Abstract
Objective:
Dealing with severe blood stream infections (BSI) is one of the intractable conditions in hospitals. The empirical treatment given remains pertinent in determining patient outcome, which becomes evidence based when substantiated by knowledge of susceptibility patterns of prevalent pathogenic organisms in the set up. This study was undertaken to determine the occurrence, species prevalence, and antibiotic susceptibility pattern of laboratory confirmed BSI (LCBSI) in patients admitted to our multi-specialty sanatorium.
Materials and Methods:
Eight hundred and forty-six blood samples from 829 patients suspected of having BSI were cultured as per standard microbiological procedures. Antimicrobial susceptibility testing was done for bacterial isolates from positive blood cultures.
Results:
Sixty (7.2%) cases were established as LCBSI. A total of eight pathogenic bacterial genera were identified and their antimicrobial susceptibility pattern was noted. Staphylococcus spp. were most prevalent (33%), followed by Klebsiella pneumoniae (20%), Escherichia coli (13%), Acinetobacter spp. (13%), Enterococcus spp. (12%), Pseudomonas aeruginosa (3%), Proteus spp. (2%), and Citrobacter spp. (2%).
Conclusions:
The study shows the prevalence of common bacterial pathogens causing BSI and their susceptibility patterns. Such studies provide benefit of instantaneous choice of antibiotic therapy aiming at improved patient management and reduced drug resistance.
Keywords: Antimicrobial susceptibility, Drug resistance, Laboratory confirmed blood stream infections
INRODUCTION
Despite many advances in patient care, blood stream infections (BSI) remain important causes of morbidity and mortality in hospitals especially in developing countries like India. These are among top 10 leading causes of death worldwide and most significant challenges in critical care. Laboratory confirmed BSI (LCBSI) have been classified into community acquired or hospital acquired/healthcare associated; of which, the latter is of particular concern, as the patient acquires BSI during the course of receiving treatment for other conditions within a healthcare setting. Among the other healthcare associated infections, which mainly include urinary tract infections, surgical site infections, and lung infections; BSI constitute for about 14% and are one of the leading causes of death globally.[1] Besides considerably affecting the mortality, LCBSI add significantly to the morbidity and economic burden of patients and hospitals. The direct and indirect costs that are affected principally include increased hospital stay, drug treatment, medical, and surgical procedures as well as patient's lost salary and illness.[2]
Where an early diagnosis and administration of accurate antibiotics based on patient's blood culture report is the ideal approach of managing BSI, the empirical treatment given remains pertinent in determining patient outcome. The increasing antimicrobial resistance among pathogenic bacteria causing BSI has been reported in many studies conducted in India, and is of major concern.[3,4] Antimicrobial resistance is a biological phenomenon acquired by microbes in response to the selective pressure of antimicrobial agents. The cock-tail of antimicrobials, which the patient is put on, for empirically managing severely ill patients such as those having BSI, further escalates the development of drug resistance, which can be reduced if the empirical treatment is made evidence based by substantiating it with the knowledge of prevalent pathogenic organisms and their antibiograms.
Therefore, there is a dearth of observational studies, which look into local epidemiology of BSI. The lack of such updated data from central India prompted us to conduct this study, to establish the leading pathogens causing BSI in our cohort and know their antimicrobial susceptibility patterns.
MATERIALS AND METHODS
Study population
It was a prospective study conducted over a period of 13 months, starting from June 1 to June 30, 2012 in a multi-specialty sanatorium of Bhilai, Chhattisgarh, Central India. A total of 829 patients clinically suspected to have BSI, admitted in the wards of different specialties of the hospital; were included in the study.
Collection and processing of samples
Venous blood, 10 ml from adults and 2-5 ml from children was obtained aseptically and inoculated into brain heart infusion broth. Blind subcultures were done on 5% sheep blood agar and MacConkey's agar after 24 and 48 h of incubation. A negative result was followed up by examining the broth daily and doing a final subculture at the end of the 7th day or at the appearance of turbidity or gas production, which ever was earlier. Organisms were identified by cultural characters, morphology, and standard biochemical tests. Antibiotic susceptibility testing was performed by the Kirby Bauer disc diffusion method as per Central Laboratory Standards Institute (CLSI) recommendations.[5,6]
Interpretation of culture results
Incidence of LCBSI was determined following the Centers for Disease Control and Prevention (CDC) guidelines and met one of the following criteria:
Patient had a recognized pathogen cultured from one or more blood samples,
Patient had at least one of the following signs or symptoms: Fever >38°C, chills, or hypotension
Patient <1 year of age had at least one of the following signs or symptoms: Fever >38°C, hypothermia <36°C, apnea, or bradycardia.
Furthermore, those signs and symptoms and positive laboratory results were not related to an infection at another site and a commensal, when isolated — was considered pathogenic if cultured from two or more blood samples drawn on separate occasions.[7]
The patients’ details, pathogen isolated, and the susceptibility patterns of all isolates were entered in excel sheets.
Patients with LCBSI were followed for the treatment given and the outcome was recorded in terms of patient discharge/mortality.
RESULTS
Study population
A total of 829 subjects were enrolled in the study, out of which 438 (53%) were males. Fourteen percent (n = 117) of the subjects belonged to pediatric age group. From 829 patients, 846 blood samples were obtained for culture. From each suspected case of BSI, a single blood specimen was obtained initially; only in cases showing growth of coagulase negative Staphylococcus (CONS) (n = 17), a repeat blood sample was obtained to confirm pathogenicity. A total of 60 (7.2%) cases were established as LCBSI including 32 males and 28 females. The cases of LCBSI belonged to age range newborn - 88 years, with a median age of 47 years for males and 56 years for females. The underlying medical conditions of the LCBSI cases were neurological illness (17%), neurosurgical condition (12%), gastroenterological illness (10%), malignancy (10%), nephrological problem (10%), and respiratory pathology (8%). Other than this, 20% were having miscellaneous medical conditions including electrolyte imbalance, hormonal imbalance, for example, hypo-/hyperthyroidism, drowning, snake/unknown bites and diabetes mellitus; while 13% were miscellaneous pediatric cases [Figure 1a].
Figure 1.
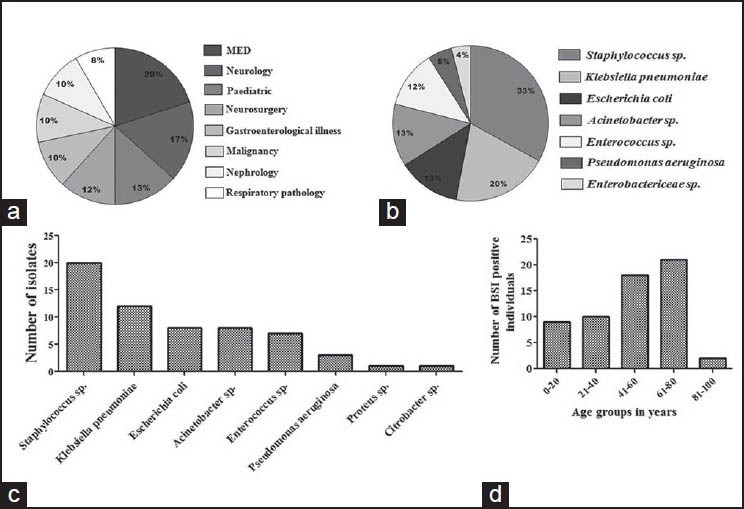
Type and distribution of bacterial pathogens causing laboratory confirmed blood stream infections (LCBSI). (a) Medical conditions for which LCBSI patients were admitted. (b, c) Frequency and distribution of pathogenic bacteria causing LCBSI. (d) Distribution of LCBSI patients in different age groups
Pathogens isolated
Of the causative pathogens, Staphylococcus spp. including Staphylococcus aureus (n = 13) and CONS (n = 7) were most prevalent (33%). Of the 17 repeat samples obtained to confirm the pathogenicity of CONS, only 7 showed the growth of the same organism on repeat blood culture, so remaining 10 were excluded as skin commensals.[7] Other pathogenic bacteria were Klebsiella pneumoniae (20%), Escherichia coli (13%), Acinetobacter spp. (13%), Enterococcus spp. (12%), Pseudomonas aeruginosa (3%), Proteus spp. (2%), and Citrobacter spp. (2%) [Figure 1b and c]. Maximum number of LCBSI cases fell in the age group of 61-80 years [Figure 1d].
Susceptibility patterns of isolates
The susceptibility pattern of different organisms is shown in Tables 1 and 2. As per the CLSI guidelines, there were different set of antibiotics that were used for different organisms, and the susceptibility patterns of different pathogens were obtained.[6] About 100% of isolates of the most common pathogen Staphylococcus spp. showed sensitivity to linezolid, teicoplanin, and vancomycin. In Klebsiella spp. 100% sensitivity was seen only to one antibiotic — imipenem. In case of E. coli however, 100% sensitivity was not seen for any antibiotic; the highest seen was 88% sensitivity for tigecycline. Hundred percent of P. aeruginosa and Acinetobacter spp. isolates were sensitive to polymixin-B and colistin. About 100% isolates of Enterococcus spp. were susceptible to linezolid, teicoplanin, tigecycline, and vancomycin; whereas in Citrobacter spp. and Proteus spp. 100% isolates were found to be sensitive to tigecycline.
Table 1.
Antibiotic susceptibility patterns of bacterial pathogens causing LCBSI
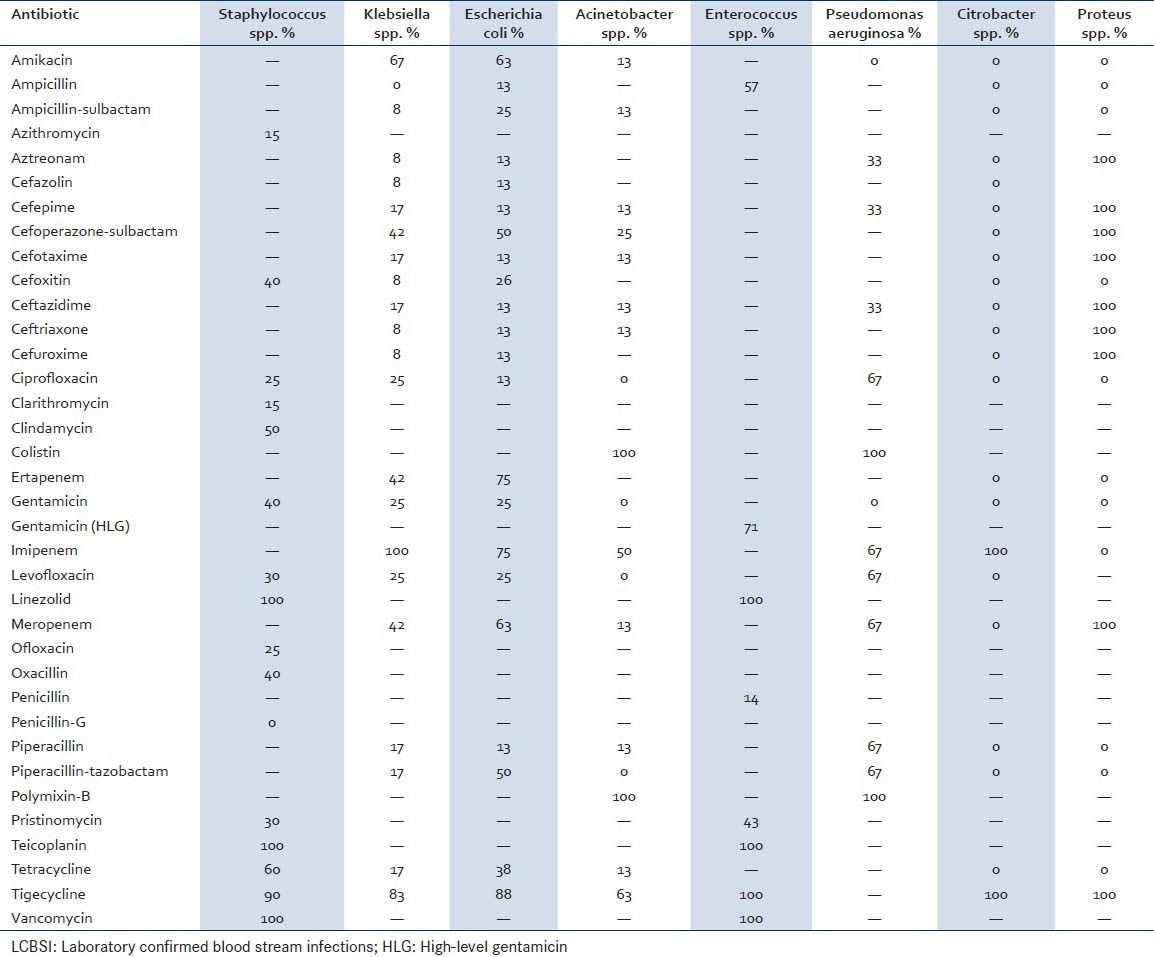
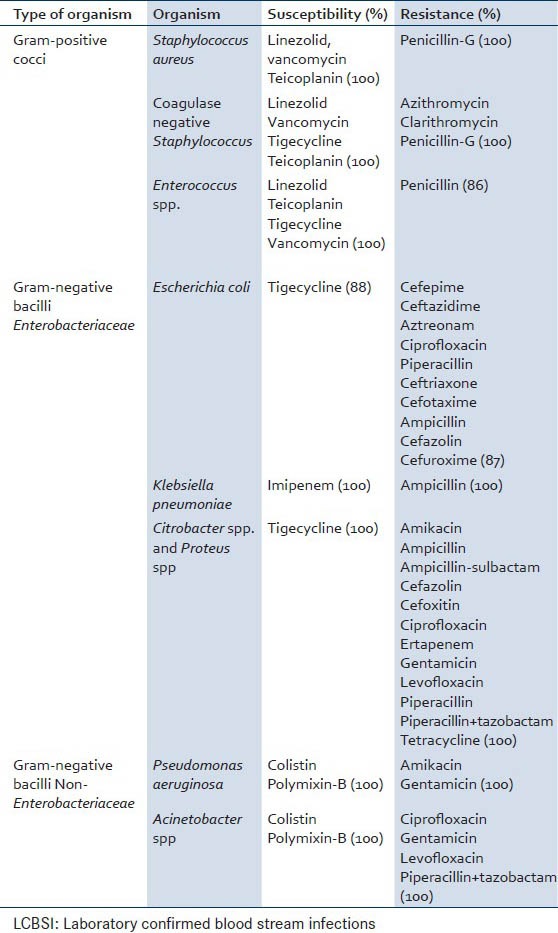
Table 2.
Highest susceptibility and highest resistance pattern of bacterial pathogens causing LCBSI
Outcome
The patients of LCBSI were followed for treatment and outcome; overall crude mortality in LCBSI was 40% (24/60 deaths). The mortality in Gram-negative LCBSI (55%) was higher than that in Gram-positive LCBSI (22%).
DISCUSSION
Blood stream infections are an important cause of morbidity and mortality in hospitalized patients. This study showed that both Gram-positive and Gram-negative bacteria were responsible for BSI, most of which were multidrug-resistant. Blood culture positivity was seen in 60/829 (7.2%) cases. In the literature, positivity in the range of 5-44% has been reported.[8,9,10,11,12,13,14] This wide range may be due to various reasons including:
The difference in the method of isolation and identification of organisms,
The difference in the type of patients recruited for the study, as in the present study patients recruited belonged to different categories, while most of the other studies were conducted on cohort belonging to a specific category of patients such as infective endocarditis and neonatal septicemia,
Difference in geographical regions, as these studies were conducted in different parts of the country and
Exposure of patients to a self-prescribed antibiotic, which is very common due to the availability of medicines over the counter without prescription in the country.
In our study, the incidence of Gram-positive organisms was 45%, while 55% were Gram-negative, this is in accordance with the study done by Arora et al. where the incidence of Gram-positive organisms was 52.67% while 47.33% isolates were Gram-negative.[8] Other studies have reported a varied incidence (11.2-25%) of Gram-positive and (15-88.8%) Gram-negative organisms. The emerging higher incidence of Gram-negative organisms especially in the hospital settings is alarming.[9,10,15,16] In our study Staphylococcus spp. were most prevalent similar to other studies with 21.66% incidence of S. aureus and 11.6% of CONS.[8,9] In a study done by Arora et al. S. aureus was isolated in 27.37% of cases and CONS in 20.16% of the cases.[8] However, Roy et al. have reported CONS 16.5% and S. aureus 14% in neonatal septicemia.[13]
Among the Gram-negative bacilli 37% belonged to Enterobacteriaceae family including E. coli, K. pneumoniae, Citrobacter spp., and Proteus spp. One more member of the family — Enterobacter spp., which is also an important nosocomial pathogen, was not observed in any of our patients; though in the literature, Enterobacter spp. have been reported in 14.19% and 22.9% LCBSI cases.[8,17] E. coli was isolated from 13% of the cases, at par with previous reports where 9.27% and 14.4% positivity has been reported.[8,18] Citrobacter spp. and Proteus spp. were very few showing only 2% positivity each, consistent with the previous findings.[8,18] K. pneumoniae was observed in 20% of the cases, which is in concert with the report by Surinder et al. who have reported 25.8% isolation, whereas it is high above than the report by Arora et al. where 5.76% positivity has been reported.[8,18]
Amongst other Gram-negative bacilli, P. aeruginosa, one of the important nil-fermenters was isolated in 5% of the cases consistent with the previous findings showing 7.62% and 5.9% positivity.[8,18] Acinetobacter spp., which is an emerging cause of late onset septicemia, was isolated in 13% of the cases, similar to study performed by Arora et al. showing 12.13% positivity.[19] In our cohort, number of BSI positive individuals increased with age until 80 years and dropped considerably for the age group >80 years. The reason for this observation could be a decrease in the immunity of patients with an increase in age resulting in making the individual more prone to infections, and due to recruitment of very few patients having >80 years of age a sharp decline in BSI positivity for 80-100 years age group was observed.
The organisms isolated from the cohort of suspected cases of BSI showed resistance to various antimicrobial agents. Among Gram-positive organisms maximum resistance was shown against azithromycin, clarithromycin, erythromycin, penicillin-G (100%). Penicillin was the common antibiotic against which all Gram-positive organisms showed resistance although percent resistance was 85% in case of Enterococcus spp. and 100% in Staphylococcus spp. In other studies, high level of resistance has been reported with ampicillin and erythromycin.[8,20,21] Most of the Gram-negative organisms showed resistance to three or more groups of antibiotics. Other workers also have reported the majority of Gram-negative isolates in their study as multidrug resistant.[13] The rapid development and spread of antibiotic resistance occurs due to nonjudicial use of antibiotics.
There are studies where prevalence of BSI in specific cohorts or by specific pathogens has been looked for. The study by Prabhash et al. reports BSI in cancer patients, where they have found Gram-negative bacteria as more common pathogens; and have reported poor activity of primary empirical agents of BSI.[22] Another study by Tak et al. reports Staphylococci as causative agents of 41% of BSI in trauma patients associated with high mortality.[23]
CONCLUSION
Our study thus provides information regarding prevalence and antibiotic susceptibility patterns of bacteria isolated from LCBSI. Such data is important in the selection of empirical antimicrobial treatment of BSI cases; in showing a path for clinical research and as an aid in educating and spreading awareness among medical personnel. This also draws our attention toward emerging trends of antibiotic resistance in pathogenic bacteria. However, as this was a single center study of limited duration, the results cannot be generalized for guiding empirical therapy of BSI in the area. It is highly recommended that more studies, involving patients from multiple medical centers are done to throw light on the epidemiology of infectious diseases, and the resistance patterns of common pathogens; to make practicing physicians aware of the magnitude of an existing problem of antibacterial resistance, so that they join hands in fighting this deadly threat by rational prescription of drugs.
Footnotes
Source of Support: Nil.
Conflict of Interest: None declared.
REFERENCES
- 1.Digiovine B, Chenoweth C, Watts C, Higgins M. The attributable mortality and costs of primary nosocomial bloodstream infections in the intensive care unit. Am J Respir Crit Care Med. 1999;160:976–81. doi: 10.1164/ajrccm.160.3.9808145. [DOI] [PubMed] [Google Scholar]
- 2.Orsi GB, Di Stefano L, Noah N. Hospital-acquired, laboratory-confirmed bloodstream infection: Increased hospital stay and direct costs. Infect Control Hosp Epidemiol. 2002;23:190–7. doi: 10.1086/502034. [DOI] [PubMed] [Google Scholar]
- 3.Sharma R, Sharma CL, Kapoor B. Antibacterial resistance: Current problems and possible solutions. Indian J Med Sci. 2005;59:120–9. [PubMed] [Google Scholar]
- 4.Kamat U, Ferreira A, Savio R, Motghare D. Antimicrobial resistance among nosocomial isolates in a teaching hospital in Goa. Indian J Community Med. 2008;33:89–92. doi: 10.4103/0970-0218.40875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Hara CM, Weinstein MP, Miller JM. Manual of Clinical Microbiology. 8th ed. Washington, DC: ASM Press; 2003. Manual and automated systems for detection and identification of microorganisms; p. 185. [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. Wayne: CLSI; 2012. Performance Standards for Antimicrobial Susceptibility Testing. 22nd Informational Supplement. [Google Scholar]
- 7.Horan TC, Andrus M, Dudeck MA. CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control. 2008;36:309–32. doi: 10.1016/j.ajic.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Arora U, Devi P. Bacterial profile of blood stream infections and antibiotic resistance pattern of isolates. J K Sci. 2007;9:186–90. [Google Scholar]
- 9.Khanal B, Harish BN, Sethuraman KR, Srinivasan S. Infective endocarditis: Report of a prospective study in an Indian hospital. Trop Doct. 2002;32:83–5. doi: 10.1177/004947550203200208. [DOI] [PubMed] [Google Scholar]
- 10.Sharma M, Goel N, Chaudhary U, Aggarwal R, Arora DR. Bacteraemia in children. Indian J Pediatr. 2002;69:1029–32. doi: 10.1007/BF02724380. [DOI] [PubMed] [Google Scholar]
- 11.Chaudhury A, Rao TV. Bacteraemia in a tertiary care urban hospital in South India. Indian J Pathol Microbiol. 1999;42:317–20. [PubMed] [Google Scholar]
- 12.Mitra S, Panigrahi D, Narang A. Anaerobes in neonatal septicaemia: A cause for concern. J Trop Pediatr. 1997;43:153–5. doi: 10.1093/tropej/43.3.153. [DOI] [PubMed] [Google Scholar]
- 13.Roy A, Maiti PK, Adhya S, Bhattacharya A, Chakraborty G, Ghosh E, et al. Neonatal candidemia. Indian J Pediatr. 1993;60:799–801. doi: 10.1007/BF02751051. [DOI] [PubMed] [Google Scholar]
- 14.Dhawan A, Grover A, Marwaha RK, Khattri HN, Anand IS, Kumar L, et al. Infective endocarditis in children: Profile in a developing country. Ann Trop Paediatr. 1993;13:189–94. doi: 10.1080/02724936.1993.11747644. [DOI] [PubMed] [Google Scholar]
- 15.Karpuch J, Azizi I, Beer S. Bacteraemia among children in hospital — A four year prospective study. J Infect. 1984;9:139–42. doi: 10.1016/s0163-4453(84)91000-4. [DOI] [PubMed] [Google Scholar]
- 16.Chugh K, Aggarwal BB, Kaul VK, Arya SC. Bacteriological profile of neonatal septicemia. Indian J Pediatr. 1988;55:961–5. doi: 10.1007/BF02727838. [DOI] [PubMed] [Google Scholar]
- 17.Roy I, Jain A, Kumar M, Agarwal SK. Bacteriology of neonatal septicaemia in a tertiary care hospital of Northern India. Indian J Med Microbiol. 2002;20:156–9. [PubMed] [Google Scholar]
- 18.Kumar S, Rizvi M, Vidhani S, Sharma VK. Changing face of septicaemia and increasing drug resistance in blood isolates. Indian J Pathol Microbiol. 2004;47:441–6. [PubMed] [Google Scholar]
- 19.Arora U, Jaitwani J. Acinetobacter spp. An emerging pathogen in neonatal septicemia in Amritsar. Indian J Med Microbiol. 2006;24:81. doi: 10.4103/0255-0857.19911. [DOI] [PubMed] [Google Scholar]
- 20.Guha DK, Jaspal D, Das K, Guha AR, Khatri RL, Kumar RS. Outcome of neonatal septicemia: A clinical and bacteriological profile. Indian Pediatr. 1978;15:423–7. [PubMed] [Google Scholar]
- 21.Karki BM, Parija SC. Analysis of blood culture isolates from hospitalized neonates in Nepal. Southeast Asian J Trop Med Public Health. 1999;30:546–8. [PubMed] [Google Scholar]
- 22.Prabhash K, Medhekar A, Ghadyalpatil N, Noronha V, Biswas S, Kurkure P, et al. Blood stream infections in cancer patients: A single center experience of isolates and sensitivity pattern. Indian J Cancer. 2010;47:184–8. doi: 10.4103/0019-509X.63019. [DOI] [PubMed] [Google Scholar]
- 23.Tak V, Mathur P, Lalwani S, Misra MC. Staphylococcal blood stream infections: Epidemiology, resistance pattern and outcome at a level 1 Indian trauma care center. J Lab Physicians. 2013;5:46–50. doi: 10.4103/0974-2727.115939. [DOI] [PMC free article] [PubMed] [Google Scholar]


