Abstract
Here, we demonstrate that sphere formation triggers immortalization and stable reprogramming of mouse fibroblasts. Cell contact signaling in spheres causes downregulation of the EMT transcription factor Zeb1 leading to rapid mesenchymal-to-epithelial transition. And, hypoxia within spheres together with loss of Zeb1 repression synergize to cause superinduction of Hif1a, which in turn leads to induction of the DNA demethylase Aid/Aicda, demethylation of the Oct4 promoter/enhancer and multipotency. Oct4 and Nanog expression diminish when cells are removed from the hypoxic environment of spheres and placed in monolayer culture, but the cells retain multipotential capacity, demonstrating stable reprogramming and a gene expression pattern resembling adult stem cells. Oct4 has been shown to induce Dnmt1 in mesenchymal stem cells, and we link Oct4 and Dnmt1 to silencing of cell cycle inhibitory cyclin dependent kinase inhibitors and Arf, and immortalization of the reprogrammed fibroblasts. Sphere formation then represents a novel and rapid protocol for immortalization and stable reprogramming of fibroblasts to multipotency that does not require exogenous expression of a stem cell factor or a lineage-specifying transcription factor.
INTRODUCTION
Fibroblasts can be reprogrammed by fusion to embryonic stem cells (ESC), nuclear transfer or forced expression of stem cell specification factors Oct4, Sox2, Klf4, and c-myc (1). For this reprogramming, inhibitory CpG methylation must be removed from the Oct4 promoter/enhancer. In induced pluripotent stem cell (iPS) reprogramming, exogenous Oct4 autoactivates the endogenous gene, and it is thought that CpG methylation is lost passively from the endogenous Oct4 promoter/enhancer as the reprogramming cells divide multiple times, thereby at least partially accounting for the protracted time required for iPS reprogramming (1, 2, 3). By contrast, when fibroblasts are fused to ESC, demethylation of the Oct4 promoter/enhancer occurs rapidly in the fibroblast nucleus in the absence of DNA replication (4). These results demonstrate an active demethylation pathway in ESC, and thus suggest a fundamentally different mechanism for Oct4 promoter demethylation in nuclear transfer and cell fusion compared to iPS reprogramming. Surprisingly, activation-induced cytidine deaminase (Aid or Acida), which classically mediates antibody class-switching, has recently been shown to be involved in DNA demethylation. Some 5-methylcytosines become converted to 5-hydroxymethylcytosine by 5-methylcytosine hydroxylase (TET), but deamination by Aid converts both 5-methylcytosine to thymidine and 5-hydroxymethylcytosine to 5-hydroxymethyluracil. This change in base then signals base excision-repair by a complex containing thymidine DNA glycosylase and Gadd45a, which replaces the mismatched thymidine or 5-hydroxymethyl uracil with an unmethylated cytosine (2, 5, 6, 7). Aid localizes to the Oct4 promoter in fibroblast nuclei following fusion to ESC, and knockdown of Aid prevented demethylation of the promoter thereby blocking reprogramming (4), implying that this active demethylation pathway is important in establishing/maintaining Oct4 demethylation in vivo.
There has been debate as to whether Oct4 and its target gene Nanog have a functional role in adult stem cells. However, recent studies show expression of both Oct4 and Nanog in freshly isolated mesenchymal stem cells (MSC), and demonstrate that their expression is lost when the cells are placed in culture (8). Knockdown of Oct4 and Nanog led to decreased differentiation capacity and rapid senescence, and overexpression maintained the MSC in an undifferentiated state and led to immortalization. MSC reside in hypoxic niches in vivo, and it was shown that expression Oct4 and Nanog in MSC is dependent upon hypoxia, thereby accounting for their loss of expression in culture. Induction of cyclin dependent kinase (cdk) inhibitors and the p53 regulatory protein Arf triggers cell cycle arrest and senescence, and is a major barrier to iPS reprogramming (9). An important and novel function of Oct4 and Nanog in MSC was shown to be induction of Dnmt1, which led to CpG methylation and thus stable silencing of promoters for p21 and Ink4 (encodes both p16 and Arf), allowing for immortalization of the cells in culture (8).
Early embryogenesis occurs under hypoxic conditions and induction of hypoxia inducible factor (Hif) has been shown to be critical for expression of Oct4 in the early embryo and in primordial germ cells (10, 11). Likewise, hypoxia and Hif induce Oct4 in cancer stem-like cells, which also reside in hypoxic niches (12, 13). As noted above, Oct4 expression in MSC also depends upon hypoxia (8). By contrast, iPS cells and ESC do not require hypoxia to maintain Oct4 and Nanog expression in culture, but hypoxia has been shown to augment iPS reprogramming (14).
Interestingly, iPS reprogramming of fibroblasts requires early mesenchymal-to-epithelial transition (MET), which is driven by downregulation of transcription factors such as Zeb1 that cause the opposing epithelial-to-mesenchymal transition (EMT) (15). Upon cell-cell contact, activation of the Hippo pathway causes the closely related Yap1 and Taz Hippo pathway transcription factors to bind Angiomotin (Amot) within tight junction complexes leading to their sequestration in the cytoplasm (16, 17). Nuclear translocation of Taz is required for induction of EMT factors including Zeb1, and it thereby drives a mesenchymal phenotype (16). But, it is unclear whether cytoplasmic retention of Yap1/Taz in tightly packed colonies arising during reprogramming might initiate MET by causing downregulation of EMT factors.
As noted above, cdk inhibitors and Arf inhibit reprogramming. And, gradual induction of these genes by serum growth factor activation of Ras signaling is responsible for senescence of fibroblasts in culture (17). Likewise, cdk inhibitors and Arf are also important in establishing cell cycle arrest initiated by cell-cell contact (17, 19). Consistent with silencing of cdk inhibitors and Arf during iPS reprogramming, as noted above, reprogrammed iPS cells are immortal in culture and they are not contact-inhibited. Although Oct4 induces Dnmt1 in MSC causing CpG methylation and stable silencing of cdk inhibitors such as p21 and the Ink4 locus, it is unclear if Dnmt1 has a corresponding role in silencing these genes during iPS reprogramming (3).
A hallmark of adult stem cells as well as ESC and iPS is their ability to form spheres, which survive in suspension culture. Beyond simply survival, sphere formation is thought to mimic cell-cell contacts and hypoxic conditions in the early embryo and in solid tumor outgrowth. Such embryoid body formation signals changes in gene expression leading to generation of multiple lineages from ESC and iPS, but surprisingly it also may contribute to reprogramming of mutant somatic cells to cells with properties of stem cells and cancer stem cells (20, 21). Here, we forced mouse fibroblasts to form spheres in suspension culture. Sphere formation resulted in rapid loss of Zeb1, which we link to cell contact inhibition and cytoplasmic retention of Taz. And, this loss of Zeb1 was associated with rapid MET. Subsequently, hypoxia in the interior of the spheres and loss of Zeb1 repression synergized to cause superinduction of Hif1a and in turn induction of Aid, Oct4, and Dnmt1. Dissociated spheres were sorted to isolate Oct4+ cells, and even though Oct4 and Nanog expression was reduced as these cells were passaged in culture, cdk inhibitors and Arf repression was maintained and the cells were immortal in culture allowing for their unlimited expansion. Importantly, the cells were stably reprogrammed to multipotency and they gave rise to cells representative of all three embryonic lineages when exposed to standard differentiation conditions. These results demonstrate a rapid protocol for immortalization and stable reprogramming of fibroblasts to multipotency that does not require forced expression of stemness genes or lineage-specifying transcription factors.
MATERIALS AND METHODS
Cells and cell culture
Mouse adult fibroblasts (MAFs) and mouse embryonic fibroblasts (MEFs) were isolated as described (20, 22). Cells were cultured in DMEM with 10% heat-inactivated fetal bovine serum. Mouse ESC (W9.5) (23) and sphere-derived colonies cells were maintained in DMEM medium containing 20% fetal bovine serum (FBS, HyClone), and 1000 units/ml leukemia inhibition factor (Lif, Chemicon) on Matrigel (BD Biosciences) or an irradiated fibroblast feeder as described (20). To induce hypoxia, cells under normoxia (21% O2, 5% CO2) were placed in a sealed container in 3% O2, 5% CO2 at 37°C for 3 days.
Sphere formation and cell sorting
For sphere formation, monolayer fibroblasts were dissociated to single cells by pipetting with a P200 pipetman, and transferred to culture dishes that do not permit adhesion. The cells rapidly associated in suspension to form small spheres. Spheres were allowed to attach to Matrigel-coated plates, and all cells in the spheres migrated onto the plate to form a monolayer. Monolayer cells were trypsinized and sorted using a MoFlo cell sorter for Abcg2, Hoechst dye exclusion and a diameter ≤ 10 μm as we described previously (20).
Cell differentiation
See Supplemental Methods.
Knockdown of Zeb1 and Taz
Lentiviral shRNA knockdown of Zeb1 and Taz was described previously (20). Also see Supplemental Fig. 1.
ChIP assays
ChIP assays were performed as we have described previously (20). Primers and antibodies are shown in Supplemental Tables 1 and 2.
Immunohistochemistry
Slides were immunostained as described previously (20). Primary and secondary antibodies are described in Supplemental Table 1.
Detection of hypoxia in spheres
After three days in suspension culture, spheres were transferred to a gelatin-coated chamber slide, treated with 200 μM of pimonidazole HCl (EF5) for 2 hrs, fixed with 4% paraffinaldehyde and immunostained with Hypoxyprobe™-1, an antibody to EF5. See www.hypoxyprobe.com for further details.
Real-time PCR
Real time PCR was described previously (20). Briefly, the 2(-Delta Delta C(T)) method was used (24). Fold change of a testing gene = testing gene(POWER(2,(Ct(Control)−Ct(testing sample))))/normalizer gene(POWER(2,(Ct(Control)−Ct(testing sample)))). The normalizer gene for our studies was β-actin (Actb). PCR primers are shown in Supplemental Table 2. PCR primers used to detect Oct4 mRNA were designed so they do not recognize Oct4 pseudogenes http://www.pseudogene.org/site_map.html. A mouse stem cell differentiation real time PCR Array was also analyzed (SuperArray) as described (20). At least three independent samples, each in triplicate, were analyzed for each real time PCR condition. All PCR products were analyzed for size by electrophoresis.
Promoter methylation analysis
Bisulfite sequencing was performed as described previously (25). PCR primers matching the bisulfite-converted sequence were used to amplify the 5′ end, promoter and proximal enhancer region of the Oct4 gene (26), and amplified sequences were cloned into the TOPO TA cloning vector (Invitrogen). Inserts were sequenced using M13 forward and reverse primers. For methylation analysis of p21 and Ink4 CpG islands, DNA was treated with bisulfite (25) and methylation-sensitive PCR primers were designed using the MethPrimer program (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi. See Supplemental table 2 for primers.
Affymetrix microarray analyses
See Supplemental Methods.
RESULTS
Fibroblasts efficiently form spheres where cells are resistant to anoikis
It has been demonstrated previously that sphere formation in conjunction with Rb1 family mutation and Ras mutation can lead to induction of Oct4 and Nanog and reprogramming to cancer stem-like cells (20, 21). We then wondered if sphere formation might provide a reprogramming signal in wild-type cells. To begin testing this hypothesis, we assessed the ability of primary cultures of mouse embryo fibroblasts (MEFs) and mouse adult fibroblasts (MAFs) to form spheres in suspension culture. MAFs at passage four in monolayer culture were pipetted to single cells and placed in suspension culture in dishes that do not permit cell adhesion. The cells rapidly and efficiently formed small spheres (Fig. 1A), and cells in these spheres remained viable in suspension for more than one month (Fig. 1B). Cells that did not form spheres underwent anchorage-dependent cell death (anoikis), losing viability after 24 hr—trypsinizing cells from monolayers prevented sphere formation and the cells also underwent anoikis (Fig. 1B inset panel). Similar results were seen with MEFs, demonstrating that primary fibroblasts readily form spheres that remain viable in suspension culture.
Figure 1.
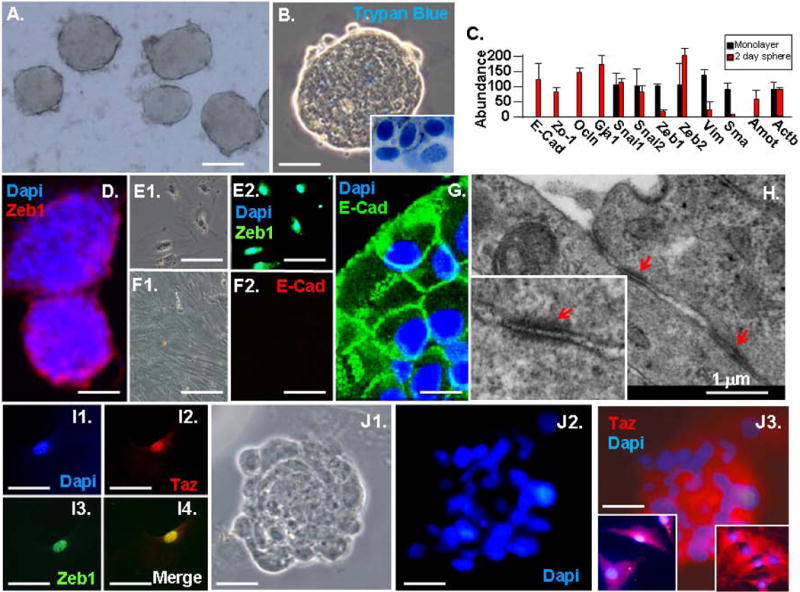
Sphere formation protects fibroblasts from anoikis, and it triggers cytoplasmic retention of Taz, loss of Zeb1 and rapid MET. A. MAF spheres after three days in suspension. B. Trypan blue exclusion demonstrates viability of cells in spheres. Trypan blue+ cells in the insert did not form spheres. C. Real time PCR showing repression of Zeb1 and mesenchymal mRNAs, and induction of epithelial specification mRNAs in spheres. D. Zeb1 becomes restricted to cells loosely attached to the exterior of the spheres. E. Monolayer fibroblast show nuclear expression of Zeb1. F. Monolayer fibroblasts do not express E-cadherin (E-Cad). G. Immunostaining of a section of a three-day-old sphere for E-Cad. H. EM of three-day-old spheres. Arrows indicate tight junctions. I. Zeb1 and Taz colocalize in the nucleus of fibroblasts in monolayer culture. J. Taz becomes sequestered in the cytoplasm in three-day-old spheres. The inset on the right shows a higher power view of cells within the sphere, and the inset on the left shows cells mechanically dislodged from sphere by pipetting. The scale bar represents 200 μm in panel A; 100 μm in panels B, D and J; 30 μm in panels E and I; 40 μm in panel F; 20 μm in panel G.
Sphere formation leads to rapid MET
Mesenchymal-to-epithelial transition (MET) is a critical early event in reprogramming of fibroblasts (15). After three days in suspension culture, real time PCR showed induction of epithelial specification genes and downregulation of mesenchymal genes, demonstrating MET in the spheres (Fig. 1C). mRNAs for the EMT transcription factors Snai1, Snai2 and ZEB2 were not downregulated in spheres, but Zeb1 was significantly downregulated (Fig. 1C). β-actin (Actb) mRNA was used as a control. We have demonstrated previously that mutation of Zeb1 in fibroblasts is sufficient for induction of epithelial specification genes including E-cadherin, and for epithelial morphology, and that Zeb1 is important in maintaining epithelial-mesenchymal balance in vivo (27). Consistent with mRNA levels, Zeb1 was expressed uniformly in the nucleus of monolayer fibroblasts, but its expression was lost in the spheres—it was restricted to cells loosely associated with the spheres (Fig. 1D and E). By contrast, E-cadherin was not expressed on monolayer fibroblasts, but it was evident on cells displaying epithelial morphology in the spheres (Fig. 1F and G). Beyond induction of E-cadherin and transition to epithelial morphology, remarkably electron microscopy (EM) showed that cells in the spheres had assembled tight junctions between the epithelial-like cells (Fig. 1H).
Translocation of the Hippo cascade transcription factor Taz to the nucleus induces expression of Zeb1 (16, 28). And, knockdown of Taz in MEFs led to downregulation of Zeb1, demonstrating that Zeb1 expression is dependent upon Taz in fibroblast monolayer culture (Supplemental Fig. 1). We found that Taz co-localized with Zeb1 in the nucleus of monolayer fibroblasts (Fig. 1I), but it was retained in the cytoplasm of cells in spheres, where Zeb1 was downregulated (Fig. 1J). Consistent with this cytoplasmic retention of Taz in the spheres, the Taz-binding factor in tight junctions, Amot, was induced in the spheres (Fig. 1C). Taken together, these results suggest that cytoplasmic retention of Taz in spheres is responsible for downregulation of Zeb1and in turn MET in the spheres.
Induction of Oct4, Nanog and Oct4-EGFP in spheres
Sections of spheres after three weeks in suspension culture were immunostained for Oct4 and its target gene product Nanog to determine whether these stem cell factors were induced. As expected, no immunostaining was evident in monolayer cultures of MAFs or MEFs prior to sphere formation (Fig. 2A and C), but nuclear immunostaining for Oct4 and Nanog was evident in approximately 5% of the cells in the spheres (Fig. 2B, D and O). We also isolated tail tip fibroblasts at birth from Oct4-EGFP mice, where EGFP is knocked into the Oct4 locus (29). These mice then express EGFP under control of the endogenous Oct4 promoter. No EGFP was evident in fibroblasts in monolayer culture, but EGFP was induced in spheres after two weeks in suspension culture—EGFP expression was confined to cells in the inner region of the spheres and was excluded from peripheral cells that expressed Zeb1 (Fig. 2F). This pattern of Oct4-EGFP matched that of Oct4 within the spheres (Fig. 2G). These results demonstrate that stem cell factors are induced in the interior of the spheres.
Figure 2.
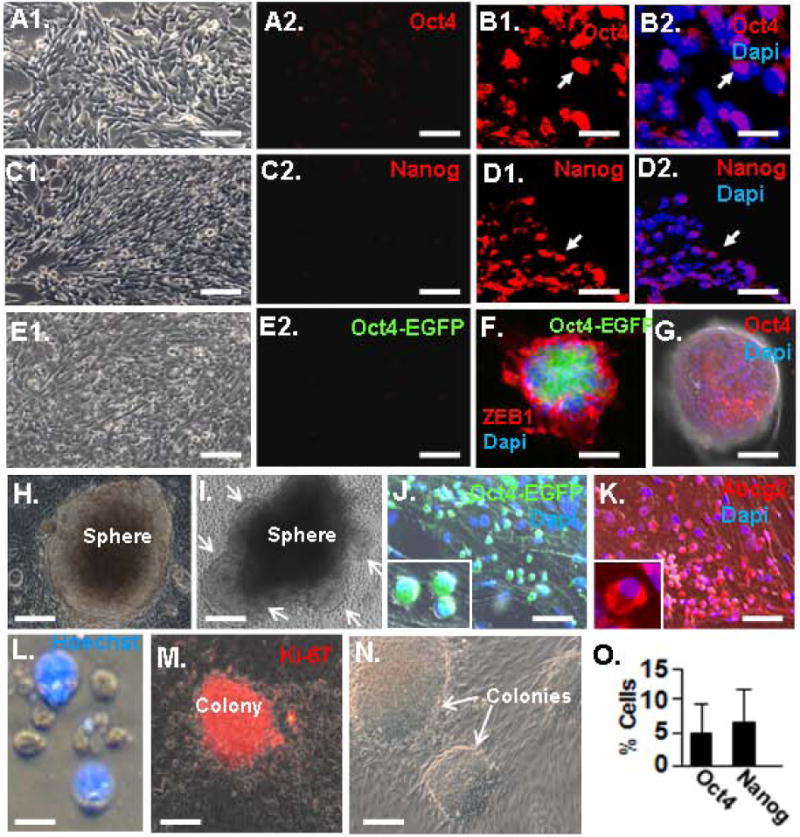
Sphere-induced induction of Oct4 and Nanog. A. Immunostaining for Oct4 in MEF monolayer culture. A1 is a phase image. B. Three-week-old spheres were sectioned and immunostained for Oct4. Note nuclear Oct4 expression. Arrows indicate the same position in the panels. C. Monolayer cultures of MEFs immunostained for Nanog. C1 is a phase image. D. Immunostaining of sphere section for Nanog as in panel B. Note nuclear Nanog expression. E. No EGFP expression is evident in monolayer cultures of tail tip fibroblast isolated at birth from Oct4-EGFP mice. E1 is the phase image. F. Sphere fromed from Oct4-EGFP fibroblast after two weeks in suspension culture immunostained for Zeb1 and showing EGFP immunofluorescence. G. Similar sphere to that in panel G immunostained for Oct4. H. Three-week-old sphere bound to a Matrigel-coated plate. I. Sphere similar to that in panel H, three days after adhesion. Arrows show cell migrating out of the sphere. J. Small, round and poorly adherent Oct4-EGFP+ cells evident at the site of sphere dissociation, one week after sphere adhesion to Matrigel. K. Similar cells to those in panel J immunostained for Abcg2. L. Cell rinsed from plates in panels K and J are enriched in a population of small, Hoechst- cells. M. Cells from panel L from colonies that immunostain for the proliferation marker Ki-67. N. Phase image of colonies. O. Quantification of Oct4+ and Nanog+ cells in three-week-old spheres. The bar is 10 μm in panels B and D, 15 μm in panel L, 20 μm in panels A, C, E, J, K, M an N, and 100 μm in panels F and G.
Isolation of Oct4+ cells from spheres as a side population
We attempted to dissociate spheres to isolate and characterize the Oct4+ cells. However, we were unable to efficiently dissociate the spheres enzymatically or mechanically without causing a loss in cell viability. Spheres were then allowed to adhere to Matrigel-coated plates (Fig. 2H), and we found that cells migrated from spheres onto the plate to form a monolayer (Fig. 2I). Among these sphere-derived cells were small poorly adherent cells that expressed Oct4-EGFP and the Abc transporter Abcg2 (Fig. 2J and K). These cells could be dislodged from culture plates by gently tapping the plate and rinsing with media, and consistent with their expression of the Abcg2 transporter (which pumps Hoechst dye out of cells), the small sphere-derived cells rinsed from the plate were enriched in cells that excluded Hoechst dye (Fig. 2L). The cells could be replated on Matrigel coated dishes or on irradiated feeder layers, where they proliferated to form colonies (Fig. 2M and N).
We sorted the sphere-derived population for Hoechst dye exclusion and size (≤10 μm), which has been used with both stem cells and tumor cells to separate a side population (SP) of small Hoescht− cells from a major population (MP) of lager Hoechst+ cells (Fig. 3A), as we have described (20). The SP cells showed uniform nuclear staining for Oct4 (Fig. 3B). EM of these SP cells showed that they are small, contain relatively uncondensed chromatin, and have a high nuclear-to-cytoplasmic ratio (Fig. 3C). Similar cells can be seen in EM of spheres prior to dissociation (Fig. 3D).
Figure 3.
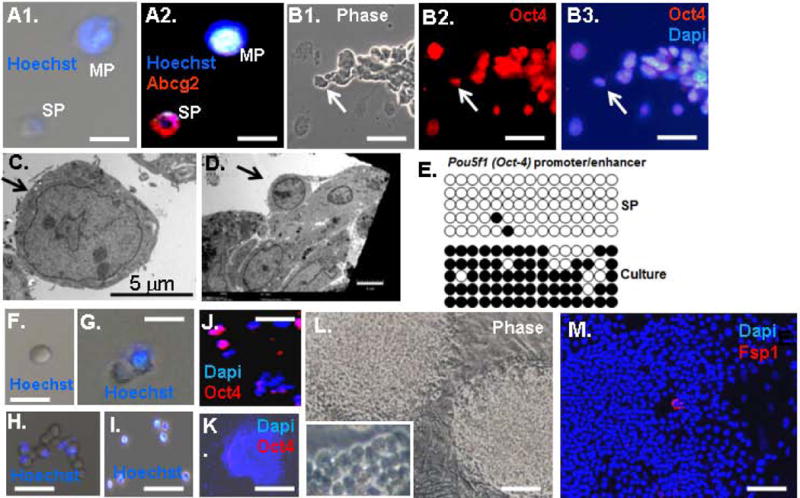
Isolation of a side population of sphere-derived cells that have demethylated the Oct4 promoter/enhancer and express Oct4. A. Cells from dissociated spheres are comprised of a side population (SP) of small cells that exclude Hoechst dye and a major population (MP) of larger cells that retain Hoechst dye. B. Sphere-derived cells were sorted for Hoechst dye exclusion and small size (<10 μm) (Materials and Methods), and the sorted cells were immunostained for Oct4. Arrows indicate the same position in the panels. C. EM of cells in panel B. D. EM of a sphere showing a cell (arrow) resembling the sorted SP cells in panel C. E. Methylation analysis of the Oct4 promoter/enhancer (Materials and methods). Circles indicate known CpG methylation sites, filled circles are methylated and open circles are unmethylated. “SP” indicates sorted cells from panel B, and “culture” indicates colonies of SP cells at passage (P)2. F–I. Phase images of sorted SP cells placed in culture. Panel F shows a Hoechst- cell after sorting. Panel G shows this same cell the next day when it has given rise to a Hoechst+ cell. By day 4, Panel H shows that the cell has given rise to a mixture of Hoechst+ and Hoechst− cells. In Panel I, after one week most of the cells are Hoechst+. J. Sorted SP cells from Panel B immunostained for Oct4 after 4 days in culture. K. A colony of SP cells immunostained for Oct4. L. Phase image of SP colonies. M. SP colony immunostained for Fsp1. The bar is 15 μm in Panels A, B, F and G, 25 μm in Panel H, 40 μm in Panels I, L and M, and 50 μm in Panel K.
Sphere-derived SP cells have demethylated the Oct4 promoter/enhancer, but it is remethylated when cells are placed in culture
Oct4 is silenced by CpG methylation of the promoter/enhancer, and this methylation must be removed for its transcription and stable reporgramming. The Oct4 promoter/enhancer region in the SP cells was amplified by PCR, and bisulfite sequencing was done to assess CpG methylation in 5 independent clones, as described (25). Consistent with expression of Oct4 and activation of the Oct4 promoter in Oct4-EGFP, the promoter/enhancer was demethylated in the sphere-derived SP cells (Fig. 3E).
Next, we placed the isolated SP cells in culture. As the cells proliferated to form colonies, they lost exclusion of Hoechst dye and Oct4 expression (Fig. 3F–L), and consistently the Oct4 promoter/enhancer was remethylated (Fig. 3E). A similar loss of Oct4 expression and Hoechst exclusion was also seen in colonies that formed directly from dissociated spheres and cells rinsed from plates. Taken together, these results suggest that sphere formation is causing reprogramming of fibroblast to a SP where the Oct4 promoter/enhancer is somehow demethylated and Oct4 is induced. But, methylation is restored and Oct4 expression is diminished when the cells are removed from the environment of the sphere and placed in monolayer culture. However, cells in these colonies continued to be very small and rounded and thus they did not resemble the parent fibroblasts, and few cells expressed the fibroblast marker Fsp1 (Fig. 3M).
Sphere-derived SP colonies express markers of adult stem cells
Microarrays were used to compare gene expression profiles in three sphere-derived colonies, and a very similar pattern was evident (Fig. 4A). Such results suggest that proliferating cells arising from the spheres represent a relatively uniform population. Gene expression in these sphere-derived colonies was then compared to parent fibroblast monolayers prior to sphere formation and to ESC. And, these results were then compared to gene expression profiling data published for mesenchymal and neural stem cells (Fig. 4A). The sphere-derived colonies were distinct from the other populations, but they were more closely related to mesenchymal stem cells in overall gene expression than either the parent fibroblasts or ESC (Supplemental Fig. 2).
Figure 4.
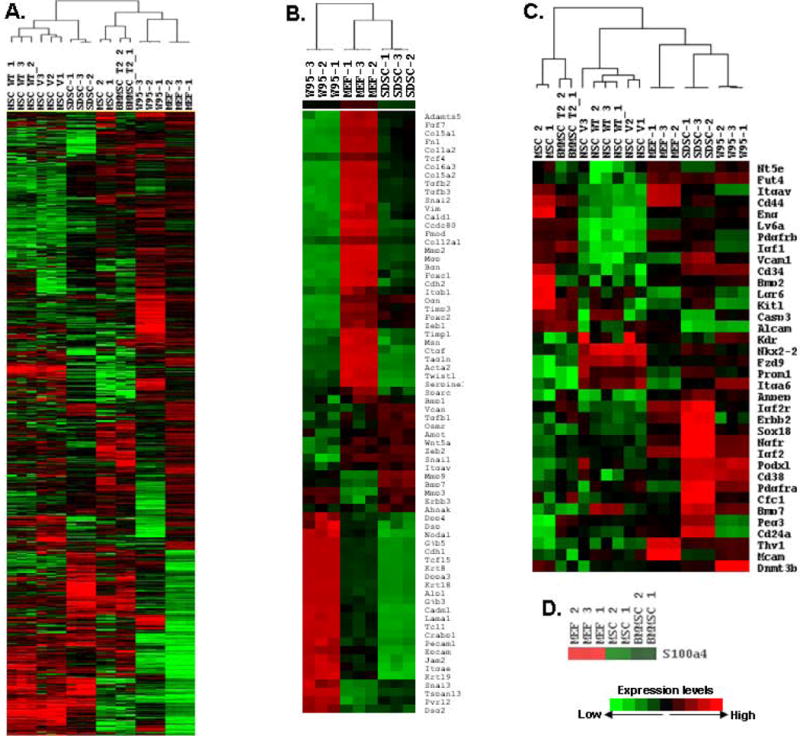
Sphere-derived colonies resemble progenitors in their gene expression. Microarrays of three independent colonies of SP sphere-derived stem-like cells (SDSC) were compared to monolayer fibroblasts prior to sphere formation, mouse ESC (W9.5) and published results for mesenchymal stem cells (MSC), neural stem cells (NSC) and bone marrow-derived mesenchymal stem cells (BMMSC) (Materials and Methods; Supplemental Methods and Supplemental Fig. 2). A. Heat map showing that the overall pattern of gene expression in sphere-derived stem cell-like colonies is most similar to MSC, but the pattern is clearly distinct from MSC. B. Heat map comparing expression of mesenchymal and epithelial genes. C. Heat map comparing expression of specific markers seen in adult stem cells, MSC, NSC and endothelial progenitors. Note that the sphere-derived stem-like cells are closest to the ESC in expression of these genes. D. Heat map showing lack of Fsp1/S100a4 expression in MSC and BMMSC compared to MEFs.
Few markers of pluripotency were induced in the sphere-derived colonies, and although many mesenchymal markers were downregulated compared to MEFs, the induction of epithelial genes seen early within the spheres was no longer evident in the colonies (Fig. 4B). Accordingly, Zeb1 was induced in the colonies compared to spheres, but its level was still below that in MEFs (Figs. 4B and 5B).
Figure 5.
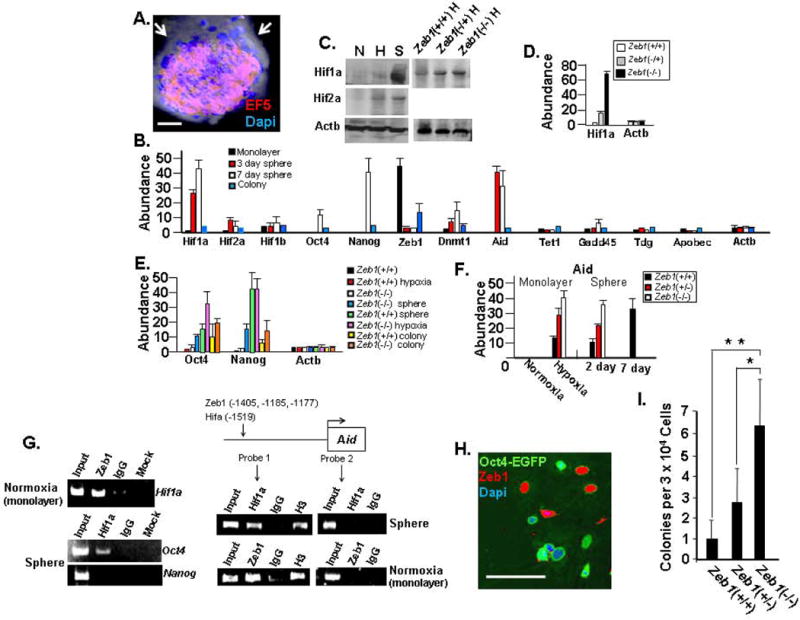
Interplay between Hif1a and Zeb1 regulates Aid, Oct4 and Dnmt1. A. After three days in suspension culture, MAF spheres were treated with the hypoxia sensor EF5 and immunostained (Material and methods). Note the hypoxic interior of the sphere. B. Real time PCR comparing mRNA levels in spheres and colonies of sphere-derived SP cells. C. Western blot showing the effects of sphere formation, hypoxia and Zeb1 on Hif expression. “N” indicates normoxic monalayer culture, “H” Hypoxic monolayer culture, and “S” three-day-old sphere. D. Real time PCR showing the effect of Zeb1 mutation on Hif1a mRNA in MEFs. E. Real time PCR showing expression of Oct4 and Nanog mRNA. F. Real time PCR showing the effects of hypoxia, sphere formation and Zeb1 on Aid mRNA expression. G. ChIP assays showing binding of Zeb1 and Hif1a to promoters. The location of Zeb1and Hif binding sites in the Aid promoter is shown along with the location of the two probes used in the assays. “IgG” indicates control normal serum and “Mock” indicates no antibodies. H. Co-immunostaining of cells dissociated spheres after adhesion to Matrigel for Oct4 and Zeb1. I. The three indicated MEF genotypes at P1 were used for spheres formation, and then after 7 days in suspension spheres were allowed to adhere to Matrigel and colony formation was assessed two weeks later. The 30,000 cells used for the assays gave rise to an average of 8.5±3.6 spheres. The bar is 50 μm in Panels A, 20 μm in Panel H.
We then examined the sphere-derived colonies for expression of adult stem cell markers (Fig. 4C). Podxl and Peg3/Pw1, which have been described as stem cell markers (30), were induced. Ngfr and Nkx2-2 which are evident in neural stem cells and are important for neural differentiation (Nkx2-2 is also critical for pancreatic β-cell specification) (31, 32) were elevated, as was Lgr6, a marker of epithelial and hair follicle stem cells (33). Another neural stem cell marker linked to tumor initiating cells in a variety of tumor types, Prominin/CD133 (34), was also elevated. Erbb2 (Her-2) was elevated—it is classically induced in breast cancer and in breast stem cells, it also expressed on neural stem cells where it is important for glial differentiation, and it is required for specification of cardiac progenitors (35). Cfc-1, a Cripto-related gene important for cardiac progenitor formation and evident in a variety of stem cells (36), was elevated. Pdgfrb, which is present on adult stem cells and is required for cardiac and hematopoietic differentiation (37), was elevated. BMP-7 was also elevated, and it is required in MSC for specification of cartilage differentiation (36). Igf-1, Igf2, Igf2r and Igf2bp2 were elevated. Igf signaling is important for specification and differentiation of muscle satellite stem cells (38). Interestingly, we also noted induction of genes expressed in endothelial progenitors and required for endothelial cell specification including Vcam-1, Sox18, and CD13 (Anpep) (39). We concluded from this expression pattern that the sphere-derived colonies do not display a pluripotent gene expression profile, but they do express a number of markers of adult stem cells and progenitors.
Hypoxia and loss of Zeb1 repression in spheres synergize to superinduce Hif1a, which in turn binds the Oct4 promoter
We initiated experiments to begin examining the pathway leading to reprogramming of fibroblasts in spheres. Hif is important for induction of Oct4 early in embryogenesis and in MSC (8, 15), and we found an outside-to-inside gradient of hypoxia in the spheres using the substituted 2-nitroimidazole EF5 (Materials and Methods), which forms adducts with proteins under hypoxic conditions that can be detected by immunostaining (Fig. 5A). We compared mRNA levels of Hif1a, Hif2a and Hif1b in fibroblast monolayers and spheres by real time PCR. Sphere formation led to a dramatic increase in Hif1a mRNA in 3-day-old spheres and a further increase was evident in 7-day-old spheres (Fig. 5B). A more modest effect was seen on Hif2a, and there was little change in Hif1b (Fig. 5B). Although hypoxia-induced stabilization is required for Hif protein expression, it has been demonstrated that transcriptional activation of Hif1a by NFκB is essential along with hypoxic stabilization to reach a functional level of Hif1a expression in hypoxic brain and liver (40). Indeed, we found by Western blot that Hif1a was induced in spheres to a level well beyond that seen simply in hypoxic monolayer culture (Fig. 5C), suggesting that transcriptional activation in the spheres synergizes with hypoxic stabilization to cause superinduction of Hif1a. Hif1a was diminished in colonies derived from dissociated spheres, which are maintained in monolayer culture under normoxic conditions (Fig. 5B). Hypoxia and Hif are required for Oct4 expression during embryogenesis and in primordial germ cells, and we noted that Oct4 and Oct4-EGFP were induced specifically in the hypoxic interior of spheres (Fig. 2F and G), but Oct4 and Nanog diminished along with Hif1a when sphere-derived SP cells were placed in monolayer culture under normoxic conditions (Fig. 5B). PCR primers that do not recognize Oct4 pseudogenes were used for these real time PCR assays (Supplemental Table 1). Oct4 was further induced when spheres were placed in hypoxic conditions (Fig. 5E). Consistent with Hif having a role in induction of Oct4, we found using chromatin immunoprecipitation (ChIP) assays that it was bound to the Oct4 promoter under hypoxic conditions (Fig. 5G).
Our results provide evidence that, as with NFκB induction of Hif1a mRNA in the liver and brain, induction of Hif1a mRNA in spheres is important for high level protein expression following hypoxic stabilization. Thus, we wondered why Hif mRNA was induced in spheres. We noted that Hif1a expression was inversely related to that of Zeb1—while Zeb1 was repressed in spheres and reexpressed in sphere-derived colonies in culture, Hif1a showed the opposite pattern (Fig. 5B). We then compared Hif1a mRNA levels in Zeb1(+/+), Zeb1(+/−) and Zeb1(−/−) MEFs, and found that Hif1a mRNA was induced in a concentration-dependent fashion with loss of Zeb1 (Fig. 5D). Inspection of the Hif1a promoter revealed a number of Zeb1 consensus E-box binding sites, and Zeb1 was bound to the Hif1a promoter in vivo in ChIP assays (Fig. 5G). Together, these results suggest that Hif1a is repressed by Zeb1 and downregulation of Zeb1 in spheres together with the hypoxic conditions in the interior of the spheres synergize to cause superinduction of Hif1a, which in turn binds the Oct4 promoter. Consistent with this pathway, we found that expression of Zeb1 and Oct4 was mutually exclusive in cells that dissociate from spheres (Fig. 5H). We then reasoned that placing Zeb1(−/−) MEFs under hypoxic condition in monolayer culture would lead to similar induction of Hif1a seen in spheres. Although Hif1a was further induced in Zeb1(−/−) MEFs compared to Zeb1(+/+) MEFs under hypoxic conditions, the level of Hif1a was still well below that seen in spheres (Fig. 5C). And, the level of Oct4 induction seen in hypoxic Zeb1(−/−) MEF monolayers also did not rise to the level seen in spheres (Fig. 5E). These results suggest that hypoxic conditions imposed in the sphere may be more severe than in monolayer culture leading to more effective stabilization of Hif1a. But, it is also possible that other conditions within the spheres, beyond downregulation of Zeb1, also contribute to induction of Hif1a.
Hif1a and Zeb1 regulate Aid
CpG methylation has been shown to block binding of Hif to target gene promoters (41), thus we reasoned that transactivation of Oct4 by Hif1a might require prior Oct4 promoter/enhancer demethylation. Therefore, we examined expression of components of the active DNA demethylation pathway. We detected little or no induction of TET1, which hydroxylates 5-methylcytosine, thymidine DNA glycosylase (Tdg) or Gadd45a, which act together in base excision and repair, or the Aid-related deaminase Apobec upon sphere formation (Fig. 5B). But, Aid was dramatically induced in spheres, and it was induced when MEFs in monolayer were placed under hypoxic conditions (Fig. 5B and F). When sphere-derived SP cells were placed in monolayer culture, Aid was diminished along with Hif1a (Fig. 5B), and CpG methylation at the Oct4 promoter was reestablished (Fig. 3E). These results then link hypoxia and Hif1a to induction of Aid, and downregulation of Aid to remethylation of the Oct4 promoter/enhancer. Consistent with a direct role for Hif in induction of Aid, we found that Hif1a bound to the Aid promoter under hypoxic conditions in ChIP assays (Fig. 5G).
Because of the link between Zeb1 and Hif1a expression, we also examined the effect of Zeb1 mutation on Aid expression. Mutation of Zeb1 in MEFs did not lead to induction of Aid in normoxic monolayer culture (Fig. 5F); however, mutation of Zeb1 did augment induction of Aid when MEFs formed spheres or when the cells were placed under hypoxic conditions in monolayer culture (Fig. 5F). These results demonstrate that mutation of Zeb1 is not sufficient alone to cause induction of Aid, but under hypoxic conditions, removal of Zeb1 augments induction of Aid. One explanation for these findings is that superinduction of Hif1a resulting from a combination of hypoxic stabilization and loss of Zeb1 repression is responsible for this further induction of Aid seen with Zeb1 mutation. But, it was also possible that Zeb1 actively represses Aid such that loss of repression in conjunction with Hif transactivation is important for full Aid induction. Consistent with this second possibility, we found multiple consensus E-box binding sites for Zeb1 in the Aid promoter in the region of the consensus Hif binding site located at position -1519 bp (Fig. 5G). And, using primers covering this upstream region of the Aid promoter in ChIP assays, we found binding of Zeb1 as well as Hif1a, as noted above (Fig. 5G). However, using primers overlapping the 5′ end of the Aid gene, which is well downstream of the consensus binding sites, we did not detect binding of either Hif1a or Zeb1. Based on these findings, we propose that Zeb1 represses Aid, but transactivation by Hif1a is required for Aid induction once Zeb1 is lost. But, Zeb1 figures into Aid induction in a second way because its loss is also important for superinduction of Hif1a.
Zeb1 presents a barrier to sphere-induced reprogramming
Based on our findings, we then hypothesized that Zeb1 serves as a barrier to generation of reprogrammed SP cells in spheres. To test this hypothesis, we compared the number of SP colonies arising from Zeb1(+/+), Zeb1(+/−) and Zeb1(−/−) MEF spheres. Consistent with elevated Oct4 expression in Zeb1 mutant spheres (Fig. 5E), we found that mutation of Zeb1 led to an increase in the number of colonies (Fig. 5I), providing evidence that it is an important barrier. A colony arising at the site of sphere adhesion to the plate is actually comprised of multiple colonies (Supplemental Fig. 3). Thus, Fig. 5I is more precisely measuring the ability of spheres to produce colonies.
Induction of Dnmt1 is linked to stable repression of cdk inhibitors and Arf and immortalization of sphere-derived colonies
In MSC, it was demonstrated that Oct4 induces Dnmt1 leading to methylation and silencing of p21 and Ink4 (8). But, loss of Oct4 as MSC are placed in culture prevents this induction of Dnmt1, leading to expression of cdk inhibitors, loss of differentiation potential and passage-dependent senescence (8). Consistent with these findings in MSC, we found induction of Dnmt1 along with Oct4 following sphere formation (Fig. 5B), and Dnmt1 was diminished along with Oct4 when sphere-derived SP cells formed colonies in culture (Figs. 5B and 6A). However, Dnmt1 levels in the colonies were still above that seen in monolayer culture of MEFs, and Dnmt1 was elevated in colonies derived from Zeb1(−/−) MEFs, where Oct4 is also elevated (Fig. 6A). Despite the fact that Dnmt1 diminished along with Oct4 as SP cells grew out into colonies in monolayer culture, p21, p16 and Arf were never induced with cell passage, as occurs with MEFs and MAFs prior to sphere formation, and the colonies were immortal (Fig. 6B and D). Beyond its well-established role in regulation of epithelial-mesenchymal balance, Zeb1 also binds the promoters of p21 and the Ink4 locus, to repress both p16 and Arf (27, 42). And, mutation of Zeb1 leads to gene-dosage-dependent induction of these genes in MEFs causing premature senescence in culture (Fig. 6B and C; ref. 27). We were then surprised that, as with colonies derived from spheres of wild-type MEFs and MAFs, similar colonies derived from Zeb1(−/−) MEFs also failed to induce p21, p16 or Arf and were likewise immortal in culture (Fig. 6B and D). We hypothesized that expression of Oct4 and the resulting induction of Dnmt1 was causing methylation and thus silencing of p21 and Ink4, thereby preventing their induction when Zeb1 is repressed in spheres and allowing for immortalization as the sphere-derived cells are passaged in culture. Consistent with this hypothesis, CpC islands located 5′ of p21 and Ink4 were fully methylated in sphere-derived colonies at passage 15 (Fig. 6E).
Figure 6.
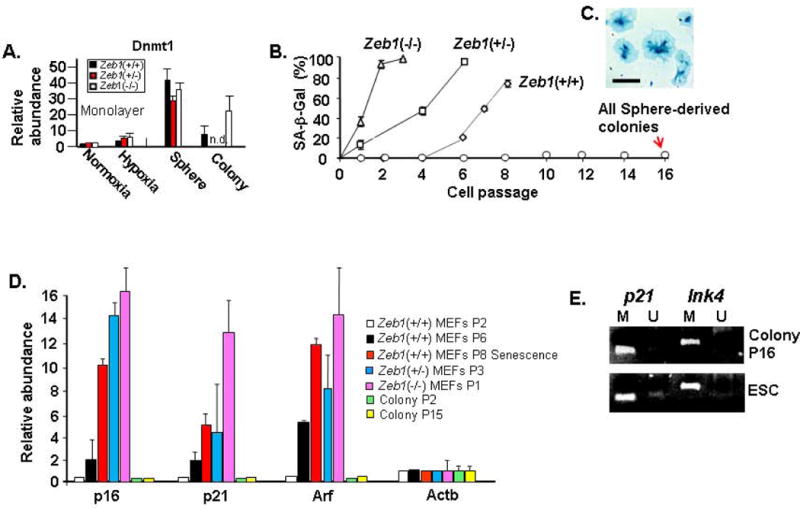
Sphere formation leads to induction of Dnmt1, and p21 and Ink4 are methylated and silenced in sphere-derived colonies, leading to immortalization. A. Real time PCR showing expression of Dnmt1 mRNA. “n.d.” not determined. B. Mutation of Zeb1 leads to premature senescence of MEFs, but sphere-derived colonies are immortal. “SA-β-Gal” indicates senescent β-galactosidase staining for cell senescence. C. Phase image showing examples of large, flat senescent cells stained for SA-β-Gal. D. Real time PCR showing expression of mRNAs for cdk inhibitors and Arf. E. Methylation-specific PCR (Materials and Methods) showing methylation (M) of CpG islands 5′ of p21 and Ink4. “U” indicates unmethylated. The bar is 30 μm in Panels A, 30 μm in Panel B and C, and E, 10 μm in Panel D and E, 15 μm in Panel F, 200 μm in Panel G, 30 μm in Panel H, 15 μm in Panel I, 20 μm in Panel J and K.
Sphere-derived colonies are multipotent
Despite the fact that Oct4 and Nanog expression diminished as sphere-derived SP cells proliferated to form colonies, their gene expression pattern resembles adult stem cells (Fig. 4A). In this regard, it is of note that MSC also rapidly lose Oct4 and Nanog expression when placed in culture, yet they retain multipotential capacity (8). Therefore, we subjected sphere-derived colonies to standard differentiation protocols for endodermal, mesodermal and ectodermal derivatives and immunostained for expression of markers of differentiation three weeks later. By this time, the cells were post-mitotic and they expressed markers representative of all three embryonic lineages (Fig. 7A–D). Ectodermal differentiation was evident by expression of Tubb3 (neurons) and Gfap (glia), endodermal differentiation was characterized by AFP (hepatic cells), and mesodermal differentiation by CD31 (endothelial cells), myogenin (skeletal muscle) and cardiac troponin I (heart). Expression of lineage markers is quantified in Fig. 7D. Beyond expression of lineage markers, staining of the differentiated cells with oil red, alcian blue and alizarin red demonstrated the presence of adipose, cartilage and calcified bone cells, respectively (Fig, 7C). As a negative control, the colonies did not express these lineage markers prior to differentiation (Fig. 7B). Additionally, the lineage markers were not induced when fibroblasts in monolayer culture prior to sphere formation were exposed to these differentiation protocols (results not shown).
Figure 7.
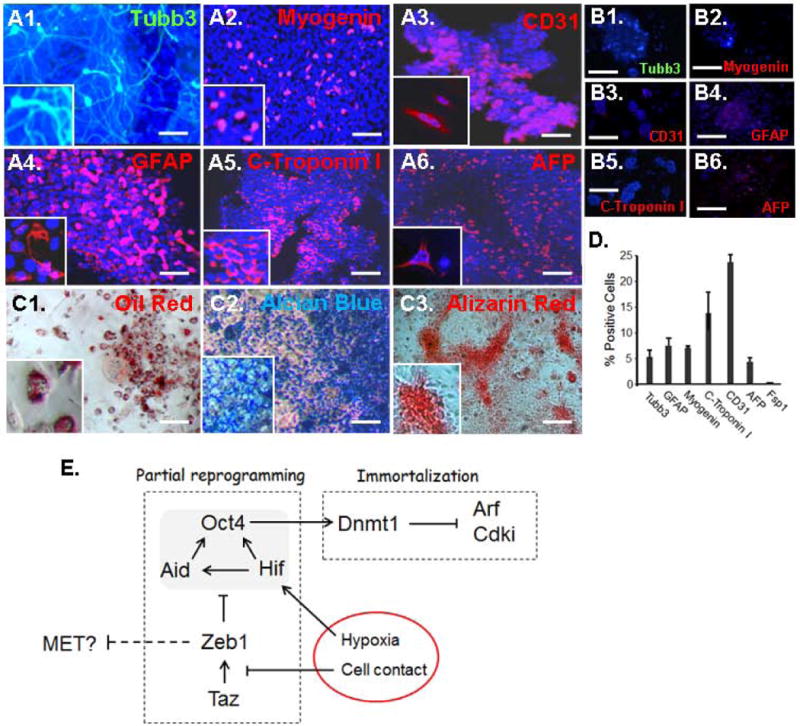
Sphere-derived colonies are multipotential. A. Sphere-derived colonies were subjected to differentiation protocols (Materials and Methods; Supplemental Methods) and three weeks later the cells were immunostained for the indicated lineage markers. Tubb3 and GFAP demonstrate ectodermal lineage, Myogenin and C-Troponin I mesodormal lineage, and CD31 and AFP endodermal lineage. B. As a control, no immunostaining was evident for any of the markers prior to differentiation. C. Differentiated colonies stained with oil red (adipose), alcian blue (cartilage) or alizarin red (calcified bone). D. Percentages of differentiated cell expressing each of the lineage markers. Standard deviations are shown. E. Model summarizing the pathway leading to reprogramming and immortalization of fibroblasts in spheres (shown as red circle). See text for discussion. The scale bar represents 20 μm.
Beyond this differentiation in monolayer culture, the sphere derived colonies also formed embryoid-like bodies where differentiation was also evident (Supplemental Figs. 4 and 5). Also, we found that post-mitotic differentiated cells begin to appear in the spheres, implying that the Oct4+ SP cells are spontaneously giving rise to a variety of lineages in situ within the sphere (Supplemental Figs. 6 and 7).
We have also specifically marked fibroblasts with LacZ prior to sphere formation in order to trace the lineage of these sphere-derived colonies from fibroblasts, to Oct4+ SP cells and finally to multipotent colonies (Supplemental Fig. 8). We conclude that the sphere-derived colonies arise from reprogrammed fibroblasts and they resemble adult stem cells in their multipotency.
DISCUSSION
A pathway to sphere-initiated reprogramming of fibroblasts
We present evidence of a molecular pathway leading to reprogramming of fibroblasts in spheres (Fig. 7E). Soon after sphere formation, Amot is induced and Taz becomes sequestered in the cytoplasm, Zeb1 expression is diminished and the cells undergo MET. Zeb1 represses transcription of Hif1a, and the hypoxia in the interior of the spheres in conjunction with loss of Zeb1 repression synergizes to cause superinduction of Hif1a. Loss of repression by Zeb1 and transactivation by Hif1a combine to induce Aid, which coincides with demethylation of the Oct4 promoter/enhancer and induction of Oct4. Hif1a also binds the Oct4 promoter, suggesting that it further contributes a transactivation function once inhibitory CpG methylation is removed. These results then link hypoxia to Oct4 promoter/enhancer demethylation and thus Oct4 expression. By contrast to ESC and iPS cells, this DNA demethylation and Oct4 and Nanog expression is not stable—it is rapidly lost when spheres are dissociated and cells are placed in cultured. In this way the Oct4 promoter/enhancer demethylation and Oct4 induction resulting from sphere formation resembles that seen in MSC where remethylation of the Oct4 promoter/enhancer and loss of Oct4 expression also occurs when the cells are placed in culture (8). A recent study has demonstrated that arachidonic and pluronic acids can promote aggregation and stable differentiation of fibroblasts to cells with properties of stem cells (43). It will then be of interest to determine how such lipid-mediated signaling might be linked to reprogramming of fibroblast in spheres.
A paradox for hypoxia in EMT and MET
Hypoxia promotes iPS reprogramming and it facilitates proliferation and prevents differentiation of ESC (12, 44), and as noted above, Hif is required to maintain Oct4 expression in primordial germ cells and in ECS during embryogenesis. And, here we provide evidence that hypoxia is also important for Oct4 induction in sphere-initiated reprogramming. iPS reprogramming is characterized by an early and essential MET with corresponding downregulation of EMT factors including Zeb1. Yet, in cancer hypoxia classically induces EMT transcription factors including Zeb1, and through the resulting EMT it is thought to trigger transition to invasive, metastatic cancer (45, 46). It is then unclear how Zeb1 becomes downregulated in association with MET under hypoxic conditions, which promote iPS reprogramming. Likewise, it is unclear and how ESC, which display a similar epithelial phenotype with downregulation of Zeb1, prevent Zeb1 induction and EMT in the hypoxic environment of the early embryo and in hypoxic culture. Our results suggest that activation of the cell-cell contact inhibition signaling pathway upon sphere formation leads to retention of the EMT-inducing transcription factor Taz in the cytoplasm, rapid downregulation of its target gene Zeb1 and MET. Although Zeb1 can be induced by hypoxia and Hif (26, 47), this does not occur in the hypoxic environment of spheres. We suggest that Taz is critical for Zeb1 expression and thus it is not induced by hypoxia and Hif1a when Taz is sequestered in the cytoplasm in spheres. In fact, we demonstrate that Zeb1 represses Hif1a such that Zeb1 loss synergizes with hypoxia to cause superinduction of Hif1a. These results imply that Zeb1 negatively feedbacks to repress Hif1a. Although fibroblast sphere formation could be thought of as mimicking early solid tumor outgrowth (where hypoxia is linked to EMT, as opposed to MET), it is of note that all solid tumors have a disrupted cell-cell contact inhibition signaling pathway, which is required for unrestricted outgrowth of cancer cells. Thus, we suggest that this lack of cell contact signaling allows for induction of Zeb1 by hypoxia and Hif under the hypoxic conditions of tumor outgrowth, leading in turn to the EMT that is important for tumor invasion. It is also interesting that cells with properties of cancer stem cells show evidence of EMT, and fibroblasts and epithelial cells can be reprogrammed to cancer stem-like cells via sphere formation if the Rb1 pathway is disrupted (20, 21). The Rb1 pathway is essential for cell contact inhibition, and it represses expression of Zeb1 (20). Thus in the absence of Rb1 Zeb1 expression is induced in sphere-reprogrammed cells as opposed to being repressed in wild-type sphere-reprogrammed cells (20). And, the resulting EMT is linked to cancer initiation in nude mice, whereas the sphere-reprogrammed wild-type cells Rb1 family expression does not change (Supplemental Fig. 9) and the cells do not form such tumors.
Sphere-induced immortalization
A key barrier to iPS reprogramming is the induction of cdk inhibitors and Arf. The cells must silence these genes to prevent senescence, and in mouse cells (where telomeres are maintained) this is sufficient for immortalization. It has been presumed that induction of p21, p16 and Arf during iPS reprogramming results in part from DNA damage. But, these cells also undergo an early MET with downregulation of Zeb1, raising the possibility that loss of Zeb1 might also contribute to induction of these genes and inhibitory senescence. The pathway to silencing the cdk inhibitors and Arf in iPS reprogramming is still unclear. However, in MSC, recent studies demonstrate that Oct4 induces Dnmt1, which in turn silences the genes by causing CpG methylation (8). In the spheres, we also link induction of Oct4 and Dnmt1 to methylation and silencing of p21 and Ink4. And, these genes remained silenced in culture, leading to immortalization. Remarkably, even Zeb1(−/−) MEFs, which have elevated levels of cdk inhibitors and Arf and thus undergo premature senescence in monolayer culture, were immortalized following sphere formation.
In summary, we present evidence of a molecular pathway for stably reprogramming and immortalizing fibroblasts to a multipotential phenotype resembling adult stem cells that does not require exogenous expression of a stem cell factor or a lineage-inducting transcription factor.
Supplementary Material
Supplemental Fig. 1. A. Sequence comparison of the Zeb1 promoter in different species showing conserved Taz/Yap1 binding site (boxed). B. Chromatin immunoprecipitation assay showing binding of Taz and to a lesser degree Yap1 to the Zeb1 promoter. “IgG” is control anti-serum, and H3 is a positive control of histone H3. C. Knockdown of Taz in MEFs leads to down regulation of ZEB1 mRNA by real time PCR. D. Western blot showing loss of Taz but not Yap1 with Taz knockdown. See ref. 19 for additional details of different Taz shRNA lentiviral constructs.
Supplemental Fig. 2. All 45101 spots on the mouse whole genome arrays for MEF, W9.5, NSC, MSC, BMMSC data in Fig. 4 were compared to the sphere derived stem cell like colonies (SDSC). The LOG-transformed data were first normalized to the mean of all the spots, then the value of each spot for SDSC was divided by the value of the same spot for MEF, W9.5, NSC, MSC, BMMSC, respectively. A student T-test of SDSC vs. other samples for each spot is calculated against ‘1’, and the probability (p value) then is assigned for all the tests. Based on the p value the number of differentially expressed spots were counted and plotted on the attached two figures. On right, the y-axis is actual spot number, whereas in the figure on left the y-axis is LOG10 spot number. The fewest significant gene expression changes are seen in the MSC and BMMSC, suggesting that the SDSC are more closely related to MSC.
Supplemental Fig. 3. After 4 days in suspension culture, MEF spheres were allowed to adhere to Matrigel-coated plates. Cells in the spheres migrated out onto the plate, and a colony was evident at the site of sphere adhesion after 10 days. Note that this colony is actually composed of a number of tightly packed colonies, which are outlined in the panel on the right. The bar is 30 μm.
Supplemental Fig. 4. Embryoid body formation with sphere-derived colonies. Three week old embryoid bodies were sectioned for H&E and immunostaining. A. Keratin pearls (KP), cartilage (C) and pigmented cells (PC). B. Region of cartilage stained with alcian blue. The insert shows a higher power view prior to alcian blue staining. C. Higher power view of a keratin pearl. D. Higher power view of pigmented cells. E. A region resembling secretory epithelium. F. Immunostaining of a section adjacent to that in panel E for E-cadherin (E-Cad). G. Low power view of an aggregate of spheres immunostained for the neuronal marker Tubb3. H. Higher power view of Tubb3 immunostaining. I. Edge of an embryoid body containing a region resembling primitive endoderm. J–K. Immunostaining of sections adjacent to that in panel I for AFP and CD31. Arrows indicate the edge of the embryoid body. The bar represents 200 mm in panel A; 75 mm in panel B–C; 400 mm in panel G; 50 mm in panels D and H–K; 30 mm in panels E–F. L. Real time PCR array comparing express in three week old embryoid bodies formed from sphere-derived colonies and ESC (Liu cell stem cell). Results are normalized to b-actin (Actb) mRNA and both samples are compared to fibroblasts maintained in monolayer culture whose mRNA levels are set to 1.0. An average of the values from three independent experiments is shown along with standard deviations. The bar is 150 μm in Panel A and B.
Supplemental Fig. 5. Real time PCR array as in Supplemental Fig. 2 comparing expression of genes in signaling pathway important for reprogramming in embryoid bodies (EB) formed from sphere-derived colonies and ESC. Expression in MEFs in monolayer culture is normalized to 1.0.
Supplemental Fig. 6. Real time PCR array showing induction of mRNA in 8 day old MEFs spheres compared to monolayer MEFs as in ref 20. Results are normalized to Actb.
Supplemental Fig. 7. After 8 days in suspension culture, MEF spheres were allowed to adhere to Matrigel coated plates. Cells migrated from the spheres, and a colony arose at the site of the sphere attachment. The colony expressed the proliferation marker Ki67, but most of the cells migrating away from the sphere did not express Ki67, and did not exclude Hoechst dye (these are MP cells). These MP cells were then immunostained for markers of differentiation three days after plating of the spheres. The bar is 20 μm in Panel A, B, C, and D, 25 μm in Panel E, 15 μm in Panel F, 10 μm in Panel G, 20 μm in Panel H.
Supplemental Fig. 8. Marking fibroblasts to assess the cell origin of sphere-derived colonies. Microarray analysis showed expression of Fsp1 in MEFs but not in MCS or bone marrow derived MSC (Fig. 4D), and accordingly immunostaining for Fsp1 showed uniform expression in MAFs and MEFs, but no expression on adipose MSC. Fibroblasts were isolated from tail tips of Rosa26 mice at birth and the cells were infected with a lentivirus expressing Cre driven by the Fsp1 promoter. This construct has been shown to drive fibroblast-specific Cre expression in vivo and in culture. We followed LacZ recombination efficiency in the infected Rosa26 fibroblasts by immunostaining for LacZ. Many of the cells expressed LacZ demonstrating that they had efficiently recombined the LacZ locus, which then permanently marked the fibroblasts. These LacZ+ fibroblast where then used to generate spheres, and Oct4+ SP cells were sorted. These Oct4+ SP cells co-expressed LacZ, demonstrating that they were derived from fibroblasts. Then colonies of these cells were subjected to differentiation protocols as in Fig. 7, and the differentiated cells were co-immunostained for LacZ and lineage markers. LacZ-positive cells were evident among those expressing each marker. We conclude that these various lineages also arise from fibroblasts. Bars are 20 mm.
Supplemental Fig. 9. Real time PCR comparing the levels of Rb1 family mRNAs in MEFs in monolayer culture to cells in four-day old spheres. Results are normalized to Actb.
Acknowledgments
These studies were supported in part by NIH grants EY019113 (DCD) and EY018603 (DCD), DE018215 (MMP), and an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences (P20GM103453) (YL), and Research to Prevent Blindness. We thank Ewa Zuba-Surma for cell sorting and Antonio Postigo for valuable comments on the studies.
Footnotes
Author contribution:
Yongqing Liu: Conception and design; collection/assembly of data; data analysis and interpretation; manuscript writing; final approval of manuscript
Partha Mukhopadhyay: collection/assembly of data; data analysis and interpretation; final approval of manuscript
Michele Pisano: data analysis and interpretation; final approval of manuscript
Xiaoqin Lu: collection/assembly of data; data analysis and interpretation; final approval of manuscript
Li Huang: collection/assembly of data; data analysis and interpretation; final approval of manuscript
Qingxian Lu: Conception and design; data analysis and interpretation; final approval of manuscript
Douglas C. Dean: Conception and design; financial support; data analysis and interpretation; manuscript writing; final approval of manuscript
References
- 1.Yamanaka S, Blau HM. Nuclear reprogramming to a pluripotent state by three approaches. NATURE. 2010;465:704–712. doi: 10.1038/nature09229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buhtani N, Burns DM, Blau HM. DNA demethylation dynamics. CELL. 2011;146:866–72. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meissner A. Epigenetic modifications in pluripotent and differentiated cells. NAT BIOTECH. 2010;28:1079–1088. doi: 10.1038/nbt.1684. 2010. [DOI] [PubMed] [Google Scholar]
- 4.Bhutani N, Brady JJ, Damian M, et al. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. NATURE. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cortellino S, Xu J, Sannai M, et al. Thymidine DNA glycosylation is essential for active DNA demethylation by linked deamination-base excision repair. CELL. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalton SR, Bellacosa A. DNA demethylation by TDG. EPIGENOMICS. 2012;4:459–467. doi: 10.2217/epi.12.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. CELL. 2011;145:422–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai C-C, Su P-F, Huang Y-F, Yew T-L, Hung S-C. Oct4 and nanog directly regulate dnmt1 to maintain self-renewal and undifferentiated state in mesenchymal stem cells. MOL CELL. 2012;47:169–182. doi: 10.1016/j.molcel.2012.06.020. [DOI] [PubMed] [Google Scholar]
- 9.Banito A, Rashid ST, Acosta JC, et al. Senescence impairs successful reprogramming to pluripotent stem cells. GENES DEV. 2009;23:2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohyeldin A, Garzon-Muvdi T, Quinones-Hinojosa A. Oxygen in stem cell biology: a critical component of the stem cell niche. CELL STEM CELL. 2010;7:150–161. doi: 10.1016/j.stem.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Covello KL, Kehler J, Yu H, et al. HIF-2a regulates Oct-4: effects of hypoxia on stem cell function embryonic development and tumor growth. GENES DEV. 2006;20:557–570. doi: 10.1101/gad.1399906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathieu J, Zhang Z, Zhou W, et al. Hif induces human embryonic stem cell markers in cancer cells. CANCER RES. 2011;71:4640–4652. doi: 10.1158/0008-5472.CAN-10-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Liu Y, Malek SN, Zheng P, Liu Y. Targeting HIF1a eliminates cancer stem cells in hematological malignancies. CELL STEM CELL. 2011;8:399–411. doi: 10.1016/j.stem.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaks S. Hypoxia enhances the generation of induced pluripotent stem cells. CELL STEM CELL. 2009;5:237–243. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Li R, Liang J, Ni S, et al. Mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. CELL STEM CELL. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 16.Lei QY, Zhang H, Zhao B, et al. Taz promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the Hippo pathway. MOLEC CELL BIOL. 2008;28:2426–2436. doi: 10.1128/MCB.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao B, Li L, Lu Q, et al. Angiomotin is a novel hippo pathway component that inhibits Yap oncoprotein. GENES DEV. 2011;25:51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. CELL. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 19.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. NAT REV CANCER. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y, Clem B, Zuba-Surma EK, et al. Mouse fibroblasts lacking RB1 function form spheres and undergo reprogramming to a cancer stem cell phenotype. CELL STEM CELL. 2009;4:336–347. doi: 10.1016/j.stem.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mani S, Gou W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. CELL. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Powers JT, Hong S, Mayhew CN, et al. E2F1 uses the ATM signaling pathway to induce p53 and Chk2 phosphorylation and apoptosis. MOL CANCER RES. 2004;2:203–214. [PubMed] [Google Scholar]
- 23.Lorico A, Rappa G, Flavell RA, Sartorelli AC. Double knockout of the MRP gene leads to increased drug sensitivity in vitro. CANCER RES. 1996;56:5351–5355. [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantative PCR and the 2(-delta delat C(T)) method. METHODS. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Takahashi K, Yamanaka S. Induction of pluoripotent stem cells from mouse embryonic and adult fibroblasts cultures by defined factors. CELL. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 26.Bui T, Sequeira J, Wen TC, et al. Zeb1 links p63 and p73 in a novel neuronal survival pathway rapidly induced in response to cortical ischemia. PLOS ONE. 2009;4:e4373. doi: 10.1371/journal.pone.0004373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, El-Naggar S, Darling DS, Higashi Y, Dean DC. Zeb1 links epithelial-mesenchymal transition and cellular senescence. DEVELOPMENT. 2008;135:579–588. doi: 10.1242/dev.007047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Xin Y, Ye F, et al. Taz-tead1 links cell-cell contact to zeb1 expression proliferation and dedifferentiation in retinal pigment epithelial cells. INVEST OPHTHALMOL VIS SCI. 2010;51:3372–378. doi: 10.1167/iovs.09-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hockemeyer D, Soldner F, Beard C, et al. Efficient targeting of expressed and silent genes in human ESCs and IPSCs using zinc-finger nucleases. NAT BIOTECH. 2009;27:851–7. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robin C, Ottersbach K, de Brujin M, et al. Developmental origins of hematopoietic stem cells. ONCOL RES. 2003;13:315–321. doi: 10.3727/096504003108748519. [DOI] [PubMed] [Google Scholar]
- 31.Micera A, Lambiase A, Stampachiacciere B, et al. Nerve growth factor and tissue repair remodeling: trkA. NGFR and p75 INTR two receptors one fate CYTOKINE GRWOTH FACTOR REV. 2007;18:245–56. doi: 10.1016/j.cytogfr.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 32.Papizan JB, Singer RA, Tschen SI, et al. Nkx2.2 repressor complex regulates islet-beta-cell specification and prevents beta-to-alpha cell reprogramming. GENES DEV. 2011;25:2291–2305. doi: 10.1101/gad.173039.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leushacke M, Barker N. Lgr5 and Lgr6 as markers to study adult stem cell roles in self-renewal and cancer. ONCOGENE. 2011;31:3009–3022. doi: 10.1038/onc.2011.479. [DOI] [PubMed] [Google Scholar]
- 34.Prestegarden L, Ender PO. Cancer stem cells in the central nervous system-a critical review. CANCER RES. 2010;77:8255–8258. doi: 10.1158/0008-5472.CAN-10-1592. [DOI] [PubMed] [Google Scholar]
- 35.Bertos NR, Park M. Breast cancer – one term many entities? J CLIN INVEST. 2011;121:3789–3796. doi: 10.1172/JCI57100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Castro NP, Rangel MC, Nagaoka T, Salomon DS, Bianco C. Cripto-1: an embryonic gene that promotes tumorigenesis. FUTURE ONCOL. 2010;6:1127–1142. doi: 10.2217/fon.10.68. [DOI] [PubMed] [Google Scholar]
- 37.Ball SG, Shuttleworth CA, Kielty CM. Platelet-derived growth factor receptors regulate mesenchymal stem cell fate: implications for neovascularization. EXPERT OPIN BIOL THER. 2010;10:57–71. doi: 10.1517/14712590903379510. [DOI] [PubMed] [Google Scholar]
- 38.Ten Broek RW, Grefte S, von den Hoff JW. Regulatory factors and cell populations involved in skeletal muscle regeneration. J CELL PHYSIOL. 2010;224:7–2216. doi: 10.1002/jcp.22127. [DOI] [PubMed] [Google Scholar]
- 39.Strunk D. Endothelial progenitor cells: quod erat demonstrandum? CURR PHARM DES. 2011;17:3245–3251. doi: 10.2174/138161211797904127. [DOI] [PubMed] [Google Scholar]
- 40.Ruis J, Guma M, Schachtrup C, et al. NF-kappaB links innate immunity to the hypoxia response through transcriptional regulation of HIF-1alpha. NATURE. 2008;453:807–811. doi: 10.1038/nature06905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rossler J, Stolze I, Frede S, et al. Hypoxia-induced erythropoietin expression in human neuroblastoma requires a methylation free Hif-1 binding site. J CELL BIOCHEM. 2004;93:153–161. doi: 10.1002/jcb.20133. [DOI] [PubMed] [Google Scholar]
- 42.Postigo A. Opposing functions of ZEB proteins in the regulation of the TGFb/BMP signaling pathway. EMBO J. 2003;22:2443–2452. doi: 10.1093/emboj/cdg225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rajanahalli P, Meyer K, Zhu L, et al. Conversion of mouse fibroblasts to sphere cells induced by albuMAXI-containing medium. FRONT BIOSCI. 2012;4:1813–1822. doi: 10.2741/e502. [DOI] [PubMed] [Google Scholar]
- 44.Westfall SD, Sachdev S, Das P, et al. Identification of oxygen-sensitive transcriptional programs in human embryonic stem cells. STEM CELLS. 17:869–881. doi: 10.1089/scd.2007.0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu MZ, Tsai YP, Yang MH, et al. Interplay between HDAC3 and WDR5 is essential for hypoxia-induced epithelial-mesenchyaml transition. MOL CELL. 43:811–822. doi: 10.1016/j.molcel.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 46.Wu CY, Tsai YP, Wu MZ, Teng SC, Wu KJ. Epigenetic reprogramming and post-transcriptional regulation during epithelial-mesenchymal transition. TRENDS GENET. 28:454–463. doi: 10.1016/j.tig.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 47.Krishnamachary B, Zagzag D, Nagasawa H, et al. Hypoxia-inducible factor-1 dependent repression of E-cadherin in von hippel-lindau tumor suppressor-null renal cell carcinoma mediated by TCF3, ZFHX1a and ZFHX1B. CANCER RES. 66:2725–2731. doi: 10.1158/0008-5472.CAN-05-3719. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1. A. Sequence comparison of the Zeb1 promoter in different species showing conserved Taz/Yap1 binding site (boxed). B. Chromatin immunoprecipitation assay showing binding of Taz and to a lesser degree Yap1 to the Zeb1 promoter. “IgG” is control anti-serum, and H3 is a positive control of histone H3. C. Knockdown of Taz in MEFs leads to down regulation of ZEB1 mRNA by real time PCR. D. Western blot showing loss of Taz but not Yap1 with Taz knockdown. See ref. 19 for additional details of different Taz shRNA lentiviral constructs.
Supplemental Fig. 2. All 45101 spots on the mouse whole genome arrays for MEF, W9.5, NSC, MSC, BMMSC data in Fig. 4 were compared to the sphere derived stem cell like colonies (SDSC). The LOG-transformed data were first normalized to the mean of all the spots, then the value of each spot for SDSC was divided by the value of the same spot for MEF, W9.5, NSC, MSC, BMMSC, respectively. A student T-test of SDSC vs. other samples for each spot is calculated against ‘1’, and the probability (p value) then is assigned for all the tests. Based on the p value the number of differentially expressed spots were counted and plotted on the attached two figures. On right, the y-axis is actual spot number, whereas in the figure on left the y-axis is LOG10 spot number. The fewest significant gene expression changes are seen in the MSC and BMMSC, suggesting that the SDSC are more closely related to MSC.
Supplemental Fig. 3. After 4 days in suspension culture, MEF spheres were allowed to adhere to Matrigel-coated plates. Cells in the spheres migrated out onto the plate, and a colony was evident at the site of sphere adhesion after 10 days. Note that this colony is actually composed of a number of tightly packed colonies, which are outlined in the panel on the right. The bar is 30 μm.
Supplemental Fig. 4. Embryoid body formation with sphere-derived colonies. Three week old embryoid bodies were sectioned for H&E and immunostaining. A. Keratin pearls (KP), cartilage (C) and pigmented cells (PC). B. Region of cartilage stained with alcian blue. The insert shows a higher power view prior to alcian blue staining. C. Higher power view of a keratin pearl. D. Higher power view of pigmented cells. E. A region resembling secretory epithelium. F. Immunostaining of a section adjacent to that in panel E for E-cadherin (E-Cad). G. Low power view of an aggregate of spheres immunostained for the neuronal marker Tubb3. H. Higher power view of Tubb3 immunostaining. I. Edge of an embryoid body containing a region resembling primitive endoderm. J–K. Immunostaining of sections adjacent to that in panel I for AFP and CD31. Arrows indicate the edge of the embryoid body. The bar represents 200 mm in panel A; 75 mm in panel B–C; 400 mm in panel G; 50 mm in panels D and H–K; 30 mm in panels E–F. L. Real time PCR array comparing express in three week old embryoid bodies formed from sphere-derived colonies and ESC (Liu cell stem cell). Results are normalized to b-actin (Actb) mRNA and both samples are compared to fibroblasts maintained in monolayer culture whose mRNA levels are set to 1.0. An average of the values from three independent experiments is shown along with standard deviations. The bar is 150 μm in Panel A and B.
Supplemental Fig. 5. Real time PCR array as in Supplemental Fig. 2 comparing expression of genes in signaling pathway important for reprogramming in embryoid bodies (EB) formed from sphere-derived colonies and ESC. Expression in MEFs in monolayer culture is normalized to 1.0.
Supplemental Fig. 6. Real time PCR array showing induction of mRNA in 8 day old MEFs spheres compared to monolayer MEFs as in ref 20. Results are normalized to Actb.
Supplemental Fig. 7. After 8 days in suspension culture, MEF spheres were allowed to adhere to Matrigel coated plates. Cells migrated from the spheres, and a colony arose at the site of the sphere attachment. The colony expressed the proliferation marker Ki67, but most of the cells migrating away from the sphere did not express Ki67, and did not exclude Hoechst dye (these are MP cells). These MP cells were then immunostained for markers of differentiation three days after plating of the spheres. The bar is 20 μm in Panel A, B, C, and D, 25 μm in Panel E, 15 μm in Panel F, 10 μm in Panel G, 20 μm in Panel H.
Supplemental Fig. 8. Marking fibroblasts to assess the cell origin of sphere-derived colonies. Microarray analysis showed expression of Fsp1 in MEFs but not in MCS or bone marrow derived MSC (Fig. 4D), and accordingly immunostaining for Fsp1 showed uniform expression in MAFs and MEFs, but no expression on adipose MSC. Fibroblasts were isolated from tail tips of Rosa26 mice at birth and the cells were infected with a lentivirus expressing Cre driven by the Fsp1 promoter. This construct has been shown to drive fibroblast-specific Cre expression in vivo and in culture. We followed LacZ recombination efficiency in the infected Rosa26 fibroblasts by immunostaining for LacZ. Many of the cells expressed LacZ demonstrating that they had efficiently recombined the LacZ locus, which then permanently marked the fibroblasts. These LacZ+ fibroblast where then used to generate spheres, and Oct4+ SP cells were sorted. These Oct4+ SP cells co-expressed LacZ, demonstrating that they were derived from fibroblasts. Then colonies of these cells were subjected to differentiation protocols as in Fig. 7, and the differentiated cells were co-immunostained for LacZ and lineage markers. LacZ-positive cells were evident among those expressing each marker. We conclude that these various lineages also arise from fibroblasts. Bars are 20 mm.
Supplemental Fig. 9. Real time PCR comparing the levels of Rb1 family mRNAs in MEFs in monolayer culture to cells in four-day old spheres. Results are normalized to Actb.


