Huang et al. demonstrate that the Drosophila KAT6 Enok acetylates histone H3 Lys 23 (H3K23). Mutants lacking functional Enok lost germline formation and abdominal segments in the embryo. Enok is localized to and acetylates H3K23 at the spire (spir) and maelstrom (mael) genes, and expression of spir in the germline can largely rescue the defective Osk localization in enok mutant ovaries.
Keywords: H3K23 acetylation, Oskar localization, germ plasm
Abstract
KAT6 histone acetyltransferases (HATs) are highly conserved in eukaryotes and have been shown to play important roles in transcriptional regulation. Here, we demonstrate that the Drosophila KAT6 Enok acetylates histone H3 Lys 23 (H3K23) in vitro and in vivo. Mutants lacking functional Enok exhibited defects in the localization of Oskar (Osk) to the posterior end of the oocyte, resulting in loss of germline formation and abdominal segments in the embryo. RNA sequencing (RNA-seq) analysis revealed that spire (spir) and maelstrom (mael), both required for the posterior localization of Osk in the oocyte, were down-regulated in enok mutants. Chromatin immunoprecipitation showed that Enok is localized to and acetylates H3K23 at the spir and mael genes. Furthermore, Gal4-driven expression of spir in the germline can largely rescue the defective Osk localization in enok mutant ovaries. Our results suggest that the Enok-mediated H3K23 acetylation (H3K23Ac) promotes the expression of spir, providing a specific mechanism linking oocyte polarization to histone modification.
In a wide range of metazoan species, primordial germ cells (PGCs) are determined by a specialized cytoplasm called germ plasm (Extavour and Akam 2003). The Drosophila germ plasm is localized at the posterior pole of the embryo and is enriched with mitochondria and ribonucleoprotein complexes. In addition to specifying the identity of PGCs, germ plasm is required to establish the gradient of the posterior determinant Nanos in embryos and therefore plays a critical role in abdominal segmentation (Williamson and Lehmann 1996). Germ plasm assembly is organized by Oskar (Osk) during oogenesis (Ephrussi and Lehmann 1992). As the critical determinant of germ plasm, the localization and translation of osk mRNA are highly regulated. More than 50 proteins have been identified to regulate the localization of osk mRNA (St Pierre et al. 2014), and among them are the osk mRNA-binding protein Staufen (Stau) and cytoskeleton regulators, including Spire (Spir), Cappuccino (Capu), and Maelstrom (Mael) (Brendza et al. 2000; Dahlgaard et al. 2007; Sato et al. 2011). The transportation of osk mRNA toward the posterior pole of the oocyte depends on a polarized microtubule (MT) network. Mael interacts with the MT-organizing center (MTOC) and regulates the formation of this polarized MT network inside the oocyte (Sato et al. 2011). In addition, the polarized MT network is maintained during stages 8–10A of oogenesis by an ooplasmic actin mesh organized by Spir and Capu (Dahlgaard et al. 2007).
During transportation of osk mRNA, the eIF4E–Cup–Bru complex binds to it and inhibits its translation (Nakamura et al. 2004). This translational inhibition is relieved by Vasa (Vas), Aubergine (Aub), and Orb once the osk mRNA reaches the posterior pole of the oocyte (Wilkie et al. 2003). Moreover, the continuous anchoring of posteriorly localized osk mRNA throughout oogenesis requires Osk protein, resulting in a positive feedback loop for Osk localization that promotes germ plasm organization (Rongo et al. 1995). Although many gene products regulating the Osk localization have been intensively studied, the specific mechanisms governing the upstream transcriptional regulation of each of these genes are still largely unknown.
KAT6 histone acetyltransferases (HATs) are conserved between yeast and metazoans and are generally involved in transcriptional regulation (Yang 2004). In yeast, Sas3 targets histone H3 Lys 9 (H3K9) and H3K14 in vivo and plays a role in transcriptional silencing and cell cycle progression (Howe et al. 2001). Human MOZ, the homolog of Sas3, was first identified as a fusion partner of CREB-binding protein (CBP) in acute myeloid leukemia. It acetylates H3K9 in vivo and facilitates transcriptional activation, with its interacting partners, p53 and Runx2, contributing to regulation of the cell cycle and hematopoiesis (Perez-Campo et al. 2013). Compared with the yeast and human homologs, much less is known about the Drosophila KAT6 Enok. Enok has been shown to play roles in neuroblast proliferation and maintenance of germline stem cells (Scott et al. 2001; Xin et al. 2013). However, its enzymatic activity and transcriptional targets are unknown. In this study, we identify H3K23 as an in vitro and in vivo substrate for Enok. We show that spir and mael are among the specific set of genes requiring Enok for expression in the ovary. Furthermore, loss of functional Enok resulted in defective Osk localization in the oocyte without affecting the overall development of nurse cells, and this defect was rescued by exogenous expression of spir. Therefore, Enok is required for Osk-dependent germline cell formation and abdominal segmentation in embryos.
Results
Enok acetylates H3K23 in vitro and in vivo
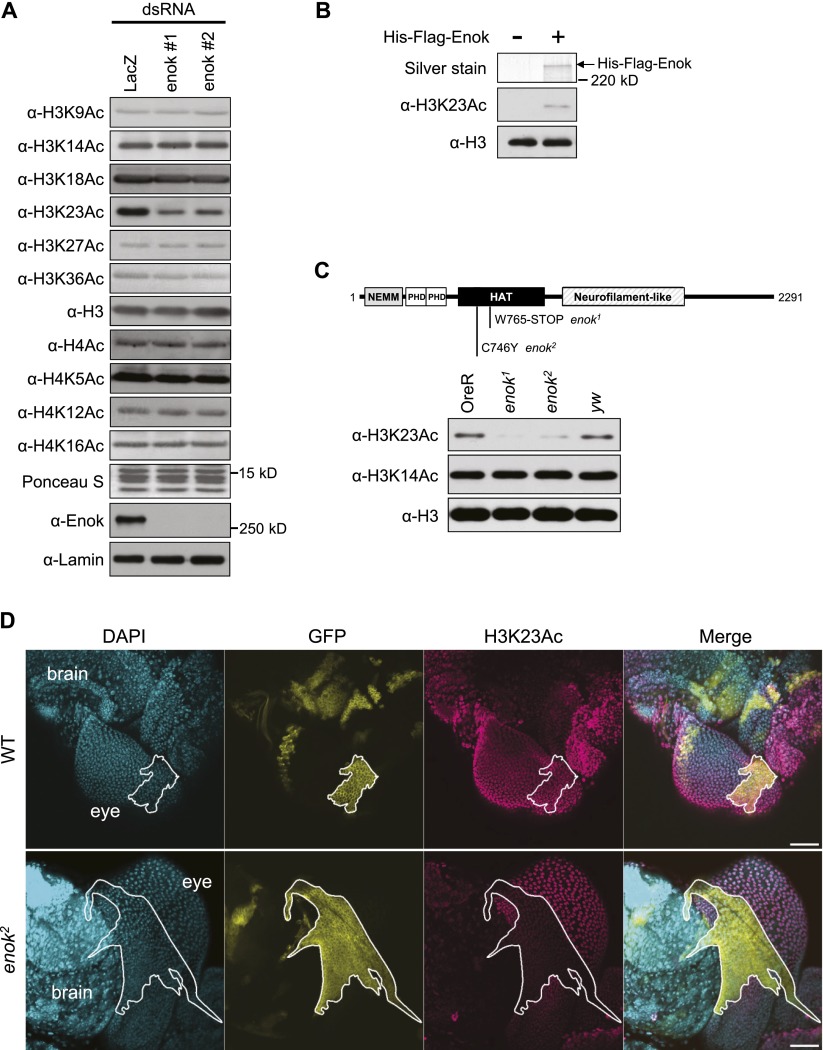
To identify the histone target for Enok, endogenous enok was knocked down in S2 cells using dsRNAs against enok, and the levels of different histone acetylation marks were examined by Western blotting. RT-qPCR analysis showed that dsRNA treatment for 4–8 d resulted in 60%–80% reduction in the enok mRNA levels (Supplemental Fig. S1A). Among the 10 histone acetylation marks examined in Figure 1A, the levels of H3K23 acetylation (H3K23Ac) were decreased in S2 cells treated with dsRNAs against enok compared with the control dsRNA, while the other lysine acetylation levels remained largely unaffected (Fig. 1A; Supplemental Fig. S1B). Furthermore, recombinant Enok purified using the baculovirus expression system showed HAT activity toward H3K23 in recombinant nucleosomes in vitro (Fig. 1B), indicating that Enok can directly acetylate histone H3 at the K23 residue. Interestingly, in vitro, recombinant Enok did not acetylate H3K9 or H3K14 (Supplemental Fig. S1C), which are the targets of its mammalian and yeast homologs (Howe et al. 2001; Ullah et al. 2008; Voss et al. 2009), suggesting that Enok uniquely prefers H3K23 as the substrate among these three residues. Nevertheless, we cannot exclude the possibility that Enok may have additional targets or that other factors may interact with Enok to expand its substrate specificity in vivo.
Figure 1.
Enok acetylates H3K23 in vitro and in vivo. (A, top 12 panels) Histones were acid-extracted from S2 cells treated with control dsRNA (LacZ) or two nonoverlapping dsRNAs against enok and subjected to Western blot analysis. (Bottom two panels) Nuclear extracts were prepared from dsRNA-treated S2 cells and subjected to Western blotting. (B) In vitro HAT assay was performed using recombinant His-Flag-tagged Enok and nucleosomes reconstituted with a 601 positioning sequence and recombinant histones. (C) Histones were acid-extracted from embryos 20–24 h old and subjected to Western blotting. Oregon-R (OreR) and yw were used as wild-type controls. Homozygous mutant embryos were obtained by picking the GFP-negative embryos from the following strains: enok1/CyO, P{GAL4-twi.G}2.2, P{UAS-2xEGFP}AH2.2 (enok1) and enok2/CyO, P{GAL4-twi.G}2.2, P{UAS-2xEGFP}AH2.2 (enok2). (NEMM) N-terminal region of Enok, MOZ, or MORF; (PHD) plant homeodomain. (D) MARCM analysis was performed in the peripodial membrane of the eye disc. Clones homozygous for the wild-type (WT) control or enok2 are marked by white lines. Genotypes were as follows: hs-Flp, USA-mCD8-GFP/+; FRTG13/FRTG13, tubP-GAL80; tubP-GAL4/+ (wild type) and hs-Flp, USA-mCD8-GFP/+; FRTG13, enok2/FRTG13, tubP-GAL80; tubP-GAL4/+ (enok2). Bars, 50 µm.
Next, we tested whether the H3K23Ac levels were affected in the two enok loss-of-function mutants enok1 and enok2 (Scott et al. 2001). Indeed, the global levels of H3K23Ac were decreased in 20- to 24-h embryos (Fig. 1C) and third instar larvae (Supplemental Fig. S1D) homozygous for the mutant alleles enok1 or enok2 compared with the wild-type controls. Also, since the point mutation in the enok2 allele changes a conserved cysteine residue in the HAT domain (Fig. 1C), we concluded that bulk H3K23Ac levels are dependent on the HAT activity of Enok. To minimize genetic background differences, we performed the mosaic analysis with a repressible cell marker (MARCM) (Lee and Luo 1999). This approach also permits analysis of the effects of enok mutants in adult tissues, since homozygotes do not survive to adulthood. In the MARCM analysis, cell clones homozygous for enok2 or the wild-type control were generated by FLP/FRT-mediated mitotic recombination and marked by the expression of GFP. As shown in Figure 1D, cells in the peripodial membrane of the eye disc in a wild-type homozygous clone showed H3K23Ac staining similar to that of cells outside the clone (top panel). However, the H3K23Ac staining signals were weaker in cells from a homozygous enok2 clone compared with cells outside the clone (Fig. 1D, bottom panel). Similar results were observed in the adult follicle cells, with reduced H3K23Ac staining in a follicle cell clone homozygous for enok1 compared with follicle cells outside the clone (Supplemental Fig. S1E). Taken together, these results indicate that Enok functions as the major acetyltransferase for H3K23 in vivo.
Enok is enriched in the ovary and embryo
To investigate the biological function of Enok, we first examined the subcellular localization of Enok in S2 cells. The cell fractionation analysis showed that Enok is present in both the nucleus and cytosol (Supplemental Figs. S2, S3A). The cytoplasmic pool of Enok may have nonhistone substrates, and the transportation of Enok between the nucleus and the cytoplasm may play a role in regulating its HAT function. We further examined the Enok levels at different stages of development (Supplemental Fig. S3B). Consistent with the previous study on the developmental transcriptome (Graveley et al. 2011), Enok protein levels are higher during the embryonic stages and in the ovary.
Maternal Enok is important for abdominal segmentation in embryos
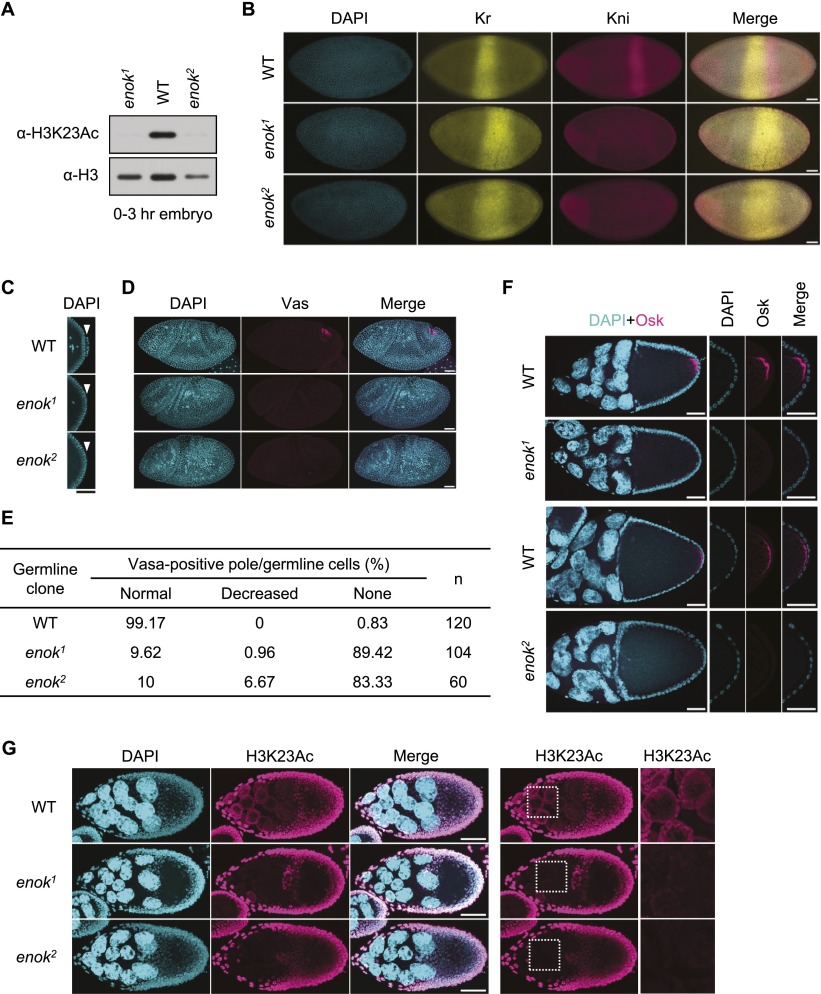
The abundance of Enok in ovaries and embryos prompted us to examine whether the maternal loading of Enok plays a role in embryonic development. To this end, a germline clone analysis using the FLP/FRT/ovoD system was performed. In the ovoD system, only egg chambers with homozygous mutant germline cells can develop beyond stage 4 of oogenesis (Chou and Perrimon 1996), and the resulting embryos will lack any maternally provided gene product. Consistent with the presence of ovoD, germline clone females without heat-shock-induced FLP expression rarely showed egg chambers developing beyond stage 4 (Supplemental Fig. S4). Unfertilized eggs (Supplemental Fig. S5A) and 0- to 3-h embryos (Fig. 2A) from enok mutant germline clones, therefore lacking the maternal contribution of functional Enok, showed reduced levels of H3K23Ac as compared with the wild-type control. The global H3K23Ac levels were also greatly decreased in 6- to 24-h embryos lacking both the maternal and the zygotic contribution of functional Enok (Supplemental Fig. S5B). These results suggest that Enok is critical for establishing the H3K23Ac mark in the mature oocyte and during embryogenesis.
Figure 2.
Enok is critical for the Osk-mediated germ plasm formation. (A) Whole-cell extracts (WCEs) were prepared from 0- to 3-h embryos and subjected to Western blotting. Embryos were collected from crosses of the germline clone females with enok1/CyO, P{GAL4-twi.G}2.2, P{UAS-2xEGFP}AH2.2 males. (B) Stage 5 germline clone embryos were stained with DAPI, α-Kr, and α-Kni antibodies. Posterior is to the right in all figures showing embryos or egg chambers. (C) Middle sections of the posterior end of embryos shown in B are enlarged. The position of pole cells is indicated by the white arrowheads. (D) Stage 7 germline clone embryos were stained with DAPI and an α-Vas antibody. (E) Embryos were stained with DAPI and an α-Vas antibody. The staining pattern of Vas in embryos beyond stage 4 was examined, and the percentages of embryos with normal or decreased numbers of Vas-positive cells or without Vas-positive cells were then calculated, respectively. The total number of examined embryos (n) is also shown. (F) Stage 10 egg chambers were stained with DAPI and an α-Osk antibody. (G) Stage 9 egg chambers were stained with DAPI and an α-H3K23Ac antibody. The genotypes of females in A–G are as follows: hs-Flp/+; FRTG13/FRTG13, ovoD1–18 (wild-type [WT]); hs-Flp/+; FRTG13, enok1/FRTG13, ovoD1–18 (enok1); and hs-Flp/+; FRTG13, enok2/FRTG13, ovoD1–18 (enok2). Embryos used in B–E were collected from crosses of the germline clone females with OreR males. Bars: B–G, 50 µm.
We then mated the germline clone females with wild-type (Oregon-R [OreR]) males to ensure that any defects in mutant embryos are due solely to the absence of maternally loaded Enok and not the loss of zygotic expression. In wild-type stage 12 embryos, the segment polarity gene engrailed (en) is expressed in a 14-stripe pattern. Interestingly, En staining in ∼60% of enok mutant germline clone embryos showed only eight stripes instead of the normal 14 stripes (Supplemental Fig. S5C,D), suggesting loss of six segments during development. Staining of Ultrabithorax (Ubx), a homeobox protein normally expressed in abdominal segments A1–A7, revealed that abdominal segments A1–A7 are fused into one segment in germline clone embryos from enok1 and enok2 (Supplemental Fig. S5E). The defects in abdominal segmentation in these embryos were further confirmed by examining the expression of the pair rule gene even-skipped (eve). In contrast to the seven-stripe pattern seen in the wild-type control, stripes 4–6 of Eve were absent in enok1 and enok2 (Supplemental Fig. S5F). As the loss of Eve stripes 4–6 and abdominal segments A2–A7 is a typical phenotype of mutants in the gap gene knirps (kni) (Nibu et al. 1998), we examined whether the expression of kni is affected in enok mutant germline clone embryos. Indeed, the posterior band of Kni staining was severely reduced in enok1 and enok2 as compared with the wild-type control, while the anterior band was largely unaffected (Fig. 2B). Consistently, the expression pattern of Kruppel (Kr), another gap gene repressed by Kni (Hoch et al. 1992), was expanded posteriorly in the enok mutant germline clone embryos compared with the wild-type control (Fig. 2B). Therefore, we concluded that Enok is required maternally for the posterior expression of kni and downstream abdominal segmentation in embryos.
Enok plays a critical role in the localization of Osk to the germ plasm
When examining stage 5 embryos, we noticed that pole cells, the germline precursors, were not formed in the enok mutant germline clone embryos (Fig. 2C). Staining of the germline marker Vas confirmed the absence of germline cells in 80%–90% of the enok1 and enok2 germline clone embryos (Fig. 2D,E). These defects in abdominal segmentation and pole cell formation strongly suggested a role for Enok in the assembly of the germ plasm, which is organized by Osk (Ephrussi et al. 1991). Supporting this possibility, the localization of Osk to the posterior pole of the oocyte was disrupted in enok mutant germline clones (Fig. 2F). Since germ plasm formation in the oocyte depends on multiple components contributed by the germline nurse cells (Williamson and Lehmann 1996), we examined whether loss of functional Enok disrupted the overall development of nurse cells. As shown in Supplemental Figure S6A, enok mutant nurse cells showed normal ploidy levels during development. On the other hand, Enok is important for the global acetylation of H3K23 in nurse cells (Fig. 2G; Supplemental Fig. S6B). Therefore, we hypothesized that the acetylation of H3K23 by Enok may transcriptionally regulate germ plasm assembly.
The expression of spir and mael are down-regulated in enok mutant ovaries
To investigate whether Enok transcriptionally regulates genes involved in osk mRNA localization, we compared the transcriptomes of the enok mutant germline clone ovaries and wild-type control ovaries by RNA sequencing (RNA-seq). There was a 60%–75% overlap in genes down-regulated in the enok1 and enok2 ovaries and a 25%–65% overlap in the up-regulated genes (Supplemental Fig. S7A; Supplemental Table S1), indicating that a considerable portion of genes affected by mutations in enok are regulated similarly in the two mutants. The larger number (approximately fivefold) of commonly down-regulated genes compared with commonly up-regulated genes in the mutants suggests a transcription-activating role for Enok during oogenesis (Supplemental Fig. S7A).
Among the 6732 genes expressed in the wild-type ovary, only 183 genes are commonly down-regulated and only 26 genes are commonly up-regulated in both enok mutants (Supplemental Fig. S7A). Thus, loss of functional Enok specifically affects a small subset (∼3%) of genes in the ovary. To obtain more functional insights from the genes regulated by Enok, we first focused on genes known to regulate oogenesis (Supplemental Tables S2, S3; St Pierre et al. 2014). As shown in Supplemental Figure S7B, 15 oogenesis-related genes are down-regulated in both enok mutants compared with the wild-type control. Among the 15 genes, five genes (33%) are involved in germ plasm formation and/or specification of the anterior–posterior axis in the embryo: spir, mael, torso (tor), caudal (cad), and mitochondrial large ribosomal RNA (mt:lrRNA). This finding suggests that Enok may play a role in transcriptionally regulating these two related pathways. The defective Osk localization observed in the enok mutant oocyte is similar to the mutant phenotypes of spir and mael (Fig. 2F; Dahlgaard et al. 2007; Sato et al. 2011). We did not observe the loss of abdominal segment A8 characteristic of the maternal tor or cad mutants in the enok mutant germline clone embryos (Supplemental Fig. S5C,E; Ambrosio et al. 1989; Wu and Lengyel 1998), probably due to the moderate reduction in levels of tor and cad in enok mutants (25%–50%). The product of mt:lrRNA is important for germ plasm formation but functions downstream from osk localization (Iida and Kobayashi 1998). In addition, although the changes in the expression levels of mt:lrRNA in enok mutants are statistically significant, this level is reduced only 20%. Taken together, we hypothesized that Enok may play an important role in the Spir/Mael–Osk localization pathway and function with other factors/mechanisms to regulate formation of the germ plasm and specification of the anterior–posterior axis in embryos.
Next, in order to identify pathways other than oogenesis that may be regulated by Enok, we performed a gene ontology (GO) term analysis of the genes that are commonly down-regulated or up-regulated in the two enok mutant germline clone ovaries. As shown in Supplemental Figure S8, three pathways are enriched in the down-regulated genes, and one pathway is enriched in the up-regulated genes. One interesting pathway enriched in the down-regulated genes is the antibacterial humoral response. The fly NF-κB Relish (Rel) has been reported to activate the transcription of all of the other genes in the list: Def, CecA2, AttB, Dro, and AttA (Boutros et al. 2002). Therefore, Enok may contribute to the innate immune response by regulating the key transcription factor Rel.
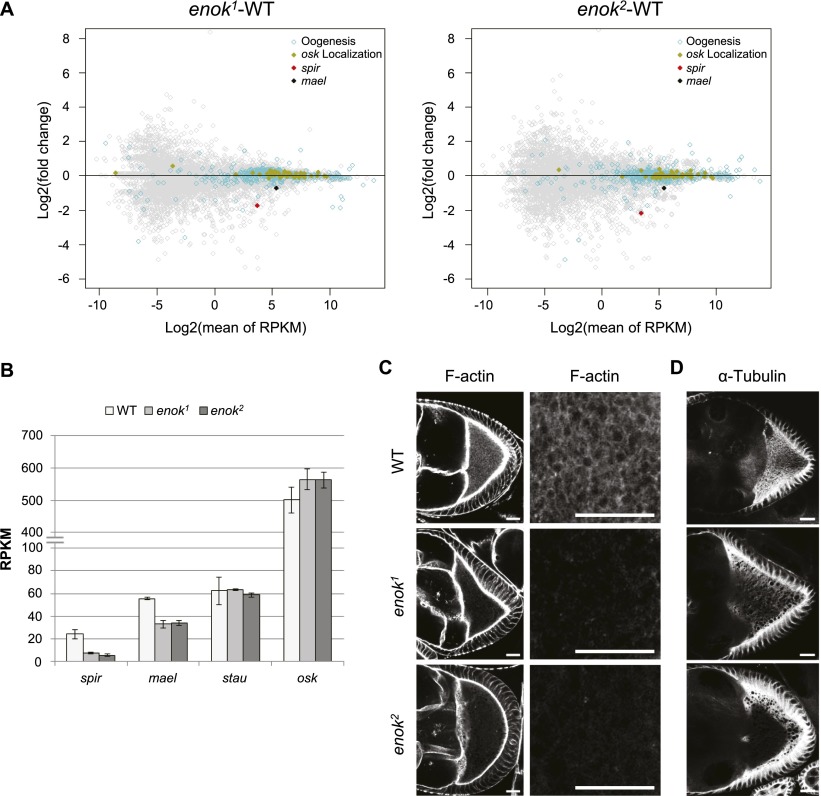
Given the defective Osk localization in oocytes observed in enok mutants, we examined the expression levels of genes known to be involved in osk mRNA localization (St Pierre et al. 2014). Among the 52 osk localization-related genes, spir and mael, the two genes mentioned earlier, were specifically down-regulated in both the enok1 and enok2 germline clone ovaries as compared with the wild-type control (Fig. 3A,B; Supplemental Fig. S9A). Mael associates with the MTOC and is required for reorganization of the MT network at stage 7 to establish oocyte polarity (Sato et al. 2011). Spir is an actin nucleation factor and functions together with the formin Capu to organize the actin mesh that regulates the polarized MT network in oocytes during stages 8–10A when the osk mRNA–protein complex is transported to and anchored at the posterior pole (Dahlgaard et al. 2007). As a result, both the spir and mael mutants display premature MT-based ooplasmic streaming starting from stage 7, which disrupts localization of the osk mRNA-binding protein Stau to the posterior pole of oocytes (Supplemental Fig. S9B; Emmons et al. 1995; Clegg et al. 1997; Dahlgaard et al. 2007). Therefore, we examined the actin mesh, the polarized MT network, and the localization of Stau in oocytes from enok1 and enok2 germline clone ovaries. Indeed, the actin mesh in the stage 9 wild-type oocyte was not formed in enok mutant oocytes (Fig. 3C), and the polarized MT network (anterior–posterior MT gradient in the wild-type oocyte) was disordered (Fig. 3D). In addition, the posterior localization of Stau in stage 9 oocytes was abrogated in enok1 and enok2 mutant egg chambers (Supplemental Fig. S9C). Taken together, these results suggest that Enok is important for the expression of spir and mael in ovaries, contributing to the proper localization of osk mRNA and formation of germ plasm in oocytes.
Figure 3.
spir and mael are down-regulated in the enok mutant ovaries. (A) MA plots showing fold change in expression (reads per kilobase per million [RPKM]) plotted against the expression levels for the enok1 (left panel) or the enok2 (right panel) mutant ovaries versus the wild-type (WT) control. Genes involved in oogenesis or osk mRNA localization are marked with cyan circles or gold dots, respectively. The red and black dots represent spir and mael, respectively. (B) The expression levels of spir, mael, stau, and osk in the wild-type control and the mutant ovaries are shown in RPKM obtained by RNA-seq. Data represent the mean of three biological replicates ± SD. (C) Stage 9 egg chambers were stained with phalloidin to label the actin filaments (F-actin). Magnifications of the actin mesh in the oocyte cytoplasm are shown in the right panel. (D) Stage 9 egg chambers were stained with an α-α-Tubulin antibody to label the MT network in the oocyte. Bars: C,D, 20 µm. Genotypes of females in A–D are as indicated in Figure 2.
Up-regulation of bruno (bru) was reported in enok mutant germline stem cells and cystoblasts, while its levels were apparently similar in region 2 of the germarium between wild-type control and enok mutants (Xin et al. 2013). Since Bru binds to and inhibits the translation of osk mRNA (Kim-Ha et al. 1995), we examined Osk localization in bru-overexpressing ovaries (Supplemental Fig. S10A,B). The original study using the UASp-Bru transgene reported that overexpression of bru resulted in one extra round of cystoblast divisions in ∼70% of the egg chambers, and 15% of the embryos showed the typical posterior group phenotype (Filardo and Ephrussi 2003). Consistent with the previous study, we observed that ∼70% of egg chambers had extra nurse cells, and 12.5% of the stage 9–10 egg chambers had defective posterior Osk localization in bru-overexpressing ovaries (Supplemental Fig. S10A,B). This result suggests that overexpression of bru may affect Osk localization in the ovary. However, the RNA-seq analysis showed that bru expression levels were not significantly changed in stage 5–14 enok mutant germline clone egg chambers (Supplemental Fig. S9A), and the protein levels of Bru were not increased in stage 9 enok mutant nurse cells (Supplemental Fig. S10C). Furthermore, bulk Bru levels were similar in stage 5–14 wild-type and enok2 germline clone egg chambers (Supplemental Fig. S10D). Taken together, we concluded that bru is not up-regulated in egg chambers beyond stage 4 of oogenesis in the enok mutants. Enok has also been reported to affect the nucleolar size in Kc cells (Neumuller et al. 2013). However, enok mutant nurse cells showed staining of the nucleolar marker Fibrillarin (Fib) similar to the wild-type control (Supplemental Fig. S10C), indicating that the nucleolar size of enok mutant nurse cells was not altered.
Enok acetylates H3K23 at spir and mael and regulates their expression
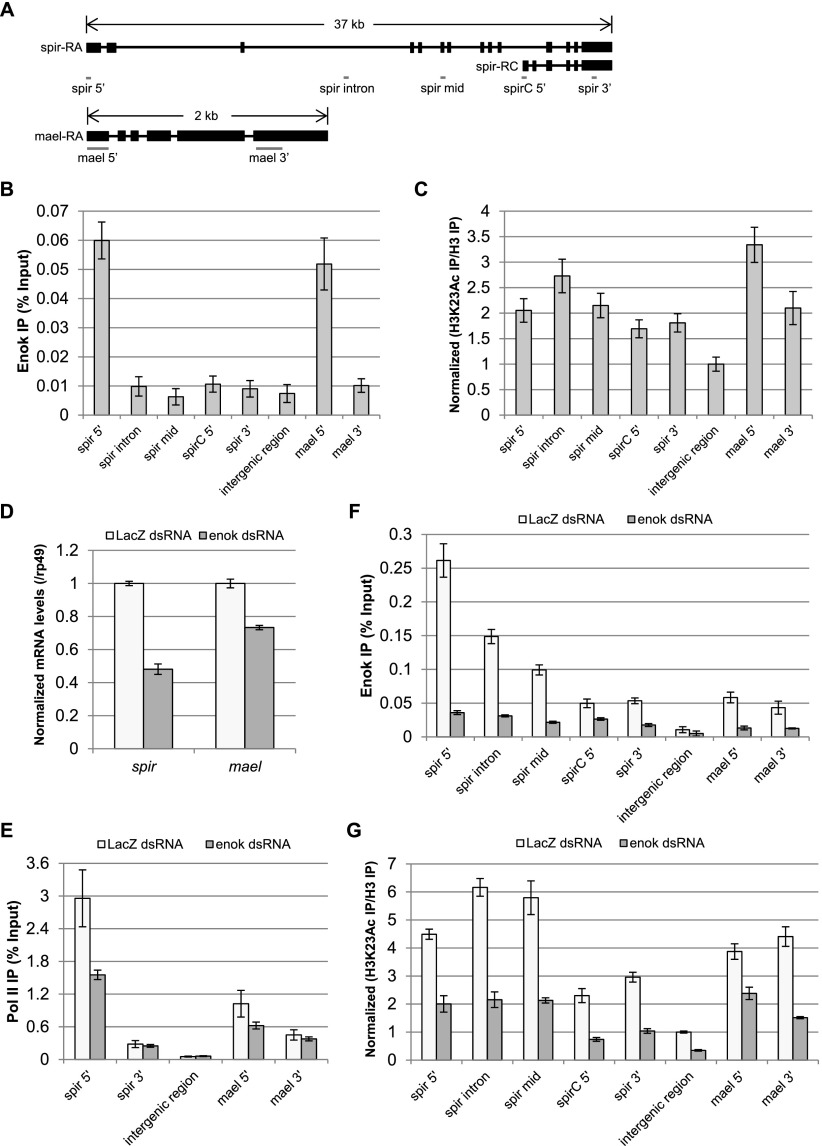
We next asked whether Enok physically localizes to and acetylates H3K23 at the spir and mael genes. To answer this question, a chromatin immunoprecipitation (ChIP) assay was undertaken to assess Enok occupancy and H3K23Ac levels across spir and mael in the wild-type ovary (Fig. 4A). Compared with the intergenic region, Enok occupancy at the 5′ ends of both spir and mael was enriched sevenfold to eightfold (Fig. 4B). Consistently, the H3K23Ac mark was enriched twofold to threefold across the spir and mael genes relative to the intergenic region (Fig. 4C). To further confirm that the acetylation of H3K23 at spir and mael is dependent on Enok, we performed the ChIP analysis in S2 cells treated with control dsRNA (LacZ) or dsRNA against enok. Knocking down enok by dsRNA resulted in a 50% and 30% reduction in the mRNA levels of spir and mael, respectively (Fig. 4D). Occupancies of RNA polymerase II (Pol II) were also decreased in the 5′ regions of spir and mael upon depletion of Enok (Fig. 4E), suggesting that Enok plays a role in activating these two genes in S2 cells similar to that in the ovary. ChIP analysis showed that dsRNA treatment against enok efficiently reduced the occupancies of Enok as well as the H3K23Ac mark across both spir and mael (Fig. 4F,G). Therefore, we concluded that Enok acetylates H3K23 at the spir and mael genes and contributes to their activation.
Figure 4.
Enok acetylates H3K23 at the spir and mael genes and regulates their expression. (A) Schematic representation of the amplicon location at the spir and mael genes used in the ChIP analysis. Exons are shown as black rectangles. The amplicon for the intergenic region is located at chr2L: 18,415,855–18,415,978. (B) The occupancies of Enok at spir and mael in the ovary of OreR were examined by ChIP analysis. (C) The H3K23Ac levels at spir and mael in the ovary of OreR were analyzed by ChIP. The H3K23Ac immunoprecipitation (IP) signals were first normalized to the histone H3 immunoprecipitation signals (H3K23Ac/H3), and then the H3K23Ac/H3 values were normalized to the mean of H3K23Ac/H3 values obtained for the intergenic region, which was set as 1. (D) The expression levels of spir and mael were examined by RT-qPCR in the S2 cells treated with control dsRNA (LacZ) or dsRNA against enok. The mRNA levels of spir and mael were normalized to the levels of rp49. (E) The occupancies of Pol II at spir and mael in the S2 cells treated with LacZ dsRNA or enok dsRNA were analyzed by ChIP. (F) The occupancies of Enok at spir and mael in the S2 cells treated with LacZ dsRNA or enok dsRNA were analyzed by ChIP. (G) Changes in H3K23Ac levels at spir and mael in the S2 cells treated with LacZ dsRNA or enok dsRNA were analyzed by ChIP. The H3K23Ac/H3 values were normalized to the mean of H3K23Ac/H3 values obtained for the intergenic region in the LacZ dsRNA-treated S2 cells, which was set as 1. In B–G, data represent the mean of triplicate experiments ± SD.
A motif analysis in the promoter regions of genes commonly down-regulated in both enok mutants revealed four candidate transcription factors that may bind to the promoters of the down-regulated genes (Supplemental Fig. S11A). However, knocking down any of these four factors in S2 cells had no obvious effect on the spir mRNA levels (Supplemental Fig. S11B,C), suggesting that there may be other yet unknown factors/mechanisms that recruit Enok to activate spir.
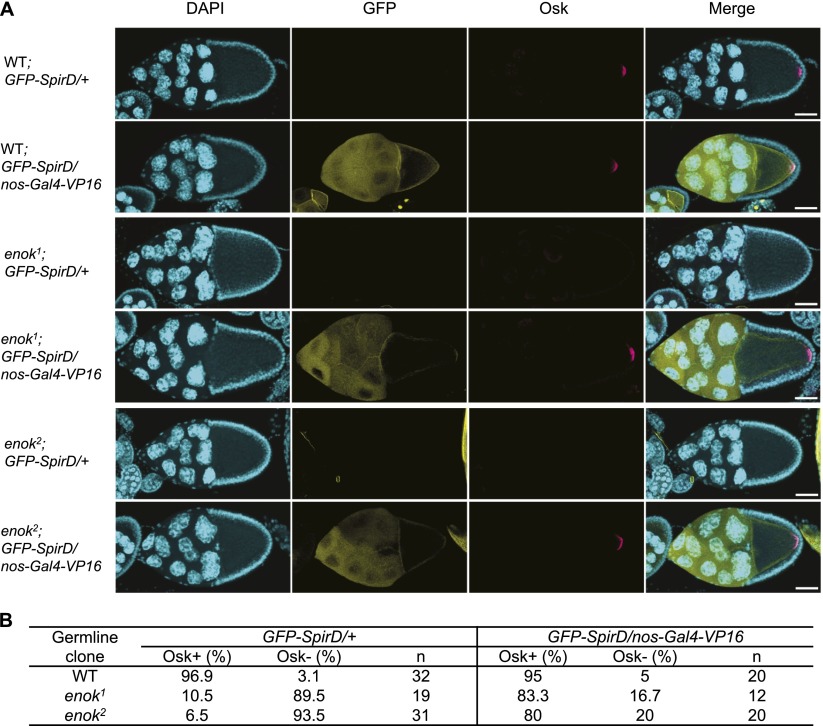
Spir isoform D (SpirD) rescues the defective Osk localization in the enok mutant egg chambers
Notably, compromising the function of Enok by genetic mutation or dsRNA had greater effects on the expression levels of spir than on those of mael (Figs. 3B, 4D). Therefore, we hypothesized that down-regulation of spir may be the major cause of the Osk localization defects in enok mutant germline clones. To test this hypothesis, an UASp-GFP-spirD transgene was expressed in the germline under the control of nos-Gal4-VP16, which has been shown to rescue the osk localization defects in spir mutant ovaries (Rosales-Nieves et al. 2006; Dahlgaard et al. 2007). Indeed, while ∼90% of enok mutant egg chambers without GFP-SpirD lacked posteriorly localized Osk at stages 9–10, the expression of GFP-spirD restored normal localization of Osk in ∼80% of the enok mutant egg chambers (Fig. 5). Taken together, our results strongly suggest that the acetyltransferase Enok plays an important role in the Osk-dependent embryonic development by stimulating the expression of spir in the female germline (Fig. 6).
Figure 5.
Rescue of enok mutant germline by GFP-SpirD. (A) Stage 9 (wild-type [WT] and enok2) or 10 (enok1) egg chambers were stained with DAPI and an α-Osk antibody. Bars, 50 µm. (B) Stage 9–10 egg chambers stained with an α-Osk antibody were examined for the presence (Osk+) or absence (Osk−) of posteriorly localized Osk in the oocytes. The percentages of Osk+ or Osk− egg chambers with or without GFP-SpirD expression were then calculated, respectively. The total number of examined egg chambers (n) is also shown. The genotypes of females in A and B are as follows: hs-Flp/+; FRTG13/FRTG13, ovoD1–18; UASp-GFP-spirD/+ (wild type; GFP-SpirD/+); hs-Flp/+; FRTG13/FRTG13, ovoD1–18; UASp-GFP-spirD/nos-Gal4-VP16 (wild type; GFP-SpirD/nos-Gal4-VP16); hs-Flp/+; FRTG13, enok1/FRTG13, ovoD1–18; UASp-GFP-spirD/+ (enok1; GFP-SpirD/+); hs-Flp/+; FRTG13, enok1/FRTG13, ovoD1–18; UASp-GFP-spirD/nos-Gal4-VP16 (enok1; GFP-SpirD/nos-Gal4-VP16); hs-Flp/+; FRTG13, enok2/FRTG13, ovoD1–18; UASp-GFP-spirD/+ (enok2; GFP-SpirD/+); and hs-Flp/+; FRTG13, enok2/FRTG13, ovoD1–18; UASp-GFP-spirD/nos-Gal4-VP16 (enok2; GFP-SpirD/nos-Gal4-VP16).
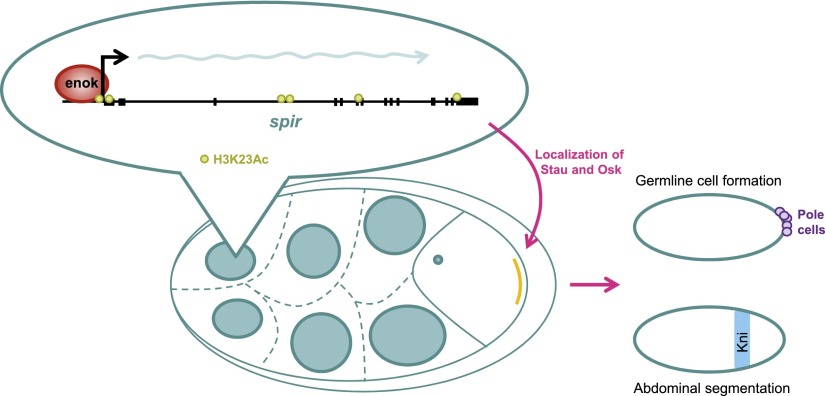
Figure 6.
Model of the regulation of Osk-dependent developmental processes by Enok in the female germline.
Expression of spir is required for assembly of an actin mesh that maintains the polarized MT network in the oocyte during stages 8–10A of oogenesis but must be down-regulated after stage 10B to allow reorganization of the MT network and fast ooplasmic streaming to take place (Dahlgaard et al. 2007; Quinlan 2013). This down-regulation does not occur when spir is expressed exogenously using the UAS-Gal4 system, and the fast ooplasmic streaming that normally occurs inside the oocyte during stages 10B–13 is blocked. Fast streaming is required in the oocyte for proper mixing of the nurse cell-derived cytoplasm and yolk. Consequently, egg chambers expressing GFP-spirD developed into nonfertile oocytes, with clear cytoplasm from nurse cells at the anterior and white yolk at the posterior (Dahlgaard et al. 2007). Thus, although we could observe rescue of the posterior Osk localization in oocytes at stages 9–10, both wild-type and enok mutant oocytes with GFP-SpirD at later stages (11–13) showed a similar stratified appearance of cytoplasm and yolk. As a result, both wild-type and the enok mutants with exogenous GFP-spirD expression do not produce fertile eggs.
Discussion
Our findings reveal a previously unknown transcriptional role for Enok in regulating the polarized localization of Osk during oogenesis through promoting the expression of spir and mael. Spir and Mael are required for the properly polarized MT network in oocytes from stages 8 to 10A (Dahlgaard et al. 2007; Sato et al. 2011). However, protein levels of both decreased at later stages of oogenesis (Clegg et al. 1997; Quinlan 2013), allowing reorganization of the MT network and fast ooplasmic streaming. The persistent presence of Spir extending into stage 11 led to loss of ooplasmic streaming and resulted in female infertility (Dahlgaard et al. 2007; Quinlan 2013). These findings suggest that the temporal regulation of spir expression is crucial for oogenesis, and, interestingly, Enok protein levels were also reduced in egg chambers during stages 10–13 compared with stages 1–9 (Supplemental Fig. S3B). While the stability of Spir or the translation of spir mRNA may also be a target for regulation, our results suggest that Enok is involved in the dynamic modulation of spir transcript. Furthermore, our results demonstrate the importance of Enok for expression of spir and mael in both ovaries and S2 cells, suggesting that Enok may play a similar role in other Spir- or Mael-dependent processes such as heart development (Xu et al. 2012).
Notably, Mael is also important for the piRNA-mediated silencing of transposons in germline cells (Sienski et al. 2012). Mutations in genes involved in the piRNA pathway, including aub and armitage (armi), result in axis specification defects in oocytes as well as persistent DNA damage and checkpoint activation in germline cells. The activation of DNA damage signaling is suggested to cause axis specification defects in oocytes, as the disruption of Osk localization in piRNA pathway mutants can be suppressed by mutations in mei-41 or mnk, which encode ATR or checkpoint kinase 2, respectively (Khurana and Theurkauf 2010). However, mutation in mnk cannot suppress the loss of posteriorly localized Osk in the mael mutant oocyte, indicating that the oocyte polarization defect in the mael mutant is independent of DNA damage signaling (Sato et al. 2011). Therefore, although we cannot exclude the possibility that the piRNA pathway is affected in enok mutants due to down-regulation of mael, the Osk localization defect in the enok mutant oocyte is likely independent of mei-41 and mnk.
In addition to the osk mRNA localization defect, both spir and mael mutants affect dorsal–ventral (D/V) axis formation in oocytes (Manseau and Schupbach 1989; Clegg et al. 1997). However, no defects in the D/V patterning were observed in the eggshells of enok mutant germline clone embryos (data not shown). Interestingly, among the spir mutant alleles that disrupt formation of germ plasm, only strong alleles result in dorsalized eggshells and embryos, while females with weak alleles produce eggs with normal D/V patterning (Manseau and Schupbach 1989). Since the enok1 and enok2 ovaries still express ∼25% of the wild-type levels of spir mRNA, enok mutants may behave like weak spir mutants. Similarly, the ∼40% reduction in mael mRNA levels in enok mutants as compared with the wild-type control may not have significant effects on the D/V axis specification.
Redundancy in HAT functions has been reported for both Moz and Sas3, the mammalian and yeast homologs of Enok, respectively. In yeast, deletion of either GCN5 (encoding the catalytic subunit of ADA and SAGA HAT complexes) or SAS3 is viable. However, simultaneously deleting GCN5 and SAS3 is lethal due to loss of the HAT activity of the two proteins, suggesting that Gcn5 and Sas3 can compensate for each other in acetylating histone residues. Indeed, while deleting SAS3 alone had no effect on the global levels of H3K9Ac and H3K14Ac, disrupting the HAT activity of Sas3 in the gcn5Δ background greatly reduced the bulk levels of H3K9Ac and H3K14Ac in yeast (Howe et al. 2001). Also, mammalian Moz targets H3K9 in vivo and regulates the expression of Hox genes, but the global H3K9Ac levels are not significantly affected in the homozygous Moz mutant (Voss et al. 2009), indicating that other HATs have overlapping substrate specificity with Moz. In flies, a previous study had reported that the H3K23Ac levels were reduced 35% in nejire (nej) mutant embryos, which lack functional CBP/p300 (Bodai et al. 2012). However, knocking down nej by dsRNA in S2 cells severely reduced levels of H3K27Ac but had no obvious effect on global levels of H3K23Ac (Tie et al. 2009). In this study, we showed that the global H3K23Ac levels decreased 85% upon enok dsRNA treatment in S2 cells (Fig. 1A; Supplemental Fig. S1B). We also showed that the H3K23Ac levels are highly dependent on Enok in early and late embryos (Figs. 1C, 2A; Supplemental Fig. S5B), larvae (Fig. 1D; Supplemental Fig. S1D), adult follicle cells and nurse cells (Fig. 2G; Supplemental Figs. S1E, S6B), and mature oocytes (Supplemental Fig. S5A). Therefore, although Nej may also contribute to the acetylation of H3K23, our results indicate that, in contrast to its mammalian and yeast homologs, Enok uniquely functions as the major HAT for establishing the H3K23Ac mark in vivo.
The H3K23 residue has been shown to stabilize the interaction between H3K27me3 and the chromodomain of Polycomb (Fischle et al. 2003). Therefore, acetylation of H3K23 may affect the recognition of H3K27me3 by the Polycomb complex. Another study showed that the plant homeodomain (PHD)–bromodomain of TRIM24, a coactivator for estrogen receptor α in humans, binds to unmodified H3K4 and acetylated H3K23 within the same H3 tail (Tsai et al. 2010). Also, the levels of H3K23Ac at two ecdysone-inducible genes, Eip74EF and Eip75B, have been shown to correlate with the transcriptional activity of these two genes at the pupal stage, suggesting the involvement of H3K23Ac in ecdysone-induced transcriptional activation (Bodai et al. 2012). In this study, we further provided evidence for the activating role of the Enok-mediated H3K23Ac mark in transcriptional regulation.
In mammals, MOZ functions as a key regulator of hematopoiesis (Perez-Campo et al. 2013). Interestingly, one of the genes encoding mammalian homologs of Spir, spir-1, is expressed in the fetal liver and adult spleen, indicating the expression of spir-1 in hematopoietic cells (Schumacher et al. 2004). Thus, it will be intriguing to investigate whether the Drosophila Enok–Spir pathway is conserved in mammals and whether Spir-1 functions in hematopoiesis. Taken together, our results demonstrate that Enok functions as an H3K23 acetyltransferase and regulates Osk localization, linking polarization of the oocyte to histone modification.
Materials and methods
Fly strains and culture
Drosophila melanogaster was crossed and grown on standard medium at 25°C unless stated otherwise. For MARCM and germline clone analyses, larvae were heat-shocked twice for 40 min at 37°C, with a 30-min break at 25°C in between, for two consecutive days starting from the second day after hatching. Females were conditioned with wet yeast for 2–3 d before ovary dissection. Embryos were collected using standard apple juice plates with wet yeast.
Antibody production
The recombinant peptide encompassing residues 1813–2197 of Enok was purified from Escherichia coli and injected into guinea pigs following the 28-d mighty quick protocol carried out by Pocono Rabbit Farm and Laboratory. For generating the α-Kr antibody, a recombinant peptide encompassing the first 200 residues of Kruppel was purified from E. coli and injected into rabbits following the 49-d mighty quick protocol conducted by Pocono Rabbit Farm and Laboratory. The α-H4K16Ac antiserum was obtained by injecting rabbits with the peptide GKGGA-K(Ac)-RHRKC conjugated to ovalbumin following the standard protocol (Covance).
RNAi knockdown in S2 cells
dsRNAs targeting LacZ or enok were synthesized as described previously (Lin et al. 2008). S2 cells were seeded at 1 × 106 cells per milliliter in serum-free Schneider’s insect medium for RNAi treatment. dsRNA was added to the S2 cell culture to a final concentration of 10 µg/mL. After incubation for 45 min at 25°C, Schneider’s insect medium containing 10% FBS (Sigma) was added to reach a final concentration of 5% FBS. S2 cells were then incubated for 4–8 d at 25°C before being subjected to acid extraction of histone or RNA isolation.
RNA purification and RT-qPCR
RNA was isolated using TRIzol (Invitrogen) and subjected to cDNA synthesis using SuperScript III reverse transcriptase (Invitrogen). Quantitative PCR (qPCR) was performed in triplicate for each sample, and the data were analyzed using the standard curve method. The expression levels of rp49 were used as the normalization control.
Western blot analysis
S2 cells or dechorionated embryos were lysed in buffer A (10 mM HEPES at pH 7.5, 1.5 mM MgCl2, 10 mM KCl, 1% NP-40, 1 mM PMSF, 5 mM sodium butyrate, 1 µg/mL pepstatin A, 1 µg/mL leupeptin, 1 µg/mL aprotinin), and the lysates were centrifuged at 1500g for 3 min at 4°C to separate the cytosol fraction and the nuclei. After being washed once with buffer A, nuclei were lysed in 1× laemmli sample buffer (Bio-Rad) for nuclear extract preparation. For acid extraction of histone, nuclei were resuspended in 0.4 N HCl and incubated for 5 min on ice. Following centrifugation at 14,000 rpm for 5 min at 4°C, supernatants were collected and neutralized with 2 M Tris (0.2× vol). Protein concentration was measured using Bio-Rad protein assay, and an equal amount of protein was subjected to Western blotting. The following antibodies were used: H3K9Ac (1:500; 06-942, Millipore), H3K14Ac (1:1000; 07-353, Millipore), H3K18Ac (1:1000; ab1191, Abcam), H3K23Ac (1:100,000; 07-355, Millipore), H3K27Ac (1:10,000; 07-360, Millipore), H3K36Ac (1:1000; 07-540, Millipore), H3 (1:5000; ab1791, Abcam), H4Ac (1:2500; 06-866, Millipore), H4K5Ac (1:1000; 06-759, Millipore), H4K12Ac (1:1000; 06-761, Millipore), H4K16Ac (1:500), β-Tubulin (1:5000; E7, Developmental Studies Hybridoma Bank), Bru (1:5000; gift from Mary A. Lilly) (Sugimura and Lilly 2006), Enok (1:1000), and Lamin (1:2000; ADL67.10, Developmental Studies Hybridoma Bank).
Expression and purification of recombinant Enok in Sf9 insect cells
Full-length cDNA of enok was cloned into vector pBacPAK8 carrying a N-terminal His-Flag tag. Purification of the recombinant enok was performed essentially as described previously with minor modifications (Lin et al. 2008). Briefly, baculovirus-infected Sf9 insect cells were cultured at 27°C in the Sf-900 III SFM (Invitrogen) supplemented with 10% FBS and penicillin—streptomycin (Gibco). After lysis and centrifugation, clarified cell lysates were incubated with α-Flag (M2) agarose beads (Sigma) for 3 h at 4°C followed by washes and elution with triple Flag peptide.
HAT assay
Purification of yeast SAGA complex was performed as described previously (Dutta et al. 2014). Recombinant histones were purified from E. coli and reconstituted into octamers. Mononucleosomes were then reconstituted on a biotin-conjugated DNA fragment containing a 601 positioning sequence (Yun et al. 2012). Nucleosomes were immobilized onto streptavidin-coupled Dynabeads (Invitrogen) in HAT buffer (50 mM Tris-HCl at pH 8.0, 50 mM KCl, 100 µM EDTA, 10 mM sodium butyrate, 5% glycerol, 1 mM DTT, 100 µg/mL BSA, 1 µM acetyl-CoA, 1 mM PMSF) and incubated with or without recombinant Enok for 2 h at 27°C. The immobilized nucleosomes were then heated in 1× SDS sample buffer (62.5 mM Tris-HCl at pH 6.8, 10% glycerol, 2% SDS, 0.01% bromophenol blue, 143 mM β-mercaptoethanol) for 6 min at 98°C and subjected to Western blotting.
Immunofluorescent staining
Embryos were fixed and processed as described previously (Fogarty et al. 1994). The following primary antibodies were used: En (1:10; 4D9, Developmental Studies Hybridoma Bank), Ubx (1:10; FP3.38, Developmental Studies Hybridoma Bank), Eve (1:10; 3C10, Developmental Studies Hybridoma Bank), Vas (1:100; sc-30210, Santa Cruz Biotechnology), Kni (1:100; Asian Distribution Center for Segmentation Antibodies) (Kosman et al. 1998), and Kr (1:1000).
Ovaries or larval eye discs were dissected in Shields and Sang M3 insect medium and processed as described previously (Shcherbata et al. 2004). The following primary antibodies were used: H3K23Ac (1:2500; 07-355, Millipore), Osk (1:3000; gift from Anne Ephrussi) (Yoshida et al. 2004), Bru (1:1000; gift from Mary A. Lilly) (Sugimura and Lilly 2006), Fib (1:100; MCA-38F3, EnCor Biotechnology), and Stau (1:200; sc-15823, Santa Cruz Biotechnology). The following secondary antibodies were used: Alexa 488 goat anti-mouse, Alexa 488 goat anti-rabbit, Alexa 568 goat anti-guinea pig, Alexa 488 donkey anti-goat (1:300; Molecular Probes), and Cy3 donkey anti-rabbit (1:300; Jackson ImmunoResearch). Cell nuclei were stained using DAPI dye (Invitrogen).
Staining of the cytoskeleton in the oocyte was performed as described previously with minor modifications (Dahlgaard et al. 2007). Briefly, ovaries were dissected in 1× PBS containing 0.01% Tween-20 and 10% formaldehyde and fixed for 10 min. After three washes with 1× PBS, ovaries were permeabilized in 1× PBS with 1% Triton X-100 for 2 h. Ovaries were then washed twice with PBST (1× PBS containing 0.2% Triton X-100) and blocked in PBSTB (PBST containing 5% normal horse serum) for 1 h followed by cytoskeleton staining overnight at 4°C. The following staining reagents were used: Alexa 568-phalloidin (1:10; Molecular Probes) and FITC-conjugated mouse anti-α-Tubulin antibody (1:200; Sigma). Phalloidin-stained actin mesh in the oocyte cytoplasm was imaged 10 µm below the cortex. Images were taken by Zeiss LSM 510 confocal microscope and processed using Zeiss LSM Image Browser or OMERO. Adobe Photoshop was used to assemble images into the figures.
Flow cytometry analysis
Ovaries were dissected from 100–170 germline clone females for each sample in Shields and Sang M3 insect medium. After one wash with EBR (10 mM HEPES at pH 7.5, 130 mM NaCl, 5mM KCl, 2mM CaCl2) (Calvi and Lilly 2004), ovaries were digested with 5 mg/mL collagenase (C9891, Sigma) in EBR for 15 min followed by two washes with EBR. The dissociated cells were then resuspended in 1× PBS and filtered through a 100-µm nylon mesh. After centrifugation at 700g for 2 min, cells were fixed in ice-cold 70% ethanol and stored overnight at −20°C. The next day, cells were rehydrated in 1× PBS containing 0.5% BSA. Cells were then stained in 1× PBS containing 0.5% BSA, 100 µg/mL RNase A, and 50 µg/mL propidium iodide for 30 min at 37°C and subjected to flow cytometry analysis of DNA content by MACSQuant Analyzer (Miltenyi Biotec, Inc.).
RNA-seq analysis
Ovaries were dissected from 4- to 6-d-old flies. Total RNA was isolated from egg chambers of stages 5–14 using TRIzol (Invitrogen) followed by DNase treatment (Invitrogen) and subjected to RNA-seq analysis.
Reads generated were 51-base-pair (bp) single-end, poly-A-selected, directional using the Illumina protocol. Fastq files were aligned to University of California at Santa Cruz dm3 with TopHat 2.0.10 (Bowtie 2.1.0) with additional parameters “-library-type fr-firststrand.” TopHat BAM files were split into stranded BAM files based on the bit flag, and strand identities were reversed, as the Illumina directional protocol yields the reverse complement of the original read. Nonredundant exonic regions of genes were made by passing bed files of Ensembl 74 (FlyBase 5.46) exons per gene through mergeBed and then using intersectBed on the BAM files to count reads aligning to these nonredundant exonic spaces per gene. Resulting sense strand reads per gene were analyzed in R with the edgeR package using default methods. Differentially expressed genes (DEGs) were identified using the cutoff of P-value < 0.05 and reads per kilobase per million (RPKM) > 5. The RPKM cutoff was applied to the wild-type data sets for identifying down-regulated genes and to the mutant data sets for identifying up-regulated genes.
RNA-seq data are available in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-2521.
GO term analysis and motif analysis
GO term analysis was performed using GOrilla with a cutoff of P-value < 0.001 (Eden et al. 2009). The genes expressed in wild-type ovaries were used as the background gene set, and the DEGs identified in the RNA-seq analysis were used as the target gene set.
The promoter regions of DEGs were defined as 500 bp upstream of the transcriptional start sites of DEGs. Motif analysis in the DEG promoters was performed with MEME using a cutoff of E-value < 0.001 (Bailey et al. 2009). TOMTOM was then used to search for transcription factors that have a DNA-binding motif matching the motifs identified by MEME in the DEG promoters (Gupta et al. 2007). Among the top 20 hits identified by TOMTOM for each motif, we defined the candidate transcription factors as the hits that fit the following criteria: (1) The candidate has a homolog in flies, and (2) the RPKM value of its fly homolog in the wild-type ovary is greater than five.
ChIP analysis
ChIP assay was carried out as described previously with minor modifications (Weake et al. 2009). Sixty pairs of ovaries were dissected from OreR females in Shields and Sang M3 insect medium and fixed in 950 µL of PBS containing 1.8% formaldehyde for 10 min at room temperature. Cross-linking was stopped by adding 50 µL of 2.5 M glycine and incubating for 5 min at room temperature. After two washes with PBS, ovaries were homogenized in buffer A1 (15 mM HEPES at pH 7.5, 15 mM NaCl, 60 mM KCl, 4 mM MgCl2, 0.5% TritonX-100, 0.5 mM DTT, 1 mM PMSF, 5 mM NaF, 5 mM sodium butyrate, 1 µg/mL pepstatin A, 1 µg/mL leupeptin, 1 µg/mL aprotinin). The chromatin pellet was washed three times with buffer A1 and once with buffer A2 (15 mM HEPES at pH 7.5, 140 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1% TritonX-100, 0.1% sodium deoxycholate, 0.1% SDS, 0.5% N-lauroyl sarcosine, 1 mM PMSF, 5 mM NaF, 5 mM sodium butyrate, 1 µg/mL pepstatin A, 1 µg/mL leupeptin, 1 µg/mL aprotinin) and was sonicated in 400 µL of buffer A2 six times for 10 sec at 30% power. After centrifugation at 14,000 rpm for 10 min at 4°C, 250 µL of clarified soluble chromatin was incubated with 5 µL of guinea pig α-Enok, 2 µL of rabbit α-H3K23Ac (Millipore), or 2 µL of rabbit α-H3 (Abcam) overnight at 4°C. Antibody-bound chromatin was pulled down by protein A-conjugated Dynabeads (Invitrogen), and the beads were washed five times with RIPA buffer (50 mM HEPES at pH 7.5, 1 mM EDTA, 0.7% sodium deoxycholate, 1% NP-40, 0.5 M LiCl, 1 mM PMSF) and once with TE buffer containing 50 mM NaCl. Chromatin was eluted twice in TE buffer containing 1% SDS and 250 mM NaCl for 20 min at 65°C. After RNase A and proteinase K treatment, cross-linking was reversed overnight at 65°C. DNA was purified by phenol-chloroform extraction and ethanol precipitation. The qPCR data obtained from input and immunoprecipitated DNA samples were analyzed using the standard curve method.
ChIP of S2 cells was performed with 1 × 107 cells. Cells were fixed in Schneider’s insect medium containing 5% FBS and 1% formaldehyde for 10 min at room temperature, and the cross-linking was stopped by adding glycine to a final concentration of 125 mM. Following two washes with PBS, fixed cells were sonicated in 180 µL of buffer A2 by Bioruptor (Diagenode) for 15 min (30 sec on/30 sec off) at the high power setting. After centrifugation, 150 µL of soluble chromatin was incubated with 2 µL of α-Enok, 1 µL of α-Pol II (ab5408, Abcam), 1 µL of α-H3K23Ac, or 1 µL of α-H3 overnight at 4°C and processed as the ovary ChIP described above.
Supplementary Material
Acknowledgments
We thank A. Ephrussi, M.A. Lilly, and the Asian Distribution Center for Segmentation Antibodies for antibodies. We also thank A. Ephrussi and the Bloomington Stock Center for fly stocks, and L. Shelton for making the Kr antibody. The α-En and α-Eve antibodies developed by C. Goodman, the α-β-Tubulin antibody developed by M. Klymkowsky, the α-Lamin antibody developed by P.A. Fisher, and the α-Ubx antibody developed by R. White were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the The Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) and maintained by the Department of Biology, The University of Iowa, Iowa City, IA. This study was funded by the Stowers Institute and the National Institutes of Health (grant GM099945) to S.M.A. and J.L.W.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.249730.114.
References
- Ambrosio L, Mahowald AP, Perrimon N. 1989. l(1)pole hole is required maternally for pattern formation in the terminal regions of the embryo. Development 106: 145–158. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME suite: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodai L, Zsindely N, Gaspar R, Kristo I, Komonyi O, Boros IM. 2012. Ecdysone induced gene expression is associated with acetylation of histone H3 lysine 23 in Drosophila melanogaster. PLoS ONE 7: e40565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros M, Agaisse H, Perrimon N. 2002. Sequential activation of signaling pathways during innate immune responses in Drosophila. Dev Cell 3: 711–722. [DOI] [PubMed] [Google Scholar]
- Brendza RP, Serbus LR, Duffy JB, Saxton WM. 2000. A function for kinesin I in the posterior transport of oskar mRNA and Staufen protein. Science 289: 2120–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvi BR, Lilly MA. 2004. Fluorescent BrdU labeling and nuclear flow sorting of the Drosophila ovary. Methods Mol Biol 247: 203–213. [DOI] [PubMed] [Google Scholar]
- Chou TB, Perrimon N. 1996. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics 144: 1673–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg NJ, Frost DM, Larkin MK, Subrahmanyan L, Bryant Z, Ruohola-Baker H. 1997. maelstrom is required for an early step in the establishment of Drosophila oocyte polarity: posterior localization of grk mRNA. Development 124: 4661–4671. [DOI] [PubMed] [Google Scholar]
- Dahlgaard K, Raposo AA, Niccoli T, St Johnston D. 2007. Capu and Spire assemble a cytoplasmic actin mesh that maintains microtubule organization in the Drosophila oocyte. Dev Cell 13: 539–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A, Gogol M, Kim JH, Smolle M, Venkatesh S, Gilmore J, Florens L, Washburn MP, Workman JL. 2014. Swi/Snf dynamics on stress-responsive genes is governed by competitive bromodomain interactions. Genes Dev 28: 2314–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmons S, Phan H, Calley J, Chen W, James B, Manseau L. 1995. Cappuccino, a Drosophila maternal effect gene required for polarity of the egg and embryo, is related to the vertebrate limb deformity locus. Genes Dev 9: 2482–2494. [DOI] [PubMed] [Google Scholar]
- Ephrussi A, Lehmann R. 1992. Induction of germ cell formation by oskar. Nature 358: 387–392. [DOI] [PubMed] [Google Scholar]
- Ephrussi A, Dickinson LK, Lehmann R. 1991. Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell 66: 37–50. [DOI] [PubMed] [Google Scholar]
- Extavour CG, Akam M. 2003. Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development 130: 5869–5884. [DOI] [PubMed] [Google Scholar]
- Filardo P, Ephrussi A. 2003. Bruno regulates gurken during Drosophila oogenesis. Mech Dev 120: 289–297. [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. 2003. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev 17: 1870–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty P, Kalpin RF, Sullivan W. 1994. The Drosophila maternal-effect mutation grapes causes a metaphase arrest at nuclear cycle 13. Development 120: 2131–2142. [DOI] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. . 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471: 473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. 2007. Quantifying similarity between motifs. Genome Biol 8: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch M, Gerwin N, Taubert H, Jackle H. 1992. Competition for overlapping sites in the regulatory region of the Drosophila gene Kruppel. Science 256: 94–97. [DOI] [PubMed] [Google Scholar]
- Howe L, Auston D, Grant P, John S, Cook RG, Workman JL, Pillus L. 2001. Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes Dev 15: 3144–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida T, Kobayashi S. 1998. Essential role of mitochondrially encoded large rRNA for germ-line formation in Drosophila embryos. Proc Natl Acad Sci 95: 11274–11278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana JS, Theurkauf W. 2010. piRNAs, transposon silencing, and Drosophila germline development. J Cell Biol 191: 905–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Ha J, Kerr K, Macdonald PM. 1995. Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell 81: 403–412. [DOI] [PubMed] [Google Scholar]
- Kosman D, Small S, Reinitz J. 1998. Rapid preparation of a panel of polyclonal antibodies to Drosophila segmentation proteins. Dev Genes Evol 208: 290–294. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22: 451–461. [DOI] [PubMed] [Google Scholar]
- Lin CH, Li B, Swanson S, Zhang Y, Florens L, Washburn MP, Abmayr SM, Workman JL. 2008. Heterochromatin protein 1a stimulates histone H3 lysine 36 demethylation by the Drosophila KDM4A demethylase. Mol Cell 32: 696–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manseau LJ, Schupbach T. 1989. cappuccino and spire: two unique maternal-effect loci required for both the anteroposterior and dorsoventral patterns of the Drosophila embryo. Genes Dev 3: 1437–1452. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Sato K, Hanyu-Nakamura K. 2004. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev Cell 6: 69–78. [DOI] [PubMed] [Google Scholar]
- Neumuller RA, Gross T, Samsonova AA, Vinayagam A, Buckner M, Founk K, Hu Y, Sharifpoor S, Rosebrock AP, Andrews B, et al. . 2013. Conserved regulators of nucleolar size revealed by global phenotypic analyses. Sci Signal 6: ra70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibu Y, Zhang H, Levine M. 1998. Interaction of short-range repressors with Drosophila CtBP in the embryo. Science 280: 101–104. [DOI] [PubMed] [Google Scholar]
- Perez-Campo FM, Costa G, Lie-a-Ling M, Kouskoff V, Lacaud G. 2013. The MYSTerious MOZ, a histone acetyltransferase with a key role in haematopoiesis. Immunology 139: 161–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan ME. 2013. Direct interaction between two actin nucleators is required in Drosophila oogenesis. Development 140: 4417–4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongo C, Gavis ER, Lehmann R. 1995. Localization of oskar RNA regulates oskar translation and requires Oskar protein. Development 121: 2737–2746. [DOI] [PubMed] [Google Scholar]
- Rosales-Nieves AE, Johndrow JE, Keller LC, Magie CR, Pinto-Santini DM, Parkhurst SM. 2006. Coordination of microtubule and microfilament dynamics by Drosophila Rho1, Spire and Cappuccino. Nat Cell Biol 8: 367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Nishida KM, Shibuya A, Siomi MC, Siomi H. 2011. Maelstrom coordinates microtubule organization during Drosophila oogenesis through interaction with components of the MTOC. Genes Dev 25: 2361–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher N, Borawski JM, Leberfinger CB, Gessler M, Kerkhoff E. 2004. Overlapping expression pattern of the actin organizers Spir-1 and formin-2 in the developing mouse nervous system and the adult brain. Gene Expr Patterns 4: 249–255. [DOI] [PubMed] [Google Scholar]
- Scott EK, Lee T, Luo L. 2001. enok encodes a Drosophila putative histone acetyltransferase required for mushroom body neuroblast proliferation. Curr Biol 11: 99–104. [DOI] [PubMed] [Google Scholar]
- Shcherbata HR, Althauser C, Findley SD, Ruohola-Baker H. 2004. The mitotic-to-endocycle switch in Drosophila follicle cells is executed by Notch-dependent regulation of G1/S, G2/M and M/G1 cell-cycle transitions. Development 131: 3169–3181. [DOI] [PubMed] [Google Scholar]
- Sienski G, Donertas D, Brennecke J. 2012. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell 151: 964–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Pierre SE, Ponting L, Stefancsik R, McQuilton P. 2014. FlyBase 102—advanced approaches to interrogating FlyBase. Nucleic Acids Res 42: D780–D788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura I, Lilly MA. 2006. Bruno inhibits the expression of mitotic cyclins during the prophase I meiotic arrest of Drosophila oocytes. Dev Cell 10: 127–135. [DOI] [PubMed] [Google Scholar]
- Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, Harte PJ. 2009. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 136: 3131–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WW, Wang Z, Yiu TT, Akdemir KC, Xia W, Winter S, Tsai CY, Shi X, Schwarzer D, Plunkett W, et al. . 2010. TRIM24 links a non-canonical histone signature to breast cancer. Nature 468: 927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah M, Pelletier N, Xiao L, Zhao SP, Wang K, Degerny C, Tahmasebi S, Cayrou C, Doyon Y, Goh SL, et al. . 2008. Molecular architecture of quartet MOZ/MORF histone acetyltransferase complexes. Mol Cell Biol 28: 6828–6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss AK, Collin C, Dixon MP, Thomas T. 2009. Moz and retinoic acid coordinately regulate H3K9 acetylation, Hox gene expression, and segment identity. Dev Cell 17: 674–686. [DOI] [PubMed] [Google Scholar]
- Weake VM, Swanson SK, Mushegian A, Florens L, Washburn MP, Abmayr SM, Workman JL. 2009. A novel histone fold domain-containing protein that replaces TAF6 in Drosophila SAGA is required for SAGA-dependent gene expression. Genes Dev 23: 2818–2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie GS, Dickson KS, Gray NK. 2003. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem Sci 28: 182–188. [DOI] [PubMed] [Google Scholar]
- Williamson A, Lehmann R. 1996. Germ cell development in Drosophila. Annu Rev Cell Dev Biol 12: 365–391. [DOI] [PubMed] [Google Scholar]
- Wu LH, Lengyel JA. 1998. Role of caudal in hindgut specification and gastrulation suggests homology between Drosophila amnioproctodeal invagination and vertebrate blastopore. Development 125: 2433–2442. [DOI] [PubMed] [Google Scholar]
- Xin T, Xuan T, Tan J, Li M, Zhao G. 2013. The Drosophila putative histone acetyltransferase Enok maintains female germline stem cells through regulating Bruno and the niche. Dev Biol 384: 1–12. [DOI] [PubMed] [Google Scholar]
- Xu P, Johnson TL, Stoller-Conrad JR, Schulz RA. 2012. Spire, an actin nucleation factor, regulates cell division during Drosophila heart development. PLoS ONE 7: e30565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ. 2004. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res 32: 959–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Muller HA, Wodarz A, Ephrussi A. 2004. PKA-R1 spatially restricts Oskar expression for Drosophila embryonic patterning. Development 131: 1401–1410. [DOI] [PubMed] [Google Scholar]
- Yun M, Ruan C, Huh JW, Li B. 2012. Reconstitution of modified chromatin templates for in vitro functional assays. Methods Mol Biol 833: 237–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.