Abstract
To understand the evolutionary histories and conservation potential of wild animal species it is useful to assess whether taxa are genetically structured into different populations and identify the underlying factors responsible for any clustering. Landscape features such as rivers may influence genetic population structure, and analysis of structure by sex can further reveal effects of sex-specific dispersal. Using microsatellite genotypes obtained from noninvasively collected fecal samples we investigated the population structure of 261 western lowland gorillas (WLGs) (Gorilla gorilla gorilla) from seven locations spanning an approximately 37,000km2 region of mainly continuous rain forest within Central African Republic (CAR), Republic of Congo and Cameroon. We found our sample to consist of two or three significantly differentiated clusters. The boundaries of the clusters coincided with courses of major rivers. Moreover, geographic distance detoured around rivers better-explained variation in genetic distance than straight line distance. Together these results suggest that major rivers in our study area play an important role in directing WLG gene flow. The number of clusters did not change when males and females were analyzed separately, indicating a lack of greater philopatry in WLG females than males at this scale.
Keywords: Gorilla gorilla gorilla, population structure, sex-specific dispersal, philopatry
INTRODUCTION
Although many researchers have investigated the pattern of genetic variation in organisms living in multiple disjunct populations or in fragmented habitats [e.g., Haag et al., 2010; Martínez-Cruz et al., 2007; Ruell et al., 2012], relatively few studies have investigated the structure exhibited by animal populations living in mainly continuous, apparently uniform habitat [Neaves et al., 2009; Piertney et al., 1998; reviewed in Storfer et al., 2010]. Even in areas of continuous habitat, rivers may hinder the movement of terrestrial animals [Storfer et al., 2010], thus potentially inhibiting gene flow and affecting patterns of genetic differentiation. However, because water levels vary by season and river courses can change over time, it can be difficult to assess whether a river serves as complete barrier to gene flow or acts as an occasionally permeable hindrance. The effects of rivers on the genetic structure of large mammal populations have accordingly varied. For example, indications that rivers reduce gene flow were found in white-tailed deer (Odocoileus virginianus) [Blanchong et al., 2008] and lowland tapir (Tapirus terrestris) [De Thoisy et al., 2010], whereas in bobcats (Lynx rufus) the large Mississippi River was not significantly associated with genetic differentiation [Reding et al., 2012]. In red deer (Cervus elaphus) from the Scottish Highlands, rivers were even suggested to facilitate gene flow [Pérez-Espona et al., 2008]. Studies of great apes found that in chimpanzees (Pan troglodytes) [Gonder et al., 2006] and Bornean orangutans (Pongo pygmaeus) [Arora et al., 2010; Jalil et al., 2008], rivers apparently act as barriers to individual movement and thereby increase genetic differentiation between subpopulations on either side of the river. No barrier effect of rivers was detected in other smaller-bodied, more arboreal primates like spider monkeys (Ateles) [Collins & Dubach, 2000], long-tailed macaques (Macaca fascicularis) [Ruiter & Geffen, 1998] and a number of lemur species (Lemuriformes) [Craul et al., 2008].
In addition to rivers, postglacial expansions from forest refugia that became isolated during the Pleistocene [Haffer, 1969] may be expected to influence the distribution of present-day genetic variation in geographically continuous populations of forest dwelling species. For example, although forest elephants (Loxodonta cyclotis) were found to be only weakly structured [Roca et al., 2001], it was suggested they were restricted to isolated patches during the Pleistocene [Brandt et al., 2012]. While an early study in bonobos (Pan paniscus) attributed patterns of genetic variation to the effect of river barriers on migration [Eriksson et al., 2004], a recent study by Kawamoto et al. [2013] suggested that present day rivers have only a weak effect on gene flow among bonobo populations and that their current population genetic structure was shaped by Pleistocene forest refugia [Haffer, 1969; Hewitt, 1996]. For gorillas (Gorilla) it has been suggested that both Pleistocene refugia and rivers played a role in structuring genetic diversity [Anthony et al., 2007]. This study found signs of demographic expansion consistent with postglacial expansion from formerly isolated refugia as well as admixture along the borders of mtDNA haplogroups. Nonetheless, some rivers acted as barriers between different haplogroups of gorillas.
If population genetic structure is found to be mainly caused by rivers or other hindrances to movement then these barriers will have a greater impact on the dispersing sex. Dispersal is male-biased in most mammals [Greenwood, 1980; Lawson Handley & Perrin, 2007]. However, there are also male-philopatric species (e.g., chimpanzee, bonobo) and species in which both sexes disperse (e.g., African wild dog (Lycaon pictus), gray wolf (Canis lupus)). Differences in dispersal distance are typical for species in which both sexes disperse [reviewed in Mabry et al., 2013]. If the rate of dispersal is similar, then the sex that travels further is less likely to be structured into differentiated clusters.
Gorillas are among the relatively few mammal species for which both sexes usually disperse from their natal groups [Stoinski et al., 2009; Stokes et al., 2003]. The typical reproductive unit in western lowland gorillas (WLGs) (Gorilla gorilla gorilla) is composed of a single adult silverback male accompanied by several females [Breuer et al., 2010; Gatti et al., 2004; Parnell, 2002; Robbins et al., 2004]. Adult females transfer directly between breeding groups during inter-group encounters and do not range on their own [Stokes et al., 2003]. Males, in contrast, leave their natal groups after maturation and range several years as solitaries before eventually obtaining a harem [Breuer et al., 2010]. Bradley et al. [2004] found evidence that WLG silverbacks leading social groups were often related to nearby males, suggesting low-distance dispersal of many males. In contrast, results of a recent study by Inoue et al. [2013] suggested that closely related WLG males do not stay close to each other after maturation arguing for relatively long dispersal distances. Thus, dispersal patterns of male WLGs may vary between different sites, as the former study was in the Central African Republic and the latter was in Gabon.
Research on WLGs as well as on mountain gorillas (Gorilla beringei beringei) suggested greater population genetic structure in females as compared to males, which was attributed to a tendency of females to disperse shorter distances than males [Douadi et al., 2007; Guschanski et al., 2008]. However, these studies were conducted over limited spatial scales (WLGs: 70km×85 km; mountain gorillas: 28km×18 km). In addition, the study of WLGs relied on sex-specific markers, namely mitochondrial DNA and the Y-chromosome. Although rapidly evolving and hence likely to reveal any underlying genetic structure, it is difficult to directly compare results from these female-and male-transmitted systems. This is because the Y-chromosome is likely to exhibit lower variation than mtDNA [Burrows & Ryder, 1997; Hellborg, 2003] and differences in genetic differentiation estimates between mtDNA and the non-recombining portion of the Y-chromosome cannot be tested for significance. In addition, differences in reproductive skew between the sexes will alter the relative effective population sizes of males and females. Thus it is difficult to make inferences about dispersal rate from the extent of genetic differentiation observed from sex-specific markers [Lawson Handley & Perrin, 2007].
For the present study WLG samples from key locations across a large expanse of their range were collected. Genetic characterization of those samples may effectively address questions on (i) population structure including the effects of rivers and (ii) the relative roles of male and female dispersal in shaping genetic structure. Samples were collected in a relatively large, roughly 160km×230km region consisting of continuous rainforest with several notable rivers and few roads (Fig. 1).
Fig. 1.
Sampled region with red dots depicting sampling locations. Rivers mentioned in the text are in blue, national parks in yellow and roads in white. The three major sampling areas are: (i) Odzala: two sites east of Odzala NP (Libonga and Liouesso); (ii) Lobéké: three sites west of and within LobékéNP (Lobéké-West, Lobéké-Central, and Lobéké-East); and (iii) Ndoki forest: two sites within or close by Dzanga Ndoki and Nouabalé-Ndoki NP (southern Ndoki and Bai Hokou).
METHODS
Study Area and Sample Collection
Fecal samples from wild WLGs were obtained from three areas within Central Africa (Fig. 1): (i) two sampling sites east of Odzala NP, Republic of Congo; (ii) three sampling sites within and west of Lobéké NP, Cameroon; and (iii) various locations (southern Ndoki and Bai Hokou) within and close to Nouabalé-Ndoki NP, Republic of Congo and Dzanga Ndoki NP, Central African Republic. The latter two national parks are adjacent and only separated by the national border with no physical barrier between them. We will summarize that area as “Ndoki forest”. The Ngoko River may serve as natural barrier between Odzala and Lobéké, and the Sangha River separates the Ndoki forest from Lobéké and Odzala. The whole region of approximately 37,000km2 is covered by continuous rainforest without notable elevation changes and has few public roads. One road runs between the two sampling sites east of Odzala NP, another is located between Odzala NP and the two other major areas. Both are regularly crossed by WLGs (F. Maisels, personal communication). Samples were collected noninvasively between 2003 and 2012 using Nsubuga et al.’ s [2004] two-step ethanol-silica storage method (samples from Odzala and the Ndoki forest) or were stored in RNAlater (samples from Lobéké).
DNA Extraction, Quantification and Amplification
The QIAamp Stool kit (QIAGEN) was used to extract DNA from fecal samples. To estimate the amount of amplifiable DNA in the extracts we used a real-time quantitative PCR with Maxima SYBR® Green (Thermo Scientific, Waltham, MA, USA) as Master Mix and c-myc_E3_F1U1 plus c-myc_ E3_R1U as primers [Morin et al., 2001] to amplify a portion of the c-myc gene. Standard curves were constructed from serially diluted human genomic DNA (BIO-35025, Bioline, London, UK). Initially, three independent amplifications from each extract were performed at 12 microsatellite loci (D10s1432, D8s1106, D6s474, D1s2130, D5s1470, D4s1627, D7s817, D1s550, D16s2624, D6s1056, D2s1326, D3s2459), using a two-step multiplex PCR [Arandjelovic et al., 2009]. For sex determination we also amplified a segment of the X-Y homologous amelogenin gene in a one-step PCR [Bradley et al., 2001].
The ABI PRISM 3100 Genetic Analyzer was used for electrophoresis of the PCR products. The sizes of the alleles relative to an internal size standard were determined using GeneMapper Software version 3.7 (Applied Biosystems, Foster City, CA, USA). We calculated allelic dropout rates to assess how many observations from independent reactions are necessary to confirm homozygosity [Morin et al., 2001]. We found that three observations from independent reactions are sufficient to confirm homozygous genotypes with 99% certainty even in the extracts with the lowest DNA concentrations. Heterozygote genotypes were confirmed with 99% certainty by observing each allele two or more times [Taberlet et al., 1996]. For each extract up to six independent reactions were analyzed per locus.
Discrimination of Individuals and Distinguishing Gorilla and Chimpanzee Samples
We used CERVUS 3.0 [Kalinowski et al., 2007] to calculate PIDsib [Waits et al., 2001], the probability that samples with matching genotypes come from siblings rather than from the same individual. Matching genotypes were combined into a consensus genotype when the PIDsib was <0.01. In the rare cases where two genotypes matched but the PIDsib was >0.01 the less complete genotype was removed from further analyses. That approach resulted in a dataset in which the least completely genotyped individuals were typed at a minimum of four loci, because genotypes consisting of fewer loci were in no case informative enough to be found unique.
In the field, fecal samples coming from chimpanzees are occasionally mistakenly identified as gorilla feces, and vice versa. We therefore checked for incorrectly identified samples by genotyping 10 samples of known chimpanzee origin at the 12 microsatellite loci typically analyzed in gorillas (see above) and performing STRUCTURE analysis of them in combination with the purported gorilla samples as in Arandjelovic et al. [2010]. After attributing samples to the correct species, we added those samples to our dataset that were genetically identified as WLGs and removed samples that were genetically identified as chimpanzees.
Data Analysis
Using SPAGeDi 1.3 [Hardy & Vekemans, 2002] we tested via permutations (equivalent to carrying out a Mantel test) for a significant correlation between genetic distance and two different measures of geographic distance: (i) straight line Euclidean distance between sampling sites (also called isolation by distance, IBD) and (ii) detoured distance from one sampling site to another around the headwaters of the Sangha and the Ngoko rivers. We chose seven geographically distinct sampling sites (Fig. 1): Odzala- South (Libonga), Odzala-North (Liouesso), Lobéké-West, Lobéké-Central, Lobéké-East, Bai Hokou, and southern Ndoki. We plotted for each of the resulting 21 pairs the natural logarithm of the geographic distance in meters against the genetic differentiation measured as FST/(1−FST) and plotted a regression line. The slope of this line was considered significantly positive if the slope was >0 and the probability in 20,000 permutations of obtaining by chance a slope larger than the observed one is less than 5%.
To assess which measure of geographic distance better explains variation in genetic distance we ran general linear models in R (R Development Core Team 2012) for calculations of the Akaike Information Criterion (AIC). The response variable for each model was genetic distance and the predictor variables were straight distance and detoured distance. The intercept was excluded from the models. The predictor variable of the model with the smaller AIC better explains the variation in the response variable. ΔAIC is the difference of the AIC values of the two models and the larger it is the less empirical support the model with the higher AIC receives. Due to the non-independence of the data points we ran 5,040 permutations of the genetic distances (total number of possible permutations) and declared ΔAIC to be significant if the proportion of cases in which ΔAIC from permuted genetic distances was larger than that from the original data was <0.05.
Most programs that output summary statistics require a priori delineation of populations which is a conflicting issue if the main goal of the study is to find the previously unknown number of clusters within the area of interest. However, software like Geneland 4.0.3 [Guillot, 2005, 2008; Guillot et al., 2005, 2008] and STRUCTURE 2.2 [Falush et al., 2003, 2007; Pritchard et al., 2000] are based on Bayesian clustering algorithms for inferring genetic structure and do not need a priori assignment of populations. STRUCTURE employs a nonspatial cluster algorithm that assumes an independent identically distributed (iid) mixture model. The iid mixture model puts equal prior weights on all partitions, whereas the hidden model of spatial organization through Voronoi tessellation implemented in Geneland favors partitions that are spatially organized. This leads to better classification with limited data due to a more informative prior [Guillot, 2005]. We therefore first investigated our data from the whole study area with Geneland to assess the most likely number of underlying clusters for each of the following different data sets: (i) all individuals; (ii) only females; and (iii) only males. To account for differences in sample size we conducted the male-specific analysis with the same number of samples as the female-specific analysis (N=89). For each dataset we conducted five independent runs using the following parameters: maximum rate of Poisson process: 100; uncertainty on coordinate: 5; minimum and maximum number of populations: 1 and 8, respectively; maximum number of nuclei: 300; number of iterations: 100,000; thinning: 100; allele frequency model: correlated; with spatial model; without null allele model. From the five runs we always present the results of the one with the highest mean posterior density.
For comparison with results generated by Geneland we additionally analyzed our data from the whole study area in STRUCTURE and BAPS 5.3 [Corander et al., 2007; Corander &Marttinen, 2006]. As with Geneland, BAPS also makes use of the geographic origin of the genotypes. However, inferences in BAPS are based on another model and different computational techniques [Corander et al., 2007]. BAPS assumes that all individuals have unique coordinates. Therefore we added random noise of up to 100m to each UTM_X and each UTM_Y coordinate that was not unique. We ran the spatial clustering of individuals with 10 replicates for each number of clusters K (K=3, 5, 10, and 20, with the numbers specifying the upper limit of expected clusters). In STRUCTURE we varied K from 1 to 8 clusters and conducted 10 independent iterations at each K using the admixture model, correlated allele frequencies, a burn-in period of 100,000 steps and then 500,000 steps of data collection. Subsequently we used Harvester Web v0.6.93 [Earl & vonHoldt, 2011] to calculate ΔK [Evanno et al., 2005], a measure of second-order rate of change in the likelihood of K which is widely used to determine the optimal value of K. For that calculation we removed runs with extraordinarily high variance of Ln likelihood compared to other runs within the same K.
It is important to check that the inferred groups are significantly differentiated from each other and in Hardy–Weinberg equilibrium (HWE). Geneland further assumes that marker loci are at linkage equilibrium within populations. Using CERVUS 3.0 [Kalinowski et al., 2007] we tested for the existence of null alleles within each cluster inferred by the Geneland analysis of all individuals. GENEPOP 4.2 [Raymond & Rousset, 1995] was used to perform the exact tests of Guo and Thompson [1992] for deviations from HWE as well as the test of composite linkage disequilibrium [Weir, 1996] within each cluster. Once clusters were identified, pairwise FST estimates of population differentiation with associated P-values (using 9999 permutations) were obtained using GenAlEx 6.5 [Peakall & Smouse, 2006, 2012].
Because our study did not involve animal testing and the samples were collected noninvasively we did not violate any regulations of the Deutsches Tier-schutzgesetz and the US Public Health Service Policy on Humane Care and Use of Laboratory Animals. The study complied with ASP principles for the ethical treatment of non-human primates. All research was conducted in accordance with the laws of Republic of Congo, Cameroon, Central African Republic, and Germany.
RESULTS
Genotype Analysis
We genotyped 411 putative WLG fecal samples at 12 autosomal microsatellite loci. These samples yielded 339 usable genotypes representing 272 unique individuals. An analysis of these genotypes using STRUCTURE in comparison with known chimpanzee DNAs typed at the same loci, showed that 12 of the newly generated genotypes were actually from chimpanzees, leaving a total of 260 different WLGs. Field researchers thus correctly identified WLG dung in 327/339=96% of cases. In a reciprocal analysis of genotypes purportedly from 134 chimpanzees and typed as part of a complementary study, we found three samples that derived from WLGs. These three samples were secondarily genotyped at microsatellite loci used here. One sample failed to produce a usable genotype, one sample was identical to an already existing genotype and one represented an additional individual. The final dataset therefore contained 261 different WLG genotypes, which were on average 84% complete. Two hundred and twenty-six out of these 261 individuals (=86.6%) had complete genotypes at more than eight loci and only 18/261 (=6.9%) had complete genotypes at six or fewer loci. The 261 individuals consisted of 89 females (F) and 168 males (M) while the sex could not be determined for four individuals. Forty-two individuals were from Odzala (20F, 21M, 1 unknown sex), 24 from Lobéké (8F, 16M) and 195 (61F, 131M, 3 unknown sex) from the Ndoki forest.
The average DNA concentration of the 298 quantified extracts was 247.3 pg/μl (SD: 548.8 pg/μl; range: 3.6–5682.5 pg/μl). Considering results from individual PCRs, allelic dropout rate was 0.06 over all loci and all samples and 0.22 over all loci for extracts with the lowest DNA amounts (≤5 pg/μl). Three independent observations of homozygosity were therefore sufficient to confirm homozygous genotypes with 99% certainty (0.223=0.01) even in samples with very low DNA concentrations.
Using the clusters defined by Geneland (see below), we found null allele frequency estimates >0.05 at one locus within the Ndoki forest cluster, at three loci within the Lobéké cluster and at one locus within the Odzala cluster. However, it is possible that null allele frequency is overestimated when sample size is small, which is the case for the Lobéké and the Odzala cluster (24 and 42 samples, respectively). We tested for HWE both before and after removal of the above mentioned loci from the respective clusters. In no case did any of the three clusters deviate significantly from HWE (P>0.05). We found no indication for linkage disequilibrium (P>0.05 after Bonferroni correction for multiple tests) between any pair of loci within any of the inferred clusters.
Geographic Distribution of Genetic Variation
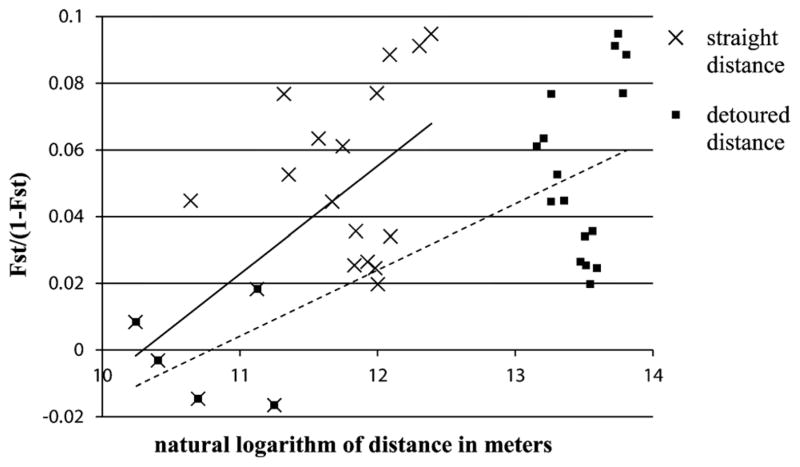
Pairwise comparisons between the seven sampling sites showed that genetic variation increased both with straight line geographic distance and detoured distance (Fig. 2). However, the correlation coefficient of the regression line was smaller in the straight distance (r=0.60) than in the detoured distance plot (r=0.73). Additionally, the GLM with detoured distance as predictor variable was found to better explain variation in genetic distance (AIC=−83.6) than the GLM with straight distance as predictor variable (AIC=−81.7). The difference in AIC was highly significant (P=0.0032).
Fig. 2.
Genetic distances measured as Fst/(1−Fst) plotted against natural logarithm of straight distance and detoured distance in meters, respectively. Each point represents one pairwise comparison between the seven sampling sites. The linear equations for the regression lines were y=0.0324x−0.3341 with P=0.0343 for straight distance (solid line) and y=0.0199 x−0.2142 with P=0.0096 for detoured distance (dashed line).
In an analysis of all genotypes, the Geneland run with the highest mean posterior density (−6399.11) depicted three clusters. All Ndoki forest samples were grouped into the first, all Lobéké samples into the second and all Odzala samples into the third cluster (Fig. 3). The boundaries of the inferred clusters roughly correspond to naturally occurring barriers: the Ngoko River between Odzala and Lobéké and the Sangha River between Ndoki forest and the two other sites. Considering each of the clusters as populations, we found significant pairwise FST estimates between all three localities with the highest differentiation between Odzala and Ndoki forest, an intermediate level between Lobéké and Ndoki forest and the lowest between Odzala and Lobéké (Table I).
Fig. 3.
A Voroni tesssellation map depicting the sorting of all genotypes into three clusters. The clusters are represented by the colors white (Odzala), green (Lobéké) and yellow (Ndoki forest). The map was produced by the Geneland run with the highest mean posterior density. GPS points at which samples were collected are depicted as black dots.
TABLE I.
FST Estimates (Below the Diagonal) and Associated P-values After 9999 Permutations (Above the Diagonal) for Pairwise Comparisons Between Odzala, Lobéké, and Ndoki Forest Including All Genotypes (Number of Included Samples From Each Locality in Brackets)
| Odzala (42) | Lobéké (24) | Ndoki forest (195) | |
|---|---|---|---|
| Odzala (42) | — | <0.0001 | <0.0001 |
| Lobéké (24) | 0.027 | — | <0.0001 |
| Ndoki forest (195) | 0.042 | 0.031 | — |
In contrast to the results from Geneland, the STRUCTURE analysis inferred two clusters of individuals. Nearly all individuals (244/261) had ≥75% ancestry from one population. One cluster contained individuals from Odzala and Lobéké and the other contained the individuals from the Ndoki forest. However, there were also three individuals (one female, two males) that were sampled in Lobéké, but assigned to the Ndoki forest cluster and three others (two females, one male) that were sampled in the Ndoki forest but assigned to the Odzala/Lobéké cluster (Fig. 4).
Fig. 4.

STRUCTURE plot for all genotypes at K=2. Analysis included WLGs sampled in Odzala (bars 1–42), Lobéké (bars 43–66) and the Ndoki forest (bars 67–261). The 17 individuals marked by a yellow asterisk have less than 75% ancestry from a single population. Individuals indicated by numbers from 1 to 6 were assigned to another cluster than the one in which they were sampled.
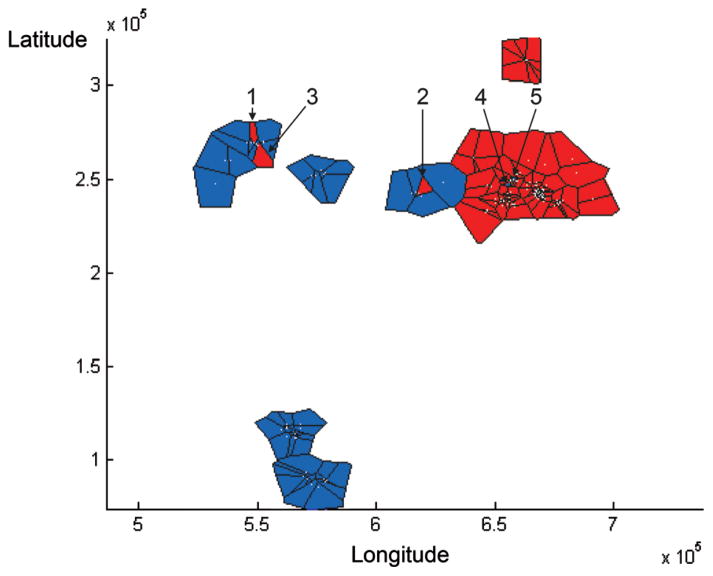
Analysis with BAPS also grouped individuals into two clusters, which can be seen on a map of Voronoi tessellation in which each tile represents one individual (Fig. 5). Of the 244 genotypes that could be allocated to a single cluster by STRUCTURE, BAPS assigned 243 genotypes into the same clusters (99.6% agreement). Of the six individuals assigned by STRUCTURE to a cluster other than the one where they were sampled, five (three females, two males) were also flagged by BAPS.
Fig. 5.
The spatial genetic structure of all generated genotypes of WLGs from Odzala, Lobéké, and the Ndoki forest depicted as a Voronoi tessellation by BAPS. Each tile refers to one genotype. The tiles indicated by numbers belong to individuals that were sampled in another cluster than the one to which they were allocated. Note that equal numbers in Figure 4 and Figure 5 refer to the same individuals.
We also conducted genetic cluster analysis using just male or just female genotypes. Using Geneland, analysis of only females produced three clusters in the run with the highest mean posterior density (−2026.85). However, in that analysis the two females from Lobéké-Central formed one cluster together with the Odzala samples. The second cluster contained all remaining Lobéké females and the third all individuals from the Ndoki forest. A Geneland analysis of only males also showed three clusters in the run with the highest mean posterior density (−1649.14). The three clusters were identical with the ones that were found by Geneland when all genotypes were included: the Ndoki forest males formed the first, the Lobéké males the second and the Odzala males the third cluster.
DISCUSSION
Effect of Rivers on Population Structure
The area under study consists of continuous rain forest and swamps with high uniformity and low fragmentation through human impact. Nevertheless, we found that our WLG sample could be divided into two (STRUCTURE and BAPS) or three (Geneland) different clusters. Apart from very few exceptions, individuals from the Ndoki forest formed one and the remaining individuals the other subpopulation when two clusters were found. That pattern could be explained by the Sangha River which runs between the Ndoki forest and the other areas and might form a substantial although not absolute impediment for migration. The Sangha is around 1,300km long and up to hundreds of meters wide [Stauch, 1963], potentially forming a considerable natural barrier. Although elephants and duikers have been observed crossing the Sangha, WLGs avoid it and do not swim [A. Todd, personal communication]. As all analyses grouped the Ndoki forest into a distinct cluster the Sangha can be considered the greatest obstacle in the studied area. However, using simulations it has been argued that Geneland is more successful in detecting barriers than STRUCTURE and other similar software [Blair et al., 2012]. Analyzing the data with Geneland indeed revealed three instead of two clusters (Odzala, Lobéké and the Ndoki forest). Lobéké and Odzala are separated from each other by the Ngoko River which hence might also function as a barrier to gene flow, albeit probably to a lesser extent than the Sangha. The lesser effect of the Ngoko is also in agreement with its smaller dimensions of approximately 700km length and 120m width [Balarin, 1985]. The varied results obtained using different software highlight the importance of not relying on the output of a single program. Only the comparison of results from Geneland with results from STRUCTURE and BAPS gave us a comprehensive view: If we would have used only Geneland we would not have been able to infer that the Sangha River is a larger barrier to gene flow than the Ngoko. Conversely, using only STRUCTURE or BAPS would have resulted in not recognizing the effect of the Ngoko River at all.
Information about whether a population is continuous or not is also of key importance for conservation. In an action plan for the conservation of chimpanzees and gorillas in western equatorial Africa Tutin et al. [2005] defined seven areas of exceptional priority. One of them was the Sangha Trinational Complex comprising the Lobéké NP and the Ndoki forest and another one a complex including Odzala NP. As we found less evidence for a clustering between Lobéké and Odzala than between Lobéké and the Ndoki forest it might be more reasonable to group Lobéké and Odzala into one area of exceptional priority and the Ndoki forest into another. However, our results indicate that some individuals also migrated across the Sangha. Therefore one should generally be careful in defining distinct conservation units within our study site.
Might there be additional causes for the population structure we observe besides the occurrence of riverine barriers? The Pleistocene refuge theory [Haffer, 1969] posits that present-day continuous rain forests have repeatedly been fragmented due to the cold and arid climate occurring during glacial maxima. Fragmentation might have led to isolation of populations of taxa that are restricted to forests. As western gorillas (Gorilla gorilla) are one of such taxon and do not occur in savannah habitats [Tutin et al., 1997], their widespread present day occurrence could be the consequence of recolonization during periods of warmer climate. Examining western gorilla populations across their entire range, Anthony et al. [2007] found highly different mtDNA haplogroups with the boundaries of these haplogroups only rarely occurring along the courses of major rivers. However, both the Sangha and the Ngoko Rivers demarcated different haplogroups, consistent with these rivers acting as barriers to gene flow. They concluded that both rivers and the occurrence of Pleistocene refugia shaped present-day genetic variation in WLGs.
It is difficult to know the exact locations of putative Pleistocene refugia in west central Africa. Although we cannot rule out that the patterns we observe are partially due to sampling of populations corresponding to different Pleistocene refugia we suggest that our results strongly support the occurrence of a barrier effect on gene flow by major rivers in our study area. This is consistent not only with the results from the clustering analyses, but also with the correlations of genetic distance with geographic distances. We found significant increase of genetic distance both when correlated with straight line and detoured distance. However, the significantly lower AIC of the GLM that used detoured distance as predictor variable suggests that detoured distance better explains the variation in genetic distance than straight line distance. Considering our results in combination with those from Anthony et al. [2007], suggests that gorillas temporarily restricted to certain refugia began to admix when their formerly fragmented patches were reconnected. The admixture, however, seems to have been impeded by certain riverine barriers. The Sangha and to a lesser extent the Ngoko apparently constituted such barriers.
While the rivers within our study site have only a relatively subtle barrier effect, other watercourses have been found to act as borders between subspecies or even species of great apes. The most striking example comes from the genus Pan, consisting of two species, the common chimpanzee (P. troglodytes) and the bonobo (P. paniscus) which range on either side of the Congo River [Osman-Hill, 1969]. That river probably existed since well before the Pleistocene [Beadle, 1981] and possibly constituted an impermeable barrier to gene flow thereby facilitating allopatric speciation in the genus Pan. Rivers as barriers between subspecies can be found within the common chimpanzees. They consist of the four subspecies P.t.verus, P.t.ellioti, P.t.troglodytes and P.t.schweinfurthii [Schwarz, 1934; Gonder et al., 1997] which are separated from each other through the Dahomey Gap, the Sanaga River and the Ubangi River, respectively. These examples illustrate that gene flow of great ape populations is sometimes profoundly affected through the existence of rivers acting as obstacles to individual movement.
Patterns of Female and Male Population Structuring
Clustering analyses in Geneland revealed three clusters of WLGs, irrespective if all individuals, only females or only males were incorporated. Therefore our results do not support a quantitative sex-bias in migration. Observational studies report that both sexes disperse when reaching sexual maturity [Breuer et al., 2009; Robbins et al., 2004; Stokes et al., 2003]. However, it is much more difficult to use observational data for drawing conclusions about the distance that each sex travels at dispersal. Adult females do not range on their own and mostly appear to transfer directly between breeding groups, whereas most males live solitarily after leaving their natal group and before creating their own group [Breuer et al., 2010; Robbins et al., 2004]. It has been suggested that at least some males travel very long distances at this phase [Levréro, 2005; Douadi et al., 2007]. Indirect evidence for long-distance dispersal in WLG males also comes from a recent study [Inoue et al., 2013] which did not find kin relationships between neighboring silverbacks. Given these points one would expect to find females to be more philopatric and therefore also genetically more structured than males, as has been previously reported for WLGs and mountain gorillas [Douadi et al., 2007; Guschanski et al., 2008]. However, the study by Douadi et al. [2007] utilized sex-specific markers which are difficult to compare, represent single loci and might be biased by sex-specific differences in effective population size. Therefore, corroboration of their results using the same markers for both sexes is necessary for confirmation of their findings. Our study fills that gap and is the first that utilizes autosomal markers to look at sex-specific differences in WLG population clustering over a large scale. BAPS and STRUCTURE both detected possible migrants that originated from another cluster than the one where they were sampled. Three of these migrants were females. Also the individual that was sampled most far away from the cluster to which it was assigned was a female. Thus our results do not support previous findings of females dispersing less far than males. Instead, it seems that both sexes migrate over similar distances. Although females may only transfer to neighboring groups, multiple dispersal events can occur through adulthood [Stokes et al., 2003], which increases the total potential distance that a female may migrate. Such a pattern of dispersal resembles a stepping-stone model and can produce a pattern of isolation by distance (IBD). Our test of IBD by sex was significant for both females and males. This supports the previously proposed pattern of migration for females, but raises questions about the impact of any long-distance dispersal by males, because it means that males are more related to other males born nearby than to males born further away. This is in line with Bradley et al. [2004] who described dispersed male networks in WLGs in which silverbacks were usually related to nearby males. As that study, however, was conducted on a very small scale (9km×6 km) it could not assess whether some males might have migrated large distances. In sum, it seems that the pattern of genetic variation is quite similar in male and female WLGs, pointing to similar frequencies and extent of dispersal by males and females.
Acknowledgments
Contract grant sponsor: Max Planck Society; contract grant sponsor: National Science Foundation; contract grant number: SBR-9729126; contract grant sponsor: L.S.B. Leakey Foundation; contract grant sponsor: US Fish and Wildlife Services—Great Ape Conservation Fund; contract grant sponsor: Conservation International-Margot Marsh Biodiversity Fund; contract grant sponsor: University of Rome “La Sapienza”; contract grant sponsor: Arcus Foundation; contract grant sponsor: Paul G. Allen Family Foundation; contract grant sponsor: Dunemere Foundation; contract grant sponsor: Bradley L. Goldberg Foundation; contract grant sponsor: National Institutes of Health; contract grant number: R01 AI50529; contract grant sponsor: Agence Nationale de Recherches sur le SIDA, France; contract grant number: ANRS 12255.
For permission to carry out research in Central African Republic the authors thank the Ministries of Eaux et Forets and Recherches Scientifiques. For permission to carry out research in Republic of Congo the authors thank the Ministère de l’Économie Forestière et Dévelopement Durable; the Ministère de la Recherche Scientifique et de l’Innovation Technologique; and the Ministère de la Santé. For permission to collect samples in Cameroon the authors thank the Cameroonian Ministries of Health and Research. We thank the staff from PRESICA, the Wildlife Conservation Society’s Congo Program, Bai Hokou and the Dzanga-Sangha Project for crucial logistical and administrative support. The authors also thank African Parks Network for facilitation of the work. The authors thank A. Abraham for laboratory assistance, C. Stephens and R. Mundry for statistical assistance and F. Maisels for information regarding the Sangha and the Ngoko River. The authors thank three anonymous referees for helpful comments. The authors thank A. Bassouama, D. Mangola, R. Ngotto, Y. Sylvain, P. Gombo, F. Dodonou, J. Dzohi-Ebeni, S. Kaba, M.J. Akongo, E.B. Ngouembe, and G. Bounga for sample collection. Special thanks go to the local Ba’Aka trackers. The laboratory work was funded by the Max Planck Society and sample collection was funded by the National Science Foundation (D.M.D-S. SBR-9729126), L.S.B. Leakey Foundation (D.M.D-S.), US Fish and Wildlife Services -Great Ape Conservation Fund, Conservation International-Margot Marsh Biodiversity Fund, University of Rome “La Sapienza”, Arcus Foundation, Paul G. Allen Family Foundation, Dunemere Foundation, Bradley L. Goldberg Foundation, National Institutes of Health (R01 AI50529) and Agence Nationale de Recherches sur le SIDA, France (ANRS 12255). All work presented herein was conducted in compliance with appropriate animal care regulations and national laws.
References
- Anthony NM, Johnson-Bawe M, Jeffery K, et al. The role of Pleistocene refugia and rivers in shaping gorilla genetic diversity in central Africa. Proceedings of the National Academy Sciences of the United States of America. 2007;104:20432–20436. doi: 10.1073/pnas.0704816105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arandjelovic M, Guschanski K, Schubert G, et al. Two-step multiplex polymerase chain reaction improves the speed and accuracy of genotyping using DNA from noninvasive and museum samples. Molecular Ecology Resources. 2009;9:28–36. doi: 10.1111/j.1755-0998.2008.02387.x. [DOI] [PubMed] [Google Scholar]
- Arandjelovic M, Head J, Kühl H, et al. Effective noninvasive genetic monitoring of multiple wild western gorilla groups. Biological Conservation. 2010;143:1780–1791. [Google Scholar]
- Arora N, Nater A, van Schaik CP, et al. Effects of Pleistocene glaciations and rivers on the population structure of Bornean orangutans (Pongo pygmaeus) Proceedings of the National Academy Sciences of the United States of America. 2010;107:21376–21381. doi: 10.1073/pnas.1010169107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balarin JD. The status of fish farming in Zimbabwe and future prospects. Zimbabwe Agricultural Journal. 1985;83:3–9. [Google Scholar]
- Beadle LC. The inland waters of tropical Africa. London, United Kingdom: Longman; 1981. [Google Scholar]
- Blair C, Weigel DE, Balazik M, et al. A simulation-based evaluation of methods for inferring linear barriers to gene flow. Molecular Ecology Resources. 2012 doi: 10.1111/j.1755-0998.2012.03151.x. Available online at: http://onlinelibrary.wiley.com/doi/10.1111/j1755-0998.2012.03151.x/full. [DOI] [PubMed]
- Blanchong JA, Samuel MD, Scribner KT, et al. Landscape genetics and the spatial distribution of chronic wasting disease. Biology Letters. 2008;4:130–133. doi: 10.1098/rsbl.2007.0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley BJ, Chambers KE, Vigilant L. Accurate DNA-based sex identification of apes using non-invasive samples. Conservation Genetics. 2001;2:179–181. [Google Scholar]
- Bradley BJ, Doran-Sheehy DM, Lukas D, Boesch C, Vigilant L. Dispersed male networks in western gorillas. Current Biology. 2004;14:510–513. doi: 10.1016/j.cub.2004.02.062. [DOI] [PubMed] [Google Scholar]
- Brandt AL, Ishida Y, Georgiadis NJ, Roca AL. Forest elephant mitochondrial genomes reveal that elephantid diversification in Africa tracked climate transitions. Molecular Ecology. 2012;21:1175–1189. doi: 10.1111/j.1365-294X.2012.05461.x. [DOI] [PubMed] [Google Scholar]
- Breuer T, Hockemba MB-N, Olejniczak C, Parnell RJ, Stokes EJ. Physical maturation, life-history classes and age estimates of free-ranging western gorillas-insights from Mbeli Bai, Republic of Congo. American Journal of Primatology. 2009;71:106–119. doi: 10.1002/ajp.20628. [DOI] [PubMed] [Google Scholar]
- Breuer T, Robbins AM, Olejniczak C, et al. Variance in the male reproductive success of western gorillas: acquiring females is just the beginning. Behavioural Ecology and Sociobiology. 2010;64:515–528. [Google Scholar]
- Burrows W, Ryder OA. Y-chromosome variation in great apes. Nature. 1997;385:125–126. doi: 10.1038/385125a0. [DOI] [PubMed] [Google Scholar]
- Collins A, Dubach J. Biogeographic and ecological forces responsible for speciation in Ateles. International Journal of Primatology. 2000;21:421–444. [Google Scholar]
- Corander J, Marttinen P. Bayesian identification of admixture events using multilocus molecular markers. Molecular Ecology. 2006;15:2833–2843. doi: 10.1111/j.1365-294X.2006.02994.x. [DOI] [PubMed] [Google Scholar]
- Corander J, Sirén J, Arjas E. Bayesian spatial modeling of genetic population structure. Computation Statistics. 2007;23:111–129. [Google Scholar]
- Craul M, Radespiel U, Rasolofoson DW, et al. Large rivers do not always act as species barriers for Lepilemur sp. Primates. 2008;49:211–218. doi: 10.1007/s10329-008-0092-3. [DOI] [PubMed] [Google Scholar]
- De Thoisy B, da Silva A, Ruiz-García M, et al. Population history, phylogeography, and conservation genetics of the last Neotropical mega-herbivore, the lowland tapir (Tapirus terrestris) BMC Evolutionary Biology. 2010;10:278. doi: 10.1186/1471-2148-10-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douadi MI, Gatti S, Levrero F, et al. Sex-biased dispersal in western lowland gorillas (Gorilla gorilla gorilla) Molecular Ecology. 2007;16:2247–2259. doi: 10.1111/j.1365-294X.2007.03286.x. [DOI] [PubMed] [Google Scholar]
- Earl DA, vonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources. 2011;4:359–361. [Google Scholar]
- Eriksson J, Hohmann G, Boesch C, Vigilant L. Rivers influence the population genetic structure of bonobos (Pan paniscus) Molecular Ecology. 2004;13:3425–3435. doi: 10.1111/j.1365-294X.2004.02332.x. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti S, Levréro F, Ménard N, Gautier-Hion A. Population and group structure of western lowland gorillas (Gorilla gorilla gorilla) at Lookout, Republic of Congo. American Journal of Primatology. 2004;63:111–123. doi: 10.1002/ajp.20045. [DOI] [PubMed] [Google Scholar]
- Gonder MK, Oates JF, Distill TR, et al. A new west African chimpanzee subspecies? Nature. 1997;388:337. doi: 10.1038/41005. [DOI] [PubMed] [Google Scholar]
- Gonder MK, Distill TR, Oates JF. New genetic evidence on the evolution of chimpanzee populations and implications for taxonomy. International Journal of Primatology. 2006;27:1103–1127. [Google Scholar]
- Greenwood PJ. Mating systems, philopatry and dispersal in birds and mammals. Animal Behavior. 1980;28:1140–1162. [Google Scholar]
- Guillot G. A spatial statistical model for landscape genetics. Genetics. 2005;170:1261–1280. doi: 10.1534/genetics.104.033803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot G. Inference of structure in subdivided populations at low levels of genetic differentiation—the correlated allele frequencies model revisited. Bioinformatics. 2008;24:2222–2228. doi: 10.1093/bioinformatics/btn419. [DOI] [PubMed] [Google Scholar]
- Guillot G, Mortier F, Stoup A. GENELAND: a computer package for landscape genetics. Molecular Ecology Notes. 2005;5:712–715. [Google Scholar]
- Guillot G, Santos F, Stoup A. Analysing georeferenced population genetics data with Geneland: a new algorithm to deal with null alleles and a friendly graphical user interface. Bioinformatics. 2008;24:1406–1407. doi: 10.1093/bioinformatics/btn136. [DOI] [PubMed] [Google Scholar]
- Guo SW, Thompson EA. Performing exact test of Hardy–Weinberg proportion for multiple alleles. Biometrics. 1992;48:361–372. [PubMed] [Google Scholar]
- Guschanski K, Caillaud D, Robbins MM, Vigilant L. Females shape the genetic structure of a gorilla population. Current Biology. 2008;18:1809–1814. doi: 10.1016/j.cub.2008.10.031. [DOI] [PubMed] [Google Scholar]
- Haag T, Santos AS, Sana DA, et al. The effect of habitat fragmentation on the genetic structure of a top predator: loss of diversity and high differentiation among remnant populations of Atlantic Forest jaguars (Panthera onca) Molecular Ecology. 2010;19:4906–4921. doi: 10.1111/j.1365-294X.2010.04856.x. [DOI] [PubMed] [Google Scholar]
- Haffer J. Speciation in Amazonian forest birds. Science. 1969;165:131–137. doi: 10.1126/science.165.3889.131. [DOI] [PubMed] [Google Scholar]
- Hardy OJ, Vekemans X. SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molecular Ecology Notes. 2002;2:618–620. [Google Scholar]
- Hellborg L. Low levels of nucleotide diversity in mammalian y chromosomes. Molecular Biology and Evolution. 2003;21:158–163. doi: 10.1093/molbev/msh008. [DOI] [PubMed] [Google Scholar]
- Hewitt GM. Some genetic consequences of ice ages and their role in divergence and speciation. Biological Journal of the Linnean Society. 1996;58:247–276. [Google Scholar]
- Inoue E, Akomo-Okoue EF, Ando C, et al. Male genetic structure and paternity in western lowland gorillas (Gorilla gorilla gorilla) American Journal of Physical Anthropology. 2013;151:583–588. doi: 10.1002/ajpa.22312. [DOI] [PubMed] [Google Scholar]
- Jalil M, Cable J, Inyor J, et al. Riverine effects on mitochondrial structure of Bornean orang-utans (Pongo pygmaeus) at two spatial scales. Molecular Ecology. 2008;17:2898–2909. doi: 10.1111/j.1365-294X.2008.03793.x. [DOI] [PubMed] [Google Scholar]
- Kalinowski ST, Taper ML, Marshall TC. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Molecular Ecology. 2007;16:1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x. [DOI] [PubMed] [Google Scholar]
- Kawamoto Y, Takemoto H, Higuchi S, et al. Genetic structure of wild bonobo populations: diversity of mitochondrial DNA and geographical distribution. PLoS ONE. 2013;8:e59660. doi: 10.1371/journal.pone.0059660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson Handley LJ, Perrin N. Advances in our understanding of mammalian sex-biased dispersal. Molecular Ecology. 2007;16:1559–1578. doi: 10.1111/j.1365-294X.2006.03152.x. [DOI] [PubMed] [Google Scholar]
- Levréro F. PhD Thesis. France: Université de Rennes; 2005. Structure d’une population de gorilles (Gorilla gorilla gorilla) visitant une Clairière Forestière—nature et rôle des rencontres intergroupes dans sa dynamique; p. 1. [Google Scholar]
- Mabry KE, Shelley EL, Davis KE, Blumstein DT, Van Vuren DH. Social mating system and sex-biased dispersal in mammals and birds: a phylogenetic analysis. PLoS ONE. 2013;8:e57980. doi: 10.1371/journal.pone.0057980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Cruz B, Godoy JA, Negro JJ. Population fragmentation leads to spatial and temporal genetic structure in the endangered Spanish imperial eagle. Molecular Ecology. 2007;16:477–486. doi: 10.1111/j.1365-294X.2007.03147.x. [DOI] [PubMed] [Google Scholar]
- Morin PA, Chambers KE, Boesch C, Vigilant L. Quantitative polymerase chain reaction analysis of DNA from noninvasive samples for accurate microsatellite genotyping of wild chimpanzees (Pan troglodytes verus) Molecular Ecology. 2001;10:1835–1844. doi: 10.1046/j.0962-1083.2001.01308.x. [DOI] [PubMed] [Google Scholar]
- Neaves LE, Zenger KR, Prince RIT, Eldridge MDB, Cooper DW. Landscape discontinuities influence gene flow and genetic structure in a large, vagile Australian mammal, Macropus fuliginosus. Molecular Ecology. 2009;18:3363–3378. doi: 10.1111/j.1365-294X.2009.04293.x. [DOI] [PubMed] [Google Scholar]
- Nsubuga AM, Robbins MM, Roeder AD, et al. Factors affecting the amount of genomic DNA extracted from ape faeces and the identification of an improved sample storage method. Molecular Ecology. 2004;13:2089–2094. doi: 10.1111/j.1365-294X.2004.02207.x. [DOI] [PubMed] [Google Scholar]
- Osman-Hill WC. The nomenclature, taxonomy, and distribution of chimpanzees. In: Bourne GH, editor. The chimpanzee. Vol. 1. Basel, Switzerland: Karger; 1969. pp. 22–49. [Google Scholar]
- Parnell RJ. Group size and structure in western lowland gorillas (Gorilla gorilla gorilla) at Mbeli Bai, Republic of Congo. American Journal of Primatology. 2002;56:193–206. doi: 10.1002/ajp.1074. [DOI] [PubMed] [Google Scholar]
- Peakall ROD, Smouse PE. GENALEX 6: genetic analysis in excel. Population genetic software for teaching and research. Molecular Ecology Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R, Smouse P. GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research —an update. Bioinformatics. 2012 doi: 10.1093/bioinformatics/bts460. Available online at: http://bioinformatics.oxfordjournals.org/content/early/2012/07/20/bioinformatics.bts460.short. [DOI] [PMC free article] [PubMed]
- Pérez-Espona S, Pérez-Barbería FJ, Mcleod JE, et al. Landscape features affect gene flow of Scottish Highland red deer (Cervus elaphus) Molecular Ecology. 2008;17:981–996. doi: 10.1111/j.1365-294X.2007.03629.x. [DOI] [PubMed] [Google Scholar]
- Piertney SB, MacColl ADC, Bacon JP, Dallas JF. Local genetic structure in red grouse (Lagopus lagopus scoticus): evidence from microsatellite DNA markers. Molecular Ecology. 1998;7:1645–1654. doi: 10.1046/j.1365-294x.1998.00493.x. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. Journal of Heredity. 1995;86:248–249. [Google Scholar]
- Reding DM, Bronikowski AM, Johnson WE, Clark WR. Pleistocene and ecological effects on continental-scale genetic differentiation in the bobcat (Lynx rufus) Molecular Ecology. 2012;21:3078–3093. doi: 10.1111/j.1365-294X.2012.05595.x. [DOI] [PubMed] [Google Scholar]
- Robbins MM, Bermejo M, Cipolletta C, et al. Social structure and life-history patterns in western gorillas (Gorilla gorilla gorilla) American Journal of Primatology. 2004;64:145–159. doi: 10.1002/ajp.20069. [DOI] [PubMed] [Google Scholar]
- Roca AL, Georgiadis N, Pecon-Slattery J, O’Brien SJ. Genetic evidence for two species of elephant in Africa. Science. 2001;293:1473–1477. doi: 10.1126/science.1059936. [DOI] [PubMed] [Google Scholar]
- Ruell EW, Riley SPD, Douglas MR, et al. Urban habitat fragmentation and genetic population structure of bobcats in Coastal Southern California. American Midland Naturalist. 2012;168:265–280. [Google Scholar]
- Ruiter JD, Geffen E. Relatedness of matrilines, dispersing males and social groups in long-tailed macaques (Macaca fascicularis) Proceedings of the Royal Society B: Biological Sciences. 1998;265:79–87. doi: 10.1098/rspb.1998.0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz E. On the local races of chimpanzees. Annals and Magazine of Natural History, London. 1934;13:576–583. [Google Scholar]
- Stauch A. Contribution a l’etude de la peche dans le Cuvette Congolaise. Bulletin de l’ Institut des Recherches Scientifiques du Congo. 1963;2:49–85. [Google Scholar]
- Stoinski TS, Vecellio V, Ngaboyamahina T, et al. Proximate factors influencing dispersal decisions in male mountain gorillas, Gorilla beringei beringei. Animal Behavior. 2009;77:1155–1164. [Google Scholar]
- Stokes EJ, Parnell RJ, Olejniczak C. Female dispersal and reproductive success in wild western lowland gorillas (Gorilla gorilla gorilla) Behavioral Ecology and Sociobiology. 2003;54:329–339. [Google Scholar]
- Storfer A, Murphy MA, Spear SF, Holderegger R, Waits LP. Landscape genetics: where are we now? Molecular Ecology. 2010;19:3496–3514. doi: 10.1111/j.1365-294X.2010.04691.x. [DOI] [PubMed] [Google Scholar]
- Taberlet P, Griffin S, Goossens B, et al. Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Research. 1996;24:3189–3194. doi: 10.1093/nar/24.16.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutin CEG, White LJT, Mackanga-Missandzou A. The use by rain forest mammals of natural forest fragments in an equatorial African savanna. Conservation Biology. 1997;11:1190–1203. [Google Scholar]
- Tutin C, Stokes E, Boesch C, et al. Regional action plan for the conservation of chimpanzees and gorillas in western equatorial Africa. Washington, DC: IUCN/SSC Primate Specialist Group Conservation International; 2005. [Google Scholar]
- Waits LP, Luikart G, Taberlet P. Estimating the probability of identity among genotypes in natural populations: cautions and guidelines. Molecular Ecology. 2001;10:249–256. doi: 10.1046/j.1365-294x.2001.01185.x. [DOI] [PubMed] [Google Scholar]
- Weir BS. Genetic data analysis II. Sunderland, MA: Sinauer; 1996. [Google Scholar]