Rac1 is essential for generating the protrusive activity that drives the collective migration of anterior visceral endoderm (AVE) cells in the early mouse embryo. Omelchenko et al. identified β-Pix as a potential regulator of Rac1. Genetic deletion of β-Pix in mice disrupts collective AVE migration.
Keywords: AVE migration, collective cell migration, Rho GTPases, GEFs, anterior–posterior axis, mouse embryo
Abstract
Collective epithelial migration is important throughout embryonic development. The underlying mechanisms are poorly understood but likely involve spatially localized activation of Rho GTPases. We previously reported that Rac1 is essential for generating the protrusive activity that drives the collective migration of anterior visceral endoderm (AVE) cells in the early mouse embryo. To identify potential regulators of Rac1, we first performed an RNAi screen of Rho family exchange factors (guanine nucleotide exchange factor [GEF]) in an in vitro collective epithelial migration assay and identified β-Pix. Genetic deletion of β-Pix in mice disrupts collective AVE migration, while high-resolution live imaging revealed that this is associated with randomly directed protrusive activity. We conclude that β-Pix controls the spatial localization of Rac1 activity to drive collective AVE migration at a critical stage in mouse development.
Collective epithelial cell migration is used throughout embryonic development to shape tissues and organs and is also a feature associated with cancer cell invasion and wound healing in the adult (Rorth 2007; Friedl and Gilmour 2009; Cheung et al. 2013; Tatin et al. 2013). The earliest example of collective migration during mouse development involves specification of the anterior–posterior axis (Takaoka and Hamada 2012). At embryonic day 5.5 (E5.5), a subpopulation of visceral endoderm (VE) cells, the presumptive anterior VE (AVE) cells, collectively migrates over a period of 5 h from the distal end of the embryo toward the extraembryonic (ExE) border (Srinivas et al. 2004; Migeotte et al. 2010; Trichas et al. 2012). AVE cells secrete antagonists of Nodal, BMP, and Wnt, while on the opposite side of the embryo, Nodal and Wnt signaling remain active and stimulate the formation of the primitive streak, thus breaking the radial symmetry of the embryo and defining the anterior–posterior axis (Arnold and Robertson 2009).
Conserved among amniotes, AVE cell migration is relatively simple and involves two epithelial cell layers—the VE and the epiblast—separated by extracellular matrix (ECM). Migrating AVE cells display key features of collective epithelial migration: the maintenance of cell–cell junctions, unidirectional leading-edge protrusions, coordinated cell body displacements, and multicellular rosette formation (Srinivas et al. 2004; Friedl and Gilmour 2009; Migeotte et al. 2010). During migration, the actin cytoskeleton is spatially organized anterior to posterior to drive forward-facing protrusions, and this behavior is coordinated between neighbors so as to maintain tissue integrity. Members of the Rho family of small GTPases—notably, Rho, Rac, and Cdc42—are key regulators of the actin cytoskeleton, and their spatially localized activities are thought to be essential for promoting cell migration (Machacek et al. 2009). In agreement with this, Rac1−/− mouse embryos display defects in AVE migration leading to an anterior–posterior axis duplication phenotype and early embryonic lethality (Rakeman and Anderson 2006; Migeotte et al. 2010). Live imaging revealed that Rac1 generates the spatially localized protrusive activity associated with AVE cells, and this is likely mediated through Nap1 and the WAVE complex, which in turn regulate Arp2/3 and actin polymerization (Rakeman and Anderson 2006).
Mammalian Rho GTPases are regulated by 82 guanine nucleotide exchange factors (GEFs) that promote GDP/GTP exchange and GTPase activation (Rossman et al. 2005; Cherfils and Zeghouf 2013). GEFs are thought to be responsible for defining the spatial compartmentalization of active GTPases, a key feature of signal transduction pathways regulating the actin cytoskeleton, although the function of only a few GEFs has so far been examined in vivo (Heasman and Ridley 2008). To identify potential regulators of Rac1 required for AVE migration and given this complexity, we first turned to an in vitro epithelial cell migration assay and identified the Cdc42/Rac GEF β-Pix as an essential regulator of collective migration. Using a conditional knockout mouse, we show that β-Pix is essential for collective AVE migration. Live imaging revealed that this GEF controls the spatial localization of Rac1 activity to drive directional AVE migration in the mouse embryo.
Results
Cellular mechanisms of AVE collective migration
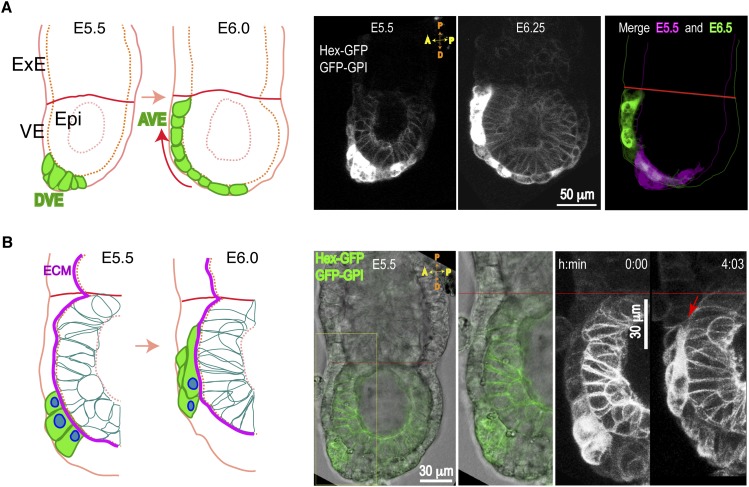
Collective AVE migration is complex and involves highly coordinated protrusive activity, cell translocation, cell division, and intercalation during a period of embryo growth and global cell movements (Srinivas et al. 2004; Migeotte et al. 2010; Takaoka et al. 2011; Trichas et al. 2011, 2012; Morris et al. 2012). To analyze AVE migration in more detail, we used high-resolution confocal video microscopy on live embryos expressing two transgenes: the AVE cell marker Hex-GFP and a ubiquitous membrane marker, GFP-GPI (Srinivas et al. 2004; Rhee et al. 2006). The expression of GFP-GPI within the VE is weak, allowing clear visualization of AVE cells (Hex-GFP) migrating over epiblast cells (GFP-GPI). AVE cells are localized at the distal end of the mouse embryo at E5.5 and translocate anteriorly over a period of ∼6 h (Fig. 1A; Supplemental Movie S1). Highly dynamic behavior can be seen in both AVE and epiblast cells, including cell divisions and changes in cell morphologies and relative positions (Supplemental Movie S2).
Figure 1.
AVE collective migration in the early mouse embryo. Transgenic embryos (E5.5–E6.5) expressing the indicated transgenes were imaged by DIC and confocal time-lapse microscopy. (A) Sagittal sections showing AVE (Hex-GFP) cells migrating over epiblast (GFP-GPI) cells. Schematic (left) and maximal projections (right) of four optical sections (6-μm thickness) taken from time-lapse snaphots (from Supplemental Movie S1). (Epi) Epiblast; (DVE) distal VE. A morphologically distinct, highly columnar subpopulation of VE cells expressing the Hex-GFP marker (green) and located at the distal end (DVE) move on the presumptive anterior side (red arrow), arrive at the embryonic/ExE border (red line), and become AVE. (B) Schematic (left) and optical projections (right) from time-lapse snapshots (10-μm thickness) of migrating Hex-GFP cells (green) moving on the ECM (magenta line). Long protrusions intercalate between front VE cells and the epiblast (red arrow).
Closer examination of AVE cells revealed long protrusions extending proximally toward the ExE border and formed in multiple cell rows (Supplemental Movies S2, S3). The VE is separated from the epiblast by the ECM, and migrating AVE cells intercalate their protrusions between the surrounding VE cells and the epiblast as they move along the ECM (Fig. 1B; Supplemental Fig. S1A,B). Time-lapse analysis of sagittal optical sections revealed the dynamics of the intercalating basal protrusions (Supplemental Fig. S1A,B; Supplemental Movie S4). Protrusions run underneath neighboring VE cells, possibly leading to competition for matrix attachment leading to displacement of adjacent non-AVE cells (Supplemental Fig. S1B), as has been observed in two-dimensional (2D) cultures of heterotypic epithelial sheets (Fetisova et al. 1990). The nonuniform levels of Hex-GFP expression allow visualization of long protrusions in back-row AVE cells in sagittal sections (Supplemental Fig. S1E).
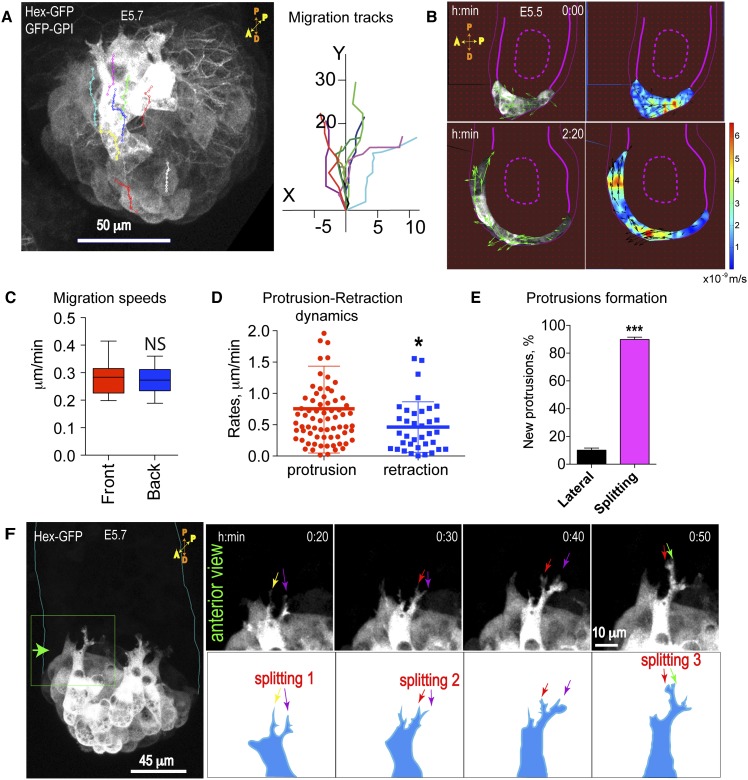
During AVE migration, cells move coordinately from the distal to the proximal end of the embryo, eventually reaching the ExE border within ∼6–7 h in dissected, cultured embryos. Once collective migration is established, “front-row” and “back-row” cells are defined relative to the direction of migration such that front-row cell bodies contact nonfluorescent VE cells. Measurements of migration trajectories (tracks) and cell migration speeds in different cell rows show that front- and back-row cells migrate directionally and with similar speeds (front row: 0.29 μm/mi ± 0.02 μm/min; back rows: 0.27 μm/mi ± 0.02 μm/min) (Fig. 2A,C; Supplemental Movie S5). Quantitative analysis of tissue flow using particle image velocimetry (PIV) revealed coordinated directional vectors in multiple cell rows as AVE cells migrate as a group toward the ExE (Fig. 2B, left panels; Poujade et al. 2007). The higher instantaneous velocities (Fig. 2B, red) seen in some back-row cells (Fig. 2B, right panels) suggest the presence of highly contractile cells within the group (Vedula et al. 2012).
Figure 2.
Morphological characterization of migrating AVE cells. Transgenic embryos (E5.5–E5.7) were imaged by DIC and confocal time-lapse microscopy. (A) Anterior view of an embryo: AVE (Hex-GFP) cells migrate unidirectionally toward the ExE border as visualized by overlay tracking of eight cells over a period of 2 h from a time-lapse movie (from Supplemental Movie S5). (B) PIV analysis showing vectors of particle flow in sagittal sections (left two panels; time is hours:minutes) and heat maps of instantaneous velocities (right two panels in meters per second). (C) Migration speeds in the front- and back-row cells. Data represent mean ± SEM of 14 front cells and 10 back-row cells from three embryos. (NS) Not significant; P = 0.663, unpaired Student’s t-test. (D) Protrusions and retractions cycle every 5–10 min, with a mean protrusion rate of 0.74 μm/min ± 0.08 μm/min (±SEM; n = 73 cells, six embryos) and mean retraction rate of 0.46 μm/min ± 0.07 μm/min (±SEM; n =36 cells, six embryos); (*) P = 0.027, unpaired Student’s t-test (from Supplemental Movies S2, S6). (E) Percentage of new protrusions formed by splitting of existing ones versus newly formed lateral protrusions (mean ± SEM; n = 14 cells; three embryos; unpaired Student’s t-test, [***] P < 0.001). (F) An anterior view of the embryo showing AVE protrusions at the front row formed by the splitting of existing ones (arrows) (from Supplemental Movie S7).
Morphometric analysis was used to characterize the properties of the highly dynamic protrusions seen in AVE cells (Fig. 2D,E; Supplemental Fig. S1C,D; Supplemental Movie S6). Extension and retraction protrusion rates in front-row cells were 0.74 μm/min ± 0.08 μm/min and 0.46 μm/min ± 0.07 μm/min, respectively. As a result, the difference in protrusion versus retraction rates correlates with cell translocation. This is similar to what has been reported for migrating epithelial leader cells observed in 2D cultures (Omelchenko et al. 2003). AVE protrusions are associated with numerous filopodia, which underlap cells in front and often run between cells at the zone of cell–cell contacts (Supplemental Fig. S1D). We previously reported the “snail-like” shape of protrusions in migrating AVE cells (Fig. 2F; Migeotte et al. 2010). Time-lapse microscopy revealed that an emerging protrusion bifurcates at its end, and this is then followed by a subsequent bifurcation in the more dominant protrusion (Fig. 2F; Supplemental Movie S7). Most of the protrusions analyzed (87.8% ± 2.8%) are formed by bifurcation rather than de novo at lateral sides (Fig. 2E), which, interestingly, is characteristic of cells migrating in shallow gradients, such as neutrophils, Dictyostelium, and fibroblasts (Andrew and Insall 2007).
β-Pix is required for collective epithelial cell migration
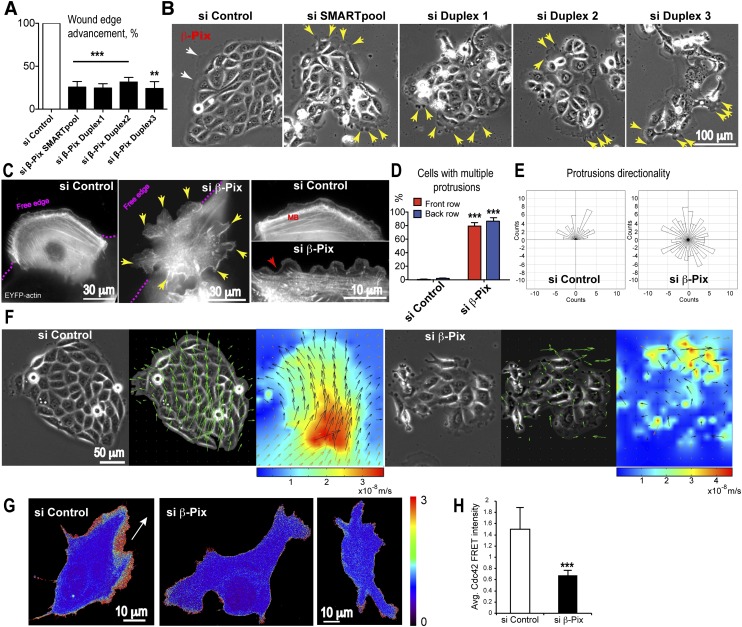
We previously established a critical role for Rac1 in collective AVE migration, but the upstream regulatory mechanisms directing Rac1 activity to produce directed migration are unknown (Migeotte et al. 2010). Given the complexity of potential Rac1 regulators encoded in the mammalian genome, we first exploited an in vitro collective epithelial cell migration assay to perform an RNAi screen of Rho family GEFs. An siRNA SMARTpool library targeting the 82 known human Rho GEFs was screened in the human bronchial epithelial cell line 16HBE14o (16HBE) using a previously described monolayer scratch migration assay (Omelchenko and Hall 2012). These cells undergo collective migration in culture and provide a simple and straightforward screen to identify important GEFs. Depletion of β-Pix (also known as ARHGEF7/Cool-1) by the SMARTpool or the four individual duplexes (Supplemental Fig. S2A) caused a strong inhibition of 16HBE cell migration compared with control cells (wound edge advancement: 25.69% ± 6.6% in SMARTpool; 24.61% ± 5% in duplex 1; 31.57% ± 5.5% in duplex 2; and 24.17% ± 7.9% in duplex 3) (Fig. 3A). As a negative control, we depleted the p114RhoGEF, which we previously showed disrupts apical junctions but is not required for collective migration (Supplemental Fig. S2G; Xu et al. 2013). Phase-contrast video microscopy of collectively migrating islands of 16HBE cells revealed that depletion of β-Pix resulted in multiple, misoriented dynamic protrusions formed at free cell edges all around the island (Fig. 3B, yellow arrows; Supplemental Movie S8). This is in contrast to control islands, which have only a few protrusions restricted to the front of the migrating group (Fig. 3B, white arrows; Supplemental Movie S8). The multiprotrusion phenotype is similar to that generated after depletion of Cdc42, whereas depletion of Rac1 leads to a complete loss of protrusive activity in these cells (Supplemental Fig. S2B).
Figure 3.
β-Pix is required for collective epithelial migration. 16HBE epitheliocytes were used in an in vitro collective epithelial migration assay as cells in either a scratched monolayer or discrete islands. An siRNA screen of the 82 mammalian Rho family GEFs performed in the scratch assay identified β-Pix as a candidate regulator of collective migration. (A) siRNA-mediated depletion of β-Pix by an siRNA SMARTpool (mixture of four siRNAs) or the four individual duplexes inhibits wound edge advancement in a monolayer scratch assay (mean ± SEM; n = 3 independent experiments; unpaired Student’s t-test, [***] P < 0.001 for duplexes 1 and 2 and SMARTpool; [**] P = 0.01 for duplex 3). (B) Phase contrast images of 16HBE cells showing multiple protrusions at the free edges of cells all around epithelial islands (yellow arrows) in β-Pix-depleted cells. In control islands, protrusion distribution is polarized (white arrows) in the direction of migration. (C) Mosaic expression of EYFP-actin in front-row cells of a scratched monolayer reveals multiple protrusions at the free edge (pink dotted line) and at the posterior and lateral sides (yellow arrows) in β-Pix-depleted cells (live epifluorescence imaging). In control cells, a single lamellipodium can be seen at the free edge, with relatively quiescent rear edges. The prominent marginal bundle (MB) seen in control cells is disassembled in β-Pix-depleted cells (inset; red arrow). (D) Quantification showing that β-Pix depletion generates multiple protrusions in both front- and back-row cells (mean ± SEM; n = 3 independent experiments; unpaired Student’s t-test, [***] P < 0.001 for both front- and back-row cells). (E) Quantification of protrusion directionality shows a random distribution after β-Pix depletion (n = 60 control protrusions; n = 164 protrusions in β-Pix-depleted cells; three independent experiments). (F) Quantitative PIV analysis was used to analyze cell flow in migrating islands. A collectively migrating control island shows unidirectional vectors (middle image) and a gradient of velocities (heat map) (taken from Supplemental Movie S9). β-Pix-depleted islands lose both directionality of migration (middle image) and a collective gradient of velocities (heat map). (G) Maximum projections of confocal sections of migrating 16HBE cells transiently expressing a dual-chain Cdc42 biosensor. Fluorescent cells are surrounded by nonfluorescent cells. Active Cdc42 is located at the leading edge in a control cell, in contrast to reduced Cdc42 activity in β-Pix knockdown cells. The white arrow shows the direction of migration. (H) Quantification of Cdc42 FRET intensity in lamellipodial regions. Data are mean ± SD; 20 control cells and 25 siRNA duplex 1 β-Pix knockdown cells; unpaired Student’s t-test, (***) P < 0.0001.
To characterize the phenotype in more detail, migrating 16HBE cells expressing EYFP-actin in a mosaic distribution were examined in scratched monolayers. Wild-type cells at the leading edge each showed a typical, single lamellipodial protrusion at the front and a relatively quiescent trailing edge associated with some retraction fibers (Fig. 3C). After β-Pix depletion, leading-edge cells each showed multiple, distorted protrusions at not only the anterior free cell edge but also the lateral sides and rear of the cell, underlapping neighboring cells (Fig. 3C, yellow arrows). In addition, the actin marginal bundle was disassembled (Fig. 3C, MB). Rescue experiments using an siRNA-resistant mouse β-Pix construct demonstrated the specificity of the phenotype (Supplemental Fig. S2C). Multiple protrusions were observed in front- and back-row cells in β-Pix-depleted cells (Fig. 3D; Supplemental Fig. S2D), and the protrusions lost their directionality (Fig. 3E; Supplemental Fig. S2E).
PIV analysis was used to assess collectivity of migration and tissue flow. Control migrating islands showed unidirectional vectors and a gradient of instantaneous velocities, with highest rates at the rear of the colony (Fig. 3F, heat map, red). This cooperation was maintained over time, allowing the colony to migrate collectively, reflecting a high level of cooperation within the migrating group (Supplemental Movie S9). Thus, the 16HBE colony is migrating as one large “supercell” (Khalil and Friedl 2010). The directionality of vectors and gradient of instantaneous velocities were lost in β-Pix knockdown cells, reflecting a loss of collective migration (Fig. 3F; Supplemental Fig. S2F; Supplemental Movie S9). Together, these results suggest that β-Pix regulates collective epithelial migration by spatially restricting Rac-mediated protrusions.
To confirm that β-Pix controls the activity of Cdc42 during collective migration, we made use of a dual-chain Cdc42 biosensor (Fig. 3G; Hodgson et al. 2010). Imaging and quantification of FRET (fluorescence resonance energy transfer) revealed a significant reduction in Cdc42 FRET intensity in the regions of protrusive activity upon β-Pix depletion (Fig. 3H). This result suggests that β-Pix regulates Cdc42 activity in migrating 16HBE cells.
β-Pix is expressed in AVE cells, and its genetic ablation leads to early embryonic lethality
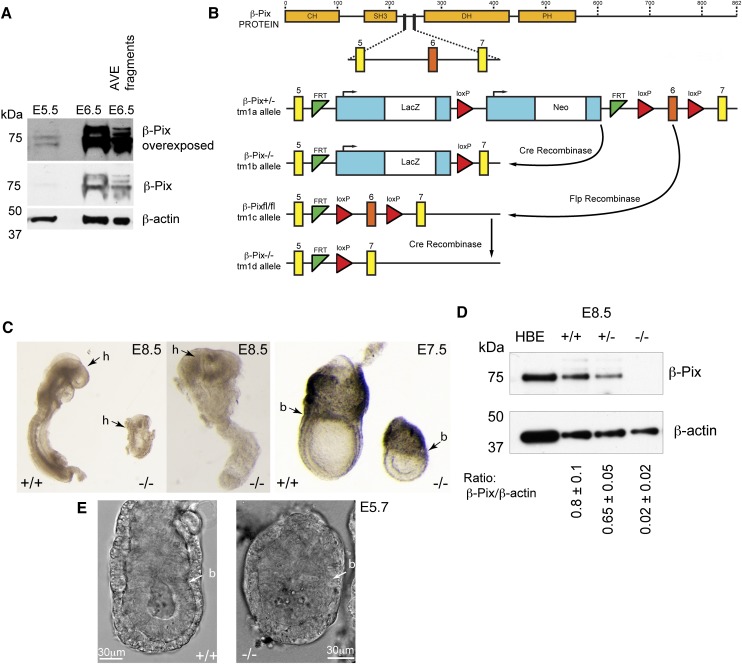
With the 16HBE result in hand, we next examined whether β-Pix might play an analogous role in controlling the localization of Rac1 activity during collective AVE migration. The murine Arhgef7 (β-Pix) gene is located on chromosome 8 and contains 19 exons encoding three isoforms of β-Pix protein (a, b, c) (Kim et al. 2000; Kim and Park 2001; Yoshizawa et al. 2003; Rhee et al. 2004). Multiple β-Pix isoforms were detected in both E5.5 and E6.5 mouse embryo extracts on Western blots (Fig. 4A). Both isolated fragments enriched for Hex-GFP (AVE cells) (Supplemental Fig. S3A), and ExE fragments showed similar multiple isoforms (Fig. 4A; data not shown).
Figure 4.
β-Pix is required for mouse embryonic development. (A) Western blot showing β-Pix protein expressed in multiple isoforms and detected at E5.5, with increasing levels at E6.5 in whole embryos and in isolated AVE fragments (representative of three independent experiment, equal amount of protein loaded) (Supplemental Fig. S3A). (B) Schematic of β-Pix protein and a portion of the Arhgef7 gene showing the organization of exons 5, 6, and 7 and the strategies used to disrupt Arhgef7. (C) Morphological analysis of normal and β-Pix−/− embryos. Images of littermates dissected at various stages (E6.5–E8.5) as indicated. (h) Head; (b) embryonic/ExE border. Note the different magnifications for the panels: E8.5, 1.25× and 3.2× (left); E7.5, 2.5×. (D) Western blot of embryos from β-Pix+/− tm1b mouse intercrossing. Equal amounts of embryo protein lysates were loaded. Data are mean ± SEM (major band); two independent experiments. Note the absence of any isoforms in the β-Pix−/− embryo sample. Uncropped blots are shown in Supplemental Figure S5. (E) Single confocal DIC sections of live β-Pix+/+ tm1b and β-Pix−/− embryos at E5.7, the earliest stage at which β-Pix−/− embryos become morphologically distinct. (b) Embryonic/ExE border.
The β-Pix protein is composed of the four conserved domains: CH (calponin homology/actin binding), SH3 (Src homology 3/proline-rich peptide binding), DH (DBL homology/Cdc42/Rac1 binding), and PH (plekstrin homology/phosphatidylinositol lipids/heterotrimeric G protein binding) (Fig. 4B). It has been reported to have GEF activity toward Rac and Cdc42 (Baird et al. 2006; ten Klooster et al. 2006; Kuo et al. 2011; Kutys and Yamada 2014). We obtained a transgenic mouse tm1a (targeted mutation 1a), generated as a part of the genome-wide study of mouse gene function and harboring a frameshift-inducing deletion in one β-Pix allele (Skarnes et al. 2011). From the tm1a mouse, β-Pix-deficient mice were generated by Cre- and Flp-mediated recombination (CAG promoter), deleting exon 6 flanked with loxP sites (producing tm1b- and tm1d-null alleles with or without lacZ reporter, respectively) (Fig. 4B). Mendelian ratios of wild-type, heterozygous, and homozygous embryos were detected. At both E6.5 and E8.5, β-Pix−/− embryos showed developmental defects in morphology and size (Fig. 4C), and β-Pix−/− embryos died at about E8.5. As expected, β-Pix−/− embryos did not express β-Pix protein, while lower levels were found in heterozygous β-Pix+/− embryos compared with wild type (Fig. 4D). The antibody used in this study recognizes the β-Pix SH3 domain, which is located before the deletion site, suggesting that no truncated proteins are produced.
During early development, at E6.5, β-Pix expression was detected in the VE by in situ hybridization of mRNA transcripts (Supplemental Fig. S3B). β-Galactosidase staining in tm1a embryos carrying the LacZ cassette revealed β-Pix expression in the VE at E5.5. At E6.0, a strong signal was found in the ExE VE; at E7.5, it was also found at the midline; later, at E8.5, histological sections showed expression in the neural tube cells (Supplemental Fig. S3B). These data suggest a possible functional role of β-Pix at numerous sites in early mouse development.
β-Pix is required for the anterior–posterior axis specification and proper AVE positioning
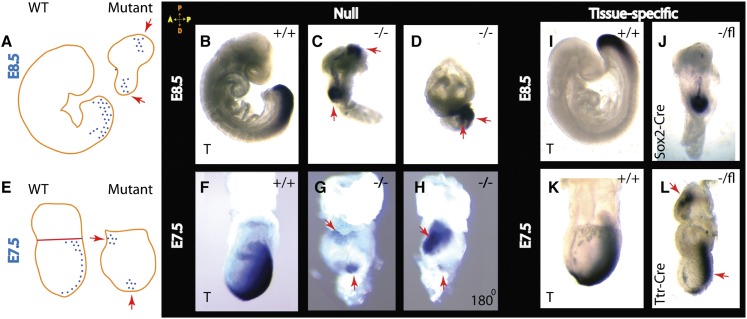
During embryogenesis, development of the anterior–posterior axis takes place through two distinct cellular events: AVE migration and primitive streak formation (Arnold and Robertson 2009). In β-Pix−/− embryos, an early primitive streak marker, Brachyury, revealed a double-axis phenotype (Fig. 5, red arrows) in contrast to a single streak in wild-type embryos at E8.5 (Fig. 5A–D) and at E7.5 (Fig. 5E–H). Another primitive streak marker, the Wnt reporter BAT-gal, showed a proximal activity across the ExE–epiblast border in β-Pix−/− embryos (Supplemental Fig. S3C; Maretto et al. 2003). To check whether the axis duplication phenotype is a consequence of loss of β-Pix in the VE or in the epiblast, β-Pix−/Sox2-Cre male mice were generated and crossed with tm1c (β-Pixflox/flox) mice to generate embryos lacking β-Pix in the epiblast (Hayashi et al. 2002). β-Pix−/epiblast-deleted embryos showed a single primitive streak marked by expression of Brachyury (Fig. 5I,J). To delete β-Pix specifically in the VE, a β-Pix−/Ttr-Cre male was generated and crossed with tm1c (β-Pixflox/flox). β-Pix−/VE-deleted embryos showed axis duplication at E7.5 (Fig. 5K,L). We conclude that β-Pix acts autonomously in the VE to promote axis specification.
Figure 5.
Deletion of β-Pix results in anterior–posterior axis duplication. In situ hybridization analysis of Brachyury (T) expression is shown schematically at E8.5 (A) and E7.5 (E) for wild-type and β-Pix-null embryos. (B–D) Whole-mount in situ hybridization of wild-type littermate control embryos (β-Pix+/+) and β-Pix−/−-null (tm1b allele) embryos at E8.5. The primitive streak marker Brachyury (T) is seen on the posterior side of the wild-type embryo, while in β-Pix−/−-null embryos, Brachyury (T) is localized in two distinct sites (red arrows), thus duplicating the anterior–posterior axis. (F–H) At E7.5, the primitive streak marker in mutant embryos is located at two different sites (snapshots of the same embryo rotated 180°). (I–L) Tissue-specific knockouts reveal axis duplication after β-Pix knockout in VE cells (Ttr-Cre; β-Pix−/VE-deleted) (L) but not after β-Pix knockout in epiblast cells (Sox2-Cre; β-Pix−/epiblast-deleted) (J). At E6.5, a Brachyury (T) signal is not clearly established (see Supplemental Fig. S3E).
β-Pix is required for directional migration of AVE cells
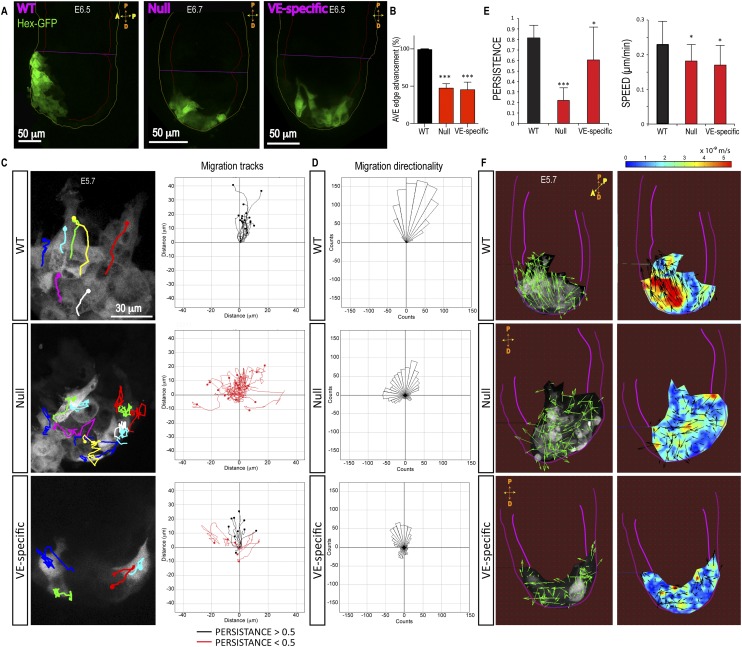
In wild-type embryos, AVE cells are localized at the ExE on the anterior side by E6.5, as visualized by Hex-GFP (Fig. 6A; Supplemental Fig. S3C,D) or the AVE marker Cer1 (Supplemental Fig. S3D). In contrast, in β-Pix−/− and β-Pix−/VE-deleted embryos, Hex-GFP cells remain distally located (Fig. 6A; Supplemental Fig. S3C,D). AVE edge advancement was inhibited by 47% ± 6% in β-Pix−/− and 45% ± 10% in β-Pix−/VE-deleted embryos relative to wild type (Fig. 6B). Cell-tracking analysis in time-lapse movies revealed a severe loss of directionality in AVE cell migration in both β-Pix−/− and β-Pix−/VE-deleted embryos (Fig. 6C,D; Supplemental Movies S5, S10). β-Pix−/− embryos showed a strong reduction in directional persistence of migration (0.22 ± 0.12), calculated as a ratio of absolute distance divided by accumulated distance traveled (wild type = 0.81 ± 0.1) (Fig. 6C,E). However, the migration speeds of individual AVE cells showed only a small reduction in β-Pix−/− embryos (0.18 μm/min ± 0.05 μm/min, ±SD) and β-Pix−/VE-deleted embryos (0.17 μm/min ± 0.06 μm/min) compared with wild-type embryos (0.23 μm/min ± 0.07 μm/min) (Fig. 6E). Tissue flow analysis by PIV showed a loss of unidirectional vectors and a disorganized pattern of instantaneous velocities in β-Pix−/− and β-Pix−/VE-deleted embryos compared with wild-type AVE cells (Fig. 6F). In wild-type embryos, high instantaneous velocity vectors (red regions) were distributed in a gradient fashion, predominantly localized in back-row cells (Fig. 6F, wild type [WT]), as observed earlier (Fig. 2B). In contrast, mutant embryos revealed a disorganized distribution of velocities, suggesting a loss of collectivity (Fig. 6F).
Figure 6.
Loss of β-Pix disrupts collective AVE migration. (A) An image from live confocal microscopy reveals AVE (Hex-GFP) cells located at the anterior side of control embryos at E6.5. AVE cells fail to migrate in β-Pix−/− (null) and β-Pix−/VE-deleted (VE-specific) mutant embryos and remain distal. (B) Quantification of AVE edge advancement toward the ExE border at E6.5 (mean ± SEM; n = 6 embryos; unpaired Student’s t-test, [***] P < 0.001 for both genotypes). (C) Cell tracking from time-lapse microscopy reveals disorganized migration of Hex-GFP cells in β-Pix−/− (null) and β-Pix−/VE-deleted (VE-specific) mutant embryos compared with wild type (from Supplemental Movie S10). (D) Quantification of migration directionality was performed in two independent experiments (n = 17 wild-type], 22 null, and 14 VE-specific cells per condition). (E) Quantification of AVE cell migration persistence (n = 20 cells in two null embryos; n = 17 cells in two wild-type embryos) and migration speeds (β-Pix−/−-null embryos: n = 20 cells, two embryos; β-Pix−/VE-deleted embryos: n = 15 cells, two embryos; wild-type embryos: n = 17 cells, two embryos). (F) PIV analysis of Hex-GFP migrating cells. Representative of three independent experiments. The left panels represent directionality of vectors, and the right panels show the spatial distribution of instantaneous velocities (heat map, meters per second). Pink lines outline the embryo and VE.
Immunostaining for apical cell–cell junctions revealed no defects in localization of E-cadherin or ZO-1 in β-Pix−/− embryos, indicating that no epithelial-to-mesenchymal (EMT)-like phenotype had occurred (Supplemental Fig. S4A). This was also confirmed in 16HBE cells (Supplemental Fig. S4B–D), where the levels of E-cadherin and N-cadherin remain unchanged after β-Pix depletion, whereas the levels of β-Pix-binding partners GIT-1 and PAK1/2 were reduced (Supplemental Fig. S4C,E).
β-Pix is required for directing protrusive activity in AVE cells
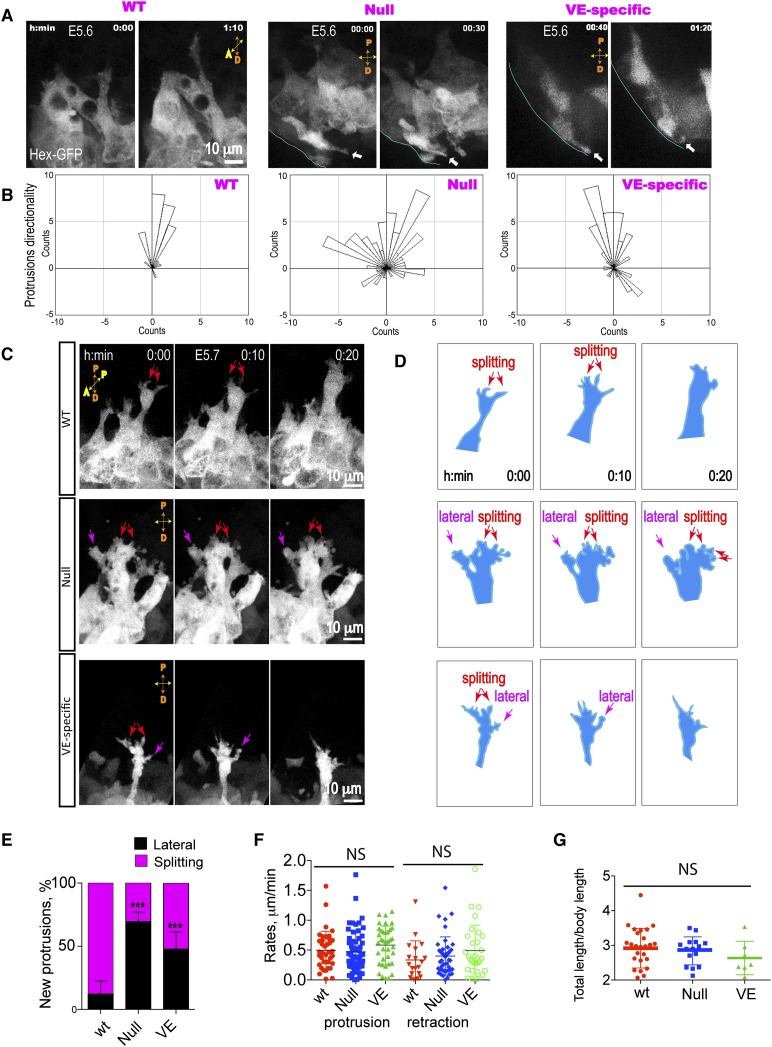
During migration, AVE cells extend long-lived (10 min) but dynamic protrusions directed toward the ExE region (Figs. 1, 7A; Supplemental Movie S3; Migeotte et al. 2010). The directionality of protrusions is uniform among multiple cells over the time of migration in wild-type embryos (Fig. 7A). In both β-Pix−/− and β-Pix−/VE-deleted embryos, however, AVE protrusions were misoriented, often facing toward the distal end (Fig. 7A [arrows], B). Morphological analysis revealed significant protrusive activity in mutant embryos; however, the shape and dynamics of protrusions were aberrant (Fig. 7C; Supplemental Movie S11). In β-Pix−/− and β-Pix−/VE-deleted embryos, many protrusions were formed de novo from lateral sides of the cells. The increase was from 12.19% ± 2.8% of lateral protrusions in wild-type cells to 69.52% ± 2.7% in β-Pix−/−-null cells and to 47.77% ± 7.8% in β-Pix−/VE-deleted cells (Fig. 7D,E). Protrusion lengths and rates of retraction and extension were similar to wild type (Fig. 7F,G), but directionality was severely disrupted in mutant embryos (Fig. 7A,B), consistent with the loss of directed AVE migration (Fig. 6C).
Figure 7.
Deletion of β-Pix disrupts the directionality of AVE protrusions. Images were obtained by maximal projections of optical sections from time-lapse confocal sequences of Hex-GFP-expressing embryos at E5.6. (A) Confocal snapshots from time-lapse sequences show protrusive activity in the proximal direction in wild-type embryos (from Supplemental Movie S3), but both proximal and distal (arrows) protrusions are seen in mutant embryos. (B) Measurements of protrusion directionality reveal the disorganized orientation of protrusions after β-Pix deletion. n = 30 protrusions in three wild-type embryos, n = 86 protrusions in three β-Pix−/− null embryos, and n = 54 protrusions in β-Pix−/VE-deleted embryos. (C) Morphological analysis of protrusion bifurcations. β-Pix+/+ (wild-type [WT]) embryos show long protrusions originating from multiple cells and oriented toward the embryonic/ExE border. New protrusions are formed by splitting (in the proximal direction; red arrows). In contrast, in β-Pix−/− (null) or β-Pix−/VE-deleted (VE-specific) mutant embryos, Hex-GFP cell protrusions are frequently formed de novo at lateral sides (magenta arrows) or by splitting but are oriented to the side rather than proximal (from Supplemental Movie S11). (D) Schematic of protrusion formation shown in C. (E) Quantitative analysis of protrusion formation (mean ± SEM; n = 14 wild-type cells; n = 8 β-Pix−/− cells; n = 3 β-Pix−/VE-deleted cells; from three embryos). (F) Protrusion dynamics of Hex-GFP cells at E5.7 (protrusion activity: unpaired Student’s t-test, P = 0.7, n = 38 protrusions in β-Pix+/+ cells, n = 60 protrusions in β-Pix−/− cells, P = 0.237, n = 38 protrusions in β-Pix−/VE-deleted cells; retraction activity: unpaired Student’s t-test, P = 0.453, n = 19 protrusions in β-Pix+/+ cells, n = 44 protrusions in β-Pix−/− cells, P = 0.174, n = 30 protrusions in β-Pix−/VE-deleted cells). (G) Length of protrusions (unpaired Student’s t-test, P = 0.646; n = 26 protrusions in β-Pix+/+ cells; n = 15 protrusions in β-Pix−/− cells; P = 0.242; n = 7 protrusions in β-Pix−/VE-deleted cells) are not significantly different in mutant and wild-type embryos.
Discussion
The migration of AVE cells along the presumptive anterior side of E5.5 mouse embryos is responsible for setting up the anterior–posterior body axis of the mouse embryo (Takaoka and Hamada 2012). Previous work has revealed that during migration, AVE cells maintain their epithelial characteristics and move collectively as a group through the VE surrounding the epiblast (Migeotte et al. 2010; Trichas et al. 2011; Bloomekatz et al. 2012). Genetic ablation studies have identified some key players required to drive this migratory behavior; namely, the small GTPase Rac1 and the Rac1 target protein Nap1 (Rakeman and Anderson 2006; Migeotte et al. 2010). Nap1 is a component of the WAVE complex, which in turn recruits the Arp2/3 complex to promote localized actin polymerization. Migrating AVE cells display extended protrusions in the direction of migration (i.e., toward the ExE border), and these are absent in Rac1- or Nap1-null cells (Rakeman and Anderson 2006; Migeotte et al. 2010). Actin-driven protrusions are therefore an essential component of AVE cell collective migration.
The mechanisms regulating directionality of AVE migration are not well understood. Rac1 activation is controlled by members of a large family of GEFs, and during directed migration, Rac1 activation must be spatially localized. In order to identify potential regulators of Rac1 in AVE cells, we first made use of an in vitro collective epithelial migration assay. Following an RNAi-based screen of the 82 known mammalian Rho family exchange factors in the bronchial epithelial cell line 16HBE, we identified a Cdc42/Rac-specific GEF β-Pix that is required for collective migration in these cells. This GEF is expressed in early mouse embryos, and so we generated conditional β-Pix knockout mice to determine whether it plays a role in collective AVE migration. As described here, in the absence of β-Pix, AVE cells are unable to move toward the ExE border in E5.5 embryos and remain at the distal end of the embryo.
The disruption of AVE migration seen in the absence of β-Pix appears similar to that seen after loss of Rac1 or Nap1; cells remain distal, leading to axis duplication and lethality at around E9.5 (Rakeman and Anderson 2006; Migeotte et al. 2010). Careful analysis of AVE cells, however, revealed significant differences. Cell tracking revealed a strong directional persistence of AVE migration toward the ExE border in control embryos, and although this is severely disrupted in the absence of β-Pix, cells still migrate with speeds similar to that of wild-type cells. Also, unlike Rac1 knockout AVE cells, which lack significant protrusive activity, β-Pix knockout cells maintain protrusive activity, but this is disorganized and not efficiently directed toward the ExE border. Finally, PIV analysis to examine tissue flow and velocity gradients within the AVE group of cells revealed high directional coordination of velocity vectors and focused gradients of velocities in wild-type embryos. Both were severely disrupted in the absence of β-Pix. We conclude that Rac1 is still active in AVE cells in the absence of β-Pix but that its activity is no longer spatially organized so as to promote directed protrusion activity and migration. This is in contrast to what has been reported in cultured fibroblasts, where β-Pix is required for protrusion formation (Cau and Hall 2005).
The external cues directing AVE migration are not clear. The predominant view is that cells navigate along prepatterned gradients and that Wnt and Nodal play important roles (Yamamoto et al. 2004; Kimura-Yoshida et al. 2005; Arnold and Robertson 2009; Rorth 2011). AVE cells send long protrusions toward the ExE border, raising the possibility that cell–cell communication also plays a role (Rorth 2003; Migeotte et al. 2010). The bifurcating nature of AVE protrusive activity that we describe here is typical of cells moving by chemotaxis, and it has been suggested that chemotactic signals may influence the maintenance and retraction of protrusions rather than inducing protrusions de novo (Andrew and Insall 2007). The PIV analysis that we describe here also reveals distinct regions of high instantaneous velocities within the AVE population, suggesting a push–pull mode of group migration, a characteristic of collective epithelial migration under narrow geometrical constraints in 2D (Vedula et al. 2012). Furthermore, the relatively low velocities seen at the leading front suggest that the contribution of protrusions to active migration may not be direct but may have a more exploratory or pathfinding role. Hex-GFP is known to mark two subpopulations of VE cells, AVE and distal VE, and the latter are essential for AVE migration (Takaoka et al. 2011). This raises the possibility that cell heterogeneity may contribute to the velocity patterns that we observed by PIV analysis.
Finally, the effect of β-Pix knockout in AVE cells is similar to that seen in the 16HBE cell culture assays after β-Pix knockdown; namely, the formation of delocalized protrusions. Interestingly, this is also similar to the phenotype that we describe after RNAi depletion of Cdc42 in 16HBE cells and that others have described after depleting Cdc42 in cultured astrocytes (Osmani et al. 2006). The use of a Cdc42 biosensor directly demonstrated β-Pix-dependent regulation of Cdc42 activity in the protrusions of migrating 16HBE cells. This raises the possibility that, in AVE cells, β-Pix also acts as a GEF (i.e., activator) for Cdc42 and that Cdc42 is required to spatially organize Rac1 activity. We are currently using a combination of in vitro collective migration assays and mouse genetics to identify the specific GEF responsible for activating Rac1 in AVE cells and the mechanism by which β-Pix is spatially localized/activated to direct protrusive activity toward the ExE border.
Materials and methods
Animals
Animal experiments were approved by the institutional committee (Memorial Sloan Kettering Cancer Center, Research Animal Resource Center). Mouse strains C3H, CD1, and C57Bl/6 were used. Mice containing the Arhgef7tm1a(EUCOMM)Wtsi allele on a C57BL/6NTac background were purchased from Wellcome Trust Sanger Institute/International Knockout Mouse Consortium/EUC. The Arhgef7tm1a(EUCOMM)Wtsi allele is a knockout first, reporter-tagged insertion with conditional potential (promoter-driven cassette) (Skarnes et al. 2011) in C57BL/6N embryonic stem cells. This strategy relies on the identification of a “critical” exon common to all transcript variants that, when deleted, creates a frameshift mutation. The β-Pix conditional allele (tm1c) or null alleles (tm1b and tm1d) were analyzed on a mixed C3H/C57Bl/6 background. Genomic information on mouse Arhgef7 was obtained from the NCBI (http://www.ncbi.nlm.nih.gov) and Ensemble (ENSMUSG00000031511; http://www.useast.ensembl.org) Web sites.
Cell culture and nucleofection
16HBE human bronchial epithelial cells were maintained in MEM with GlutaMax (Invitrogen) supplemented with 10% fetal calf serum (FCS) and the antibiotics streptomycin and penicillin (Gibco; Life Technologies) (Cozens et al. 1994). Sixty nanograms to 100 ng of siRNA (Dharmacon) was used to transfect 0.5 × 105 to 1 × 105 16HBE cells. Duplex sequences for human ARHGEF7 (NM_003899) were duplex-1 (GGAAGAAGAUGCUCAGAUU), duplex-2 (GAAGAGCCCUCCCAAAGGA), duplex-3 (UCAAAGAGCUCGAGAGACA), and duplex-4 (GGAGGGCGAUGACAUUAAA). For transient expression, 4 μg of DNA was nucleofected into 0.5 × 106 16HBE cells using kit T (Lonza) with program A-23 according to the manufacturer’s protocol. All stable lines were obtained by selection in G418.
Lentiviral-mediated generation of stable 16HBE cells with knockdown of β-Pix was performed with Sigma MISSION constructs (NM_003899): shRNA #1 (5′-CCGGGCCCTCCCAAAGGATTTGATACTCGAGTATCAAATCCTTTGGGAGGGCTTTTTG-3′), shRNA #2 (5′-CCGGGCGGATATTAGTGTCGTGCAACTCGAGTTGCACGACACTAATATCCGCTTTTTG-3′), shRNA #3, (5′-CCGGCCTGGGATGAGACCAATCTATCTCGAGATAGATTGGTCTCATCCCAGGTTTTTG-3′), and nontarget pLKO.1-puro control. For lentivirus production, HEK293T cells were transfected with shRNA, VSV-G, and pDeltaR8.9 DNA constructs using Lipofectamine 2000 (Invitrogen). 16HBE cells were infected with a viral suspension diluted in MEM/10% FCS growth medium containing 8 μg/mL polybrene. Puromycin-resistant cells were used for experiments.
Genotyping
Mouse punches were digested in 100 μL of 300 μg/mL Proteinase K (Roche) in PCR buffer (Invitrogen) overnight at +55°C an then heat-inactivated. One microliter to 3 μL was used for genotyping. Embryos were digested in 15 μL of Proteinase K/PCR buffer, and 5 μL was used for genotyping. Genotyping for GFP, LacZ, and Cre was done as described previously (Migeotte et al. 2010). To detect wild-type β-Pix/Arhgef7, tm1a, 5′FRT, tm1b, tm1c, tm1d, and LoxP, the following primer combinations were used: wild-type β-Pix/Arhgef7 (Arhgef7_40333_F, 5′-TGCTAAAACAGTGGCAGGTG-3′; Arhgef7_40333_R, 5′-ACAGAACACTGCTGCTTCCA-3′), 200 base pairs (bp); Tm1a (Arhgef7_40333_F; CAS_R_Term, 5′-TCGTGGTATCGTTATGCGCC-3′), 146 bp; 5′FRT (5′FRT_F, 5′-AGGCGCATAACGATACCACGAT-3′; 5′FRT_R, 5′-CCACAACGGGTTCTTCTGTT-3′), 204 bp; Tm1b (Tm1b_prom_F, 5′-CGGTCGCTACCATTACCAGT-3′; Floxed LR, 5′-ACTGATGGCGAGCTCAGACC-3′), 380 bp; Tm1d (tm1c_F, 5′-AAGGCGCATAACGATACCAC-3′; Floxed LR), 174 bp; Tm1c (tm1c_F; tm1c_R, 5′-CCGCCTACTGCGACTATAGAGA-3′), 218 bp; and LoxP (Floxed PNF, 5′-ATCCGGGGGTACCGCGTCGAG-3′; Floxed LR), 800–1000 bp.
Imaging of mouse embryos
Deciduae were dissected from E5.5 pregnant females killed by cervical dislocation and placed in ice-cold dissection medium containing phenol red-free DMEM/F12 and 10% heat-inactivated fetal bovine serum (FBS). Embryos were dissected in the same medium under a stereoscope and cultured in a 1:1 mixture of phenol red-free DMEM/F12 and rat serum under 5% CO2 and 95% air. For live imaging, dissected embryos were incubated in phenol red-free F12/DMEM supplemented with 50% rat serum in glass-bottomed dishes (MatTek Corporation) and imaged on Leica SP5 or Leica SP8 confocal microscopes at 37°C and 5% CO2 in a humidified chamber. High-resolution, 12-bit or 8-bit z-stacks of the embryos (pixel size: 0.123 μm [ 3× zoom] or 0.223 μm [1.7× zoom]; Z-space: 1–2 μm) were captured using a HCX PL APO CS 40× water immersion objective (1.10 NA; Leica). Green and transmitted light signals (ex., 488 nm; em., 489–550 nm) were acquired simultaneously. Imaging was performed on embryos at E5.5 with an unknown genotype obtained from crosses with a known genotype of parents, one embryo at a time, for 6 h (time interval of 10 min). For high temporal resolution, 4 h was used (time interval of 1 or 2 min). After imaging, embryos were incubated further, analyzed for AVE migration defects, and collected for genotyping at E6.5. An overview of E6.5 embryos was acquired using a Leica MZ16F fluorescence dissection stereoscope.
Image analysis
After confirmation of embryo genotypes, images were processed. Z-stacks of embryos acquired as described above were corrected for background (Statistical Correction in MetaMorph). Z-stacks were flattened by maximum intensity projection. Individual frames of video stack sequences were manually aligned in the X–Y direction using Align Stack (MetaMorph), and the rotation was fixed by the Rotate function (MetaMorph) and Adobe Photoshop. Cell tracking was performed by manually tracking cell centroids over time by the Track Points application (MetaMorph) and Manual tracking plug-in (ImageJ). (X; Y) Coordinates were generated in Excel format and used to calculate migration speeds. Protrusion–retraction mean rates were calculated as described previously (Gloushankova et al. 1995). Briefly, two still frames acquired in a 2- or 10-min interval were used to outline (Region tool) the active cell edges using MetaMorph. Two regions were merged (Supplemental Fig. S1C). Protrusive activity was measured by dividing the total area of protrusion or retraction by the shortest contour length measured in two frames over time. Protrusion formation was counted by analyzing video sequences (4 h) and sorting new protrusions based on the location site of formation (lateral sides vs. the front leading edge). Wound edge advancement was quantified from images taken at time 0 h and time 4 h by averaging the distance between the advancing wound edge and the distal edge of the field of view. Wound edge advancement delay was quantified as a distance from the position of the wound edge after 24 h of migration to the “finish” line marked on the surface substrate. For directionality of protrusions in EYFP-actin-expressing 16HBE cells, fluorescent images were thresholded, an object was generated, and Integrated Morphometry analysis was used to generate centroids and collect morphological parameters (vectors) (Supplemental Fig. S2E). For directionality of protrusions in Hex-GFP cells, all embryo positions in the video sequences were aligned along the distal–proximal axis. Aligned frames were used to manually draw vectors from the cell centroids toward the center of the protrusion. Migration directionality and persistence of migration of Hex-GFP cells were quantified using aligned time-lapse images of embryos (distal–proximal axis), where centroids of the cells were tracked manually over time (10 min per frame) and processed using the Chemotaxis plug-in (ImageJ). Directionality and persistance analyses were performed using the Chemotaxis plug-in (ImageJ), and graph generation was done using ImageJ or GraphPad Prizm 6. PIV analysis was performed by PIVlab version 1.32 (Time-Resolved Digital Particle Image Velocimetry Tool for MATLAB, developed by W. Thielicke and E. J. Stamhuis, http://pivlab.blogspot.com). Briefly, image video sequences were preprocessed by Kalman filtering, and the region of interest was selected for every individual frame. Images were calibrated and analyzed. Every fourth vector was visualized. Heat maps were generated by plotting the velocity magnitude (meters per second). The relative length of the protrusions was quantified by dividing the length of the protrusion over the diameter of the cell body (Migeotte et al. 2010).
FRET ratiometric imaging
16HBE cells were treated with siRNA. At day 2 post-treatment, cells were nucleofected with DNA constructs, plated in Mattek glass-bottomed dishes, and imaged within 16–24 h post-nucleofection. Culture medium was replaced 30 min before FRET imaging, with imaging medium containing phenol-free MEM supplemented with 10% FCS, GlutaMAX-I (1:100; Invitrogen), and OxyFluor (1:100; Oxyrase). Optimal FRET acquisition settings were determined using mYPet, mCer, and mYpet–mCer as controls. Cells were imaged with a Leica TCS SP8 confocal microscope equipped with HC PL APO 63× oil 1.4 NA lens using the SP8 SE module and allowing quantification of bleed-through coefficients, background subtractions, and FRET evaluation. To quantify FRET, we used Method 1 (Sensitized Emission). The laser was set at 3%–15% power, and images were captured in 12-bit format. The laser settings were as follows: 458-nm Argon laser at 15% power and 523-nm white light laser at 1%. Three to four optical Z sections with 1-μm step size were acquired. Quantification of FRET intensity in lamellipodia was performed by selecting a region of interest within an area of 35 μm2. The same pseudocolor intensity scale was maintained for control and knockdown conditions.
In situ hybridization and LacZ staining
Whole-mount in situ hybridization of Brachyury and Cer1 markers was performed using standard procedures (Eggenschwiler and Anderson 2000). A 282-bp β-Pix RNA probe was generated using the following primers: 5′-AAACCTTTCAGCTCAGTGTCAAG-3′ and 5′-AGCTTTGTGATTGTCATTCCTGT-3′. Embryos were fixed in 4% paraformaldehyde and processed using whole-mount in situ hybridization. LacZ staining was performed as described previously (Migeotte et al. 2010). After in situ hybridization or LacZ staining, whole-mount embryos were imaged using a Zeiss Axiocam HRC digital camera or Canon EOS camera on a Leica MZFLIII microscope. LacZ in histological sections was imaged using a 10× Plan-Neofluar NA 0.5 Zeiss Axio microscope equipped with a color Axiocam camera.
DNA constructs
Mouse pCDNA3-Flag-β-Pix was obtained from Addgene, pEGFP-βPIX was from Dr. Sandrine Etienne-Manneville, pEYFP-actin was from Clontech, and the pmCherry-actin plasmid was a generous gift from Dr. T. Svitkina (University of Pennsylvania). Human pVZR-hE-cadherin-GFP was a gift from Dr. M. Overholtzer (Memorial Sloan Kettering Cancer Center). For FRET studies, we used pmCer, pmYPet, and pmCer–mYPet (gifts from Dr. Songhi Shi, Sloan Kettering Institute, New York) and pTriex4-Cer3-Cdc42, pYPet-CBD, and pCer3 (a generous gifts from Dr. Klaus Hahn, University of North Carolina, Chapel Hill).
Immunofluorescence staining
Cell culture staining was performed following standard procedures. Briefly, cells were washed with warm PBS and fixed with 4% PFA for 10 min, permeabilized with 0.5% Triton X-100, blocked with 10% FBS, and incubated with primary mouse anti-Flag M2 (1:500; Sigma) antibody. Embryos were fixed with 4% PFA and stained with rabbit anti-ZO1 (1:200; Zymed) and rat anti-E-cadherin (1:500; Sigma) antibodies. Secondary antibodies were Alexa-conjugated (1:500; Invitrogen).
Western blotting
Embryo or cell lysates were prepared using standard lysis buffer containing 50 mM Tris (pH 7.5), 150 mM NaCl, 0.5% (v/v) NP-40, 1 mM Na3VO4, 10 mM NaF, and 1× protease inhibitor cocktail (Roche). Proteins were separated by 4%–12% NuPAGE Bis-Tris gels (Invitrigen) and analyzed using the following primary antibodies: polyclonal rabbit anti-β-Pix 07-1450 (1:2000; Millipore-Chemicon), mouse anti-β-actin AC-74 (1:10,000; Sigma), monoclonal rabbit anti-PAK-1 (1:1000; Cell Signaling), anti-PAK-2 (1:1000; Cell Signaling), rabbit anti-GIT (1:1000; Cell Signaling), rabbit anti-N-cadherin (1:1000; Zymed ), and mouse anti-E-cadherin (1:1000; BD Biosciences). HRP-conjugated secondary antibodies (Dako) were used at 1:5000 dilution.
Statistical analysis
Statistical analysis was performed using an unpaired two-tailed Student’s t-test in GraphPad Prizm 6. Details of statistical tests are indicated in the figure legends. In box-and-whiskers plots, the middle bar is the median. Embryos were carefully staged and always imaged at the same developmental stage to ensure compatibility.
Repeatability of experiments
Time-lapse images and Western blot data shown in this study are representative of the following number of analyzed cells and embryos in independent experiments (time-lapse data obtained from one embryo represent one independent experiment). Figure 2C represents n = 24 cells from three independent experiments; Figure 2D represents n = 109 cells from six independent experiments; Figure 2E represents n = 14 cells from three independent experiments; Figure 3A represents three independent experiments; Figure 3D represents three independent experiments; Figure 3E represents three independent experiments and >60 protrusions; Figure 4A represents three independent experiments; Figure 4C represents >10 independent experiments; Figure 4D represents two independent experiments and two embryos per genotype; Figure 5 represents three independent experiments; Figure 6B represents six independent experiments; Figure 6D represents two independent experiments and >17 cells per genotype, Figure 6E represents two independent experiments and >15 cells per genotype; Figure 6F represents three independent experiments; Figure 7B represents three independent experiments and >30 protrusions; Figure 7E represents n = 14 wild-type cells, n = 8 null cells, and n = 3 tissue-specific knockout cells and three independent experiments; Figure 7F represents more than four independent experiments and n = 29 cells; and Figure 7G represents three independent experiments.
Supplementary Material
Acknowledgments
We thank Dr. Dieter Gruenert for providing 16HBE cells; Dr. Sandrine Etienne-Manneville for pEGFP-βPIX; Dr. Tatiana Svitkina for pmCherry-actin; Dr. Michael Overholtzer for the E-cadherin-GFP; Dr. Songhi Shi for pmCer, pmYPet, and pmCer–mYPet; Dr. Klaus Hahn for pTriex4-Cer3-Cdc42, pYPet-CBD, and pCer3; and Dr. Phillip Niethammer and Dr. William Gault for help with PIVlab. We are grateful to members of the Hall and Anderson laboratories for helpful discussions. We thank Memorial Sloan Kettering Cancer Center Molecular Cytology Core Facilities staff and especially Dr. Vitaly Boyko for the use of the confocal microscopes and help with FRET and embryo imaging. The work was supported by National Institutes of Health (NIH) grants GM081435 and CA008748 (to A.H.) and 5R37HD035455 (to K.V.A.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.251371.114.
References
- Andrew N, Insall RH. 2007. Chemotaxis in shallow gradients is mediated independently of PtdIns 3-kinase by biased choices between random protrusions. Nat Cell Biol 9: 193–200. [DOI] [PubMed] [Google Scholar]
- Arnold SJ, Robertson EJ. 2009. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat Rev Mol Cell Biol 10: 91–103. [DOI] [PubMed] [Google Scholar]
- Baird D, Feng Q, Cerione RA. 2006. Biochemical characterization of the Cool (cloned-out-of-library)/Pix (Pak-interactive exchange factor) proteins. Methods Enzymol 406: 58–69. [DOI] [PubMed] [Google Scholar]
- Bloomekatz J, Grego-Bessa J, Migeotte I, Anderson KV. 2012. Pten regulates collective cell migration during specification of the anterior-posterior axis of the mouse embryo. Dev Biol 364: 192–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cau J, Hall A. 2005. Cdc42 controls the polarity of the actin and microtubule cytoskeletons through two distinct signal transduction pathways. J Cell Sci 118: 2579–2587. [DOI] [PubMed] [Google Scholar]
- Cherfils J, Zeghouf M. 2013. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev 93: 269–309. [DOI] [PubMed] [Google Scholar]
- Cheung KJ, Gabrielson E, Werb Z, Ewald AJ. 2013. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell 155: 1639–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozens AL, Yezzi MJ, Kunzelmann K, Ohrui T, Chin L, Eng K, Finkbeiner WE, Widdicombe JH, Gruenert DC. 1994. CFTR expression and chloride secretion in polarized immortal human bronchial epithelial cells. Am J Respir Cell Mol Biol 10: 38–47. [DOI] [PubMed] [Google Scholar]
- Eggenschwiler JT, Anderson KV. 2000. Dorsal and lateral fates in the mouse neural tube require the cell-autonomous activity of the open brain gene. Dev Biol 227: 648–660. [DOI] [PubMed] [Google Scholar]
- Fetisova EK, Ivanova OY, Potapova TV, Svitkina TM, Vasiliev JM. 1990. Competition of contacting heterotypic epithelial sheets for the territory in culture. Cell Biol Int Rep 14: 957–965. [DOI] [PubMed] [Google Scholar]
- Friedl P, Gilmour D. 2009. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol 10: 445–457. [DOI] [PubMed] [Google Scholar]
- Gloushankova NA, Krendel MF, Sirotkin VA, Bonder EM, Feder HH, Vasiliev JM, Gelfand IM. 1995. Dynamics of active lamellae in cultured epithelial cells: effects of expression of exogenous N-ras oncogene. Proc Natl Acad Sci 92: 5322–5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Lewis P, Pevny L, McMahon AP. 2002. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech Dev 119: S97–S101. [DOI] [PubMed] [Google Scholar]
- Heasman SJ, Ridley AJ. 2008. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol 9: 690–701. [DOI] [PubMed] [Google Scholar]
- Hodgson L, Shen F, Hahn K. 2010. Biosensors for characterizing the dynamics of ρ family GTPases in living cells. Curr Protoc Cell Biol 46: 14.11.1–14.11.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AA, Friedl P. 2010. Determinants of leader cells in collective cell migration. Integr Biol (Camb) 2: 568–574. [DOI] [PubMed] [Google Scholar]
- Kim T, Park D. 2001. Molecular cloning and characterization of a novel mouse β-Pix isoform. Mol Cells 11: 89–94. [PubMed] [Google Scholar]
- Kim S, Kim T, Lee D, Park SH, Kim H, Park D. 2000. Molecular cloning of neuronally expressed mouse β-Pix isoforms. Biochem Biophys Res Commun 272: 721–725. [DOI] [PubMed] [Google Scholar]
- Kimura-Yoshida C, Nakano H, Okamura D, Nakao K, Yonemura S, Belo JA, Aizawa S, Matsui Y, Matsuo I. 2005. Canonical Wnt signaling and its antagonist regulate anterior–posterior axis polarization by guiding cell migration in mouse visceral endoderm. Dev Cell 9: 639–650. [DOI] [PubMed] [Google Scholar]
- Kuo JC, Han X, Hsiao CT, Yates JR III, Waterman CM. 2011. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for β-Pix in negative regulation of focal adhesion maturation. Nat Cell Biol 13: 383–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutys ML, Yamada KM. 2014. An extracellular-matrix-specific GEF–GAP interaction regulates Rho GTPase crosstalk for 3D collagen migration. Nat Cell Biol 16: 909–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machacek M, Hodgson L, Welch C, Elliott H, Pertz O, Nalbant P, Abell A, Johnson GL, Hahn KM, Danuser G. 2009. Coordination of Rho GTPase activities during cell protrusion. Nature 461: 99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maretto S, Cordenonsi M, Dupont S, Braghetta P, Broccoli V, Hassan AB, Volpin D, Bressan GM, Piccolo S. 2003. Mapping Wnt/β-catenin signaling during mouse development and in colorectal tumors. Proc Natl Acad Sci 100: 3299–3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migeotte I, Omelchenko T, Hall A, Anderson KV. 2010. Rac1-dependent collective cell migration is required for specification of the anterior–posterior body axis of the mouse. PLoS Biol 8: e1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SA, Grewal S, Barrios F, Patankar SN, Strauss B, Buttery L, Alexander M, Shakesheff KM, Zernicka-Goetz M. 2012. Dynamics of anterior–posterior axis formation in the developing mouse embryo. Nat Commun 3: 673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omelchenko T, Hall A. 2012. Myosin-IXA regulates collective epithelial cell migration by targeting RhoGAP activity to cell–cell junctions. Curr Biol 22: 278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omelchenko T, Vasiliev JM, Gelfand IM, Feder HH, Bonder EM. 2003. Rho-dependent formation of epithelial ‘leader’ cells during wound healing. Proc Natl Acad Sci 100: 10788–10793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani N, Vitale N, Borg JP, Etienne-Manneville S. 2006. Scrib controls Cdc42 localization and activity to promote cell polarization during astrocyte migration. Curr Biol 16: 2395–2405. [DOI] [PubMed] [Google Scholar]
- Poujade M, Grasland-Mongrain E, Hertzog A, Jouanneau J, Chavrier P, Ladoux B, Buguin A, Silberzan P. 2007. Collective migration of an epithelial monolayer in response to a model wound. Proc Natl Acad Sci 104: 15988–15993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakeman AS, Anderson KV. 2006. Axis specification and morphogenesis in the mouse embryo require Nap1, a regulator of WAVE-mediated actin branching. Development 133: 3075–3083. [DOI] [PubMed] [Google Scholar]
- Rhee S, Yang SJ, Lee SJ, Park D. 2004. β-Pix-b(L), a novel isoform of β-Pix, is generated by alternative translation. Biochem Biophys Res Commun 318: 415–421. [DOI] [PubMed] [Google Scholar]
- Rhee JM, Pirity MK, Lackan CS, Long JZ, Kondoh G, Takeda J, Hadjantonakis AK. 2006. In vivo imaging and differential localization of lipid-modified GFP-variant fusions in embryonic stem cells and mice. Genesis 44: 202–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth P. 2003. Communication by touch: role of cellular extensions in complex animals. Cell 112: 595–598. [DOI] [PubMed] [Google Scholar]
- Rorth P. 2007. Collective guidance of collective cell migration. Trends Cell Biol 17: 575–579. [DOI] [PubMed] [Google Scholar]
- Rorth P. 2011. Whence directionality: guidance mechanisms in solitary and collective cell migration. Dev Cell 20: 9–18. [DOI] [PubMed] [Google Scholar]
- Rossman KL, Der CJ, Sondek J. 2005. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol 6: 167–180. [DOI] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, et al. 2011. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474: 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Rodriguez T, Clements M, Smith JC, Beddington RS. 2004. Active cell migration drives the unilateral movements of the anterior visceral endoderm. Development 131: 1157–1164. [DOI] [PubMed] [Google Scholar]
- Takaoka K, Hamada H. 2012. Cell fate decisions and axis determination in the early mouse embryo. Development 139: 3–14. [DOI] [PubMed] [Google Scholar]
- Takaoka K, Yamamoto M, Hamada H. 2011. Origin and role of distal visceral endoderm, a group of cells that determines anterior-posterior polarity of the mouse embryo. Nat Cell Biol 13: 743–752. [DOI] [PubMed] [Google Scholar]
- Tatin F, Taddei A, Weston A, Fuchs E, Devenport D, Tissir F, Makinen T. 2013. Planar cell polarity protein Celsr1 regulates endothelial adherens junctions and directed cell rearrangements during valve morphogenesis. Dev Cell 26: 31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Klooster JP, Jaffer ZM, Chernoff J, Hordijk PL. 2006. Targeting and activation of Rac1 are mediated by the exchange factor β-Pix. J Cell Biol 172: 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trichas G, Joyce B, Crompton LA, Wilkins V, Clements M, Tada M, Rodriguez TA, Srinivas S. 2011. Nodal dependent differential localisation of dishevelled-2 demarcates regions of differing cell behaviour in the visceral endoderm. PLoS Biol 9: e1001019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trichas G, Smith AM, White N, Wilkins V, Watanabe T, Moore A, Joyce B, Sugnaseelan J, Rodriguez TA, Kay D, et al. 2012. Multi-cellular rosettes in the mouse visceral endoderm facilitate the ordered migration of anterior visceral endoderm cells. PLoS Biol 10: e1001256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedula SR, Leong MC, Lai TL, Hersen P, Kabla AJ, Lim CT, Ladoux B. 2012. Emerging modes of collective cell migration induced by geometrical constraints. Proc Natl Acad Sci 109: 12974–12979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Jin D, Durgan J, Hall A. 2013. LKB1 controls human bronchial epithelial morphogenesis through p114RhoGEF-dependent RhoA activation. Mol Cell Biol 33: 2671–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Saijoh Y, Perea-Gomez A, Shawlot W, Behringer RR, Ang SL, Hamada H, Meno C. 2004. Nodal antagonists regulate formation of the anteroposterior axis of the mouse embryo. Nature 428: 387–392. [DOI] [PubMed] [Google Scholar]
- Yoshizawa M, Sone M, Matsuo N, Nagase T, Ohara O, Nabeshima Y, Hoshino M. 2003. Dynamic and coordinated expression profile of dbl-family guanine nucleotide exchange factors in the developing mouse brain. Gene Expr Patterns 3: 375–381. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.