By studying Drosophila ovarian follicle cell progenitors, Lee et al. identified lysine-specific demethylase 1 (lsd1) and CoRest as regulators of differentiation. The follicle cell progenitors in heterozygotes prematurely lose epigenetic plasticity, undergo the Notch-dependent mitotic–endocycle transition, and stop dividing before a normal number of follicle cells can be produced. Reducing the dosage of the histone H3K4 methyltransferase Trithorax reverses these effects.
Keywords: Lsd1, differentiation, epigenetic plasticity, progenitor
Abstract
Progenitors are early lineage cells that proliferate before the onset of terminal differentiation. Although widespread, the epigenetic mechanisms that control the progenitor state and the onset of differentiation remain elusive. By studying Drosophila ovarian follicle cell progenitors, we identified lysine-specific demethylase 1 (lsd1) and CoRest as differentiation regulators using a GAL4∷GFP variegation assay. The follicle cell progenitors in lsd1 or CoRest heterozygotes prematurely lose epigenetic plasticity, undergo the Notch-dependent mitotic-endocycle transition, and stop dividing before a normal number of follicle cells can be produced. Simultaneously reducing the dosage of the histone H3K4 methyltransferase Trithorax reverses these effects, suggesting that an Lsd1/CoRest complex times progenitor differentiation by controlling the stability of H3K4 methylation levels. Individual cells or small clones initially respond to Notch; hence, a critical level of epigenetic stabilization is acquired cell-autonomously and initiates differentiation by making progenitors responsive to pre-existing external signals.
Progenitors are characterized by an epigenetically flexible chromatin state (Iglesias-Bartolome et al. 2013). However, the gene expression, nucleosome organization, histone modification, and chromatin modulator activity that promote progenitor proliferation and restrain differentiation remain unclear (Badeaux and Shi 2013). Developmental gene activation or repression correlates strongly with methylation on two histone 3 residues (H3K4 and H3K27) within the promoter region of key lineage genes (Shi 2007). The dynamics of histone methylation reflect the opposing activities of multiple lysine methyltransferases and lysine demethylases (Kooistra and Helin 2012) with differing specificities. For example, lysine-specific demethylase 1 (LSD1) specifically demethylates H3K4me2 and H3K4me (Shi et al. 2004; Klose and Zhang 2007), whereas a different demethylase, LID, demethylates H3K4me3 and H3K4me2 (Lee et al. 2007). LSD1 is highly conserved through evolution and regulates gene expression, heterochromatin spreading, and epigenetic memory in both stem and differentiating cells (Di Stefano et al. 2007, 2011; Rudolph et al. 2007; Katz et al. 2009; Adamo et al. 2011; Eliazer et al. 2011, 2014; Kerenyi et al. 2013; Zhu et al. 2014).
The ovarian follicle cells of Drosophila melanogaster provide a well-characterized in vivo system for understanding the molecular mechanisms that underlie the progenitor state and regulate the onset of differentiation (Margolis and Spradling 1995; Skora and Spradling 2010). All ∼800 of the somatic cells on each mature ovarian follicle are derived from two follicle stem cells (FSCs) located midway in the initial segment of the ovariole, known as the germarium (Fig. 1A). Early follicle cell progenitors associate with a cyst of 16 germ cells and undergo four to five rounds of division before surrounding the oocyte and its 15 nurse cells to form a new follicle. They each divide five more times (DIV5–9) as a monolayer on the follicle surface before a major regulatory event, the mitotic/endocycle (M–E) transition, terminates proliferation and initiates differentiation (Fig. 1A; Deng et al. 2001; Sun and Deng 2005, 2007). The M–E transition triggers when follicle cells respond to Delta expressed from associated germ cells to activate Notch signaling, inducing hindsight (hnt) and repressing cut and M-phase genes. During the three endocycles that follow, follicle cells differentiate into multiple cell types in response to well-understood JAK/STAT, Ras/MAPK, and other signals (Klusza and Deng 2011). The onset of germ cell Delta expression precedes the M–E transition (Lopez-Schier and St Johnston 2001), so the rate-limiting step controlling follicle cell progenitor differentiation remains uncertain.
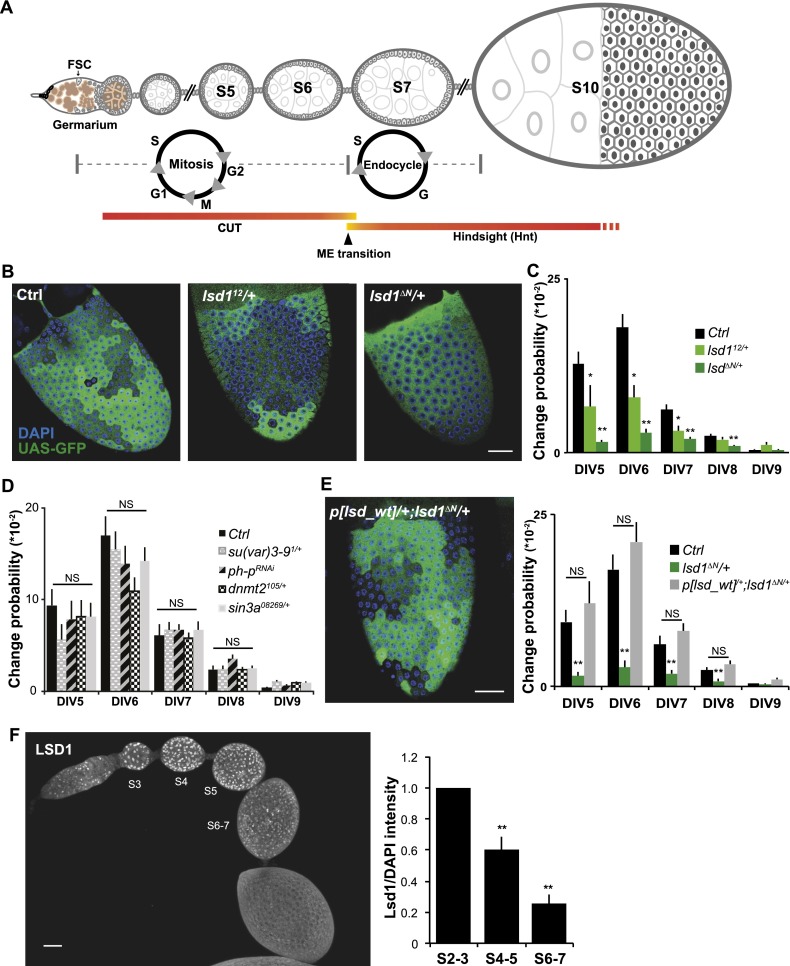
Figure 1.
Lsd1 mediates epigenetic plasticity in developing follicle cells. (A) Schematic of ovarian follicle development and the M–E transition. (B) GAL4∷GFP variegation in a control stage 10 follicle (Ctrl) is much greater than in follicles heterozygous for the hypomorphic lsd12 allele or the null lsd1ΔN allele. (C) Variegation levels in B are quantified as the probability that expression will heritably change at follicle cell DIV5–9 (see the Materials and Methods) to indicate changes in epigenetic plasticity. N = 44 control, 45 lsd112/+, and 51 lsd1ΔN/+ ovarioles. (D) Reducing the gene dosage or activity of genes tested shows no effect on variegation. N = 20 control, 20 Su(Var)3-91/+, 17 ph-pRNAi/+, 32 dnmt21/+, and 22 sin308269/+ ovarioles. (E) Variegation suppression by lsd1ΔN/+ is reversed by an additional copy of wild-type lsd1: p[lsd1_wt]. N = 47. (F) Endogenous Lsd1 expression assayed by immunostaining is gradually down-regulated from stage 3 to stage 7 follicles. N = 10. Bars, 20 μm. Error bars indicate standard errors. (*) P < 0.05; (**) P < 0.01; (NS) nonsignificant difference, Student’s t-test.
Recently, a new approach to identifying genes involved in the epigenetic regulation of follicle cell progenitors was developed (Skora and Spradling 2010). Unlike genes introduced on simple transposons, the binary gene expression systems based on the yeast GAL4 protein and its target UAS promoter show variegated expression in many tissues. The variegation of GAL4∷GFP transgenes in follicle cell progenitors was shown to reveal an ongoing epigenetic maturation process that precedes progenitor differentiation. During eight mitotic divisions (DIV2–9), follicle cell progenitors gradually reduce their propensity to alter the heritable GFP expression state of such transgenes (i.e., epigenetic plasticity). Although the instability of GAL4∷UAS expression may reflect the artificial nature of these constructs, the epigenetic inheritance of their gene expression levels likely relies on normal Drosophila epigenetic machinery. Consequently, identifying genes that modify variegation in follicle cell progenitors should reveal important components acting in these cells at the onset of differentiation.
Here we identify lsd1 and CoRest as dominant suppressors of GAL4∷GFP variegation during ovarian follicle progenitor divisions. These proteins form a complex that not only influences epigenetic plasticity but maintains progenitor proliferation and times the Notch-dependent M–E transition. Lsd1 likely mediates epigenetic stability by controlling H3K4 methylation levels, since reducing the dosage of the histone H3K4 methyltransferase Trithorax reverses its effects. Interestingly, as differentiation begins, Notch signaling becomes active initially in small groups of epigenetically related cells. Thus, an ongoing epigenetic program mediated by Lsd1/CoRest ultimately controls when progenitors differentiate by modulating their ability to respond autonomously to external signals.
Results
Lsd1 mediates epigenetic plasticity in developing follicle cells
We assayed genes for their effects on epigenetic plasticity during the late follicle progenitor divisions (DIV5–9) by examining the effects of heterozygous mutants on GAL4∷GFP variegation. Epigenetic changes were scored quantitatively by identifying distinct GFP patches (Fig. 1B) and then calculating the specific division at which each patch arose based on its size. Next, the change probability at that division (Fig. 1C) was calculated by dividing the frequency of epigenetic changes arising at a given division by the number of cells that underwent that division (see the Materials and Methods; Skora and Spradling 2010). To make sure that the size of distinct GFP expression patches accurately reflected the timing of epigenetic changes and was not affected by the fluorescent protein movement through follicle cell intercellular bridges (Airoldi et al. 2011; McLean and Cooley 2013), we compared follicle cell variegation patterns using a normal diffusible GFP with tethered derivatives (UAS-mGFP, UAS-nlsRFP, or UAS-yps-mRFP) (Airoldi et al. 2011) that are unlikely to move through intercellular bridges. Very similar variegation patterns were observed (Supplemental Fig. S1), showing that intercellular protein movement does not preclude using GAL4∷GFP variegation as an assay indicating epigenetic plasticity. Consequently, we searched for genetic enhancers and suppressors of GAL4∷GFP variegation to further understand the molecular machineries mediating progenitor differentiation. To aid in interpreting changes in variegation patterns caused by modifier genes, we also simulated the observed variegation mathematically (Supplemental Fig. S2).
We found that lsd1 acts as a strong dominant suppressor of GAL4∷GFP variegation in a preliminary screen of chromatin-related genes (Fig. 1B, C). In follicles heterozygous for a hypomorphic allele (lsd112) or a null allele (lsd1ΔN), variegation was greatly reduced during the later follicle cell divisions (DIV5–9), as indicated by nearly homogeneous GFP expression patterns with many fewer clonal patches (Fig. 1B). Dose-sensitive suppression of GAL4∷GFP variegation was a rare and highly specific property of lsd1; other tested chromatin genes had no significant effect (Fig. 1D; Skora and Spradling 2010). A systematic screen for variegation modifier loci on the Drosophila autosomes is in progress. Adding a wild-type copy of lsd1 (p[lsd_wt]) to lsd1ΔN/+ flies restored variegation, showing that suppression was Lsd1-specific and not due to genetic background (Fig. 1E). Driving lsd1 RNAi specifically in follicle progenitors (R10H05∷lsd1RNAi) suppressed variegation, indicating that Lsd1 function in maintaining plasticity is needed only in follicle cells (Supplemental Fig. S3). The average GFP intensity within large or small patches was not affected when lsd1 function was reduced; hence, variegation was not suppressed due to a general enhancement of GFP expression (Supplemental Fig. S4). Prior to the M–E transition in vivo, we found by immunostaining that Lsd1 expression in cell nuclei decreases gradually as progenitors proliferate (Fig. 1F; Supplemental Fig. S5). Therefore, Lsd1 levels correlate with epigenetic plasticity not only following dosage reduction (Fig. 1C) but also during normal follicle cell progenitor development.
Lsd1 functions with CoRest as a H3K4 demethylase mediating epigenetic plasticity
Lsd1 functions in a complex with the transcription factor CoREST in many tissues (Tsai et al. 2010; Ceballos-Chavez et al. 2012; Zhang et al. 2013). When we lowered CoREST expression using RNAi or reduced CoRest dosage using CoRestEY14218/+ flies (Fig. 2A), follicle cell GAL4∷GFP variegation was significantly suppressed during DIV5–9 (Fig. 2A,B). We carried out coimmunoprecipitation experiments using extracts from young ovaries enriched in follicle cells prior to the M–E transition and consistently found strong interaction between LSD1 and CoREST (Fig. 2C), suggesting that LSD1 and CoREST act in a complex to maintain the epigenetic plasticity of follicle cell progenitors.
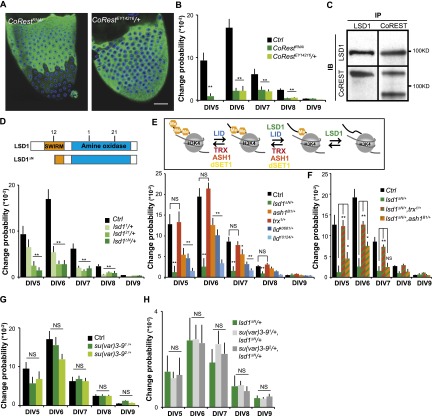
Figure 2.
Lsd1 functions with CoREST as a H3K4 demethylase. (A) GAL4∷GFP variegation is suppressed by knocking down CoRest expression (left) or reducing CoRest gene dosage (right). For control, see Figure 1B. (B) Quantitation of variegation levels in A. N = 20 CoRestRNAi and 20 CoRestEY14216/+ ovarioles). (C) LSD1 and CoREST form a complex in adult ovaries. Immunoprecipitates (IP) were prepared from ovarian lysates using anti-LSD1 or anti-CoREST antibodies (Dallman et al. 2004) and then immunoblotted (IB) with the indicated antibodies. N = 3 experiments. (D) Point mutations in the Lsd1 enzymatic domain (i.e., lsd11 and lsd121; see the diagram above) suppress GAL4 variegation. N = 44 control, 35 lsd11/+, 22 lsd21/+, and 51 lsd1ΔN/+ ovarioles. (E) Schematic (shown above) of methylases (red) and demethylases (green and blue) that control histone H3K4 methylation. The effects of reducing the dosage of these genes on GAL4∷GFP variegation is shown. N = 51 lsd1ΔN/+, 29 ash1B1/+, 27 Trx1/+, 51 (LidK0681/+, and 33 Lid10424/+ ovarioles. (F) Trx and ash1 dominantly counteract lsd1-mediated suppression of GAL4∷ GFP variegation. N = 46 lsd1ΔN/+, Trx1/+ and 29 lsd1ΔN/+, ash1B1/+ ovarioles. (G,H) The su(var)3-9 gene encoding an H3K9 methylase does not affect GAL4∷GFP variegation (G) or suppress the effects of lsd1ΔN/+ (H). N = 20 su(var)3-91, 20 su(var)3-92, 34 su(var)3-91, lsd1ΔN/+, and 36 su(var)3-92, lsd1ΔN/+ ovarioles. Bars, 20 μm. Error bars indicate standard errors. (*) P < 0.05; (**) P < 0.01; (NS) nonsignificant difference, Student’s t-test.
Lsd1 likely acts via its demethylase activity, since point mutants in conserved enzymatic domains (lsd11 and lsd121) scored as dominant suppressors modifying GAL4∷GFP variegation patterns (Fig. 2D). If changes in methylated H3K4 pools are critical, we reason that lsd1-mediated suppression of variegation might be reversed by lowering the function of genes encoding histone H3K4 methyltransferases, such as Trithorax (Trx), dSET1, or ash1 (Fig. 2E). Indeed, lowering Trx dosage but not dSET1 expression (Supplemental Fig. S6) was sufficient to restore the variegation of lsd1ΔN/+ follicles to wild-type level (lsd1ΔN/+ Trx1/+; P < 0.01, Student’s t-test) (Fig. 2F). Reducing the dosage of ash1 also caused a smaller but statistically significant restoration of epigenetic plasticity (Fig. 2F). Further support came from the observation that a second H3K4 demethylase, Lid, also strongly suppressed GAL4:GFP variegation in follicle progenitors (Fig. 2E). In contrast, genetic interaction tests did not support lsd1 action as an H3K9 demethylase in this context. We reduced the dosage of two different Su(var)3-9 alleles encoding a H3K9 methylase in lsd1ΔN/+ follicles but observed no restoration of GAL4∷GFP variegation (Fig. 2G,H). These results support the idea that lsd1 functions as an H3K4 demethylase to mediate epigenetic plasticity.
Early epigenetic stabilization leads to a premature M–E transition
Ovarian follicles do not develop normally when epigenetic maturation is accelerated. lsd1ΔN/+ and CoRestEY14218/+ follicles are smaller than normal and contain fewer follicle cells (Fig. 3A,B). In wild type, 253 ± 18 cells could be readily visualized per side on mounted stage 10 follicles, whereas lsd1ΔN/+ follicles only had 170 ± 20 follicle cells per side, a reduction of 30% (N = 51; P < 0.001) (Fig. 3A,B). Normal follicle cell number was restored in lsd1ΔN/+ follicles by the same second site mutations (Trx1/+ or ash1/+) that restore epigenetic plasticity (Fig. 3A,B). Compared with control follicles, the frequency of S-phase-specific EdU incorporation (Fig. 3D) and the number of M-phase-specific phosphohistone-positive (PH3+) nuclei (Supplemental Fig. S7) were reduced in lsd1ΔN/+ follicles during stages 5 and 6, indicating that cell cycling terminated earlier than normal. The fecundity of lsd1ΔN/+ females was only ∼30% of wild type and was restored to wild-type level in lsd1ΔN/+ Trx1/+ females, indicating that the changes in epigenetic plasticity are detrimental to efficient egg production (Fig. 3E). Finally, increased lsd1 dosage had the reverse effect (Fig. 3C), indicating that lsd1 plays a rate-limiting, dose-dependent role in controlling the proliferation of follicle progenitor cells.
Figure 3.
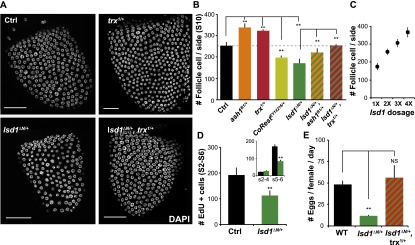
Epigenetic plasticity correlated with total follicle cell numbers. (A) Follicle cell nuclei visualized by DAPI staining in control (Ctrl), ash1B1/+, trx1/+, lsd1ΔN/+, ash1B1/+, lsd1ΔN/+, and trx1/+, lsd1ΔN/+ stage 10 follicles. (B) Follicle cell number (mean ± SEM per visual field) in stage 10 follicles from controls and genotypes with altered GAL4∷GFP variegation. Follicles from genotypes with reduced epigenetic plasticity contain fewer cells. N = 44 control, 29 ash1B1/+, 27 trx1/+, 51 lsd1ΔN/+, 34 CoRestEY14216/+, 27 ash1B1/+, lsd1ΔN/+, and 46 trx1/+, lsd1ΔN/+ ovarioles. (C) Follicle cell number (mean ± SD per visual field) in stage 10 follicles from genotypes with 1× (lsd1ΔN/+), 2× (Ctrl), 3× (p[lsd1_wt]/+), and 4× (p[lsd1_wt]/p[lsd1_wt]) copies of lsd1. N = 51, 39, 42, and 52 ovarioles respectively. (D) Follicle cell cycle is reduced in lsd1ΔN/+ follicles based on the reduced number of cells that underwent EdU incorporation. (Inset) Significant differences are only observed at stages 5–6. N = 23 control and 19 lsd1ΔN/+. (E) Female fecundity is affected by epigenetic plasticity. Reduced fecundity in lsd1ΔN/+ females and the restoration of fecundity in lsd1ΔN/+, Trx1/+ were quantified by measuring the number of eggs laid per female per day compared with wild-type controls. N = 3 and 3, respectively. Error bars indicate standard errors. (*) P < 0.05; (**) P < 0.01; (NS) nonsignificant difference, Student’s t-test.
To determine whether the premature termination of mitotic cycles was caused by a precocious M–E transition, we assayed the timing of Notch activation in wild-type and lsd1ΔN/+ follicles. Normally, Notch signal activation in follicle cells takes place at stages 6–7 as indicated by the Notch reporter Gbe-Su(H)-LacZ (Fig. 4A,D). Consistent with this temporal profile, cut expression begins to shut down during stage 6 and turns completely off by stage 7 (Fig. 4B,E), while hnt expression is absent in stage 5 and only begins to appear in stage 6 (Fig. 4C,E). In striking contrast, the kinetics of these events were shifted earlier by an entire developmental stage in lsd1ΔN/+ follicles. Notch signaling becomes active in stage 5 (Fig. 4A,D), cut expression turns completely off in stage 6 (Fig. 4B,E), and hnt expression begins in stage 5 (Fig. 4C,E). Thus, accelerated epigenetic maturation correlates with the premature onset of the M–E transition (Fig. 4F). While Notch is necessary for the M–E transition at stages 6–7, we found that premature expression of the Notch intracellular domain in follicle cells prior to stage 6 (R10H05∷NICD) (Supplemental Fig. S8) is not sufficient to trigger a precocious M–E transition (Fig. 4G). This suggests that epigenetic maturation is independently required for the M–E transition and that it is rate-limiting under normal conditions.
Figure 4.
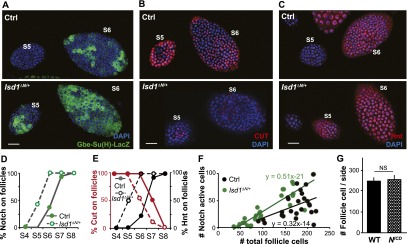
Accelerated epigenetic maturation leads to premature M–E transition. (A–C) Precocious M–E transition in lsd1ΔN/+ follicles compared with control (Ctrl). Activation of Notch signal reception [Gbe-Su(H)-lacZ; green; A] and induction of hnt (red; C) occur in stage 5 rather than stage 6 in lsd1ΔN/+ follicles, while repression of cut (red; B) is prematurely completed by stage 6. (D) Quantification of the precocious activation of Notch signal reception. N = 30 control and 36 lsd1ΔN/+. (E) Quantification showing the premature loss of cut expression (red circles) and the early acquisition of hnt (black circles). For Hnt expression, N = 14 control and 18 lsd1ΔN/+. For Cut expression, N = 16 control and 18 lsd1ΔN/+. (F) Differential timing of the M–E transition in lsd1ΔN/+ follicles (green circles) versus control follicles (green circles) was shown by staging follicles with follicle cell number rather than size. N = 25 control and 22 lsd1ΔN/+. (G) Driving the Notch intracellular domain (NICD) using R10H05-GAL4 failed to terminate mitosis precociously. N = 14. Bars, 20 μm.
Follicle cell progenitors undergo the M–E transition in epigenetically similar groups
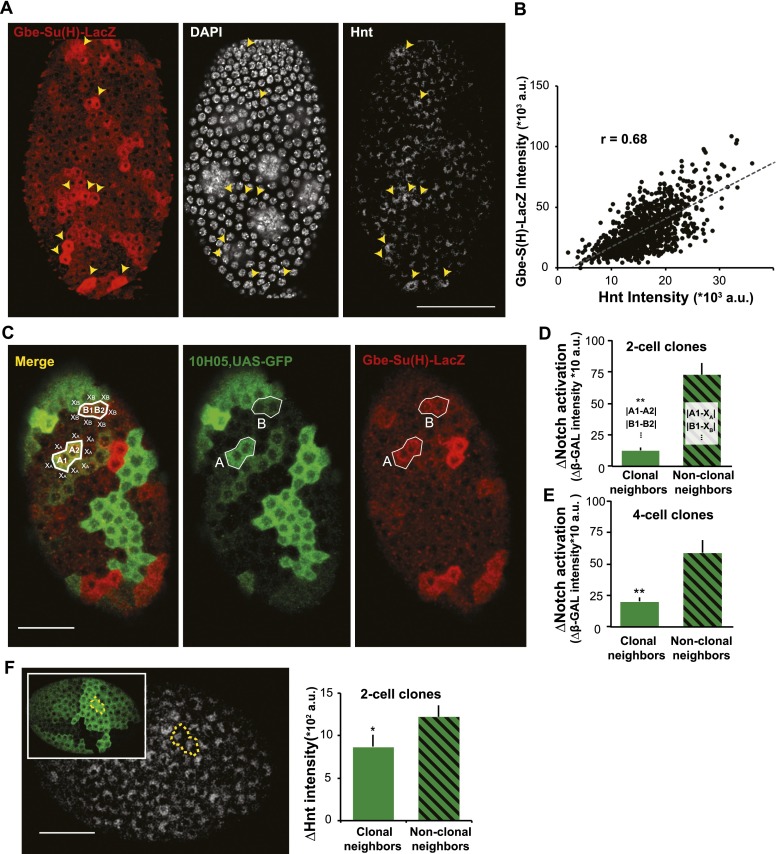
The fact that a premature decline in epigenetic plasticity leads to a precocious M–E transition strongly suggests that the state of follicle cell chromatin rather than an external signal determines when cell division shuts off. The idea that intrinsic epigenetic states gate the M–E transition was further supported by our observation that a Notch reception marker, Gbe-Su(H)-lacZ, was not induced uniformly in stage 6 follicle cells but sporadically in patches of one to four cells (Fig. 5A). The patchy expression of the Notch reporter was unlikely to be an artifact because its expression level correlated significantly with the expression of the key downstream gene hnt as measured by antibody staining (Fig. 5A,B). To determine whether epigenetically similar cell neighbors that share a common GFP expression level (clonal neighbors) were likely to induce Notch reception together, we analyzed two-cell GFP clones, which correspond to daughters of a common precursor. Indeed, clonal neighbors express very similar levels of Gbe-Su(H)-lacZ, whereas cell neighbors that express different levels of GFP (nonclonal neighbors) show much greater differences in Notch reception activity (Fig. 5C,D). Cells within four-cell clones are also more similar to each other than to their nonclonal neighbors but are not as similar as two-cell clones (Fig. 5D,E). Significantly greater concordance between the Hnt levels in two-cell GFP clones was also observed compared with Hnt levels in the nonclonal neighbors (Fig. 5F). Thus, progenitor proliferation terminates due to an epigenetic process acting during the final progenitor cell cycles that allows Notch signaling to proceed.
Figure 5.
Follicle cell behavior at the M–E transition depends on epigenetic relationships. (A) The timing and the degree of Notch activation [Gbe-Su(H)-LacZ; red] predicts endogenous Hnt expression in individual cells. Cells with higher Notch activation (yellow arrowheads) showed higher Hnt expression. (B) The degree of Notch activation [Gbe-S(H)-LacZ intensity] and endogenous Hnt expression is well correlated among stage 7–8 follicle cells. N = 973 cells from eight ovarioles. (C) Notch activation (red) and UAS-GFP variegation (green) driven by G10H05-GAL4 in stage 6 follicles undergoing the M–E transition. Individual cells were analyzed quantitatively to determine whether Notch reception values are concordant in clonally related cells sharing equal GFP expression (clonal neighbors; for example, A1, A2 and B1, B2) or cell neighbors that express different levels of GFP (nonclonal neighbors; for example, A1 vs. X1 and B1 vs. X1). Notch activation was significantly more similar in clonal neighbor in two-cell clones (D) and four-cell clones (E). N = 26 two-cell clones and 19 four-cell clones. (F) hnt expression level was also more similar between clonal neighbors among all of the two-cell clones measured. N = 32 for two-cell clones from 15 cells. Data are presented as mean ± SEM. Bars, 20 μm. Error bars indicate standard errors. (*) P < 0.05; (**) P < 0.01; (NS) nonsignificant difference, Student’s t-test.
Discussion
Lsd1 and CoREST function in multiple protein complexes as major epigenetic regulators during cell differentiation within the nervous system and many other tissues (Adamo et al. 2011; Domanitskaya and Schupbach 2012; Whyte et al. 2012; Kerenyi et al. 2013; Rudolph et al. 2013; Zhu et al. 2014). By using a sensitive variegation assay, we discovered an epigenetic function mediated by Lsd1 and CoREST that suppresses the differentiation of follicle cell progenitors until they have completed an appropriate number of divisions. Our data suggest that, in progenitors, Lsd1 complexed with CoRest acts as an epigenetic eraser that prevents methylated H3K4 marks at critical sites from prematurely reaching levels that are high enough and/or stable enough for cell cycle down-regulation and cell differentiation. They may act on newly replicated DNA, which associates with Polycomb group, Trithorax group, and other chromatin proteins after fork passage to re-establish chromatin architecture and patterns of modified histones (Petruk et al. 2012, 2013).
The process of restoring chromatin structure and organization following DNA replication is likely to be complex and involve many different proteins, including some that vary between different tissues and developmental contexts. However, our results argue that Lsd1 plays a preeminent role in follicle cell and possibly other progenitors. Lsd1 is easily detected by immunofluorescence in early follicle cell progenitors, and its level gradually falls as follicles approach the M–E transition and become less epigenetically plastic. The timing of the M–E transition, as reflected in the final number of follicle cells, is directly proportional to lsd1 gene dosage (Fig. 3C). The amount of Lsd1 present in chromatin, particularly as chromatin architecture is being re-established following DNA replication, may determine the stability of chromatin states and the sensitivity of progenitors to intercellular cues that drive differentiation (Di Stefano et al. 2007, 2011; Mulligan et al. 2011). Low levels of Lsd1/CoRest activity may allow stable epigenetic states to arise and may contribute to the context dependency of developmental signaling by modulating which target genes are able to respond to pathway activation (de Groote et al. 2012).
The relationship that we observed between Lsd1/CoRest action and Notch pathway activation clarified the relative roles played by epigenetic maturation and developmental signaling in follicle cell progenitor differentiation. Although we were able to induce premature epigenetic stabilization as early as stage 2 in lsd1ΔN/+ animals, the M–E transition did not occur until stage 5 (Fig. 3). On the other hand, while we could activate Notch in early progenitors by expressing NICD, this was not sufficient to induce an early M–E transition (Fig. 4G; Supplemental Fig. S8). These results argue that both epigenetic stabilization and Notch signaling are required to induce an M–E transition. Normally, Delta expression in germ cells increases around stage 5 (Lopez-Schier and St Johnston 2001), and we detected NICD production in overlying follicle cells before stage 6 (Supplemental Fig. S8). Thus, the initial steps of Notch signaling in follicle cells likely begin in stage 5, prior to the M–E transition, but follicle cells do not respond until they have completed Lsd1/CoRest-dependent epigenetic steps during stage 6. These results emphasize the importance of cell competency in determining the effects of intercellular signaling events and show that epigenetic competency is the rate-limiting step in the differentiation of at least some progenitors.
What process causes Lsd1 levels to fall during stages 2–6 as follicle cell progenitors proliferate and approach the time of the M–E transition? Intercellular signaling has been extensively studied during Drosophila oogenesis, but pathways other than Notch that can impact the transition are thought to act indirectly by affecting the ability of FSCs to generate normal daughters or functioning downstream from Notch signaling (Klusza and Deng 2011). A checkpoint based on normal follicle size at stage 6 is unlikely, since our experiments showed that the M–E transition can be accelerated and take place in undersized follicles. However, regulated changes in Lsd1 protein stability may play a role, since a specific ubiquitin ligase, Jade 2, targets Lsd1 for degradation via the proteasome pathway in mammalian neural progenitors (Han et al. 2014), while Lsd1 turnover is attenuated by expression of the ubiquitin-specific protease Usp28 in breast cancer cells (Wu et al. 2013). Perhaps the most interesting possibility is that gradual changes in the cellular chromatin state mediated by Lsd1/CoREST activity act as a “differentiation clock.” One or more genes encoding an Lsd1-negative regulator as well as Notch target genes would eventually become active, leading to Lsd1 down-regulation and competence to undergo the M–E transition.
How similar is the action of Lsd1 in follicle cell progenitors and the stem and differentiating cells of other tissues where it has been shown to function by regulating gene expression, heterochromatin formation, and epigenetic memory (Di Stefano et al. 2007, 2011; Rudolph et al. 2007; Katz et al. 2009; Adamo et al. 2011; Eliazer et al. 2011, 2014; Kerenyi et al. 2013; Zhu et al. 2014)? Lsd1 contributes to the maintenance and differentiation of pluripotent stem cells (Adamo et al. 2011) by controlling the activity of pluripotency gene enhancers (Whyte et al. 2012) and maintaining the silencing of key developmental genes (Adamo et al. 2011). In follicle progenitors, Lsd1 represses differentiation genes, but whether it positively regulates genes that promote the progenitor state is unknown. Lsd1 influences the number of germline stem cells in the Drosophila ovary (Eliazer et al. 2011) by affecting the differentiation of escort cells (Eliazer et al. 2014), one of the somatic cell types contributing to niche function. The decision of escort cell progenitors to differentiate occurs prior to adulthood and has not been well characterized, but without Lsd1, gene repression is abnormal, and the niche signal Dpp is overproduced. Lsd1’s action on Notch signaling in follicle progenitors is also shared by developing mouse pituitary cells (Wang et al. 2007) and in Drosophila wing and tissue culture cells, where it binds to and represses target genes (Di Stefano et al. 2011; Mulligan et al. 2011). Clearly, the role played by Lsd1 in follicle progenitors is likely to be relevant to understanding how this key chromatin regulator acts in progenitors and during differentiation in many cell types and organisms.
Our results are also likely to inform the roles that Lsd1 plays in some cancers, including both leukemias and solid tumors, where it may be expressed at high levels (Rotili and Mai 2011; Harris et al. 2012; Amente et al. 2013). Cancers are genetically heterogeneous due to ongoing mutation and genomic instability, and cell subgroups also diverge epigenetically due to different environmental interactions. Consequently, groups of cancer cells frequently lose differentiation markers and come to resemble tissue progenitors. Learning more about the evolution of chromatin states in normal tissue progenitors as they begin to differentiate will likely provide valuable insights into the properties of cancer cells that transition back toward the progenitor state due to genetic or epigenetic changes. It would be particularly interesting to learn whether Lsd1, as a key gene controlling progenitor differentiation, plays a role in allowing a subset of reverting cancer cells to function as cancer stem cells (Wang et al. 2011; Wu et al. 2013).
Finally, this study supports the utility of identifying genes based on their ability to modify GAL4∷UAS variegation as a reporter of epigenetic plasticity. The identification of genes that modify a different type of variegation, position effect variegation, has proved to be a powerful way to identify genes and pathways involved in gene silencing (Elgin and Reuter 2013). Lsd1 is found in a variety of complexes with other chromatin modifiers whose variety and dynamic behavior are challenging to characterize by purely biochemical means. Evidence is growing that long noncoding RNAs contribute to the composition and localization of such complexes (Fatica and Bozzoni 2014). Screening for genes throughout the genome that modify follicle cell GAL4∷GFP variegation represents an unbiased, genome-wide approach to identifying genes encoding proteins or RNAs that modulate epigenetic plasticity and inheritance during differentiation.
Materials and methods
Drosophila stocks
Flies were reared on standard cornmeal agar yeast food at 22°C. Fly strains used in this study were from Bloomington, Vienna Drosophila RNAi Center (VDRC), or the fly community as indicated: 179y-Gal4, c768-Gal4, and R10H05-Gal4 (a gift from Gerald Rubin and Todd Laverty); lsd1 RNAi (#1 was from VDRC, 25218; #2 was from Bloomington, #32853); lsd1ΔN (a gift from Nick Dyson); lsd11, lsd112, and lsd121 (gifts from Gunter Reuter); Sin3A08269 (Bloomington, #12350); ph-pTRiP.HMS00082 (Bloomington #33669); dnmt2105 (a gift from Gunter Reuter); dSET1 RNAi (#1 was from Bloomington, #33704; #2 was from Bloomington, #40931); Su(Var)3-91 (Bloomington, #6209); Su(Var)3-92 (Bloomington, #6210); ash1B1 (Bloomington, #5045); CoRestEY14216 (Bloomington, #20793); CoRestTRiP.HM04053 (Bloomington, #31743); Lidk0681 (Bloomington, #10403); Lid10424 (Bloomington, #12367); trx1 (Bloomington, #2114); UAS-NICD and Gbe-Su(H)-lacZ (gifts from Jianjun Sun); and lsd1Df(3L)ED4858 (Bloomington, #8088). The p[lsd_wt] transgenic fly lines were generated by gremlin transformation of P[acman] BAC CH322-21I03 DNA using a ϕC31 integrase (Rainbow Transgenic). Oregan-R was used as a control strain in this study.
Epigenetic plasticity assay
Candidate gene mutations were combined with three different GAL4 lines (R10H05-Gal4, c768-Gal4, and 179y-Gal4) to ensure that they acted globally and did not simply affect a particular GAL4 construct. R10H05 drives GFP expression beginning in FSCs, while c768 and 179y only initiate GFP expression after the M–E transition. All three Gal4 lines showed indistinguishable post-mitotic GFP variegation patterns (Skora and Spradling 2010), and line-specific behavior was not observed in these experiments.
To quantitatively measure epigenetic plasticity during the final five follicle cell divisions, we calculated the frequency with which a change in GFP expression took place at a given division in GAL4∷GFP variegating follicles (Skora and Spradling 2010). Briefly, using stained and mounted stage 10B follicles, we measured the post-mitotic sizes of individual GFP patches (known to be clonal in origin), indicating the division at which particular events occurred during progenitor growth (by rounding to the nearest power of 2). For instance, one-cell clones derive from the last mitotic division (ninth division), while two-cell clones originate at DIV8. By measuring the size of every GFP patch within a known amount of follicular surface (∼250 cells per follicle scored), the number of epigenetic changes that produced an expression level change could be determined during each division from DIV5 to DIV9 for a known total number of scored follicle cells. By dividing the number of such epigenetic events by the total number of divisions required to produce the scored cells, the probability of an epigenetic change at each division could be calculated (“change probability”). Because of the low number of changes at DIV9 and because changes in single-cell clones might have occurred post-mitotically, the conclusions of the study were based on epigenetic change probabilities in DIV5–8. All of the change probability measurements presented were calculated from at least two independent experiments.
Immunostaining and microscopy
Ovaries were dissected in Grace’s solution. Dissected ovaries were fixed in 3.7% formaldehyde in 1× PBS for 15 min at room temperature. Primary antibodies were used overnight at 4°C. Antibodies and dilutions used in this study were rabbit anti-GFP (1:1000; Invitrogen), mouse anti-Hnt (1:20; Developmental Studies Hybridoma Bank [DSHB]), mouse anti-cut (1:20; DSHB), chicken anti-βgal (pre-absorbed, 1:1000; Abcam), rabbit anti-PH3 (1:100; Cell Signaling), mouse anti-PH3 (1:1000; Cell Signaling), mouse anti-Lsd1 (1:1000; Abmart), and mouse anti-NICD (1:20; DSHB). Secondary antibodies from Invitrogen included goat anti-rabbit 488, goat anti-mouse 488, goat anti-rat 568, goat anti-chicken 568, and goat anti-mouse 633 (1:500; Molecular Probes). Stained ovaries were mounted in VectaShield on glass slides. Images were taken on an Sp5 confocal microscope and processed with ImageJ software or metamorph. For EdU experiments, ovaries were dissected in Grace’s solution and then incubated with 20 μM EdU in PBS for 1 h at room temperature. After washing with PBS three times at room temperature, EdU was detected according to the manufacturer’s instructions (Click-it EdU imaging kit, Invitrogen).
Generation of anti-Lsd1 antisera
The mouse monoclonal antibody recognizing a unique Drosophila Lsd1 peptide (KPIPAKKLGK) was generated using Abmart SEAL service. On top of testing the specificity by ELISA against its epitope, the specificity of the antibody was further confirmed by the loss of nuclear Lsd1 staining in lsd1ΔN/Df(3L)ED4858 mutant ovarioles (Supplemental Fig. S5). This mouse anti-Lsd1 monoclonal antibody (1:1000; Abmart) was used to visualize endogenous Lsd1 expression. The Lsd1 expression was quantified by measuring the integrated intensity of endogenous Lsd1 and dividing by its DNA content (Lsd1/DAPI) for every follicle nucleus throughout the follicle stages (stage 3–stage 7). For each ovariole, the stage 4–5 and stage 6–7 Lsd1/DAPI measurements were normalized to its stage 2–3 measurements.
Western blotting and immunoprecipitation
The methylation levels of H3K4 were assayed by immunoblotting ovarian lysate with the following methyl-H3K4 antibodies: rabbit anti-H3 (Abcam, #ab-1791), rabbit anti-H3Me (Millipore, #07-436), rabbit anti-H3K4Me2 (Abcam, #ab-32356), and rabbit anti-H3K4Me3 (Cell Signaling Technology, #9751). For collecting lysates, 20 pairs of ovaries were dissected from well-fed 2- to 3-d old female flies for each genetic background. Dissection was performed in Grace’s solution with proteasome inhibitors (Roche, #11836170001). Ovaries were first homogenized in 120 μL of ice-cold lysis buffer (20 mM Tris at pH 7.4, 100 mM NaCl, 0.5 mM EDTA, 0.9 M glycerol, 0.5 mM DTT, 0.25% NP-40/IGEPAL-CA630, Complete protease inhibitor cocktail [Roche, #11836170001]) using blue pestles and incubated for 5 min on ice. Samples were prepared by supplementing each lysate with sample buffer/DDT. Five microliters to 10 μL of each sample was loaded onto the precast gel (Bio-Rad, #161-1104) and then transferred onto PVDF membranes (Invitrogen, #lc2005) for Western blotting. Transferred membranes were incubated with primary antibodies overnight at 4°C and HRP-conjugated secondary antibodies for 2 h at room temperature. The blots were finally visualized using ECL Western blotting kit (GE Healthcare, #RPN2236).
For immunoprecipitation experiments, ∼150 pairs of ovaries from well-fed 4- to 5-d-old female flies were manually dissected in Grace’s solution with proteasome inhibitors (Roche, #11836170001). Ovaries were then homogenized in 0.5–1 mL of ice-cold lysis buffer (20 mM Tris at pH 7.4, 100 mM NaCl, 0.5 mM EDTA, 0.9 M glycerol, 0.5 mM DTT, 0.25% NP-40/IGEPAL-CA630, Complete protease inhibitor cocktail [Roche#11836170001]). The lysate was spun at 12,000 rpm for 15 min at 4°C to remove tissue debris. Collected supernatant was then precleaned by adding A/G beads (50–100 μL/lysis buffer; Pierce) and incubated for 30 min at 4°C. Next, 200–300 μL of lysate was used for each immunoprecipitation reaction by adding 1–5 μg of antibodies. Immunoprecipitation was performed on a rotating plate for 3 h (or overnight) at 4°C, and 20 μL of washed A/G beads (Pierce) was added and incubated for 2 h at 4°C. These were quickly washed four times with wash buffer (50 mM TrisCl at pH 7.9, 10% glycerol, 100 mM KCl, 0.2 mM EDTA, 5 mM MgCl2, 10 mM β-ME, 0.1% NP-40/IGEPAL-CA630). The Lsd1–CoRest interaction was then assayed by immunoblot using either guinea pig anti-Lsd1 (1:1000; a gift from Dr. Michael Buszczak) or rabbit anti-CoRest (a gift from Gail Mandel) (Dallman et al. 2004).
Modeling the effects of epigenetic plasticity changes during follicle cell progenitor divisions
In order to visualize the expected patterns of follicle cell variegation under an arbitrary regime of change probabilities, we simulated the appearance of the follicle surface following eight mitotic divisions in which random shifts among 10 discontinuous expression levels occur, with specified probability values at each division. The code was written using MATLAB based on the following spatial model: A single 2 × 2 square cell underwent first horizontal and then vertical divisions with expression changes as described above until eight divisions were completed (256 “cells”) (Supplemental Fig. S2A).
When the empirically measured change probabilities for all eight divisions were used in the model, the predicted pattern of GFP variegation strongly resembled that observed in vivo (Supplemental Fig. S2B, top row). We then used the model to predict the variegation patterns expected in the case of modifier genes that greatly enhanced or suppressed epigenetic plasticity during progenitor development. High epigenetic plasticity throughout progenitor divisions predicts extensively mosaic “pepper and salt” variegation patterns (Supplemental Fig. S2B, middle row), while greatly accelerated loss of plasticity will result in relatively homogeneous GFP expression, broken only rarely by variant clonal patches (Supplemental Fig. S2B, bottom row).
Supplementary Material
Acknowledgments
We thank G.M. Rubin and Todd Laverty for the R10H05 stock. We are grateful to Jack Bateman and Ting Wu for sending ϕC31 vectors. We thank the Transgenic RNAi Project (TRiP) at Harvard Medical School (National Institutes of Health/National Institute of General Medical Sciences R01-GM084947) for providing transgenic RNAi fly stocks and/or plasmid vectors used in this study. We thank Shih-Chieh Lin for his feedback and comments in developing the mathematical model of variegation. We are grateful to members of the Spradling laboratory for comments on the manuscript and helpful discussions. A.C.S is an Investigator of the Howard Hughes Medical Institute. M.-C.L. designed and performed research, analyzed data, and wrote the paper. A.C.S. designed research, analyzed data, and wrote the paper.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.252692.114.
References
- Adamo A, Sesé B, Boue S, Castaño J, Paramonov I, Barrero MJ, Izpisua Belmonte JC. 2011. LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat Cell Biol 13: 652–659. [DOI] [PubMed] [Google Scholar]
- Amente S, Lania L, Majello B. 2013. The histone LSD1 demethylase in stemness and cancer transcription programs. Biochim Biophys Acta 1829: 981–986. [DOI] [PubMed] [Google Scholar]
- Airoldi SJ, McLean PF, Shimada Y, Cooley L. 2011. Intercellular protein movement in syncytial Drosophila follicle cells. J Cell Sci 124: 4077–4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badeaux AI, Shi Y. 2013. Emerging roles for chromatin as a signal integration and storage platform. Nat Rev Mol Cell Biol 14: 211–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceballos-Chavez M, Rivero S, Garcia-Gutierrez P, Rodriguez-Paredes M, Garcia-Dominguez M, Bhattacharya S, Reyes JC. 2012. Control of neuronal differentiation by sumoylation of BRAF35, a subunit of the LSD1-CoREST histone demethylase complex. Proc Natl Acad Sci 109: 8085–8090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallman JE, Allopenna J, Bassett A, Travers A, Mandel G. 2004. A conserved role but different partners for the transcriptional corepressor CoREST in fly and mammalian nervous system formation. J Neurosci 24: 7186–7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groote ML, Verschure PJ, Rots MG. 2012. Epigenetic Editing: targeted rewriting of epigenetic marks to modulate expression of selected target genes. Nucleic Acids Res 40: 10596–10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng WM, Althauser C, Ruohola-Baker H. 2001. Notch–Delta signaling induces a transition from mitotic cell cycle to endocycle in Drosophila follicle cells. Development 128: 4737–4746. [DOI] [PubMed] [Google Scholar]
- Di Stefano L, Ji JY, Moon NS, Herr A, Dyson N. 2007. Mutation of Drosophila Lsd1 disrupts H3-K4 methylation, resulting in tissue-specific defects during development. Curr Biol 17: 808–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano L, Walker JA, Burgio G, Corona DF, Mulligan P, Naar AM, Dyson NJ. 2011. Functional antagonism between histone H3K4 demethylases in vivo. Genes Dev 25: 17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domanitskaya E, Schupbach T. 2012. CoREST acts as a positive regulator of Notch signaling in the follicle cells of Drosophila melanogaster. J Cell Sci 125: 399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin SC, Reuter G. 2013. Position-effect variegation, heterochromatin formation and gene silencing in Drosophila. Cold Spring Harb Perspect Biol 5: a017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliazer S, Shalaby NA, Buszczak M. 2011. Loss of lysine-specific demethylase 1 nonautonomously causes stem cell tumors in the Drosophila ovary. Proc Natl Acad Sci 108: 7064–7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliazer S, Palacios V, Wang Z, Kollipara RK, Kittler R, Buszczak M. 2014. Lsd1 restricts the number of germline stem cells by regulating multiple targets in escort cells. PLoS Genet 10: e1004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Bozzoni I. 2014. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 15: 7–21. [DOI] [PubMed] [Google Scholar]
- Han X, Gui B, Xiong C, Zhao L, Liang J, Sun L, Yang X, Yu W, Si W, Yan R, et al. 2014. Destabilizing D1 by Jade-2 promotes neurogenesis: an antibraking system in neural development. Mol Cell 55: 482–494. [DOI] [PubMed] [Google Scholar]
- Harris WJ, Huang X, Lynch JT, Spencer GJ, Hitchin JR, Li Y, Ciceri F, Blaser JG, Greystoke BF, Jordan AM, et al. 2012. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell 21: 473–487. [DOI] [PubMed] [Google Scholar]
- Iglesias-Bartolome R, Callejas-Valera JL, Gutkind JS. 2013. Control of the epithelial stem cell epigenome: the shaping of epithelial stem cell identity. Curr Opin Cell Biol 25: 162–169. [DOI] [PubMed] [Google Scholar]
- Katz DJ, Edwards TM, Reinke V, Kelly WG. 2009. A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell 137: 308–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerenyi MA, Shao Z, Hsu YJ, Guo G, Luc S, O’Brien K, Fujiwara Y, Peng C, Nguyen M, Orkin SH. 2013. Histone demethylase Lsd1 represses hematopoietic stem and progenitor cell signatures during blood cell maturation. eLife 2: e00633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Zhang Y. 2007. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol 8: 307–318. [DOI] [PubMed] [Google Scholar]
- Klusza S, Deng WM. 2011. At the crossroads of differentiation and proliferation: precise control of cell-cycle changes by multiple signaling pathways in Drosophila follicle cells. BioEssays 33: 124–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra SM, Helin K. 2012. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol 13: 297–311. [DOI] [PubMed] [Google Scholar]
- Lee N, Zhang J, Klose RJ, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. 2007. The trithorax-group protein Lid is a histone H3 trimethyl-Lys4 demethylase. Nat Struct Mol Biol 14: 341–343. [DOI] [PubMed] [Google Scholar]
- Lopez-Schier H, St Johnston D. 2001. Delta signaling from the germ line controls the proliferation and differentiation of the somatic follicle cells during Drosophila oogenesis. Genes Dev 15: 1393–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis J, Spradling A. 1995. Identification and behavior of epithelial stem cells in the Drosophila ovary. Development 121: 3797–3807. [DOI] [PubMed] [Google Scholar]
- McLean PF, Cooley L. 2013. Protein equilibration through somatic ring canals in Drosophila. Science 340: 1445–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan P, Yang F, Di Stefano L, Ji JY, Ouyang J, Nishikawa JL, Toiber D, Kulkarni M, Wang Q, Najafi-Shoushtari SH, et al. 2011. A SIRT1–LSD1 corepressor complex regulates Notch target gene expression and development. Mol Cell 42: 689–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Johnston DM, Hodgson JW, Black KL, Kovermann SK, Beck S, Canaani E, Brock HW, Mazo A. 2012. TrxG and PcG proteins but not methylated histones remain associated with DNA through replication. Cell 150: 922–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S, Black KL, Kovermann SK, Brock HW, Mazo A. 2013. Stepwise histone modifications are mediated by multiple enzymes that rapidly associate with nascent DNA during replication. Nat Commun 4: 2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotili D, Mai A. 2011. Targeting histone demethylases: a new avenue for the fight against cancer. Genes Cancer 2: 663–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph T, Yonezawa M, Lein S, Heidrich K, Kubicek S, Schafer C, Phalke S, Walther M, Schmidt A, Jenuwein T, et al. 2007. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Mol Cell 26: 103–115. [DOI] [PubMed] [Google Scholar]
- Rudolph T, Beuch S, Reuter G. 2013. Lysine-specific histone demethylase LSD1 and the dynamic control of chromatin. Biol Chem 394: 1019–1028. [DOI] [PubMed] [Google Scholar]
- Shi Y. 2007. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet 8: 829–833. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119: 941–953. [DOI] [PubMed] [Google Scholar]
- Skora AD, Spradling AC. 2010. Epigenetic stability increases extensively during Drosophila follicle stem cell differentiation. Proc Natl Acad Sci 107: 7389–7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Deng WM. 2005. Notch-dependent downregulation of the homeodomain gene cut is required for the mitotic cycle/endocycle switch and cell differentiation in Drosophila follicle cells. Development 132: 4299–4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Deng WM. 2007. Hindsight mediates the role of notch in suppressing hedgehog signaling and cell proliferation. Dev Cell 12: 431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. 2010. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329: 689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I, et al. 2007. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature 446: 882–887. [DOI] [PubMed] [Google Scholar]
- Wang J, Lu F, Ren Q, Sun H, Xu Z, Lan R, Liu Y, Ward D, Quan J, Ye T, et al. 2011. Novel histone demethylase LSD1 inhibitors selectively target cancer cells with pluripotent stem cell properties. Cancer Res 71: 7238–7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. 2012. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 482: 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Wang Y, Yang XH, Kang T, Zhao Y, Wang C, Evers BM, Zhou BP. 2013. The deubiquitinase USP28 stabilizes LSD1 and confers stem-cell-like traits to breast cancer cells. Cell Rep 17: 224–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Bonasio R, Strino F, Kluger Y, Holloway JK, Modzelewski AJ, Cohen PE, Reinberg D. 2013. SFMBT1 functions with LSD1 to regulate expression of canonical histone genes and chromatin-related factors. Genes Dev 27: 749–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Holz S, Metzger E, Pavlovic M, Jandausch A, Jilg C, Galgoczy P, Herz C, Moser M, Metzger D, et al. 2014. Lysine-specific demethylase 1 regulates differentiation onset and migration of trophoblast stem cells. Nat Commun 5: 3174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.