distribution of H4K16ac across the genome was similar for proliferating and RS cells and, interestingly, was reminiscent of HIRA and histone H3.3. H4K16ac was most abundant at the coding regions and was enriched at CpG islands, promoters, and 5′ UTRs (cf. Supplemental Figs. 4A,B and 3A,C). However, both antibodies reported a twofold to threefold increase in the number of H4K16ac peaks and the number of base pairs covered in RS cells (Fig. 4A; Supplemental Tables 5, 6). Indeed, ∼45,000 regions showed an increase in H4K16ac in RS, while <10,000 pre-existing peaks showed a decrease in RS (Fig. 4B). On a larger scale, in RS cells, H4K16ac was depleted from late replicating regions of the genome and SAHF (Supplemental Fig. 4C,D), in line with incorporation of late replicating regions into compacted SAHF, H4K16ac’s ability to inhibit chromatin compaction, and a previous report correlating global decreased H4K16ac to the formation of SAHF (Shogren-Knaak et al. 2006; Chandra et al. 2012; Contrepois et al. 2012).
Figure 4.
HIRA controls H4K16ac, a marker of expressed genes in senescent cells. (A) The amount of genome (in megabases) covered by H4K16ac peaks in proliferating and RS cells, as determined by antibody 1 (Ab1), Ab2, and the intersection between the two Abs. (B) The number of peaks that gain (up) or lose (down) H4K16ac in RS cells compared with proliferating cells. (C) The percentage of base pairs covered by both HIRA and H3.3 peaks that overlap with base pairs covered by H4K16ac peaks for proliferating and RS cells. (D) HIRA, H3.3, and H4K16ac enrichment profiles centered on the midpoint of intersected HIRA and H3.3 peaks (±3 kb). The ChIP-seq signal was normalized to input (HIRA, H3.3) or H4 (H4K16ac). (E) Genome browser representative region (Chr11: 65,602,592–65,707,036, hg19) showing HIRA, H3.3, and H4K16ac profiles in proliferating and RS cells. (F) H4K16ac enrichment profiles around (±3 kb) the TSSs of genes separated into quartiles according to expression in proliferating (left) and RS (right) (from high to low expression [Q1–Q4]). ChIP-seq signal was normalized to H4. (G) Box plots showing H4K16ac normalized to H4 ChIP-seq at promoters of up-regulated (up), down-regulated (down), unchanged, and unexpressed coding genes in RS cells compared with proliferating cells. The bottom and top of the boxes correspond to the 25th and 75th percentiles, respectively, and the internal band is the median. The plot whiskers correspond to the most extreme value within 1.5× interquartile range.
There was a marked increase in overlap of HIRA/H3.3 and H4K16ac in RS cells (Fig. 4C), and H4K16ac peaks acquired in RS mirrored HIRA/H3.3 peaks (Fig. 4D), indicating a substantial increase in H4K16ac in RS at regions bound by HIRA/H3.3. At H4K16ac peaks unique to RS cells, the HIRA and H4K16ac enrichments by ChIP-seq correlated strongly (Pearson correlation coefficient [PCC] = 0.64). Indeed, in RS cells but much less so in proliferating cells, H4K16ac was enriched at early replicating gene-rich regions (Fig. 4E; Supplemental Fig. 4C), particularly on either side of the TSS (Fig. 4F; Supplemental Fig. 4E). In senescent cells, ∼12,500 genes were bound by H4K16ac, compared with <5000 genes in proliferating cells (Supplemental Fig. 4F). Remarkably, in RS cells, H4K16ac was enriched at all but the lowest expressed genes (Fig. 4F; Supplemental Fig. 4E,G), and expressed genes gained H4K16ac in RS cells compared with proliferating cells regardless of whether their expression increased, decreased, or did not change (Fig. 4G). These results show that in RS cells, H4K16ac is an especially prominent feature at promoters of expressed genes, where it colocalizes with HIRA and newly synthesized H3.3. However, rather than closely correlating with level of expression, an abundance of H4K16ac registers a binary pattern, either “high” or “low,” linked to expressed or very low expressed/off genes, respectively.
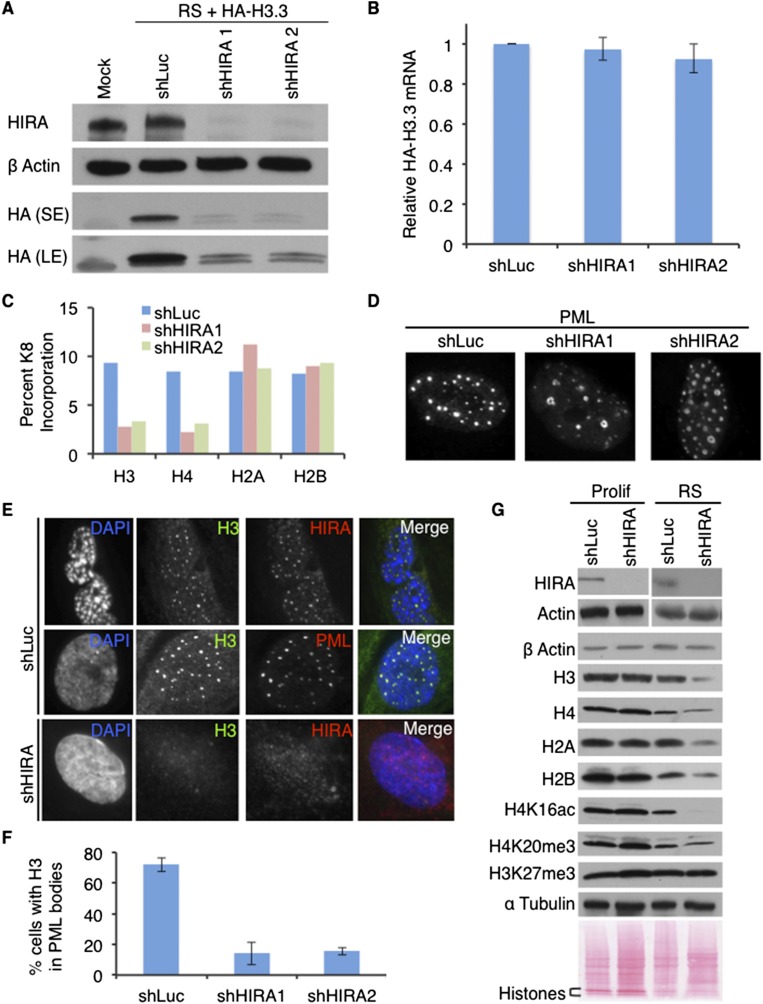
To directly test a requirement for HIRA in the deposition of histones and chromatin control in nonproliferating senescent cells, we ectopically expressed HA-tagged histone H3.3 and H4 in senescent cells infected with a lentivirus encoding a control shRNA or shRNAs to knock down HIRA. HIRA was knocked down in senescent cells using two distinct lentivirus-encoded shRNAs (Fig. 5A). HIRA blocked incorporation of histone H3.3 and H4 into chromatin (Supplemental Fig. 5A,B), but this was apparently largely due to a block to stable production of the histone (Fig. 5A). Since knockdown of HIRA did not affect histone HA-H3.3 mRNA expression (Fig. 5B), binding to the HIRA chaperone is most likely required for stability of the unincorporated histone substrate. We also confirmed by liquid chromatography-mass spectrometry (LC-MS) analysis of histones purified from heavy lysine pulse-labeled cells, either control or HIRA knockdown, that HIRA is required for the incorporation of newly synthesized endogenous histones H3 and H4 into chromatin but not H2A and H2B (Fig. 5C). Furthermore, while knockdown of HIRA did not affect the number of PML bodies in senescent cells (Fig. 5D), it did block recruitment of H3 protein to PML bodies (Fig. 5E,F). Finally, proliferating and RS cells expressing shRNAs to HIRA or a control were Western-blotted to compare the abundance of core histones and a panel of histone modifications. Previously, we and others reported that cells held in a senescent state exhibit a progressively decreased histone content (O’Sullivan et al. 2010; Ivanov et al. 2013), as do aged yeast (Feser et al. 2010). Loss of total core histones was exacerbated in cells lacking HIRA (Fig. 5G; Supplemental Fig. 5C), consistent with a role for HIRA in histone deposition during a cyclical eviction/deposition process. More strikingly, compared with total core histone, cells lacking HIRA showed a marked decrease in H4K16ac but no substantial change in a number of other modifications, including H4K20me3 and H3K27me3 (Fig. 5G; Supplemental Fig. 5C). Together, these results indicate that the HIRA chaperone is required for incorporation of histones H3.3 and H4 into chromatin in senescent cells via a direct role in either histone deposition into chromatin or the stability or trafficking of non-chromatin-bound histones. Moreover, HIRA not only colocalizes with H4K16ac across the genome of senescent cells (Fig. 4) but is required for steady-state abundance of H4K16ac in these cells. Thus, HIRA is critical to control a specialized H4K16ac-decorated chromatin landscape in senescent cells.
Figure 5.
HIRA histone chaperone is required for histone and chromatin control in senescent cells. (A) RS cells were infected with lentivirus encoding HA-histone H3.3 and Western-blotted with the indicated antibodies. Mock-uninfected lysate was used as a control. (SE) Short exposure; (LE) long exposure. (B) Cells from A were analyzed for mRNA abundance of HA-histone H3.3 by qRT–PCR. The bar chart displays the mean HA-H3.3 mRNA abundance in senescent cells compared with proliferating cells normalized to GAPDH as a housekeeping control. Standard error from three independent experiments. (C) Cells infected with the indicated shRNAs were labeled for 48 h with medium containing heavy lysine (K8). Histones were purified by acid extraction and gel purification and digested with trypsin, and the percentage of incorporation of heavy lysine (K8)—of total heavy and light (K0) lysine—in histones was determined by quantitative proteomics. (D) RS cells were infected with lentivirus-encoded shRNAs to HIRA (shHIRA1 and shHIRA2) or control (shLuc) and fluorescently stained with antibodies to PML. (E) RS cells were infected with lentivirus-encoded shRNAs to HIRA (shHIRA) or control (shLuc) and fluorescently stained with antibodies to histone H3, PML, and HIRA. (F) Quantitation of cells with H3 localized to PML bodies from E. (G) Western blots of the indicated proteins in control and shHIRA-infected proliferating and RS cells. (Bottom) Ponceau S was used as an additional loading control.
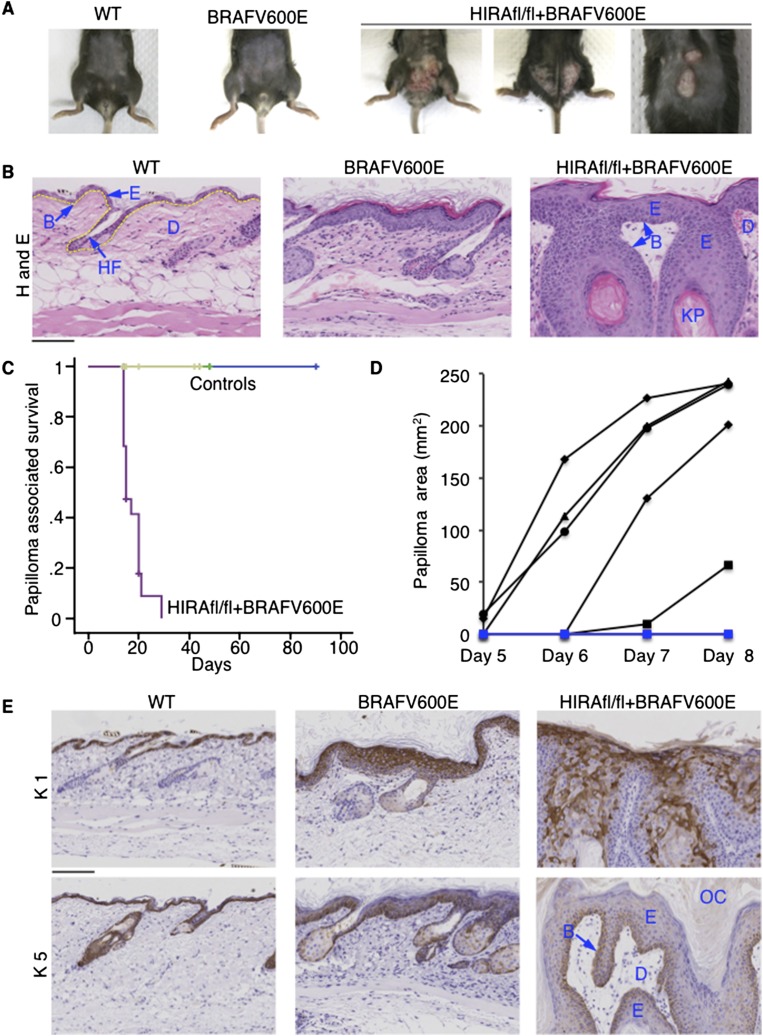
In light of these findings and the known role of senescence as a tumor suppressor mechanism, we wanted to test the requirement for HIRA in suppression of neoplasia in vivo. To do this, we employed a mouse model expressing a tamoxifen/β-naphthoflavone-induced oncogenic Braf allele (LSL-BrafV600E) (Mercer et al. 2005) controlled by an AhCreER promoter/transgene cassette (Kemp et al. 2004). BrafV600E induces senescence in mouse and human tissues, including melanocytes and intestinal epithelium (Michaloglou et al. 2005; Dhomen et al. 2009; Carragher et al. 2010). We also generated a mouse harboring a conditionally inactivated Hira allele (Hirafl.fl) in which Cre-mediated excision of loxP-flanked exons 6 and 7 generates a stop codon in exon 8 (Supplemental Fig. 6A). While this setup directs endogenous levels of expression of BRAFV600E and inactivation of HIRA in a number of tissues, in this study, we focused on one target of the AhCreER cassette: the skin (Clayton et al. 2007). On its own, expression of BRAFV600E induced hyperplasia of the skin epidermis, which was visible on microscopic analysis of H+E-stained tissues but was typically not apparent on examination of the shaved skin (Fig. 6A,B). A small proportion (two out of 11) of these mice developed papillomas (Supplemental Data Set 5). Remarkably, however, all 19 mice expressing activated BRAFV600E and lacking HIRA developed rapidly growing papillomas that reached the maximum permitted size (1.5 cm) within 2–3 wk, necessitating that they be culled (Fig. 6A–D, Supplemental Fig. 6B,C; Supplemental Data Set 5).
Figure 6.
HIRA suppresses oncogene-induced neoplasia. (A) Skin of representative wild-type (WT), AhCreER/LSL-BrafV600E, and AhCreER/LSL-BrafV600E/Hirafl.fl mice approximately 2 wk after tamoxifen treatment. (B) H+E-stained sections of skin from the indicated mice approximately 2 wk after tamoxifen treatment. (B) Basement membrane location (also indicated by dashed yellow line in wild type); (E) epidermis; (D) dermis; (HF) hair follicle; (KP) keratin pearl. (C) Papilloma-associated survival of control and AhCreER/LSL-BrafV600E/Hirafl.fl mice after tamoxifen treatment (N = 19; seven male, 12 female) for AhCreER/LSL-BrafV600E/Hirafl.fl mice. Controls include AhCreER/LSL-BrafV600E (N = 11; three male, eight female; light green), AhCreER/Hirafl.fl (N = 9; five male, four female; dark green), and AhCreER (N = 12; eight male, four female; blue). Mice culled of causes unrelated to papilloma formation were censored from analysis (indicated by “+” on the plot). See Supplemental Data Set 5 for all mouse outcomes. (D) Skin lesion area measured (length × width) at daily intervals after tamoxifen treatment. N = 5 for AhCreER/LSL-BrafV600E/Hirafl.fl (black); N = 2 for AhCreER/LSL-BrafV600E (blue), neither of which developed any skin lesion. In addition, AhCreER/LSL-BrafV600E (N = 2) and AhCreER/LSL-BrafV600E/Hirafl.fl (N = 6) were treated with corn oil and observed for 4 wk. None of the mice developed any lesions or tumors. (E) Skin from mice of the indicated genotypes ∼2 wk after tamoxifen treatment, stained with antibodies to keratin 1 and keratin 5. Histological features labeled as in B. (OC) Outer cornified layer. Bars, 100 μM.
Histological analysis revealed marked hyperplasia in the skin of induced AhCreER/LSL-BrafV600E/Hirafl.fl mice (Fig. 6B; Supplemental Fig. 6B). The epidermal keratinocytes of these mice continued to differentiate, based on overt stratification, a thickened outer cornified layer, and keratin pearls (Fig. 6B; Supplemental Fig. 6B,C). The thickest hyperplastic areas sometimes showed changes in K5 and K1 and cell enlargement. While this suggested that the papillomas could progress to carcinoma, at the age investigated, no invasion, disruption of basement membranes, or other histological signs of carcinoma were observed (Fig. 6B,E; Supplemental Fig. 6B). Inactivation of HIRA on its own had no apparent effect on skin histology or differentiation, at least over the 2- to 3-wk time frame (Supplemental Fig. 6D). In sum, combined activation of BRAFV600E and inactivation of HIRA causes rapid growth of well-differentiated nonmalignant epidermal hyperplasias. We conclude that HIRA is required for efficient suppression of oncogene-induced neoplasia in vivo.
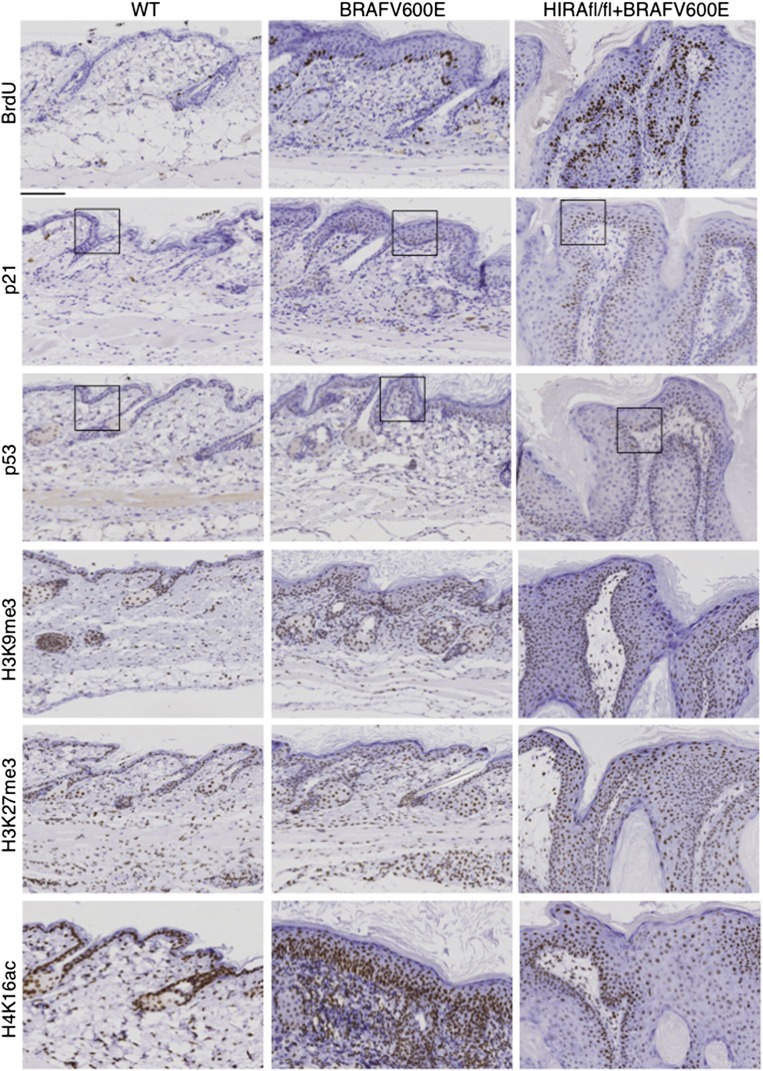
We asked whether HIRA’s role in suppression of neoplasia is linked to its regulation of chromatin structure and senescence. Inactivation of HIRA alone did not obviously affect proliferation or senescence markers (Supplemental Fig. 7A). In control and induced AhCreER/LSL-BrafV600E and AhCreER/LSL-BrafV600E/Hirafl.fl mice, the level of cell proliferation, measured by BrdU incorporation, correlated well with the degree of hyperplasia (Fig. 7). Consistent with the histologically differentiated nature of the tissues, proliferation was largely confined to the basal layer, although this layer was expanded in induced AhCreER/LSL-BrafV600E/Hirafl.fl mice (Fig. 7). Surprisingly, while we observed modest induction of senescence markers p21 and p53 in induced AhCreER/LSL-BrafV600E mice, expression of these proteins was, in regions, more marked in induced AhCreER/LSL-BrafV600E/Hirafl.fl mice (Fig. 7; Supplemental Fig. 7B). Most surprisingly, in mice harboring activated BRAFV600E and inactivation of HIRA, there was a substantial overlap of zones of proliferating cells marked by BrdU and zones of cells expressing the highest levels of p21 and p53, with all three enriched in the basal layer of the epidermis (Fig. 7).
Figure 7.
Suppression of oncogene-induced neoplasia by HIRA is linked to control of H4K16ac. Sections of skin from the indicated mice ∼2 wk after tamoxifen treatment, stained with the indicated antibodies. Boxed areas are expanded in Supplemental Figure 6B. Bars, 100 μM.
Inactivation of HIRA alone had no detectable effect on total histone H3 or histone modifications H3K9me3, H3K27me3, and H4K16ac (Supplemental Fig. 7C, cf. with wild type in Fig. 7 and Supplemental Fig. 7D). However, while inactivation of HIRA in induced AhCreER/LSL-BrafV600E/Hirafl.fl mice did not overtly affect total histone content and H3K9me3 and H3K27me3, papillomas of induced AhCreER/LSL-BrafV600E/Hirafl.fl mice were depleted of H4K16ac, particularly in the nonproliferative regions furthest from the basal layer (Fig. 7; Supplemental Fig. 7D). We conclude that, in the presence of activated BRAFV600E, inactivation of HIRA expands a population of cells that has engaged the p53–p21 senescence effector pathway yet remains in a proliferative state. Moreover, inactivation of HIRA in vivo recapitulates chromatin changes observed on HIRA inactivation in senescent cells in vitro; most notably, decreased H4K16ac.
Discussion
Cellular senescence acts as a barrier to malignant progression of potentially neoplastic cells (Salama et al. 2014), particularly those harboring activated oncogenes. This barrier function is facilitated by the exceptional stability of the senescence-associated proliferation arrest, for example, in benign human nevi (Michaloglou et al. 2005). The mechanisms by which chromatin contributes to barrier function are not fully understood. However, most efforts to date have focused on features of chromatin structure and regulation that promote compacted heterochromatin, whose repressed “closed” state is intuitively interpreted as a static barrier to proliferation and tumor progression; for example, H3K9me3, HP1 proteins, and SAHF (Braig et al. 2005; Narita et al. 2006). In contrast, here we define chromatin of senescent cells as a dynamically maintained landscape, exhibiting previously unanticipated dynamic features. We show that one critical regulator of this dynamic landscape, histone chaperone HIRA, is required for efficient suppression of oncogene-induced neoplasia.
Several lines of evidence demonstrate the dynamic nature of chromatin in senescent cells. First, histone mRNAs are expressed in senescent cells. Second, in senescent cells, histones are enriched in PML bodies. This has been previously shown to reflect trafficking of newly synthesized histones into chromatin (Chang et al. 2013; Delbarre et al. 2013; Corpet et al. 2014). Third, newly synthesized ectopically expressed and endogenous histones are actively incorporated into chromatin in senescent cells. Fourth, FRAP analysis of a fluorescent-tagged histone H3.3 demonstrates a highly mobile subpopulation of H3.3. Fifth, gene promoters in senescent cells show marked relative enrichment of H4K16ac in senescent cells. H4K16ac impedes higher-order chromatin packaging (Shogren-Knaak et al. 2006) and has also been suggested to directly promote nucleosome exchange (Elliott et al. 2013). Together, these lines of evidence indicate that chromatin of senescent cells is maintained in a state of dynamic equilibrium.
Chromatin of senescent cells exhibits at least two unanticipated specialized features, indicating its fundamentally distinct regulation from chromatin in proliferating cells. First, nonproliferating senescent cells do, surprisingly, express a subset of so-called “replication-dependent” histone genes, linked to alternative mRNA splicing to a second exon harboring a polyadenylation signal instead of a stem/loop structure. Presumably, this second exon/poly A tail bypasses the normal stem–loop-mediated degradation of these mRNAs in nonproliferating cells (Marzluff et al. 2008), facilitating their preferential expression in senescent cells. Second, H4K16ac is extraordinarily enriched at promoters of expressed genes in senescent cells, even at genes that are unchanged or decreased in expression compared with proliferating cells. As a result, in senescent cells, but not proliferating cells, H4K16ac appears to flag all expressed genes, regardless of whether those genes are expressed at a high or low level. Curiously, H4K16ac is similarly enriched at promoters and enhancers of embryonic stem cells (Taylor et al. 2013), which have been previously suggested to contain especially dynamic chromatin (Meshorer et al. 2006). H4K16ac has been reported to counter heterochromatin spreading (Wang et al. 2013). Conceivably, H4K16ac constitutes a local chromatin “barrier” to prevent promoter silencing in unusual chromatin environments in embryonic stem cells and senescent cells.
We previously implicated the DNA replication-independent histone chaperone HIRA and its binding partners (UBN1, CABIN1, and ASF1a) in chromatin regulation in senescent cells (Zhang et al. 2005; Banumathy et al. 2009; Rai et al. 2011). Here, we directly demonstrate that multiple features of the dynamic landscape in senescent cells are controlled by HIRA. HIRA colocalizes with histones in PML bodies of senescent cells, a presumptive marker of HIRA-mediated trafficking of histones into chromatin (Chang et al. 2013; Delbarre et al. 2013). HIRA is required for the accumulation of histones in PML bodies and also for the deposition and/or accumulation of newly synthesized H3 (presumably H3.3) and H4 into chromatin of senescent cells. HIRA also colocalizes with newly deposited histone H3.3 at gene promoters in senescent cells, particularly at highly expressed genes. HIRA and, to a lesser extent, H3.3 alter their target gene selection between proliferating and senescent cells, in line with changes in expression. HIRA is required for the retention of total histone content and, more specifically, H4K16ac in senescent cells. HIRA also colocalizes with H4K16ac at promoters of highly expressed genes. However, HIRA is unlikely to act alone in nucleosome dynamics of senescent cells. An alternative chaperone, DAXX, has previously been suggested to target histone H3.3 to pericentromeric heterochromatin in senescent cells (Corpet et al. 2014). A partially overlapping role for DAXX can explain why repressed genes depleted of HIRA, such as histone genes, continued to incorporate histone H3.3 in senescent cells. Nonetheless, numerous lines of evidence indicate that the HIRA chaperone complex is a critical homeostatic regulator of the dynamic, H4K16ac-flagged chromatin landscape of senescent cells.
While previous constitutive knockout animals showed that HIRA is required for early embryo development (Roberts et al. 2002; Szenker et al. 2012), these models did not allow investigation of the functions in adult tissues. Here, in a novel conditional knockout HIRA mouse model expressing an activated BRAFV600E oncogene in the skin, inactivation of HIRA dramatically enhanced oncogene-induced hyperplastic cell proliferation accompanied by p53- and p21-expressing proliferating (BrdU+) cells, consistent with impaired execution of the cellular senescence program despite p53–p21 activation. Just as in vitro, inactivation of HIRA also resulted in specific depletion of H4K16ac. Inactivation of HIRA also appeared to modestly impair differentiation in BRAFV600E-expressing epidermis, based on altered K1 and K5 expression. Thus, both impaired differentiation and senescence likely contribute to epidermal hyperplasia in AhCreER/LSL-BrafV600E/Hirafl.fl mice. Regardless, these results indicate the importance of proper chromatin regulation for tumor suppression, even in cells in which prosenescence effectors p53 and p21 are activated. Remarkably, they also unambiguously demonstrate a role for HIRA in efficient proliferation arrest to suppress neoplasia in the face of an activated oncogene.
Both histone H3.3 and H4K16ac are dysregulated in cancer. The level of H4K16ac is reduced in many cancers and cancer cell lines (Fraga et al. 2005; Elsheikh et al. 2009), suggesting a potential tumor suppressor function. Recent studies have revealed recurrent missense mutations in genes encoding histone H3.3 (H3F3A) and H3.1 (HIST3H1B) in some cancers, in particular in pediatric gliomas (Schwartzentruber et al. 2012; Sturm et al. 2012; Wu et al. 2012; Yuen and Knoepfler 2013). These mutations often target specific residues and most often in histone H3.3, specifically H3.3K27M and H3.3G34R/V. The H3.3K27M protein acts as a dominant inhibitor of the EZH2 histone methyltransferase responsible for methylation of histone H3 to generate H3K27me3 (Lewis et al. 2013). While H3.3K27M therefore acts to globally reduce H3K27me3 levels, some regions do gain H3K27me3 (Chan et al. 2013; Lewis et al. 2013; Venneti et al. 2013). One such region is reported to be the CDKN2a locus, encoding the tumor suppressor p16INK4a, and expression of p16INK4a is reduced in two glioma cell lines (Chan et al. 2013). The p16INK4a/pRB pathway is a key effector of cellular senescence, and its suppression by H3.3K27M would be expected to impair senescence as a tumor suppressor mechanism (Salama et al. 2014). Therefore, H3.3K27M and inactivation of HIRA might in fact target similar senescence-related pathways to promote tumorigenesis. Perhaps more relevant to HIRA, very recent studies have indicated that transcription-coupled methylation of H3 to yield H3K36me3 and, in particular, H3.3K36me3 exerts a tumor-suppressive function (Wen et al. 2014; Zhu et al. 2014). HIRA is responsible for deposition of histone H3.3 at many transcribed genes (Goldberg et al. 2010). Results reported here also invoke HIRA in the maintenance of H4K16ac levels in senescent cells. Thus, the tumor-suppressive functions of both histone H3.3 and H4K16ac might be controlled by the HIRA chaperone complex.
In sum, our description of specialized and dynamic chromatin in senescent cells presents a new model for chromatin control in senescent cells that is substantially different from the intuitive model of more static canonical heterochromatin enforcing a barrier to proliferation and tumor progression. Histone chaperone HIRA is a key regulator of this landscape and exhibits potent neoplastic suppressor activity in a mouse model. Extending this idea, we suggest that DNA replication-independent chromatin regulators, such as HIRA, also maintain the dynamic chromatin of senescent cells to retain the tumor-suppressive nonproliferating phenotype over the long term, a process that we refer to as chromatin homeostasis or “chromostasis.” In future studies, it will be important to investigate the extent to which HIRA, histone H3.3, and H4K16ac act in concert to mediate tumor suppression.
Materials and methods
See the Supplemental Material for additional methods, details, and references.
Proliferating and senescent cells
IMR90 fibroblast cells were cultured according to American Type Culture Collection guidelines in low oxygen (3%). Proliferating fibroblasts (<30 population doublings) were cultured in Dulbecco’s modified Eagle’s medium supplemented with 20% (v/v) fetal bovine serum until they reached RS (defined in Supplemental Fig. 1A,B). To generate OIS and control cells, IMR90 cells stably expressing a fusion protein of the estrogen receptor (ER) ligand-binding domain and H-RASG12V (ER-RA-IMR90 cells [grown phenol red-free]) (Barradas et al. 2009) were supplemented with 100 nM 4-hydroxytamoxifen (4-OHT) or ethanol vehicle, respectively.
ChIP-seq
For HIRA ChIP, native ChIP was performed using benzonase endonuclease for chromatin solubilization and fragmentation. For HA-histone H3.3, H4K16ac, and H4 ChIP, cells were cross-linked with 1% formaldehyde for 5 min at room temperature. After quenching with glycine, the cells were harvested and sonicated to produce soluble chromatin with DNA fragments in the range of 300–500 base pairs. Fragmented chromatin was incubated with antibodies to HIRA, HA, H4K16ac, or H4 preimmobilized on Dynabeads. Libraries were prepared from 10–20 ng of ChIP or input DNA using New England Biolabs ChIP-seq kit according to the manufacturer’s instructions, and the resulting libraries were sequenced on an Illumina GAIIx to yield ∼30 million to 40 million raw reads.
RNA-seq
RNA was isolated using a Qiagen RNeasy kit. Ribodepletion was performed using an Epicentre Ribo Zero kit (MRZH11124C). Libraries were prepared according to the Illumina TruSeq protocol and sequenced on an Illumina GAIIx.
Massively parallel sequencing and data analysis
ChIP-seq or input reads were mapped to the human genome (hg19) using the Bowtie 2 alignment software. Only unique reads mapping to a single location were retained. The determination of enriched regions was performed using the USeq and SICER packages, and reads were visualized using the University of California at Santa Cruz browser. The results presented were analyzed from a single ChIP-seq reaction for HIRA in each of RS and OIS and H3.3 in RS, but the results are representative of two different experiments performed with two different antibodies for H4K16ac.
LC-MS analysis of extracted histones
The extracted histones were separated by SDS-PAGE, digested with trypsin, and analyzed by LC-MS on an AB-Sciex 5600 MS system as described previously (Gabrielsen et al. 2013). Proteins were identified by searching the UniProt database using Mascot 2.3 (MatrixScience), and SILAC ratios of lysine containing peptides (K0/K8) were calculated using Mascot Distiller 2.5 run on a local server.
Supplementary Material
Acknowledgments
We thank all members of the Adams laboratory for critical discussions. We thank Jesus Gil for pLNC-RAS:ER, Katrin Pritchard for LSL-BrafV600E mice, Philippe Collas for pH3.3-EGFP, and Doug Winton for AhCreER mice. The laboratory of P.D.A. is funded by National Institute on Aging program project AG031862 and Cancer Research UK program A16566. T.S.R. is supported by a start-up grant from Institute of Biomedical and Environmental Health Research, University of West Scotland, and the Carnegie Trust.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.247528.114.
References
- Acosta JC, O’Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, Fumagalli M, Da Costa M, Brown C, Popov N, et al. . 2008. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell 133: 1006–1018. [DOI] [PubMed] [Google Scholar]
- Adam S, Polo SE, Almouzni G. 2013. Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell 155: 94–106. [DOI] [PubMed] [Google Scholar]
- Ahmad K, Henikoff S. 2002. The histone variant h3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell 9: 1191–1200. [DOI] [PubMed] [Google Scholar]
- Banaszynski LA, Wen D, Dewell S, Whitcomb SJ, Lin M, Diaz N, Elsasser SJ, Chapgier A, Goldberg AD, Canaani E, et al. . 2013. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell 155: 107–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banumathy G, Somaiah N, Zhang R, Tang Y, Hoffmann J, Andrake M, Ceulemans H, Schultz D, Marmorstein R, Adams PD. 2009. Human UBN1 is an ortholog of yeast Hpc2p and has an essential role in the HIRA/ASF1a chromatin-remodeling pathway in senescent cells. Mol Cell Biol 29: 758–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barradas M, Anderton E, Acosta JC, Li S, Banito A, Rodriguez-Niedenfuhr M, Maertens G, Banck M, Zhou MM, Walsh MJ, et al. . 2009. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev 23: 1177–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamed M, Herbig U, Ye T, Dejean A, Bischof O. 2012. Senescence is an endogenous trigger for microRNA-directed transcriptional gene silencing in human cells. Nat Cell Biol 14: 266–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, Stein H, Dorken B, Jenuwein T, Schmitt CA. 2005. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436: 660–665. [DOI] [PubMed] [Google Scholar]
- Brown DT, Wellman SE, Sittman DB. 1985. Changes in the levels of three different classes of histone mRNA during murine erythroleukemia cell differentiation. Mol Cell Biol 5: 2879–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carragher LA, Snell KR, Giblett SM, Aldridge VS, Patel B, Cook SJ, Winton DJ, Marais R, Pritchard CA. 2010. V600EBraf induces gastrointestinal crypt senescence and promotes tumour progression through enhanced CpG methylation of p16INK4a. EMBO Mol Med 2: 458–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai J, Charboneau AL, Betz BL, Weissman BE. 2005. Loss of the hSNF5 gene concomitantly inactivates p21CIP/WAF1 and p16INK4a activity associated with replicative senescence in A204 rhabdoid tumor cells. Cancer Res 65: 10192–10198. [DOI] [PubMed] [Google Scholar]
- Chan KM, Fang D, Gan H, Hashizume R, Yu C, Schroeder M, Gupta N, Mueller S, James CD, Jenkins R, et al. . 2013. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev 27: 985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra T, Kirschner K, Thuret JY, Pope BD, Ryba T, Newman S, Ahmed K, Samarajiwa SA, Salama R, Carroll T, et al. . 2012. Independence of repressive histone marks and chromatin compaction during senescent heterochromatic layer formation. Mol Cell 47: 203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang FT, McGhie JD, Chan FL, Tang MC, Anderson MA, Mann JR, Andy Choo KH, Wong LH. 2013. PML bodies provide an important platform for the maintenance of telomeric chromatin integrity in embryonic stem cells. Nucleic Acids Res 41: 4447–4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, et al. . 2005. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 436: 725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton E, Doupe DP, Klein AM, Winton DJ, Simons BD, Jones PH. 2007. A single type of progenitor cell maintains normal epidermis. Nature 446: 185–189. [DOI] [PubMed] [Google Scholar]
- Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, Benguria A, Zaballos A, Flores JM, Barbacid M, et al. . 2005. Tumour biology: senescence in premalignant tumours. Nature 436: 642. [DOI] [PubMed] [Google Scholar]
- Contrepois K, Thuret JY, Courbeyrette R, Fenaille F, Mann C. 2012. Deacetylation of H4-K16Ac and heterochromatin assembly in senescence. Epigenetics Chromatin 5: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, Nelson PS, Desprez PY, Campisi J. 2008. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol 6: 2853–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet A, Olbrich T, Gwerder M, Fink D, Stucki M. 2014. Dynamics of histone H3.3 deposition in proliferating and senescent cells reveals a DAXX-dependent targeting to PML-NBs important for pericentromeric heterochromatin organization. Cell Cycle 13: 249–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosme-Blanco W, Shen MF, Lazar AJ, Pathak S, Lozano G, Multani AS, Chang S. 2007. Telomere dysfunction suppresses spontaneous tumorigenesis in vivo by initiating p53-dependent cellular senescence. EMBO Rep 8: 497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruickshanks HA, McBryan T, Nelson DM, Vanderkraats ND, Shah PP, van Tuyn J, Singh Rai T, Brock C, Donahue G, Dunican DS, et al. . 2013. Senescent cells harbour features of the cancer epigenome. Nat Cell Biol 15: 1495–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cecco M, Criscione SW, Peckham EJ, Hillenmeyer S, Hamm EA, Manivannan J, Peterson AL, Kreiling JA, Neretti N, Sedivy JM. 2013. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell 12: 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbarre E, Ivanauskiene K, Kuntziger T, Collas P. 2013. DAXX-dependent supply of soluble (H3.3–H4) dimers to PML bodies pending deposition into chromatin. Genome Res 23: 440–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhomen N, Reis-Filho JS, da Rocha Dias S, Hayward R, Savage K, Delmas V, Larue L, Pritchard C, Marais R. 2009. Oncogenic Braf induces melanocyte senescence and melanoma in mice. Cancer Cell 15: 294–303. [DOI] [PubMed] [Google Scholar]
- Di Micco R, Sulli G, Dobreva M, Liontos M, Botrugno OA, Gargiulo G, dal Zuffo R, Matti V, d’Ario G, Montani E, et al. . 2011. Interplay between oncogene-induced DNA damage response and heterochromatin in senescence and cancer. Nat Cell Biol 13: 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte LF, Young AR, Wang Z, Wu HA, Panda T, Kou Y, Kapoor A, Hasson D, Mills NR, Ma'ayan A, et al. . 2014. Histone H3.3 and its proteolytically processed form drive a cellular senescence programme. Nat Commun 5: 5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D, Ray S, Home P, Saha B, Wang S, Sheibani N, Tawfik O, Cheng N, Paul S. 2010. Regulation of angiogenesis by histone chaperone HIRA-mediated Incorporation of lysine 56-acetylated histone H3.3 at chromatin domains of endothelial genes. J Biol Chem 285: 41567–41577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott GO, Murphy KJ, Hayes JJ, Thiriet C. 2013. Replication-independent nucleosome exchange is enhanced by local and specific acetylation of histone H4. Nucleic Acids Res 41: 2228–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsheikh SE, Green AR, Rakha EA, Powe DG, Ahmed RA, Collins HM, Soria D, Garibaldi JM, Paish CE, Ammar AA, et al. . 2009. Global histone modifications in breast cancer correlate with tumor phenotypes, prognostic factors, and patient outcome. Cancer Res 69: 3802–3809. [DOI] [PubMed] [Google Scholar]
- Feldser DM, Greider CW. 2007. Short telomeres limit tumor progression in vivo by inducing senescence. Cancer Cell 11: 461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feser J, Truong D, Das C, Carson JJ, Kieft J, Harkness T, Tyler JK. 2010. Elevated histone expression promotes life span extension. Mol Cell 39: 724–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, et al. . 2005. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet 37: 391–400. [DOI] [PubMed] [Google Scholar]
- Gabrielsen M, Schuldt M, Munro J, Borucka D, Cameron J, Baugh M, Mleczak A, Lilla S, Morrice N, Olson MF. 2013. Cucurbitacin covalent bonding to cysteine thiols: the filamentous-actin severing protein Cofilin1 as an exemplary target. Cell Commun Signal 11: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. . 2010. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140: 678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove GW, Zweidler A. 1984. Regulation of nucleosomal core histone variant levels in differentiating murine erythroleukemia cells. Biochemistry 23: 4436–4443. [DOI] [PubMed] [Google Scholar]
- Ivanov A, Pawlikowski J, Manoharan I, van Tuyn J, Nelson DM, Rai TS, Shah PP, Hewitt G, Korolchuk VI, Passos JF, et al. . 2013. Lysosome-mediated processing of chromatin in senescence. J Cell Biol 202: 129–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Felsenfeld G. 2007. Nucleosome stability mediated by histone variants H3.3 and H2A. Z. Genes Dev 21: 1519–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G. 2009. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet 41: 941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang TW, Yevsa T, Woller N, Hoenicke L, Wuestefeld T, Dauch D, Hohmeyer A, Gereke M, Rudalska R, Potapova A, et al. . 2011. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature 479: 547–551. [DOI] [PubMed] [Google Scholar]
- Kari V, Karpiuk O, Tieg B, Kriegs M, Dikomey E, Krebber H, Begus-Nahrmann Y, Johnsen SA. 2013. A subset of histone H2B genes produces polyadenylated mRNAs under a variety of cellular conditions. PLoS ONE 8: e63745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp R, Ireland H, Clayton E, Houghton C, Howard L, Winton DJ. 2004. Elimination of background recombination: somatic induction of Cre by combined transcriptional regulation and hormone binding affinity. Nucleic Acids Res 32: e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J. 2001. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc Natl Acad Sci 98: 12072–12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, Aarden LA, Mooi WJ, Peeper DS. 2008. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell 133: 1019–1031. [DOI] [PubMed] [Google Scholar]
- Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD. 2013. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340: 857–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CJ, Conti M, Ramalho-Santos M. 2013. Histone variant H3.3 maintains a decondensed chromatin state essential for mouse preimplantation development. Development 140: 3624–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loppin B, Bonnefoy E, Anselme C, Laurencon A, Karr TL, Couble P. 2005. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature 437: 1386–1390. [DOI] [PubMed] [Google Scholar]
- Lujambio A, Akkari L, Simon J, Grace D, Tschaharganeh DF, Bolden JE, Zhao Z, Thapar V, Joyce JA, Krizhanovsky V, et al. . 2013. Non-cell-autonomous tumor suppression by p53. Cell 153: 449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin N, Popov N, Aguilo F, O’Loghlen A, Raguz S, Snijders AP, Dharmalingam G, Li S, Thymiakou E, Carroll T, et al. . 2013. Interplay between Homeobox proteins and Polycomb repressive complexes in p16INK(4)a regulation. EMBO J 32: 982–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ. 2008. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet 9: 843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer K, Giblett S, Green S, Lloyd D, DaRocha Dias S, Plumb M, Marais R, Pritchard C. 2005. Expression of endogenous oncogenic V600EB-raf induces proliferation and developmental defects in mice and transformation of primary fibroblasts. Cancer Res 65: 11493–11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. 2006. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell 10: 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, Majoor DM, Shay JW, Mooi WJ, Peeper DS. 2005. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 436: 720–724. [DOI] [PubMed] [Google Scholar]
- Narita M, Nunez S, Heard E, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113: 703–716. [DOI] [PubMed] [Google Scholar]
- Narita M, Narita M, Krizhanovsky V, Nunez S, Chicas A, Hearn SA, Myers MP, Lowe SW. 2006. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell 126: 503–514. [DOI] [PubMed] [Google Scholar]
- O’Sullivan RJ, Kubicek S, Schreiber SL, Karlseder J. 2010. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat Struct Mol Biol 17: 1218–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantazis P, Bonner WM. 1984. Specific alterations in the pattern of histone-3 synthesis during conversion of human leukemic cells to terminally differentiated cells in culture. Differentiation 28: 186–190. [DOI] [PubMed] [Google Scholar]
- Pchelintsev NA, McBryan T, Rai TS, van Tuyn J, Ray-Gallet D, Almouzni G, Adams PD. 2013. Placing the HIRA histone chaperone complex in the chromatin landscape. Cell Reports 3: 1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Placek BJ, Huang J, Kent JR, Dorsey J, Rice L, Fraser NW, Berger SL. 2009. The histone variant H3.3 regulates gene expression during lytic infection with herpes simplex virus type 1. J Virol 83: 1416–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai TS, Puri A, McBryan T, Hoffman J, Tang Y, Pchelintsev NA, van Tuyn J, Marmorstein R, Schultz DC, Adams PD. 2011. Human CABIN1 is a functional member of the human HIRA/UBN1/ASF1a histone H3.3 chaperone complex. Mol Cell Biol 31: 4107–4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-Gallet D, Quivy J-P, Scamps C, Martini EM-D, Lipinski M, Almouzni G. 2002. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol Cell 9: 1091–1100. [DOI] [PubMed] [Google Scholar]
- Ray-Gallet D, Woolfe A, Vassias I, Pellentz C, Lacoste N, Puri A, Schultz DC, Pchelintsev NA, Adams PD, Jansen LE, et al. . 2011. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol Cell 44: 928–941. [DOI] [PubMed] [Google Scholar]
- Roberts C, Sutherland HF, Farmer H, Kimber W, Halford S, Carey A, Brickman JM, Wynshaw-Boris A, Scambler PJ. 2002. Targeted mutagenesis of the Hira gene results in gastrulation defects and patterning abnormalities of mesoendodermal derivatives prior to early embryonic lethality. Mol Cell Biol 22: 2318–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaie S, Salama R, Carroll T, Tomimatsu K, Chandra T, Young ARJ, Narita M, Perez-Mancera PA, Bennett DC, Chong H, et al. . 2013. Redistribution of the Lamin B1 genomic binding profile affects rearrangement of heterochromatic domains and SAHF formation during senescence. Genes Dev 27: 1800–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama R, Sadaie M, Hoare M, Narita M. 2014. Cellular senescence and its effector programs. Genes Dev 28: 99–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, et al. . 2012. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482: 226–231. [DOI] [PubMed] [Google Scholar]
- Shah PP, Donahue G, Otte GL, Capell BC, Nelson DM, Cao K, Aggarwala V, Cruickshanks HA, Rai TS, McBryan T, et al. . 2013. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev 27: 1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood PW, Tsang SV, Osley MA. 1993. Characterization of HIR1 and HIR2, two genes required for regulation of histone gene transcription in Saccharomyces cerevisiae. Mol Cell Biol 13: 28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. 2006. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311: 844–847. [DOI] [PubMed] [Google Scholar]
- Skene PJ, Henikoff S. 2013. Histone variants in pluripotency and disease. Development 140: 2513–2524. [DOI] [PubMed] [Google Scholar]
- Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, Pfaff E, Tonjes M, Sill M, Bender S, et al. . 2012. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22: 425–437. [DOI] [PubMed] [Google Scholar]
- Szenker E, Lacoste N, Almouzni G. 2012. A developmental requirement for HIRA-dependent H3.3 deposition revealed at gastrulation in Xenopus. Cell Reports 1: 730–740. [DOI] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116: 51–61. [DOI] [PubMed] [Google Scholar]
- Taylor GC, Eskeland R, Hekimoglu-Balkan B, Pradeepa MM, Bickmore WA. 2013. H4K16 acetylation marks active genes and enhancers of embryonic stem cells, but does not alter chromatin compaction. Genome Res 23: 2053–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban MK, Zweidler A. 1983. Changes in nucleosomal core histone variants during chicken development and maturation. Dev Biol 95: 421–428. [DOI] [PubMed] [Google Scholar]
- van der Heijden GW, Derijck AA, Posfai E, Giele M, Pelczar P, Ramos L, Wansink DG, van der Vlag J, Peters AH, de Boer P. 2007. Chromosome-wide nucleosome replacement and H3.3 incorporation during mammalian meiotic sex chromosome inactivation. Nat Genet 39: 251–258. [DOI] [PubMed] [Google Scholar]
- Venneti S, Garimella MT, Sullivan LM, Martinez D, Huse JT, Heguy A, Santi M, Thompson CB, Judkins AR. 2013. Evaluation of histone 3 lysine 27 trimethylation (H3K27me3) and enhancer of Zest 2 (EZH2) in pediatric glial and glioneuronal tumors shows decreased H3K27me3 in H3F3A K27M mutant glioblastomas. Brain Pathol 23: 558–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Tadeo X, Hou H, Tu PG, Thompson J, Yates JR 3rd, Jia S. 2013. Epe1 recruits BET family bromodomain protein Bdf2 to establish heterochromatin boundaries. Genes Dev 27: 1886–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H, Li Y, Xi Y, Jiang S, Stratton S, Peng D, Tanaka K, Ren Y, Xia Z, Wu J, et al. . 2014. ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature 508: 263–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BG, Roberts CW. 2011. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer 11: 481–492. [DOI] [PubMed] [Google Scholar]
- Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, et al. . 2012. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet 44: 251–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW. 2007. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445: 656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JH, Song Y, Seol JH, Park JY, Yang YJ, Han JW, Youn HD, Cho EJ. 2011. Myogenic transcriptional activation of MyoD mediated by replication-independent histone deposition. Proc Natl Acad Sci 108: 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Zerlanko B, Kennedy A, Banumathy G, Zhang R, Adams PD. 2007. Downregulation of Wnt signaling is a trigger for formation of facultative heterochromatin and onset of cell senescence in primary human cells. Mol Cell 27: 183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen BT, Knoepfler PS. 2013. Histone H3.3 mutations: a variant path to cancer. Cancer Cell 24: 567–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Poustovoitov MV, Ye X, Santos HA, Chen W, Daganzo SM, Erzberger JP, Serebriiskii IG, Canutescu AA, Dunbrack RL, et al. . 2005. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev Cell 8: 19–30. [DOI] [PubMed] [Google Scholar]
- Zhang R, Chen W, Adams PD. 2007. Molecular dissection of formation of senescent associated heterochromatin foci. Mol Cell Biol 27: 2343–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, He F, Zeng H, Ling S, Chen A, Wang Y, Yan X, Wei W, Pang Y, Cheng H, et al. . 2014. Identification of functional cooperative mutations of SETD2 in human acute leukemia. Nat Genet 46: 287–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.