Abstract
Histones post-translational modifications (PTMs) are crucial components of diverse processes that modulate chromatin. Among the histones PTMs, the histones phosphorylation appears such crucial since it plays a significant role into DNA repair structure, transcription and chromatin compaction during cell division and apoptosis. However, little is known about the prognostic value of the histone phosphorylation in human cancer. This point could be considerate such as an important gap in anti-cancer therapy since the use of adequate kinase inhibitors could remedy to the aberrant histone phosphorylation associated with a poor prognosis factor. To remedy at this situation, we analyzed the phosphorylation level of histone H3 at the residues T3, T6, S10, S28, Y41 and T45 in a collection of 42 glioblastoma multiformes (GBM). Our data indicated that the high level of pH3T6, pH3S10 and pH3Y41 are signatures associated with a poor prognosis of overall survival (OS) of GBM treated with the "temozolomide and irradiation standard" treatment of GBM (named TMZ+Irad treatment). Our data also showed that these signatures are correlated with the high activity of kinases already described as writers of the pH3T6, pH3S10 and pH3Y41 i.e. the PKC, Aurora-B and JAK2, respectively. Finally, our analysis revealed that the use of Enzastaurin, AZD1152, and AZD1480 abrogated the high level of pH3T6, pH3S10 and pH3Y41 while increasing the sensitivity to the “temozolomide and irradiation”-induced cell death. To conclude, it appears that this work provides biomarkers for patient stratification for a therapy including kinase inhibitors.
Keywords: phosphorylation, histone, epigenetic, glioma, apoptosis
Introduction
Histone H3 is one of the five main histone proteins and is composed by a principal globular domain and a long N-terminal tail 1. Histone H3 is involved with the structure of the nucleosome i.e. the basic unit of DNA packaging in eukaryotes which is composed by a segment of DNA wound in sequence around four histone protein cores 2. Thus, with histone H2A, histone H2B and histone H4, histone H3 is considered such as a histone protein core.
Like the other histone proteins, histone H3 is highly post-translationally modified. The N-terminal tail of histone H3 protrudes from the globular nucleosome core and can undergo several different types of post-translational modifications that influence cellular processes. Classically, 8 post-translational modifications of histone could be described: citrullination, ubiquitination, SUMOylation, phosphorylation, methylation, acetylation, ADP-ribosylation, and palmytoylation (according to the HIstome: The Histone Infobase).
Histone H3 phosphorylation mainly affect the T3, T6, S10, T11, S28, S31, Y41 and T45 residues 3-5. H3T3 phosphorylation occurs specifically during mitosis and is important for normal alignment of chromosomes on the metaphase plate 6. The protein kinase haspin and VRK1 reported such as being implicated into the H3T3 phosphorylation 7, 8. H3T6 phosphorylation via PKCβI prevents LSD1 from demethylating H3K4, thus maintaining the active methyl marks during androgen-dependent gene activation. Increased levels of PKCβI and phosphorylated H3T6 positively correlate with high Gleason scores of prostate carcinomas 9. PKCα can also phosphorylate H3T6 5. H3S10 phosphorylation is a mitosis specific modification and is essential for the onset of mitosis. Basal c-Jun N-terminal kinases (JNK) play a key role in controlling histone H3 phosphorylation for mitotic entry at G2/M phase 10. Aurora-B, PIM1, RSK2, MSK1/2, and IKKα can also phosphorylate H3S10 5, 11. H3T11 phosphorylation is a crucial chromatin mark for transcriptional regulation 12. H3T11 is phosphorylated at androgen receptor gene targets by PRK1 that acts as a co-activator of androgen receptor dependent transcription. Levels of PRK1 and phosphorylated H3T11 correlate with Gleason scores of prostate carcinomas 12. PKM2, Chk1 and Dlk/ZIP are also reported such as being able the capacity to promote the H3T11 phosphorylation 13-15. H3S28 phosphorylation is a mitosis specific modification. It appears first at prophase and extends till anaphase 11. JNK, MLTKα, Aurora-B and MSK1/2 can also phosphorylate H3S28 5. H3.3S31 phosphorylation is observed only in late prometaphase and metaphase and is absent in anaphase 16. Phosphorylation of H3T41 by JAK2 prevents the binding of Heterochromatin protein 1 alpha (HP1α) to hematopoietic oncogene lmo2 promoter which is implicated in leukaemogenesis 17. H3T45 phosphorylation induces structural change within the nucleosome to facilitate DNA nicking and/or fragmentation linking its role to apoptosis, and PKCδ is identified such as the kinase responsible for H3T45 phosphorylation in vitro and in vivo 18.
In the present report, we have analyzed the phosphorylation level of H3T3, T6, S10, S28, Y41 and T45 in a collection of 42 GBMs. Our work highlighted that the presence of a high level of pH3T6, pH3S10 or pH3Y41 identified different subpopulation of GBM patients presenting a poor OS. Our data also indicated that the high level of pH3T6 correlated with high level of PKC activity, the high level of pH3S10 correlated with high level of Aurora-B activity, while the high level of pH3Y41 correlated with high level of Jak2 activity. Finally, our data indicated that the use of: 1) Enzastaurin (PKC-beta inhibitor) increased the TMZ-irrad induced cell death in Primary Cultured Tumor Cells (PCTC) of GBM presenting an high level of pH3T3 and a resistance phenotype to the TMZ-Irrad treatment, 2) AZD1152 (Aurora-B inhibitor) increased the TMZ-irrad induced cell death in PCTC of GBM presenting an high level of pH3S10 and a resistance phenotype to the TMZ-Irrad treatment, and 3) AZD1480 (Jak2 inhibitor) increased the TMZ-irrad induced cell death in PCTC of GBM presenting an high level of pH3Y41 and a resistance phenotype to the TMZ-Irrad treatment.
Materials and methods
Proteins and histone proteins extraction
Proteins extracts were obtained by using T-PER (Tissue Protein Extraction Reagent, Thermo Scientific, France) according to the manufacturer's instructions. However, before to realize the last centrifigation, samples were sonicated by using the Bioruptor sonicator (Diagenode, France).
To extract histone proteins, we used The EpiQuik™ Total Histone Extraction Kit (Epigentek, Euromedex, France) i.e. a complete set of optimized buffers and reagents for extracting total core histone proteins (H2A, H2B, H3, and H4) from mammalian cells or tissue.
ELISA
For ELISA, plates were coated with Total Histone Extraction samples at a concentration of 3µg/ml or with protein extraction samples at a concentration of 5µg/ml in 50mM carbonate-bicarbonate buffer pH 9.6, incubated overnight at 4ºC and then washed three times with 150µl/well of PBS-tween (PBS-T). Plates were blocked with 150µl/well of PBS containing 3% BSA and incubated for 2 hour at 37ºC, then washed twice with PBS-T. Antibody, diluted at the optimal concentration (according to the manufacturer's instructions) was added in 100 µl in blocking buffer (PBS, 3% BSA). Test and control sera were diluted 1:10 in PBS-T containing 0.25 % BSA and were serially titrated in 3 fold dilutions to a final dilution of 1:270. Plates were incubated at 37ºC for 2 hour followed by one hour at 4ºC and then washed 4 times with PBS-T. Alkaline phosphatase conjugated was added at a dilution of 1:20,000 in PBS-T containing 0.25 % BSA. Plates were incubated for 2 hours at 37ºC, washed 4 times with PBS-T before substrate reaction was developed using 1 mg/ml p-nitrophenyl phosphate in 1 M diethanolamine buffer, pH 9.8. Plates were allowed to develop for 30 minutes at 37ºC and then read at 405 nm using an ELISA plate reader.
DNA methylation analyses
DNA was extracted by using the QiaAmp DNA mini Kit (Qiagen, France). Methylated DNA ImmunoPrecipitation experiments were realized by using MeDIP kit (Diagenode, France). Primers used to defined the MGMT gene methylation status are: CACCGTTTGCGACTTGG and CAGGACCACTCGAGGCTG.
Ethics Statements and Patient characteristics
Patient data were obtained and handled according to French laws and recommendations of the French National Commitee of Ethics (Comité Consultatif National d'Ethique pour les Sciences de la Vie et de la Santé).
Overall survival was measured from the date of surgical resection to the death. All patients included in this study had similar management (Complete resection) and similar treatment (temozolomide 75 mg/m2/d x 7 d/wk for 6 weeks and fractionated radiotherapy (60 Gy total dose)).
Statistical analysis
All experiments were done at least in triplicates. Significance of the differences in means were calculated using Student-t-test while correlations were determined using Pearson-test. Survival curves were plotted according to Kaplan-Meier method and compared by means of Cox Proportional Hazards Survival Regression analysis. All statistical tests were two-sided and statistical significance was defined as “p” less than 0.05.
Evaluation of cell death
Percentages of cell death were evaluated by using a Trypan Blue Stain 0.4%, and the Countess® Automated Cell Counter (Life Technology, France).
Western blot
In brief, proteins were size fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Proteins were transferred onto nitrocellulose or PVDF membrane. Saturation and blotting were realized by using SNAP i.d™ Protein Detection System (Millipore, France). The detection of proteins was performed using the FusionX7 Imager (Fisher Scientific, France).
Primary cultured tumor cells (PCTC)
Fresh brain tumor tissues obtained from the neurosurgery service of the Laennec Hospital (Nantes/Saint-Herblain) were collected and processed within 30 min after resection. The clinical protocol was approved by the French laws of ethics with informed consent obtained from all subjects. The primary cultured tumor cells were obtained after mechanical dissociation according to the technique previously described 19. Briefly, tumor tissue was cut into pieces of 1-5 mm3 and plated in a 60 mm2 tissue culture dish with DMEM with 10% FBS and antibiotics. Additionally and in parallel, minced pieces of tumor were incubated with 200 U/ml collagenase I (Sigma) and 500 U/ml DNaseI (Sigma) in PBS during 1 hr at 37°C with vigorous constant agitation. The single-cell suspension was filtered through a 70 mm cell strainer (BD Falcon), washed with PBS, and suspended in DMEM-10% FBS. Cell cultures were subsequently split 1:2 when confluent and experiments were done before passage 3-5.
Antibodies
Antibodies used in our work were: anti-Histone H3 (Abcam#Ab1791, France or Active Motif#39763, France), anti-pH3T3 (Abcam#Ab53762, France), anti-pH3T6 (Abcam#Ab14102, France), anti-pH3S10 (Abcam#14955, France), anti-pH3S28 (Santa Cruz#sc-12927, France), anti-pH3Y41 (Abcam#26310), France and anti-pH3T45 (Abcam#26127, France).
Results
Clinical and biological characteristics of GBM patients
To evaluate the global level of histone phosphorylation, we performed ELISA analyses in a collection of 42 patient-derived samples of GBMs.
The clinical characteristics of this collection of patient-derived samples of GBMs are summarized in table 1. Briefly, we noted that the median age was 57 years, OS median was 14.15 months, and the male/female sex ratio was 28/15. Five patients harbored a Karnofsky performance score of 70, 19 of 80, and 18 of 90. The prognostic relevance of these patient characteristics was analyzed by performing a Cox Proportional Hazards Survival Regression Analysis. No significant impact of the considered clinical characteristics was observed (Table 1).
Table 1.
Characteristic of GBM patients.
| Parameter value | Cox proportional Hazards Survival Regression Analysis |
|
|---|---|---|
| Age (years)* | 43/69/57 | p=0.9069++ |
| Overall survival (months)* | 4.4/56.2/14.15 | - |
| KPS (80/90)+ | 24/18 | p=0.3550 |
| Sex (M/F)+ | 15/27 | p=0.5348 |
| MGMT (Meth./Unmeth.)+ | 19/23 | p=0.0041 |
| pH3T3 | 0.124/0.705/0.3645 | p=0.5259 |
| pH3T6 | 0.105/0.654/0.265 | p=0.0170 |
| pH3S10 | 0.102/0.675/0.293 | p=0.0056 |
| pH3S28 | 0.102/0.675/0.293 | P=0.8871 |
| pH3Y41 | 0.028/0.654/0.3035 | p=0.0159 |
| pH3T45 | 0.109/0.874/0.4175 | p=0.1466 |
*min/max/median, +number,++age<median Vs. Age>median
One biological parameter was investigated the methylation level of the MGMT gene. The prognostic relevance of these patient characteristics was analyzed by performing a Cox Proportional Hazards Survival Regression Analysis. Thus, we noted that patients whose GBMs presented a methylated MGMT gene (n=19) have an OS more longer than those GBMs presented an unmethylated MGMT gene (n=23) (p=0.0041) (Table 1).
About the level of histone H3 phosphorylation in GBMs
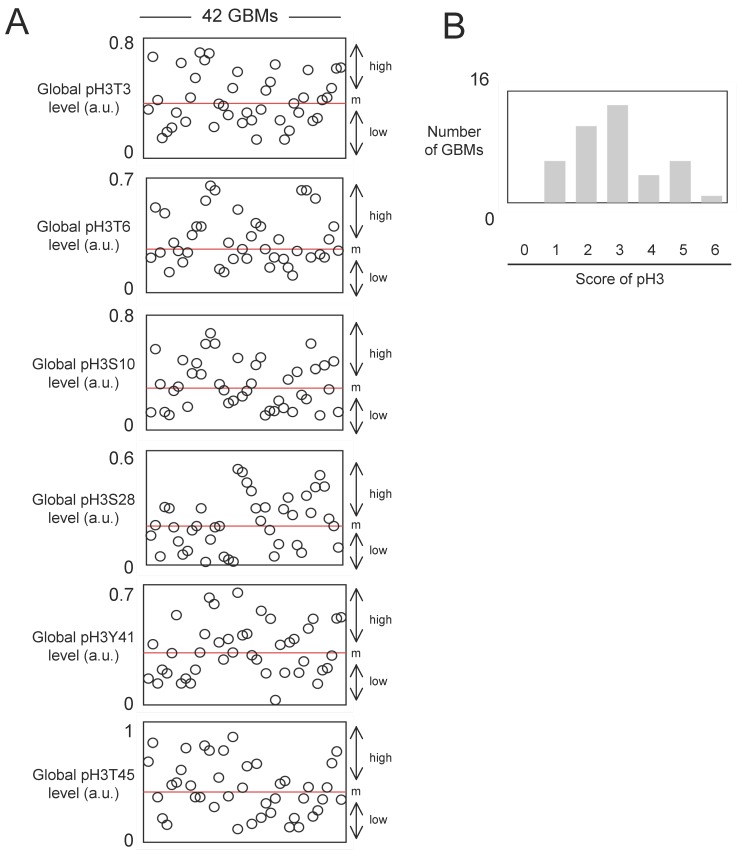
As can be seen from Figure 1A, all histone phosphorylations under investigation were presented in GBM samples. Of course, the histone phosphorylations levels were heterogeneous among GBM samples. This idea is particularly observed for the pH3Y41 since the maximal value of pH3Y41 is equal to 23 fold to the minimal value of pH3Y41.
Figure 1.
Study of the global level of pH3T3, pH3T6, pH3S10, pH3S28, pH3Y41 and pH3T45 in 42 GBMs. (A) After extraction of histone, ELISA experiments were realized to estimate the considered phosphorylation levels of histone H3. Each open circle represents a GBM. m: median. (B) The core of pH3 reflects the accumulation of several types of histone H3 phosphorylation in a same GBM. Thus, graph 1B illustrates the number of GBM presenting an accumulation of histone H3 phosphorylation.
We next asked whether there is a tendency for phospho-histones to be clustered in certain tumor specimens and not in others. For this purpose, we calculated the histone H3 phosphorylation score. Thus, when a level of histone phosphorylation is higher than the median in a considered GBM sample, this sample has one point, and the sum of this point is the histone H3 phosphorylation score. In our analysis, all of GBMs harbored at least, a score equal to 1. Only one GBM has a score of 6 (Figure 1B). The graph of pH3 score near revealing a bell curve, we postulate that the distribution of the pH3 replies a continuous probability distribution, known as the Gaussian distribution. Consequently, it appears that the pH3 were not clustered in certain GBMs.
Histone H3 phosphorylation and predictive value for overall survival of GBM patients
Next, we analyzed whether the pH3 would have predictive value for overall survival of GBM patients. For this purpose, patients were divided into two groups based on the level of each phosphorylation found on their tumors. Thus, GBM samples having a histone phosphorylation level higher than the median value obtained from all GBM samples were included in the subgroup presenting a high phosphorylation level. A contrario, GBM samples having a histone phosphorylation level lower than the median value obtained from all GBM samples were included in the subgroup presenting a low phosphorylation level (Figure 1A).
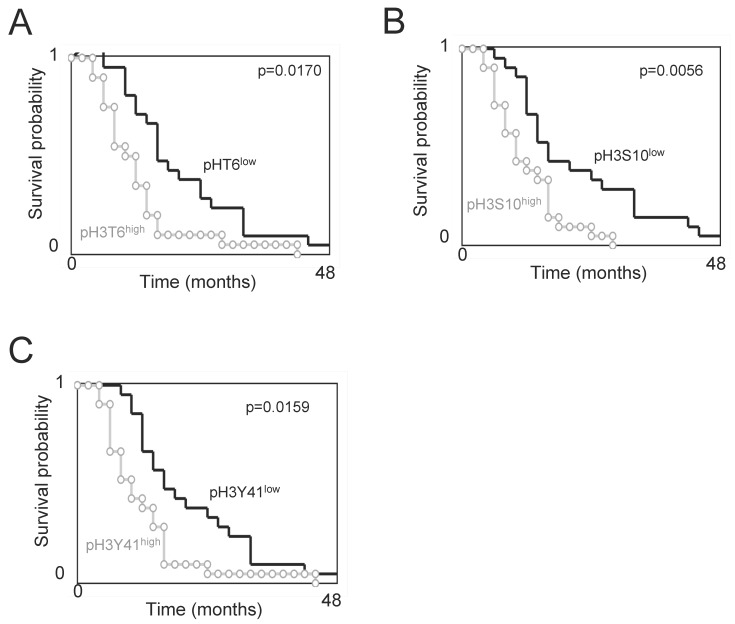
Overall survival curves were estimated by the Kaplan-Meier method and compared with the Cox Proportional Hazards Survival Regression (Figure 2). Significant differences were observed in overall survival between patients whose tumors had high levels of pH3T6, pH3S10, or pH3Y41 and those whose tumors did not (p=0.0170, p=0.0056 and p=0.0159 respectively). In other terms, these data indicate that the high levels of pH3T6, pH3S10, or pH3Y41 are markers of poor prognosis for GBM patients.
Figure 2.
Overall survival (OS) vs. subgroups of phospho-histone H3. Kaplan-Meier curves illustrate the significant difference in OS between patients with tumors harboring a low or high level of pH3T6 (A), pH3S10 (B) or pH3Y41 (C). The level of pH3T6, pH3S10 or pH3Y41 used are the ones obtained by ELISA. Significant difference was estimated via a Cox Proportional Hazards Survival Regression Analysis. For considered level of pH3, the “low” subgroup includes GBMs having a level of pH3 inferior or equal to the median value, and the “high” subgroup includes GBMs having a level of pH3 superior to the median value.
Correlation between the phosphorylation level of H3T6, H3S10 and H3Y41and methylation status of the MGMT gene
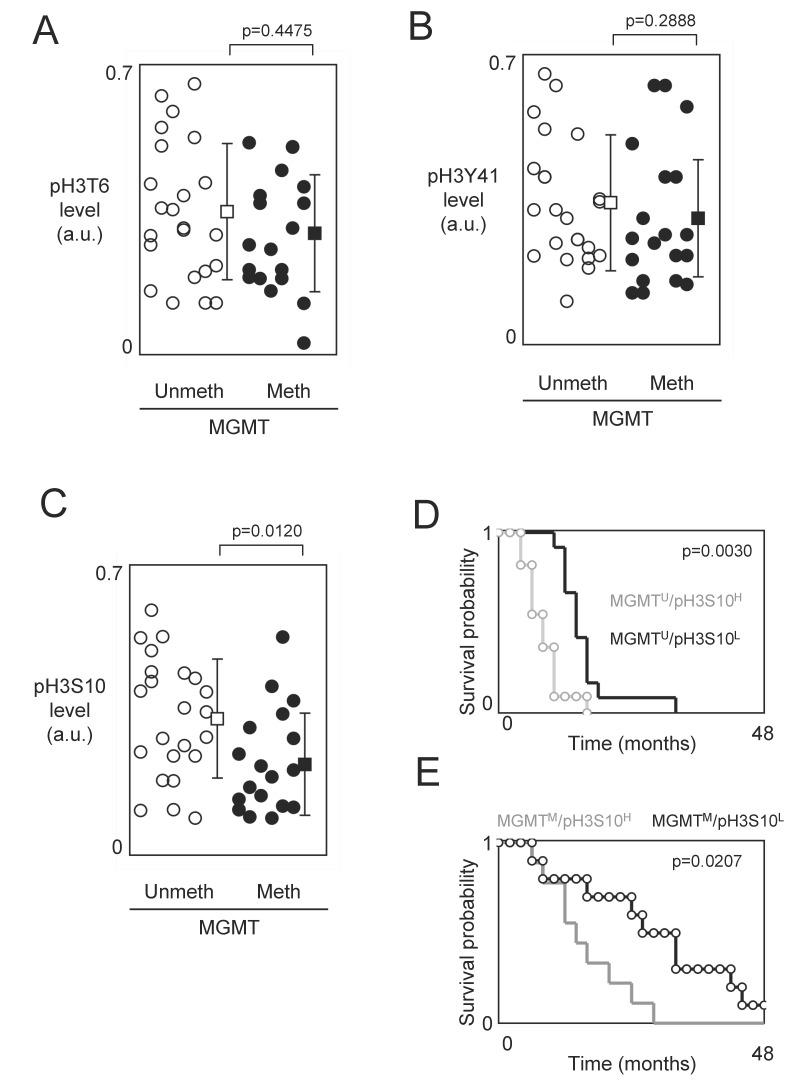
O6-methylguanine DNA methyltransferase (MGMT) promoter methylation being a predictive marker of benefit from temozolomide-irradiation treatment, we next analyzed whether the prognosis value associated with the phosphorylation level of H3T6, H3S10 and H3Y41 is influenced or not by the MGMT methylation status 20, 21. For this purpose, we analyzed whether the GBMs presenting a methylated MGMT gene also presented a high level of pH3T6, pH3S10 or pH3Y41. No statistical difference was observed between the pH3T6 and pH3Y41 in GBM presenting an unmethylated and a methylated MGMT gene (p=0.4475 and p=0.2888) (Figures 3A and 3B). Thus, the pH3T6 and pH3Y41 levels could be used as an alternative prognostic factor in GBM patients independently to the MGMT methylation status.
Figure 3.
Study of the phosphorylation level of H3T6, H3S10 and H3Y41 in function of the methylation status of the MGMT gene. (A-B-C) Graphs illustrate the level of pH3T6 (A), pH3Y41 (B) and pH3S10 (C) in GBMs previously characterized by the presence of unmethylated MGMT gene or methylated MGMT gene. Open circles represent GBMs presenting an unmethylated MGMT gene. Black circles represent GBMs presenting a methylated MGMT gene. Open square represents the average±standard deviation of GBMs presenting an unmethylated MGMT gene. Black square represents the average±standard deviation of GBMs presenting a methylated MGMT gene. (D-E) Kaplan-Meier curves illustrate the significant difference in overall survival between patients whose tumors had MGMTM/pH3S10H and those whose tumors had MGMTM/pH3S10H (E), and between patients whose tumors had MGMTM/pH3S10H and those whose tumors had MGMTM/pH3S10H (D).
On contrary, our analysis indicated that GBMs presenting a methylated MGMT gene also presented a low level of pH3S10 (Figure 3C). In other terms, this data suggest that the existence of a subpopulation of patients whose GBMs are characterized by the presence of two biomarkers (methylated MGMT gene and low level of pH3S10) associated with good prognosis of survival and/or response to the TMZ+Irrad treatment.
Supported by this last observation, we next compared the OS of patients stratified by taking into consideration the MGMT methylation status (methylated (M) or unmethylated (U)) and the pH3S10 level (low (L) or high (H)). According to these 2 parameters, we distinguished 4 subgroups of GBM patients: MGMTU/pH3S10L (n=12, median=14.75 months), MGMTU/pH3S10H (n=11, median=8.3 months), MGMTM/pH3S10L (n=10, median=29.9 months) and MGMTM/pH3S10H (n=9, median=14.1 months). As expected, we firstly noted that the MGMTM/pH3S10L subgroup (i.e. the subgroup characterized by the presence of the two biomarkers associated with good prognosis of survival and/or response to the TMZ+Irrad treatment) was the subgroup having the better median of OS, while the MGMTU/pH3S10H subgroup (i.e. the subgroup characterized by the presence of the two biomarkers associated with poor prognosis of survival and/or response to the TMZ+Irrad treatment) was the subgroup having the worst median of OS. In parallel, Kaplan-Meier curves and Cox Proportional Hazards Survival Regression analyses indicated that significant different in OS between patients whose tumors had MGMTM/pH3S10H and those whose tumors had MGMTM/pH3S10H (Figures 3E), and between patients whose tumors had MGMTM/pH3S10H and those whose tumors had MGMTM/pH3S10H (Figure 3D).
Correlation between the phosphorylation level of H3T6, H3S10 and H3Y41 and the activity of kinases implicated into these phosphorylation
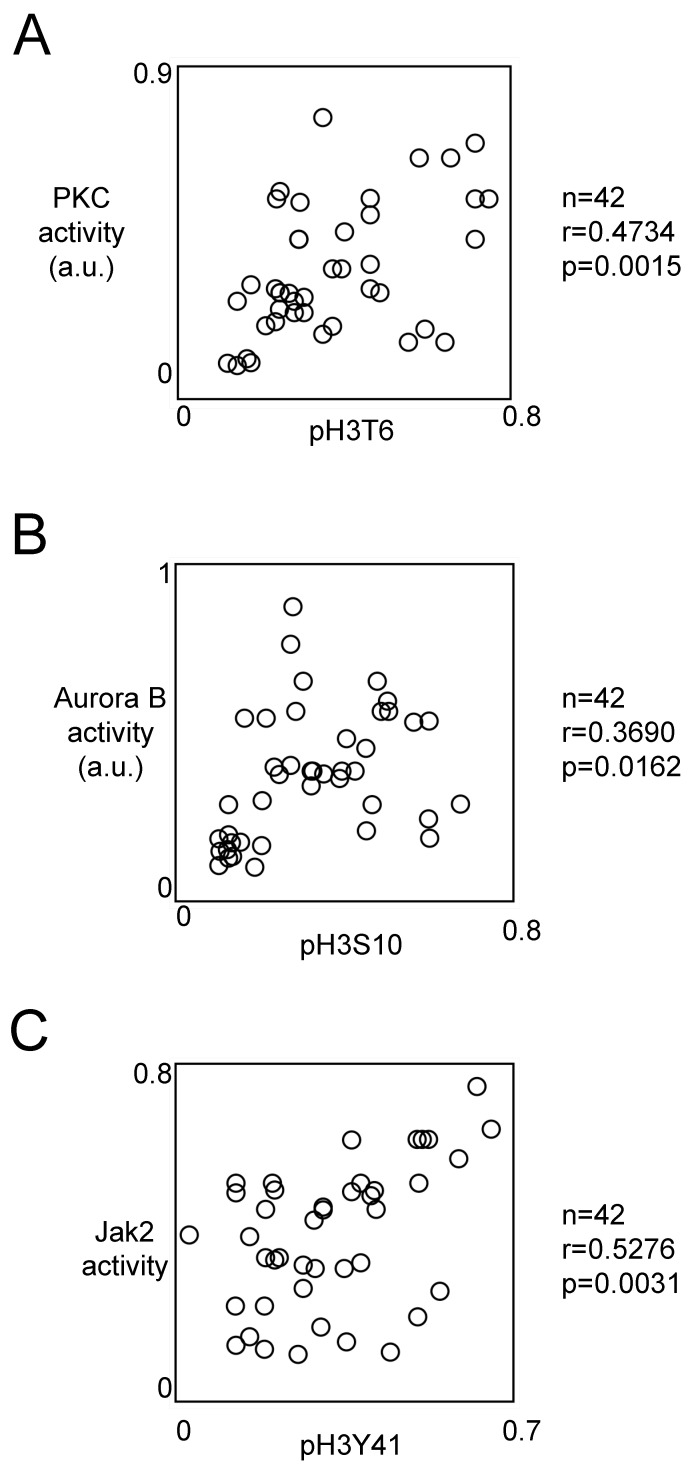
Literature reports that the phosphorylations of H3T6, H3S10 and H3Y41 can be due to the PKCβ, Aurora-B and Jak2 kinases respectively, we next analyzed whether the activity of these kinases correlated with the corresponding phoshorylation level of histone H3.
For this purpose, we used HTScan® Jak2 Kinase Assay Kit, HTScan® Aurora B Kinase Assay Kit and HTScan® PKCβ Kinase Assay Kit (Ozyme, Cell Signaling, France) according to the manufacturer's instructions. Pearson's correlation test indicated that the pH3T6 level was correlated with the PKCβ activity (p=0.0015) (Figure 4A). Similar analyses indicated that the pH3S10 level was correlated with the Aurora-B activity (p=0.0162), and the pH3Y41 level was correlated with Jak2 activity (p=0.0031) (Figures 4B and 4C).
Figure 4.
Correlation between the phosphorylation level of H3T6, H3S10 and H3Y41 and the activity of kinases implicated into these phosphorylations. Graphs illustrates the correlation existing between the PKC activity and the pH3T6 level (A), the Aurora-B activity and the pH3S10 level (B) and between the Jak2 activity and the pH3Y41 level (C). Correlation analyses were realized by using the Pearson's correlation test. The level of pH3T6, pH3S10 or pH3Y41 used are the ones obtained by ELISA. Kinase activity was obtained by using the HTScan® Jak2 Kinase Assay Kit, HTScan® Aurora B Kinase Assay Kit and HTScan® PKCβ Kinase Assay Kit (Ozyme, Cell Signaling, France) according to the manufacturer's instructions.
Effect of PKCβ, Aurora-B and Jak2 inhibitors on the TMZ+Irrad-induced cell death
The pH3T6, pH3S10 or pH3Y41 levels being markers of poor prognosis in GBM patients, we then asked the double question to know whether 1) the high level of pH3T6, pH3S10 or pH3Y41 are hallmarks associated with a phenotype of resistance to the standard treatment used in anti-GBM therapy (i.e. a treatment combining the temozolomide and the irradiation named TMZ+Irrad treatment), and 2) the presence of this phenotype can be reverted by pre-treatment abolishing the presence of high levels of pH3T6, pH3S10 or pH3Y41.
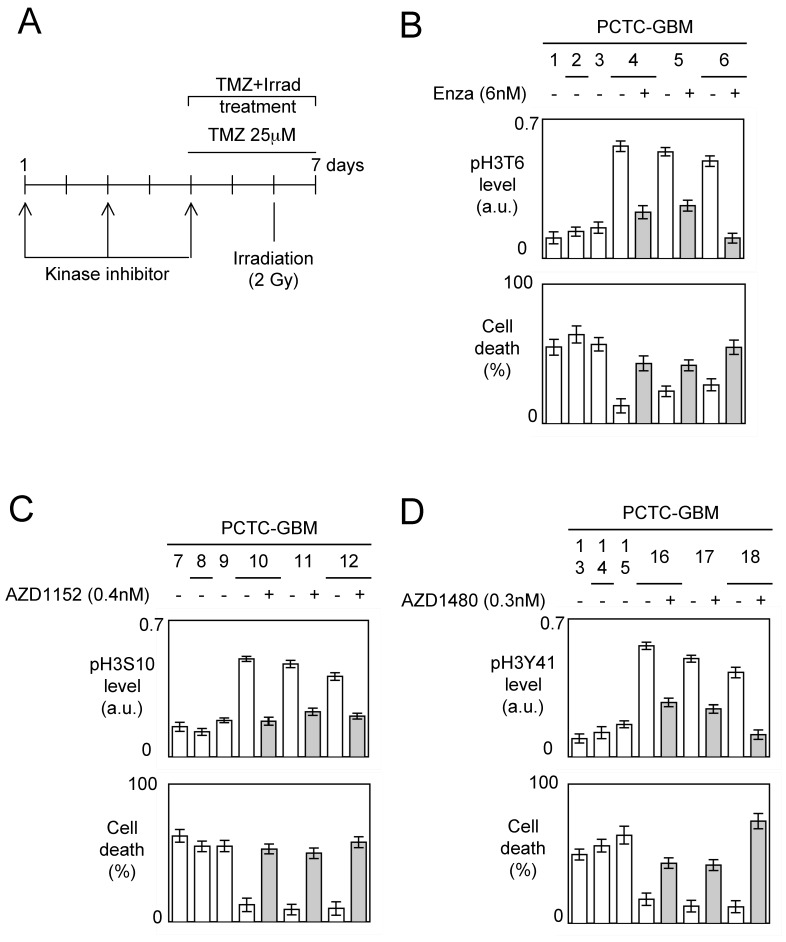
To reply at this double question, we analyzed the percentage of cell death induced by an TMZ+Irrad treatment on primary cultured tumors cells obtained from the patient-derived GBM samples (referred PCTC-GBMs) pre-treated or not by kinase inhibitors targeting the kinases implicated into the presence of high levels of pH3T6, pH3S10 or pH3Y41 in PCTC-GBM (Figure 5A).
Figure 5.
Impact of kinase inhibitors on the phosphorylation levels of histone H3 and on the TMZ+Irrad-induced cell death. (A) Design of the experiments. (B-C-D) PCTC-GBMs presenting a high level of pH3 were treated with adequate kinase inhibitors. The effects of these treatments were estimated by monitoring the pH3 level by ELISA. Cell death is next estimated by using a Trypan Blue Stain 0.4%, and the Countess® Automated Cell Counter (Life Technology, France) after the TMZ+Irrad treatment (Temozomide+Irradiation treatment).
Thus, tumor cells from different GBMs were seeded less than half an hour after tumor resection, and in less than three to five passages, we obtained enough cells to perform our experiments. For each type of histone H3 phosphorylation, we used 3 PCTC-GBMs haboring a low level of considered pH3 and 3 PCTC-GBMs harboring a high level of considered pH3.
The choice of kinase inhibitors targeting the kinases implicated into the presence of high levels of pH3T6, pH3S10 or pH3Y41 in PCTC-GBM fell on Enzastaurin (a PKCβ inhibitor), AZD1152 (an Aurora-B inhibitor) and AZD1480 (a Jak2 inhibitor) since these drugs target kinases implicated into the presence of presence of high levels of pH3T6, pH3S10 or pH3Y41 in GBM, and that these drugs are used in clinical (source: www.clinicaltrilas.gov) 22-24.
The impact of these kinase inhibitors on the considered histone phosphorylation levels have been monitored after 5 days of pre-treatment, and the percentage of cell death was determined 72h after the TMZ+Irrad treatment described in figure 5A.
A first set of experiments indicated that PCTC-GBMs having a low level of pH3T6 were more sensitive to the TMZ+Irrad treatment than the cells having a high level of pH3T6 (Figure 5B). We then noted that the Enzastaurin pre-treatment significantly decreased the pH3T6 level, and that this decrease is associated with a significant increase of the sensitivity of the TMZ+Irrad treatment (Figure 5B).
A second set of experiments (realized from a second set of PCTC-GBM) indicated that PCTC-GBMs having a low level of pH3S10 were more sensitive to the TMZ+Irrad treatment than the cells having a high level of pH3S10 (Figure 5C). We then noted that the AZD1152 treatment significantly decreased the pH3S10 level, and that this decrease is associated with a significant increase of the sensitivity of the TMZ+Irrad treatment (Figure 5C).
A third set of experiments (realized from a third set of PCTC-GBM) indicated that PCTC-GBM having a low level of pH3Y41 were more sensitive to the TMZ+Irrad treatment than the cells having a high level of pH3Y41 (Figure 5D). We then noted that the AZD1480 treatment significantly decreased the pH3Y41 level, and that this decrease is associated with a significant increase of the sensitivity of the TMZ+Irrad treatment (Figure 5D).
All these results suggest that the level of pH3T6, pH3S10 and pH3Y41 could be used to guide the administration of adequate kinase inhibitors in anti-GBM therapy.
Patients stratification and combination of kinase inhibitors
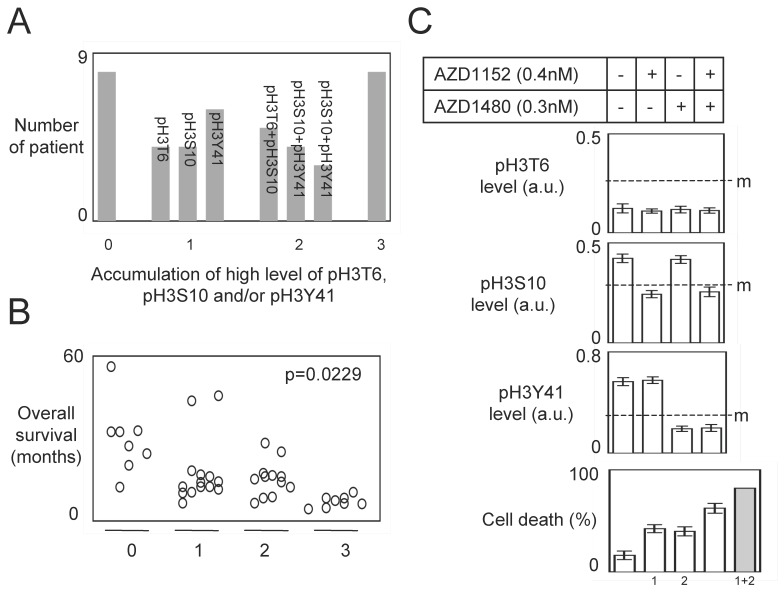
The stratification of GBM in function of the pH3T6, pH3S10 and pH3Y41 levels indicated that 8 GBM have a low level of pH3T6, pH3S10 and pH3Y41, 14 patients have a high level of one H3 phosphorylation, 12 GBM patients accumulated two high levels of H3 phosphorylation, and 8 GBM patients accumulated three high levels of H3 phosphorylation (Figure 6A).
Figure 6.
Patients stratification and combination of kinase inhibitors. (A) Graph illustrates the accumulation of high level of pH3T6, pH3S10 and pH3Y41 in 42 GBM. (B) Correlation between the accumulation of high level of pH3T6, pH3S10 and pH3Y41 and the overall survival of GBM patients. Each circle represents a GBM. (C) Effect of combinantion of kinases inhibitors on TMZ+Irrad-induced apoptosis. PCTC#19 presenting a high level of pH3 were treated with adequate kinase inhibitors. The effects of these treatments were estimated by monitoring the pH3 level by ELISA. Cell death is next estimated by using a Trypan Blue Stain 0.4%, and the Countess® Automated Cell Counter (Life Technology, France) after the TMZ+Irrad treatment (Temozomide+Irradiation treatment). m: median.
Our study also indicated that the accumulation of high level of H3 phosphorylation (among the pH3T6, pH3S10 and pH3Y41) was inversely correlated with the overall survival of GBM patients (Figure 6B).
According to our previous results and the fact that a same GBM was characterized by a 2 (or more) high levels of H3 phosphorylation ask the question to know whether the combination of kinase inhibitors could be more efficient than the use of one kinase inhibitor to increase the sensitivity of the TMZ+Irrad treatment.
To investigate this point, we analyzed the percentage of cell death induced by the TMZ+Irrad treatment on PCTC#19 since this PCT have high levels of pH3S10 and pH3Y41 (but a low level of pH3T6) (Figure 6C).
As expected, the AZD1152 (an Aurora-B inhibitor) and AZD1480 (a Jak2 inhibitor) treatments reduced the levels of pH3S10 and pH3Y41 in PCTC#19, and increased the percentage of TMZ+Irrad-induced cell death (Figure 6C). Interestingly, we also noted that the AZD1152+AZD1480 treatment induced more cell death than each single treatment. However, the percentage of cell death induced by the AZD1152+AZD1480 treatment was inferior to the percentage obtained by the addition of the percentage of cell death induced by each single treatment (Figure 6C). Thus, the effect of the AZD1152+AZD1480 treatment appears synergic and not additional. All these last points suggest that the combination of several kinase inhibitors could have a therapeutic advantage for GBM characterized by the high level of several H3 phosphorylation.
In conclusion, our data indicated that the presence of high level of pH3T6, pH3S10 and/or pH3Y41 in PCTC-GBM is a signature associated with a phenotype of resistance to TMZ+Irrad-induced cell death. Our data also indicate that this phenotype can be reverted by using adequate kinase inhibitors pre-treatment. In other terms, our data provide biomarkers (high level of pH3T6, pH3S10 or pH3Y41) for patient stratification in order to administrate to these patients the adequate kinase inhibitors having the capacity to decrease the level of histone H3 phosphorylation and to increase the TMZ+Irrad-induced cell death.
Discussion
Recent advances in the understanding of the epigenetic molecular mechanisms that govern tumorigenesis have provided meaningful progress in the development and the use of epigenetic drugs in anti-cancer treatment. Still, much more has to be done in order to improve present epigenetic therapeutic approaches: 1) identification of new epigenetic hallmarks associated with tumorigenesis, with a poor prognosis of survival, or with a poor prognosis of response to a therapeutic strategy; 2) identification of molecular actor(s) at the origin of the edification of these epigenetic hallmarks; and 3) design, development and/or use of epigenetic drugs having the double ability to specifically target the molecular actor(s) at the origin of the edification of these epigenetic hallmarks and to promote a gain of efficiency into the anti-cancer treatment.
Our study is in this context since our data identify 1) the high level of pH3T6, pH3S10 and pH3Y41 such as an epigenetic hallmark whose the presence in GBM is associated with a poor OS; 2) PKCβ, Aurora-B and Jak2 such as a molecular actors explaining the presence of high level of pH3T6, pH3S10 and pH3Y41; and 3) Enzastaurin, AZD1152, and AZD1480 such as drugs limiting the high level of pH3T6, pH3S10 and pH3Y41 in PCTC-GBM and increasing the percentage of cell death induced by the irradiation/TMZ treatment.
Thus, our data support the idea to include Enzastaurin in the standard treatment of GBMs. Besides, the promising character of this idea is also supported by different clinical trials 25. Indeed, a phase I study indicated that Enzastaurin and TMZ can be associated in a same protocol since TMZ did not appear to affect Enzastaurin exposures at the 250 mg or 500 mg dose levels 26. Literature also mentions the realization of a phase II and pharmacogenomics study of Enzastaurin plus temozolomide during and following radiation therapy in patients with newly diagnosed glioblastoma multiforme and gliosarcoma 27.
In addition to our data, the putative use of AZD1152 in the standard treatment of GBMs, is also supported by the fact that the subcutaneous administration of AZD1152 resulted in a prolongation in median survival after intracranial inoculation of U251 cells in mice 28.
Concerning the potential use of AZD1480 in anti-GBM therapy, our data found echo and are complemented by the Mac Farland's et al. results, since these authors published that AZD1480 inhibits the growth of subcutaneous tumors and increases survival of mice bearing intracranial GBM tumors 29. Thus, it appears indicating that pharmacologic inhibition of the JAK/STAT-3 pathway by AZD1480 should be considered for study in the treatment of patients with GBM tumors.
In addition to the articles supporting the use of Enzastaurin, AZD1152 or AZD1480 in anti-GBM therapy, our data provide biomarkers (the level of pH3T6, pH3S10 and pH3Y41) for the patient stratification having the most probabilities to reply to anti-GBM treatment including the Enzastaurin, AZD1152 and/or AZD1480 drugs. Our study also suggests that the combination of kinases inhibitors could be provided therapeutic advantage for the treatment of GBM harboring high level of pH3T6, pH3S10 and/or pH3Y41.
Concerning the prognosis value of the histone phosphorylation levels analyzed in this study, our work indicates that the poor prognosis associated to the presence of high level of pH3T6 and pH3Y41 is not influenced by the MGMT gene methylation status. Indeed, no correlation was observed between the level of pH3Y41 and the MGMT gene methylation status (p=0.4476) and between the level of pH3T6 and the MGMT gene methylation status (p=0.2887). Thus, our data indicated that the pH3T6 and pH3Y41 levels could be used as an alternative and independent prognosis factor to the use of the MGMT methylation status. On contrary, we noted a significant inverse correlation between the level of pH3S10 and the methylation status of MGMT (p=0.0120). In addition to this inverse correlation, our work highlights the fact that the pH3S10 level could be used in association with the MGMT gene methylation status in order to stratify the GBM patients since 1) patients whose GBM had a high pH3S10 level and a unmethylated MGMT gene have a lower overall survival than patients whose GBM had a low pH3S10 level and a unmethylated MGMT gene and 2) patients whose GBM had a high pH3S10 level and a methylated MGMT gene have a lower overall survival than patients whose GBM had a low pH3S10 level and a methylated MGMT gene. Consequently, the administration of AZD1152 to treat patients whose GBM are characterized by the unmethylated MGMT gene and a high level of pH3S10 (MGMTU/pH3S10H) appears as a promising therapeutic alternative since these patients have a poor overall survival in response to the standard anti-GBM therapy (8.3 months in our study).
To date there is only one similar and recently published observation. Indeed, high level of pH3T11 was associated with a poor prognosis in glioma patients 13. Thus, as in our case, it is an excess of phosphorylation that is used such as poor prognosis marker. In all these cases, the use of adequate kinase inhibitors appears such as a promising alternative in anti-GBM therapy.
The use of pH3T6, pH3S10 and/or pH3Y41 phosphorylation levels as biomarkers to guide the administration of kinases inhibitors in anti-cancer therapy could be discussed since histone phosphorylation could be modulated (distorded) by physiologic events such as the cell number in mitosis 30. Consequently, in our study, the detection of high level of pH3S10 in certain GBM could be due to the fact that these GBM are in mitosis. In this case, the pH3S10 level should be correlated with the level of other histone modifications (such as the H3S28 and H3T3 phosphorylations) 6, 11, 31. Now, no significant correlations were observed between the pH3S10 and pH3S28 levels (Pearson's correlation test: r =0.183, p=0.2460, n=42), pH3T3 and pH3S28 (Pearson's correlation test: r =-0.237, p=0.1307, n=42) and pH3T3 and pH3S10 (Pearson's correlation test: r =0.281, p=0.0714, n=42). We thereby conclude that the detection of high level of pH3S10 was not due to the accumulation of cells in mitosis in certain GBM.
References
- 1.Zheng C, Hayes J. Structures and interactions of the core histone tail domains. Biopolymers. 2003;68:539–46. doi: 10.1002/bip.10303. [DOI] [PubMed] [Google Scholar]
- 2.Luger K, Dechassa M, Tremethick D. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat Rev Mol Cell Biol. 2012;13:436–47. doi: 10.1038/nrm3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossetto D, Avvakumov N, Côté J. Histone phosphorylation: A chromatin modification involved in diverse nuclear events. Epigenetics. 2012;7:1098–108. doi: 10.4161/epi.21975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sawicka A, Seiser C. Histone H3 phosphorylation - A versatile chromatin modification for different occasions. Biochimie. 2012;94:2193–201. doi: 10.1016/j.biochi.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baek S. When signaling kinases meet histones and histone modifiers in the nucleus. Mol Cell. 2011;42:274–84. doi: 10.1016/j.molcel.2011.03.022. [DOI] [PubMed] [Google Scholar]
- 6.Dai J, Sultan S, Taylor S, Higgins J. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev. 2005;19:472–88. doi: 10.1101/gad.1267105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eswaran J, Patnaik D, Filippakopoulos P, Wang F, Stein R, Murray J. et al. Structure and functional characterization of the atypical human kinase haspin. Proc Natl Acad Sci U S A. 2009;106:20198–203. doi: 10.1073/pnas.0901989106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang T, Park D, Choi Y, Kim K, Yoon H, Kim K. Mitotic histone H3 phosphorylation by vaccinia-related kinase 1 in mammalian cells. Mol Cell Biol. 2007;27:8533–46. doi: 10.1128/MCB.00018-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Metzger E, Imhof A, Patel D, Kahl P, Hoffmeyer K, Friedrichs N. et al. Phosphorylation of histone H3T6 by PKCbeta(I) controls demethylation at histone H3K4. Nature. 2010;464:792–6. doi: 10.1038/nature08839. [DOI] [PubMed] [Google Scholar]
- 10.Lee K, Song K. Basal c-Jun N-terminal kinases promote mitotic progression through histone H3 phosphorylation. cell cycle. 2008;7:216–21. doi: 10.4161/cc.7.2.5155. [DOI] [PubMed] [Google Scholar]
- 11.Goto H, Yasui Y, Nigg E, Inagaki M. Aurora-B phosphorylates Histone H3 at serine28 with regard to the mitotic chromosome condensation. Genes Cells. 2002;7:11–7. doi: 10.1046/j.1356-9597.2001.00498.x. [DOI] [PubMed] [Google Scholar]
- 12.Metzger E, Yin N, Wissmann M, Kunowska N, Fischer K, Friedrichs N. et al. Phosphorylation of histone H3 at threonine 11 establishes a novel chromatin mark for transcriptional regulation. Nat Cell Biol. 2008;10:53–60. doi: 10.1038/ncb1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D. et al. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012;150:685–96. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Preuss U, Landsberg G, Scheidtmann K. Novel mitosis-specific phosphorylation of histone H3 at Thr11 mediated by Dlk/ZIP kinase. Nucleic acids Res. 2003;31:878–85. doi: 10.1093/nar/gkg176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimada M, Niida H, Zineldeen D, Tagami H, Tanaka M, Saito H. et al. Chk1 is a histone H3 threonine 11 kinase that regulates DNA damage-induced transcriptional repression. Cell. 2008;132:221–32. doi: 10.1016/j.cell.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 16.Hake S, Garcia B, Kauer M, Baker S, Shabanowitz J, Hunt D. et al. Serine 31 phosphorylation of histone variant H3.3 is specific to regions bordering centromeres in metaphase chromosomes. Proc Natl Acad Sci U S A. 2005;102:6344–9. doi: 10.1073/pnas.0502413102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dawson M, Bannister A, Göttgens B, Foster S, Bartke T, Green A. et al. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461:819–22. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurd P, Bannister A, Halls K, Dawson M, Vermeulen M, Olsen J. et al. Phosphorylation of histone H3 Thr-45 is linked to apoptosis. J Biol Chem. 2009;284:16575–83. doi: 10.1074/jbc.M109.005421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Beusechem V, Grill J, Mastenbroek D, Wickham T, Roelvink P, Haisma H. et al. Efficient and selective gene transfer into primary human brain tumors by using single-chain antibody-targeted adenoviral vectors with native tropism abolished. J Virol. 2002;76:2753–62. doi: 10.1128/JVI.76.6.2753-2762.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hegi M, Diserens A, Gorlia T, Hamou M, de Tribolet N, Weller M. et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:991–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 21.Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V. et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–4. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 22.Ma S, Rosen S. Enzastaurin. Curr Opin Oncol. 2007;19:590–3. doi: 10.1097/CCO.0b013e3282f10a00. [DOI] [PubMed] [Google Scholar]
- 23.Wilkinson R, Odedra R, Heaton S, Wedge S, Keen N, Crafter C. et al. AZD1152, a selective inhibitor of Aurora B kinase, inhibits human tumor xenograft growth by inducing apoptosis. Clin Cancer Res. 2007;13:3682–8. doi: 10.1158/1078-0432.CCR-06-2979. [DOI] [PubMed] [Google Scholar]
- 24.Ioannidis S, Lamb M, Wang T, Almeida L, Block M, Davies A. et al. Discovery of 5-chloro-N2-[(1S)-1-(5-fluoropyrimidin-2-yl)ethyl]-N4-(5-methyl-1H-pyrazol-3-yl)pyrimidine-2,4-diamine (AZD1480) as a novel inhibitor of the Jak/Stat pathway. J Med Chem. 2011;54:262–76. doi: 10.1021/jm1011319. [DOI] [PubMed] [Google Scholar]
- 25.Galanis E, Buckner J. Enzastaurin in the treatment of recurrent glioblastoma: a promise that did not materialize. J Clin Oncol. 2010;28:1097–8. doi: 10.1200/JCO.2009.26.1149. [DOI] [PubMed] [Google Scholar]
- 26.Rampling R, Sanson M, Gorlia T, Lacombe D, Lai C, Gharib M. et al. A phase I study of LY317615 (enzastaurin) and temozolomide in patients with gliomas (EORTC trial 26054) Neuro Oncol. 2012;14:344–50. doi: 10.1093/neuonc/nor221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Butowski N, Chang S, Lamborn K, Polley M, Pieper R, Costello J. et al. Phase II and pharmacogenomics study of enzastaurin plus temozolomide during and following radiation therapy in patients with newly diagnosed glioblastoma multiforme and gliosarcoma. Neuro Oncol. 2011;13:1331–8. doi: 10.1093/neuonc/nor130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diaz R, Golbourn B, Shekarforoush M, Smith C, Rutka J. Aurora kinase B/C inhibition impairs malignant glioma growth in vivo. J Neurooncol. 2012;108:349–60. doi: 10.1007/s11060-012-0835-2. [DOI] [PubMed] [Google Scholar]
- 29.McFarland B, Ma J, Langford C, Gillespie G, Yu H, Zheng Y. et al. Therapeutic potential of AZD1480 for the treatment of human glioblastoma. Mol Cancer Ther. 2011;10:2384–93. doi: 10.1158/1535-7163.MCT-11-0480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang F, Higgins J. Histone modifications and mitosis: countermarks, landmarks, and bookmarks. Trends Cell Biol. 2013;23:175–84. doi: 10.1016/j.tcb.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 31.Crosio C, Fimia G, Loury R, Kimura M, Okano Y, Zhou H. et al. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol Cell Biol. 2002;22:874–85. doi: 10.1128/MCB.22.3.874-885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]