Summary
The SWI/SNF complex plays an important role in mouse embryonic stem cells (mESCs), but it remains to be determined whether this complex is required for the pluripotency of human ESCs (hESCs). Using RNAi, we demonstrated that depletion of BRG1, the catalytic subunit of the SWI/SNF complex, led to impaired self-renewing ability and dysregulated lineage specification of hESCs. A unique composition of the BRG1-SWI/SNF complex in hESCs was further defined by the presence of BRG1, BAF250A, BAF170, BAF155, BAF53A, and BAF47. Genome-wide expression analyses revealed that BRG1 participated in a broad range of biological processes in hESCs through pathways different from those in mESCs. In addition, chromatin immunoprecipitation sequencing (ChIP-seq) demonstrated that BRG1 played a repressive role in transcriptional regulation by modulating the acetylation levels of H3K27 at the enhancers of lineage-specific genes. Our data thus provide valuable insights into molecular mechanisms by which transcriptional repression affects the self-renewal and differentiation of hESCs.
Highlights
-
•
We report a conserved but distinct role of SWI/SNF complex in hESC pluripotency
-
•
Depletion of BAF170 in SWI/SNF complex leads to impaired pluripotency of hESCs
-
•
BRG1 negatively regulates transcription of lineage-specific genes
-
•
BRG1 downregulates H3K27ac levels at enhancers of lineage-specific genes
In this article, Wang and colleagues show that a unique composition of the BRG1-SWI/SNF complex is required for maintaining self-renewal and proper differentiation of human embryonic stem cells. In addition, they demonstrate that BRG1 participates in a broad range of biological processes in human stem cells through transcriptional repression via downregulation of the acetylation levels of H3K27 at enhancers of lineage-specific genes.
Introduction
Embryonic stem cells (ESCs) are originally derived from the inner cell mass. They are pluripotent cells that can self-renew and differentiate into multiple cell types upon appropriate stimuli. Mouse ESCs (mESCs) possess a distinctive global chromatin structure that is characterized by hyperdynamic plasticity and bivalent domains marked by both active H3K4me3 (trimethylation of histone 3 lysine 4) and repressive H3K27me3 (trimethylation of histone 3 lysine 4) in the promoter regions of lineage-specific genes (Bernstein et al., 2006; Meshorer et al., 2006). In human ESCs (hESCs), H3K4me1 (monomethylation at lysine 4 of histone 3), H3K27ac (acetylation at lysine 27 of histone 3), and p300 marked chromatin loci were recently identified as active enhancers that drive gene expression (Rada-Iglesias et al., 2011). Conversely, a lack of H3K27ac but enrichment with H3K27me3 is linked to repressed but “poised” elements for genes active in early development (Rada-Iglesias et al., 2011). The switch from self-renewal to differentiation requires the participation of multiple epigenetic factors, including chromatin regulators, noncoding RNAs, and histone modifiers (Surani et al., 2007; Tay et al., 2008).
One of these factors, SWI/SNF, is a large chromatin-remodeling complex that contains either BRG1 or BRM exclusively as the catalytic ATPase subunit that drives the alteration of DNA-nucleosome structure and thus regulates gene transcription (Fryer and Archer, 1998; Trotter and Archer, 2008; Wang et al., 1996). Previous studies revealed that deletions of BRG1 or core subunits of the SWI/SNF complex, such as BAF155 and BAF47, led to peri-implantation lethality due to compromised survival of totipotent cells that give rise to both the inner cell mass and trophoblast, suggesting a requirement of the SWI/SNF complex for totipotency in vivo (Bultman et al., 2005; Kim et al., 2001; Klochendler-Yeivin et al., 2000). In addition, deficiency of SWI/SNF components also affects the developmental potential of various types of cells (e.g., neurons, hematopoietic cells, and germ cells) (Chi et al., 2003; Gebuhr et al., 2003; Griffin et al., 2008; Hang et al., 2010; Kim et al., 2012; Wang et al., 2012; Yoo and Crabtree, 2009), supporting the notion that the SWI/SNF complex plays a role in tissue development. Recently, a series of reports demonstrated that the SWI/SNF complex also plays a role in the pluripotency of mESCs. Deficiency of BRG1, BAF155, or BAF250a/b impaired the ability of mESCs to proliferate and to differentiate into three germ layers (Gao et al., 2008; Ho et al., 2009a, 2009b, 2011; Kaeser et al., 2008; Kidder et al., 2009; Schaniel et al., 2009; Yan et al., 2008). Chromatin immunoprecipitation-coupled DNA sequencing (ChIP-seq) revealed that BRG1 colocalizes with core pluripotent factors (OCT4 and SOX2) at a large number of loci in target genes to fine-tune their regulation. In addition, BRG1 also potentiates LIF/STAT3 signaling in mESCs by opposing polycomb (PcG) function via alteration of H3K27me3 levels (Ho et al., 2009a, 2009b, 2011; Kidder et al., 2009).
The mammalian SWI/SNF complex contains 15 core subunits called BRG1- or BRM-associated factors (BAFs), including several ones not found in yeast (Kadoch et al., 2013). Different assemblies of BAF components have been implicated in their tissue-specific functions (Wu et al., 2009). For instance, BAF60c, but not BAF60b, protein is selectively expressed in embryonic heart, and its deficiency results in defective cardiac development and embryonic lethality at 10–11 days postcoitum (dpc) (Lickert et al., 2004). In mESCs, BRG1 (but not BRM) and BAF155 (but not BAF170) are enriched with BAF60a to form the so-called “esBAF” complex (Ho et al., 2009b). Deficiency of either BRG1 or BAF155 leads to a loss of pluripotency in mESCs that cannot be rescued by overexpression of BAF170 (Ho et al., 2009b). Thus, the specific composition of the BAF is critical for its function in mESCs.
Although hESCs and mESCs share a number of core pluripotency-related transcription factors and cellular characteristics, hESCs differ remarkably in their clone morphology, cell-doubling time, responses to signal molecules, and culture conditions required for self-renewal. This may result in part from their distinct developmental stages and perhaps from an intrinsic human-mouse divergence. To date, only a few genes have been identified as sharing conserved roles in both hESCs and mESCs. The precise role played by the SWI/SNF complex in hESCs remains unclear. We thus sought to explore the functional relevance of the SWI/SNF complex in hESC pluripotency. Here, we demonstrated that knocking down either BRG1 or BAF170 instead of BAF155 led to the loss of pluripotency of hESCs and dysregulated lineage specification. Genome-wide arrays revealed a general upregulation of gene expression resulting from BRG1 deficiency, suggesting that BRG1 plays a repressive role in transcriptional regulation in hESCs. Further analyses showed that H3K27ac levels of lineage-specific genes were dramatically increased upon BRG1 knockdown, highlighting H3K27ac as an important marker to distinguish active from repressed gene loci in hESCs. Our study thus provides important functional and mechanistic insights into the role of the BRG1-SWI/SNF complex in regulating the pluripotency of hESCs by transcriptional repression of genes involved in early development.
Results
BRG1 Deficiency Impairs the Self-Renewal of hESCs
To investigate the importance of the SWI/SNF complex in hESCs, we established stable BRG1 knockdown lines that allowed us to evaluate the long-term effects of BRG1 depletion. At the end of the first passage after BRG1 knockdown for 7 days, the colonies with BRG1 depletion were apparently unable to maintain a normal stem cell phenotype in culture (Figure 1A). Consistent with this observation, the percentage of Tra-1-60+ or Tra-1-81+ cells was also remarkably decreased (Figure 1B; Figures S1A–S1C available online). In addition, most colonies totally lost alkaline phosphatase (AP) staining upon BRG1 depletion, and a minority of cells stained positive in the middle of the colonies (Figure 1A). By the end of the second passage, 14 days after BRG1 knockdown, there were few discernible AP+ colonies in culture, with Tra-1-60+ or Tra-1-81+ cells reduced to 20%–40% (Figures 1B and S1A–S1C). In contrast, we did not detect any obvious differentiation phenotype upon depletion of BRM (Figure S1A). Collectively, these data support an essential requirement of BRG1, but not BRM, for hESC maintenance.
Figure 1.
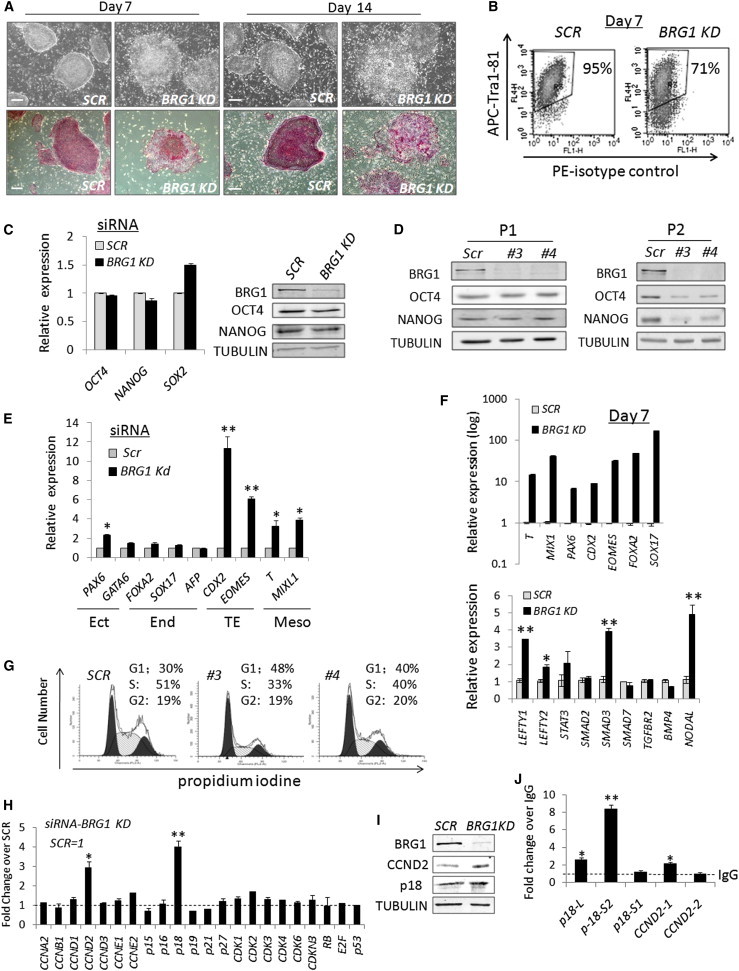
BRG1 Deficiency Impairs Self-Renewal of hESCs
(A and B) Morphology and AP staining (A) and flow-cytometry analyses (B) of Tra-1-81 in hESC colonies at day 7 or day 14 after shRNA knockdown. Scale bars, 400 μm.
(C and D) Expression of pluripotency-related transcription factors was examined on samples with transient siRNA knockdown (C) or on hESCs with shRNA knockdown at day 7 or day 14 (D) by real-time RT-PCR (histogram) or by western blots.
(E and F) Quantitative gene expression on samples with siRNA knockdown (E) or shRNA knockdown at day 7 (F) by real-time RT-PCR. Ect, ectoderm; End, endoderm; TE, trophectoderm; Meso, mesoderm.
(G) Cell-cycle profiles of control and BRG1-deficient hESCs analyzed by flow cytometry at day 14 after BRG1 knockdown.
(H) Real-time RT-PCR analyses on cell-cycle regulators upon transient siRNA BRG1 knockdown for 48 hr compared with a scrambled siRNA control.
(I) Western blots on control and BRG1-depleted hESCs with antibodies against CCND2 and p18 at day 14 after BRG1 knockdown.
(J) ChIP-PCR analyses with an antibody against BRG1 in different promoter regions of p18 and CCND2 in untreated hESCs. Three different p18 and two CCND2 primers were used. The number in the primer names indicates the distance of the amplified region upstream from the TSS. Fold changes for percentage of input are presented relative to the immunoglobulin G (IgG) control. SCR, control samples with scrambled siRNAs or shRNAs; BRG1 KD, samples with siRNAs or shRNAs against BRG1.
Four different shRNAs against BRG1 were used in this study and yielded similar results. Individual shRNA is labeled as #3 or #4. PCR data (E, H, J, and the lower panel in F) are presented as the average of six biological replicates ± 1 SEM from three independent experiments; ∗p < 0.05, ∗∗p < 0.01. Data in the upper panel of (F) are the average of biological duplicates ± 1 SEM from one representative experiment. Also see Figure S1.
Notably, although the colonies rapidly lost ESC properties upon BRG1 knockdown, the expression of pluripotency-related core transcription factors such as OCT4 and NANOG remained unchanged at both the transcript and protein levels (Figures 1C and 1D). Alterations in the expression of these pluripotency markers became apparent only after prolonged loss of BRG1 for 14 days (Figure 1D). By contrast, the levels of key genes required for early germ-layer differentiation (Wang et al., 2008), especially those involved in the mesoderm (BRAYCHURY/T and MIXL1) and trophectoderm (CDX2 and EOMES), significantly increased 48 hr after transient BRG1 knockdown (Figure 1E). By day 7, the expression of many genes required for germ-layer differentiation, as well as several important signaling factors (such as LEFTY1, SMAD3, and NODAL), was increased remarkably before any significant alteration of core pluripotency-related genes, such as OCT4 and NANOG, was observed (Figure 1F). Therefore, it is unlikely that BRG1 affects the expression of these genes in early development via downregulation of OCT4 and NANOG.
ESCs are known to lack cyclin D-associated kinase activity, which may contribute to their characteristic cell-cycle profile with a shortened G1 phase and high percentage of cells at S phase (Becker et al., 2006; Neganova et al., 2009; Stead et al., 2002). We observed that the cell-cycle profile in hESCs was significantly altered at approximately day 14 after BRG1 knockdown, with more cells arresting at G1 phase and fewer cells having progressed through S phase (Figure 1G). Consistent with these data, the expression levels of two important cell-cycle regulators, p18 and CCND2, were increased in BRG1-deficient ESCs (Figures 1H, 1I, and S1E). In addition, ChIP analyses in hESCs demonstrated that BRG1 was specifically enriched at 540 bp and 908 bp upstream of the transcription start sites (TSSs) in the promoter regions of p18 and CCND2, respectively, suggesting that BRG1 directly regulates these two genes (Figure 1J). Collectively, these results suggest that BRG1 deficiency compromises the cell-cycle progression of hESCs.
BRG1 Deficiency Compromises the Differentiation Potential of hESCs
In order to examine whether BRG1 affects the differentiation potential of hESCs, embryoid bodies (EBs) were produced following BRG1 depletion. Compared with scrambled small hairpin RNA (shRNA) controls, the BRG1-deficient EBs appeared to be smaller (Figure 2A). Gene-expression analyses demonstrated that the BRG1-depleted EBs did not differentiate properly and showed significantly increased neuron ectoderm (PAX6 and NESTIN) and moderately enhanced endoderm (SOX17, FOXA2, and CDX2) formation, whereas the mesoderm lineages (BRACHYURY/T, MIXL1, and GATA6) appeared to be hindered at different time points during EB development (Figure 2B).
Figure 2.
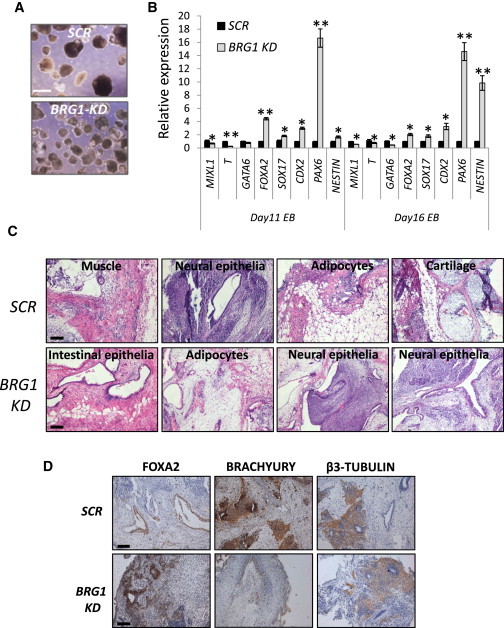
BRG1 Deficiency Compromises the Differentiation Potential of hESCs
(A) Morphology of EBs from control and BRG1 knockdown hESCs. Scale bar, 400 μm.
(B) Quantitative RT-PCR for expression of genes involved in three-germ-layer formation at different time points (day 11 and day 16) during EB differentiation from control and BRG1 knockdown hESCs. All RT-PCR data are presented as the average of six biological replicates ± 1 SEM from three independent experiments; ∗p < 0.05, ∗∗p < 0.01.
(C) Histological studies of teratoma sections from control and BRG1-depleted hESCs.
(D) IHC on teratoma sections with antibodies against the markers for endoderm (endo), mesoderm (meso), or ectoderm (ecto) derivatives. Scale bars, 200 μm.
Consistent with these in vitro results, teratomas that formed from BRG1-deficient hESCs mainly contained pigmented neural epithelia and, to a lesser degree, intestinal endothelial cells. Conversely, mesoderm-derived lineages (such as smooth muscle, bone, and cartilage) were hardly detectable (Figure 2C). Immunohistochemistry (IHC) analysis further confirmed the reduced expression of BRACHYURY (mesoderm), whereas the ectoderm (β-TUBULIN) and endoderm (FOXA2) markers were largely unaffected (Figure 2D). Taken together, these results revealed that the mesendodermal differentiation potential was markedly impaired in BRG1-deficient hESCs.
The BRG1-SWI/SNF Complex Contains both BAF170 and BAF155 in hESCs
Previous reports suggested that the unique composition of BAFs defines their specific functions (Wu et al., 2009). BAF155 (but not BAF170) forms homodimers in the mouse esBAF complex and plays an important role in the self-renewal of mESCs (Ho et al., 2009b). To determine the precise composition of the BRG1-SWI/SNF complex in hESCs, we performed gel-filtration analyses. We observed that both BAF170 and BAF155 were found with BAF250A, BAF57, BAF53A, and BAF47 in BRG1-enriched fractions/complexes in this assay (Figure 3A), constituting a different pattern compared with that reported for mESCs (Ho et al., 2009b). We further performed a coimmunoprecipitation (coIP) assay with an antibody against BRG1, followed by mass spectrometry analyses (Figure 3B). Using spectra counts as a measure for relative protein abundance, we found that BRG1, BAF155, BAF170, BAF60A, and BAF250A were enriched with BAF53A and BAF47 in this assay (Figure 3C). These proteins contributed to more than 80% of the total identified spectra counts of the SWI/SNF complex in hESCs, whereas the levels for BAF250B, BAF200, BAF180, and BAF60C, as well as BCL11A/B (the newly identified BAF subunits in human T leukemia cells; Kadoch et al., 2013), were relatively low in BRG1-immunoprecipitated fraction (Figure 3C). We confirmed these results by coIP assays. Particularly, we found that both BAF155 and BAF170 were pulled down with BRG1, and BAF170 and BRG1 were copurified with BAF155 from hESCs (Figure 3D). Taken together, these results revealed a unique composition of the BRG1-SWI/SNF complex in hESCs containing both BAF155 and BAF170.
Figure 3.
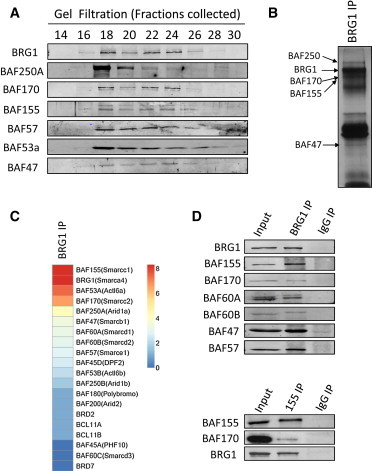
Determination of BRG1-SWI/SNF Composition in Wild-Type hESCs
(A) Gel-filtration analyses of hESCs blotted with antibodies against BRG1 or other BAF subunits.
(B) Silver-stained images of a BRG1 pull-down sample.
(C) Relative protein abundance of SWI/SNF components immunoprecipitated with BRG1, calculated according to their spectra counts. Red, the most abundant; blue, the least abundant.
(D) CoIP with antibodies against either BRG1 or other BAF subunits on hESCs. Antibodies for western blots are indicated to the left of the panels.
The Unique BRG1-SWI/SNF Composition Is Critical for Self-Renewal of hESCs
To investigate whether this unique assembly of the SWI/SNF complex is critical for maintaining the pluripotency of hESCs, we knocked down individual SWI/SNF components by RNAi. To our surprise, we observed no obvious differentiation phenotype (as indicated by morphology, AP staining, expression of OCT4, and surface antigen analyses) upon BAF155 knockdown (Figures 4A–4C, S2A, and S2B). This result was confirmed by several independent assays with different shRNAs against BAF155 (Figure S2A). Instead, BAF170 deficiency led to a detectable differentiation of hESCs (Figure 4A). The differentiation phenotype upon BAF170 knockdown was further validated by the loss of AP staining at the edges of the ESC colonies, reduced expression of OCT4, and decreased percentage of Tra-1-60+ or Tra-1-81+ cells (Figures 4A–4C and S2B).
Figure 4.
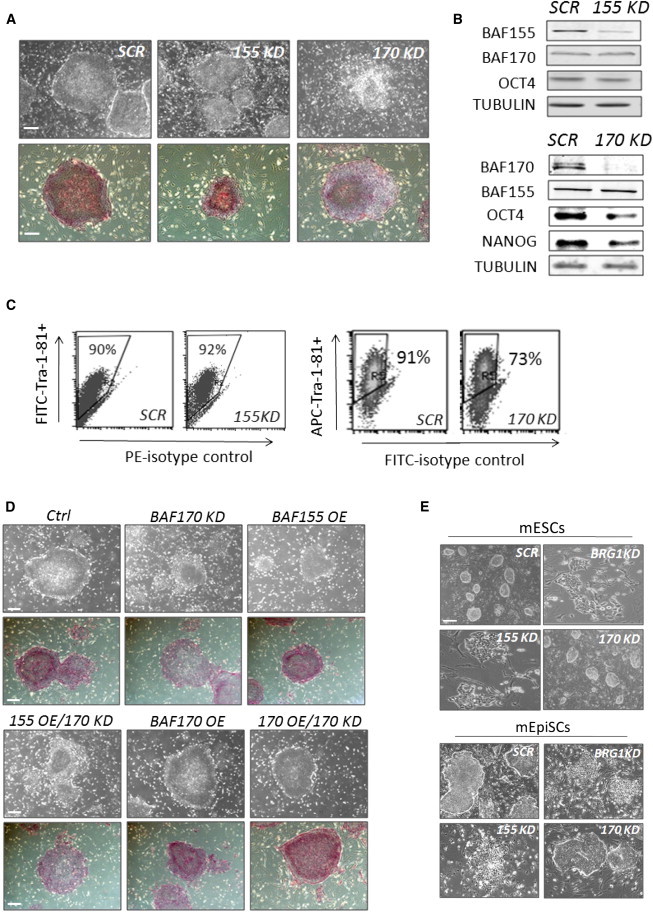
Unique BRG1-SWI/SNF Composition Is Important to Maintain the Stem Cell Phenotype of hESCs
(A) Morphology and AP staining of hESC colonies upon knockdown of BAF155 or BAF170 in hESCs.
(B) Expression of BAF components, OCT4, and NANOG was examined by western blotting upon knockdown of BAF155 or BAF170 in hESCs.
(C) Flow-cytometry analyses of the surface marker Tra-1-81 on hESCs upon depletion of BAF155 or BAF170.
(D) Morphology of hESC colonies upon BAF170 knockdown and/or expression of V5-tagged BAF155 or BAF170 cDNAs. OE, overexpression; Ctrl, empty vector control for overexpression analyses.
(E) Morphology of colonies from mESCs and mEpiSCs upon depletion of individual SWI/SNF components.
Scale bars in (A), (D), and (E), 400 μm. See also Figures S2 and S3.
To exclude the possibility that the differentiation phenomena observed upon BAF170 deficiency were caused by off-target effects of RNAi, we introduced BAF170 or BAF155 cDNAs into hESCs containing a scrambled shRNA or an shRNA against BAF170 (Figure S2C). Notably, we observed that BAF155 overexpression could not rescue the differentiation phenotype caused by BAF170 knockdown, but ectopic overexpression of BAF170 could (Figure 4D). This finding explicitly demonstrated that the BAF170-containing BRG1-SWI/SNF complex was required for the self-renewal of hESCs.
Recent studies showed that hESCs exist in a primed pluripotent stage, similarly to mouse epiblast stem cells (mEpiSCs), whereas mESCs represent a pluripotent population in a naive state (Brons et al., 2007; Tesar et al., 2007). To determine whether this unique assembly of the BRG1-SWI/SNF complex in hESCs is actually due to their more advanced developmental stage compared with mESCs, we investigated the roles of individual BRG1-SWI/SNF components in mESCs and EpiSCs by RNAi (Figures S3A–S3C). We observed that knocking down BRG1 or BAF155, rather than BAF170, led to the loss of the pluripotent features in both naive mESCs and primed mEpiSCs (Figure 4E), thereby supporting a unique roll for t BAF170 in the BRG1-SWI/SNF complex in hESCs.
BRG1 Deficiency Affects a Broad Range of Biological Processes in hESCs
To elucidate how the SWI/SNF complex functions in hESCs, we knocked down BRG1 by transient transfection of small interfering RNAs (siRNAs) against BRG1 in both mESCs and hESCs (Figure 5A), compared their gene-expression profiles with microarray analyses, and further validated the results by real-time RT-PCR assays (Figures 5B and 5C).
Figure 5.
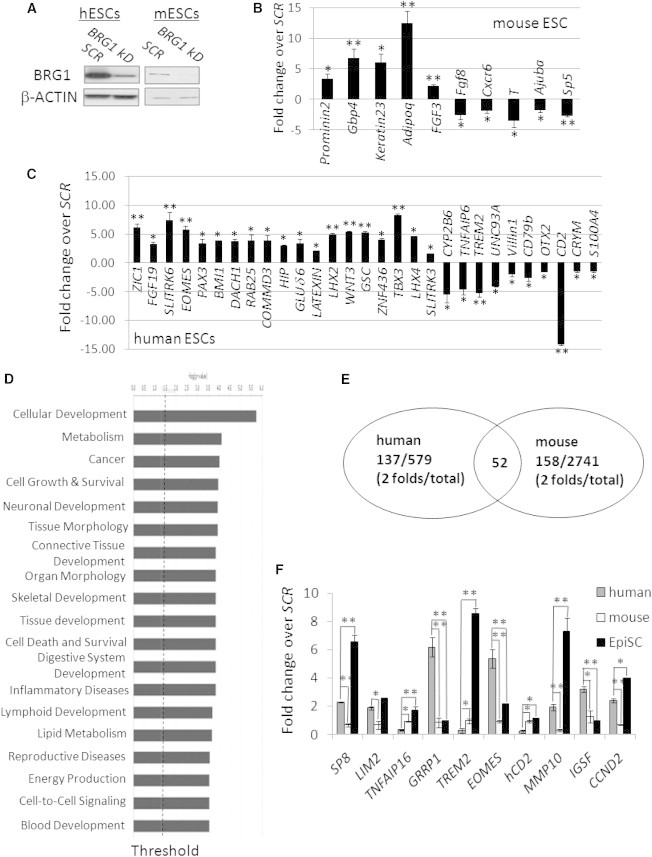
BRG1 Depletion Affects a Broad Range of Biological Processes in hESCs
(A) Knockdown of BRG1 in hESCs and mESCs was verified by western blots.
(B and C) Quantitative RT-PCR validation for genes differentially expressed in microarray analyses. RNA samples were collected from either mESCs (B) or hESCs (C) transfected with siRNAs against BRG1.
(D) Genes affected by BRG1 knockdown in hESCs were analyzed by IPA according to their functions.
(E) Venn diagram with the numbers of genes affected by BRG1 knockdown in hESCs and mESCs. The number in the overlapping circles indicates the number of common genes altered in both hESCs and mESCs upon knockdown.
(F) Expression levels of the same set of genes upon BRG1 depletion in hESCs, mESCs, or mEpiSCs examined by quantitative RT-PCR (fold changes of expression relative to their own scrambled shRNA controls).
All PCR data are presented as the average of six biological replicates ± 1 SEM from three independent experiments; ∗p < 0.05, ∗∗p < 0.01. See also Figure S4.
We identified 631 genes that were altered upon BRG1 depletion in hESCs and further classified them according to their functions and pathways using Ingenuity Pathway Analysis (IPA). These genes were associated with more than 20 categories of biological functions above the threshold, the most overrepresented of which included cancer, cellular/embryonic development, nervous system development, cell death and survival, and tissue morphology (Figures 5D and S4). Consistent with these functional distributions, the top-ranked networks were associated with neurological disorders, cellular organization, and cell-to-cell interactions and signaling (Figures 5D and S4). Specifically, genes important for cell-cycle regulation (e.g., CCND2 and CDKL3) and lineage development (e.g., EOMES, ZIC1, and OSR2) were significantly altered in BRG1-deficient hESCs, in agreement with our findings regarding the role of BRG1 in regulating the self-renewal and pluripotency of hESCs. In addition, BRG1 regulated many genes that are involved in metabolism, such as ALDH1A1, CYP2B1/2B6/2R1, LRAT, AKR1C1/C3, and RDH10 (Figure 5B; Table S1). We also noticed that several molecules involved in cell adhesion, such as cadherins (PCDH20, PCDH18, and PCDH15) and GJB5, were decreased in BRG1-deficient hESCs (Table S1). Collectively, these data demonstrated that BRG1 participated in a broad range of biological processes in hESCs in addition to its role in maintaining hESC self-renewal and differentiation.
Among the genes that were affected by BRG1 deficiency in hESCs, only a small set (52 out of 631) was also altered in mESCs upon BRG1 knockdown (Figure 5E). In addition, although most of the functional categories affected by BRG1 depletion were similar between hESCs and mESCs, the exact pathways and genes that were perturbed were quite different (Figure S4A). For example, several genes involved in embryonic development (such as SP8, EOMES, TREM2, and LIM2) and cell-cycle regulation (such as CCND2) were altered differentially upon BRG1 knockdown in hESCs and mESCs (Figure 5F). Interestingly, we further revealed that half of these genes (e.g., TNFAIP6, GRRP1, CD2, MMP10, and IGSF1) were differently affected by BRG1 depletion in hESCs from mEpiSCs (Figure 5F), indicating an intrinsic human-mouse divergence for pathways regulated by BRG1 in stem cells at the same primed stage.
BRG1 Plays a Repressive Role in Transcriptional Regulation in hESCs
From microarray analyses, we identified that 427 out of 579 genes that were differentially expressed upon BRG1 deficiency were upregulated (Figure 6A), suggesting that BRG1 primarily plays a repressive role in hESCs. Recent reports demonstrated that active enhancers (class I) in hESCs were marked by H3K4me1, H3K27ac, and p300 binding (Rada-Iglesias et al., 2011). In contrast, the “poised” enhancers (class II) for genes expressed in early development were characterized by a lack of H3K27ac (Rada-Iglesias et al., 2011). We interrogated these published ChIP-seq data and revealed that 33.5% of class II enhancers were colocalized within ±250 bp of BRG1-binding peaks (Table S3). Since a high H3K27ac level marks active gene expression, we further explored whether BRG1 could repress the expression of lineage-specific genes by affecting histone acetylation.
Figure 6.
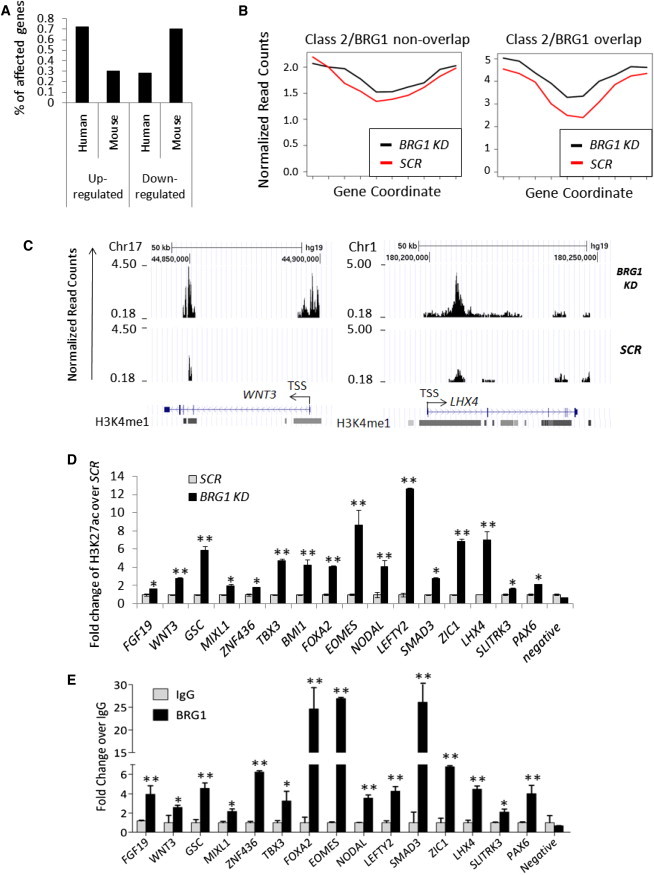
BRG1 Plays a Repressive Role in Transcriptional Regulation in hESCs
(A) Percentage of upregulated and downregulated genes upon knockdown of BRG1 in hESCs, based on microarray data.
(B) H3K27ac levels in control (SCR) and BRG1 knockdown (KD) hESCs were examined in class II enhancer regions, which were divided into two groups according to their overlap with BRG1-binding peaks. The genomic regions of the enhancers were divided equally into ten bins (shown as the x axis). The normalized read counts of H3K27ac ChIP-seq data for each bin (shown as the y axis) was calculated by the reads in that region divided by the total reads in that library (SCR versus BRG1 KD) to exclude the potential variation introduced by different sequencing coverage.
(C) The H3K27ac ChIP-seq signals from SCR control and BRG1 KD hESCs at two representative genomic loci (WNT3 and LHX4) are shown. H3K4me1 signals based on the UCSC custom track deposited by the ENCODE/Broad Institute are shown in grayscale.
(D) ChIP-PCR assays with an antibody against H3K27ac were performed on genes important for lineage differentiation from control hESCs and cells with BRG1 depletion.
(E) ChIP-PCR assays with control IgG or an antibody against BRG1 in the same regions described in (D) in wild-type hESCs.
All PCR data are presented as the average of six biological replicates ± 1 SEM from three independent experiments; ∗p < 0.05, ∗∗p < 0.01. See also Figures S5 and S6.
We performed genome-wide ChIP-seq analyses using an antibody specifically against H3K27ac on control and BRG1-depleted hESCs around day 7 after knockdown before the expression of key pluripotency factors (e.g., OCT4 and NANOG) was altered. We observed that H3K27ac levels within 5 kb of the TSS of 5,914 genes were upregulated upon BRG1 knockdown compared with scrambled control cells (Table S5). We observed that H3K27ac levels within 5 kb of the TSS of 5,914 genes were upregulated upon BRG1 knockdown compared with scrambled control cells but were downregulated in only 623 genes (Table S5), thus demonstrating that BRG1 depletion led to a general elevation of H3K27 acetylation levels. We further compared the H3K27ac status in enhancer regions of published class II genes according to the enrichment of BRG1 in control and BRG1-depleted hESCs (Rada-Iglesias et al., 2011). Interestingly, we found that the increase of H3K27ac levels was more obvious at the loci that overlapped with BRG1-binding regions (Figure 6B), suggesting that the elevated H3K27ac levels in enhancer regions were indeed BRG1 dependent.
The genes with an increased H3K27ac level in the enhancer regions included those that are important for differentiation into trophectoderm (EOMES), neurons (LHX4, PAX6, ZIC1, and SLITRK3), mesoderm (MIXL1 and GSC), and endoderm (FOXA2), and signal molecules that are important for mesendodermal development (NODAL, LEFTY1/2, and WNT3) (Figures 6C, 6D, S5E, and S5F; Table S5). Their increased H3K27ac levels were confirmed by independent ChIP-PCR analyses (Figure 6D). Notably, correlating with their elevated H3K27ac status, the expression of these genes was upregulated upon BRG1 depletion as well (Figures 1E, 1F, and 5C). We further determined the BRG1-binding intensity in these enhancer regions by performing ChIP-PCR assays with an antibody against BRG1. Indeed, we observed a significant enrichment of BRG1 in the same regions with elevated H3K27ac levels (Figure 6E), thus proving that BRG1 depletion is responsible for increased H3K27 acetylation at these enhancers of genes that are active in development, and may in turn lead to their upregulated transcriptional activation.
Discussion
Although both mESCs and hESCs are derived from the inner cell mass of blastocysts, they differ remarkably in terms of colony morphology, cell-doubling time, and the signaling pathways that maintain their self-renewal capacity. The extent to which these differences are observed at a molecular level remains only partially described. The recent generation of primed EpiSCs has added another layer of complexity (Brons et al., 2007; Tesar et al., 2007). Although hESCs share some defining features with mEpiSCs in terms of growth properties and signaling responses, they differ from mEpiSCs in several aspects (De Los Angeles et al., 2012). For instance, hESCs express REX1, a marker associated with naive mESCs, but do not express FGF5, a key mEpiSC marker (Greber et al., 2010). These differences further complicate efforts to simply extrapolate the rodent paradigm to humans. In the case of the SWI/SNF complex, the nature and importance of its role in hESCs is an important area of investigation. In this study, we found that depletion of the major catalytic subunit of the SWI/SNF complex, BRG1, compromised the self-renewal capacity of hESCs and their murine counterparts, mESCs and mEpiSCs. In addition, BRG1 deficiency impaired mesodermal development from hESCs, as revealed by both in vitro EB differentiation and in vivo teratoma formation. We further demonstrated that BRG1 participated in a broad range of biological processes in hESCs through genes and pathways distinct from those of mESCs and mEpiSCs, thus indicating that BRG1 differently regulates the pluripotency of stem cells across species. Our data define an essential and conserved role of BRG1 in ESC biology, and also provide mechanistic insights into how the BRG1-mediated SWI/SNF complex affects the maintenance of human stem cells.
The SWI/SNF complex consists of 15 core subunits whose unique composition is critical for its distinct functions in different types of cells (Kadoch et al., 2013; Wu et al., 2009). In our study, we demonstrated that both BAF170 and BAF155 existed in the human BRG1-SWI/SNF complex and were enriched with BRG1, BAF53A, and BAF47. Depletion of BAF170 instead of BAF155 led to the loss of stem cell properties in hESCs. This pattern is different from that observed in their rodent counterparts (i.e., mESCs and mEpiSCs), suggesting an intrinsic functional divergence of BAF components between human and mouse. Although BAF155 and BAF170 share more than 60% homology in their protein sequences, their functions are not interchangeable. BAF155 is unable to correct the differentiated morphology caused by BAF170 depletion in hESCs. Similarly, knockdown of BAF53A, but not BAF53B, led to obvious differentiation of hESCs (Figure S2D). It is plausible that the BAF170- and BAF53A-containing SWI/SNF complex interacts with different partners in hESCs from BAF155 and in turn regulates a distinct set of target genes that are important for pluripotency.
OCT4, SOX2, and NANOG are core pluripotency-related transcription factors that maintain undifferentiated phenotypes of ESCs (Boyer et al., 2005). Our study demonstrated that BRG1 deficiency did not lead to any immediate alteration in expression of OCT4, SOX2, or NANOG. Changes in the protein levels of these pluripotency markers became apparent only after prolonged depletion of BRG1 (Figures 1C and 1D). By contrast, significantly increased expression of genes that are important for trophectodermal and mesendodermal lineage development was observed at the early stage of BRG1 knockdown. Several of those genes (e.g., EOMES, GSC, FOXA2, NODAL, LEFTY1/2, and WNT3) were direct targets regulated by BRG1 through modulation of H3K27ac levels, thus providing insights into epigenetic regulation during human embryogenesis. In addition, our data demonstrate that BRG1 participates in a broad range of biological functions of hESCs, including cell cycle, cell-to-cell interactions, and metabolism. Therefore, it is plausible that reduced expression of these pluripotency-related factors is secondary to prolonged cellular abnormalities and differentiation. Nevertheless, we cannot exclude the possibility that BRG1 coregulates target genes of OCT4 or NANOG. We interrogated the published ChIP-seq data using antibodies against BRG1, OCT4, or NANOG (Figure S6A). About 40% of OCT4-bound loci and 18%–38% of NANOG-occupied regions were enriched with BRG1 using ±250 bp as a cutoff distance between BRG1 and OCT4- or NANOG-enriched peak centers (Figure S5A). Although BRG1 colocalizes extensively with OCT4 and NANOG in hESCs, the overlap is far less than that in mESCs (Ho et al., 2009a), highlighting the different regulatory networks between human and mouse cells.
A balance between gene activation and repression is crucial for multiple biological or pathological processes. This balance is mainly regulated through changes in chromatin structure imparted by DNA methylation, chromatin remodeling, and histone modifications. DNA methylation, histone deacetylation, and certain histone methylations (such as H3K27me3) are often associated with transcriptional repression (Perissi et al., 2010). In mESCs, BRG1 and PcG components such as EZH and SUZ12 co-occupy many of the OCT4/SOX2 target genes, and BRG1 opposes PcG activity by altering H3K27me3 levels (Ho et al., 2011). In hESCs, however, BRG1 depletion did not result in any significant changes of H3K27me3 levels in either the promoter or enhancer regions of upregulated lineage-specific genes such as EOMES and FOXA2 (Figures S5B–S5D). In contrast, we observed a significant increase of genome-wide H3K27ac levels upon BRG1 knockdown in the enhancer regions that become active in early development. This increase was detected in the majority of these enhancer regions (such as those for EOMES, FOXA2, LEFTY2, and NODAL) after transient BRG1 depletion for 3 days (Figure S6A). These data thus suggest that the elevation of H3K27ac levels is BRG1 dependent and likely a direct consequence of BRG1 depletion. We also observed an upregulation of the acetyl-H3 level in the promoter regions for about 60% of the genes we examined (Figure S6B), which was correlated with their activated transcription. However, the increase for H3K9ac was much less obvious (Figure S6C), indicating a specificity for H3K27ac in the enhancer regions regulated by BRG1. In addition, no obvious alteration in the levels of acetyl-H3, acetyl-H4, or H3K27ac modification was detected upon BRG1 depletion by western blots (Figure S6D), suggesting a regulatory specificity of BRG1 on H3K27ac in enhancer regions. Collectively, our data support the notion that BRG1 inhibits the transcription of lineage-specific genes by specifically modulating H3K27ac levels in hESCs. In this context, it is noteworthy that in several previous reports, BRG1 was shown to be associated with histone acetyltransferases (such as p300/CBP) (Huang et al., 2003) or histone deacetylases containing corepressor complexes (such as SIN3A) (Liang et al., 2008; Pal et al., 2003; Underhill et al., 2000). It will be of great interest to systematically dissect which coactivator or corepressor complex associates with BRG1 to regulate histone acetylation during early development. Nevertheless, the present study suggests a mechanism by which the BRG1-SWI/SNF complex negatively regulates gene transcription, and clearly illustrates the functional differences between the epigenetic machineries present in hESCs and mESCs that are important for pluripotency.
Experimental Procedures
ESC and Epiblast Stem Cell Culture and Differentiation
Mouse ESCs were maintained as previously described (Wang et al., 2005). hESC lines H1 and BG01 (WiCell Research Institute) were maintained in Dulbecco’s modified Eagle’s medium (DMEM)/F12 supplemented with 20% knockout serum replacement (KSR) and 4 ng/ml FGF2 (all from Invitrogen). mEpiSCs were derived from the epiblast of embryos at 5.5 dpc according to standard protocols (Brons et al., 2007; Tesar et al., 2007). The mEpiSCs were maintained routinely according to the same protocol and in the same medium used for hESCs. For EB formation, clumps of hESCs were transferred into petri dishes with DMEM/F12 containing 20% KSR. The medium was changed every other day.
AP Assay
AP staining was performed with the BCIP/NBT Alkaline Phosphatase Kit (Beyotime) according to the manufacturer’s instructions.
Flow-Cytometry Analyses
For cell-cycle analyses, cells were fixed with 70% ethanol at 4°C overnight and incubated in PBS containing 100 μg/ml ribonuclease A and 20 μg/ml propidium iodine for further analysis. Cell-cycle profiles collected by flow cytometry were processed with Modfit software. For surface antigen analyses, hESCs were stained with primary antibodies (Tra-1-60 and Tra-1-81; Santa Cruz Biotechnologies), washed with PBS, and incubated with an APC-conjugated secondary antibody (BD Biosciences) for analysis. All flow-cytometry assays were performed on the FACSCalibur system (BD Biosciences).
Plasmid Construction and RNAi
The siRNAs for transient BRG1 knockdown were purchased from Santa Cruz Biotechnologies and electroporated into ESCs with the Nucleofactor Device (Lonza). The shRNAs for knocking down BRG1 or other BAF subunits were cloned into a lentiviral vector driven by a human U6 promoter. All shRNA sequences used in this study are listed in Table S6. To construct expression plasmids, a cDNA fragment containing either V5-tagged BAF155 or BAF170 was cloned into a PiggyBac vector driven by a CMV promoter. Drug selection for hygromycin or puromycin driven by the EF1α promoter in these vectors allowed us to establish stable hESC lines for long-term shRNA knockdown or cDNA expression.
Western Blots and Gel-Filtration Analyses
Western blots were performed according to standard protocols. The final blots were incubated with fluorescence-conjugated secondary antibodies (Li-Cor Odyssey) and signals were read by the Li-Cor Odyssey system. Superose 6 gel-filtration analyses (Bio-Rad) were performed as previously described (Yan et al., 2008). The following primary antibodies were used in this study: BRG1-G7, BAF155, BAF170, BAF60A, BAF60B, BAF47, BAF57, and OCT4 (Santa Cruz Biotechnologies); NANOG, H3, and H4 (Cell Signaling Technologies); TUBULIN (Abcam); BAF53A (Bethyl Laboratories); and acetyl-H3 and acetyl-H4 (Millipore).
CoIP and Mass Spectrometry Analyses
Cells were harvested, incubated with primary antibodies for 6 hr at 4°C, and precipitated with protein A-agarose beads (Santa Cruz Biotechnologies). For mass spectrometry analysis, peptides were extracted from destained SDS-PAGE gel and analyzed on an LTQ XL (Thermo Fisher Scientific) in data-dependent acquisition mode. The spectra were analyzed using SEQUEST against the Uniprot Swiss-Prot Database and the International Protein Index (IPI) Human Database (v3.87). BRG1-H88 from Santa Cruz Biotechnologies was used for the coIP assays.
ChIP and ChIP-Seq Analyses
Cells were fixed in 1% formaldehyde for 10 min at 37°C and sonicated to shear DNA to a length of 200–1,000 bp. Lysates were precleared with protein A/G agarose beads (Millipore) and antibodies were added to immunoprecipitate targeted chromatin fragments at 4°C overnight. Enrichment of proteins at targeted DNA fragments was analyzed by real-time PCR. For ChIP-seq analyses, 1–10 ng of ChIP DNA fragments were end repaired, 5′ phosphorylated, and ligated with Illumina adaptors using the Ovation SP+ Ultralow DR system (NuGen). Adaptor-ligated ChIP DNA fragments were subjected to 12 cycles of PCR to prepare the final libraries, which were sequenced with an Illumina HiSeq-2000 Sequencer. For H3K27ac ChIP-seq data analyses, after raw sequence reads were demultiplexed, each ChIP-seq sample was aligned to the human reference genome (hg19) with BWA (Li and Durbin, 2010). Uniquely mapped reads were analyzed with SICER (Zang et al., 2009). To identify peaks in each ChIP sample, the corresponding input was used as a background control. The location and tag density of the resulting ChIP peaks were visualized by UCSC Genome Browser. The BRG1 ChIP-seq data for hESCs were downloaded from the NCBI database (GSM602297) and lifted over to hg19 with the LiftOver tool from UCSC. The class I and class II enhancer peak data were obtained from a published report (Rada-Iglesias et al., 2011) and lifted over to hg19. All ChIP-seq peaks were annotated to the nearest RefGenes (build hg19) from the UCSC genome browser. ChIP-seq peaks were considered as overlapping when the distance between the centers of loci was within ±250 bp. For comparison between BRG1 ChIP-seq data and microarray data, BRG1 ChIP-seq peaks were annotated to the nearest genes and compared with the gene list from microarray analyses. The antibodies used in these assays were BRG1 and H3K27ac (Abcam).
Microarray Hybridization and Data Processing
Expression analysis was conducted using Agilent Whole Human Genome (014850) or Whole Mouse Genome (014868) 4 × 44 multiplex format oligo arrays (Agilent Technologies) according to the Agilent one-color expression analysis protocol. For each sample, 1.65 μg of Cy3-labeled cRNAs were fragmented and hybridized for 17 hr. Slides were scanned with an Agilent Scanner. Data were obtained and adjusted using the Agilent Feature Extraction software (v9.5) and further processed with the Rosetta Resolver system (v7.1) (Rosetta Biosoftware). To identify genes that were differentially expressed, error-weighted ANOVA was performed to determine whether there was a statistical difference between group means with p value < 0.01. Function and network analyses were performed by IPA (Ingenuity Systems).
RT-PCR and Real-Time PCR
Total RNA was extracted using Trizol (Invitrogen) and cDNA was synthesized with the SuperScript II First-Strand Synthesis Kit (Invitrogen) according to the manufacturer’s protocol. Quantitative PCR was performed with SYBR Premix Ex Taq (TAKARA) on Mx3005P (Stratagene). The transcript levels were normalized to glyceraldehyde 3-phosphate dehydrogenase. All primers used for RT-PCR and real-time PCR are provided in Table S6.
Teratoma Formation, IHC, and Histological Analyses
Cells were injected subcutaneously into NOD/SCID mice (∼5 × 106 cells per site). Tumors were processed for hematoxylin-eosin staining at 8 weeks postinjection, and 4-μm-thick paraffin sections were treated with xylol and rehydrated in alcohol. The epitopes were retrieved in citrate buffer at 100°C for 30 min. IHC was performed with the Histostain-Plus IHC Kit (MRBiotech) with the following antibodies: FOXA2 (Cell Signaling Technologies), BRACHYURY (Abcam), and β-TUBULIN (Santa Cruz Technologies). All animal experimental procedures were conducted in accordance with the local Animal Welfare Act and Public Health Service Policy and approved by the animal experiment review board of the Institute of Life Sciences at East China Normal University (protocol #AR2013/05006).
Statistical Analysis
All data (except for microarray and ChIP-seq analyses) are presented as group means ± SEM, and p values were calculated using Student’s t test for comparisons as previously described (Wang et al., 2008).
Author Contributions
X.Z., B.L., W.L., Y.W., and J.Z., designed experiments; X.Z., B.L.,W.L., L.M., D.Z., L.L., W.Y., M.C., W.C., and Y.W. performed research; G.F. and R.M.B. contributed new reagents/analytic tools; X.Z., B.L., W.L., L.M., D.Z., L.L., W.Y., M.C., W.C., J.Z., and Y.W. analyzed data; and R.B.M., J.Z., T.K.A., and Y.W. wrote the paper.
Acknowledgments
We thank Dr. Jiemin Wong for his critical comments during preparation of this manuscript, Dr. Ting Ni for help with the ChIP-seq analyses, and Dr. Lei Feng for help with the mass spectrometry assays. Microarray analyses were carried out at the Sequencing Core Facility of NIEHS. This work was supported by grants from the Ministry of Science and Technology of China (2010CB945403 and 2014CB964800), the National Natural Science Foundation of China (31271589 and 30971522), the Science and Technology Commission of Shanghai Municipality (11DZ2260300 and 13JC1406402), and the Intramural Research Program of the NIH (Z01 ES071006-11 to T.K.A., ZIC HL006058 to J.Z., and ES101765 to L.L.).
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Accession Numbers
The microarray and ChIP-seq raw data reported in this work have been deposited in the GEO and Sequence Reads Archive (SRA) under accession numbers GSE57474 and SRP041744, respectively.
Supplemental Information
References
- Becker K.A., Ghule P.N., Therrien J.A., Lian J.B., Stein J.L., van Wijnen A.J., Stein G.S. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J. Cell. Physiol. 2006;209:883–893. doi: 10.1002/jcp.20776. [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons I.G., Smithers L.E., Trotter M.W., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S.M., Howlett S.K., Clarkson A., Ahrlund-Richter L., Pedersen R.A., Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Bultman S.J., Gebuhr T.C., Magnuson T. A Brg1 mutation that uncouples ATPase activity from chromatin remodeling reveals an essential role for SWI/SNF-related complexes in beta-globin expression and erythroid development. Genes Dev. 2005;19:2849–2861. doi: 10.1101/gad.1364105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi T.H., Wan M., Lee P.P., Akashi K., Metzger D., Chambon P., Wilson C.B., Crabtree G.R. Sequential roles of Brg, the ATPase subunit of BAF chromatin remodeling complexes, in thymocyte development. Immunity. 2003;19:169–182. doi: 10.1016/s1074-7613(03)00199-7. [DOI] [PubMed] [Google Scholar]
- De Los Angeles A., Loh Y.H., Tesar P.J., Daley G.Q. Accessing naïve human pluripotency. Curr. Opin. Genet. Dev. 2012;22:272–282. doi: 10.1016/j.gde.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer C.J., Archer T.K. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- Gao X., Tate P., Hu P., Tjian R., Skarnes W.C., Wang Z. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc. Natl. Acad. Sci. USA. 2008;105:6656–6661. doi: 10.1073/pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebuhr T.C., Kovalev G.I., Bultman S., Godfrey V., Su L., Magnuson T. The role of Brg1, a catalytic subunit of mammalian chromatin-remodeling complexes, in T cell development. J. Exp. Med. 2003;198:1937–1949. doi: 10.1084/jem.20030714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber B., Wu G., Bernemann C., Joo J.Y., Han D.W., Ko K., Tapia N., Sabour D., Sterneckert J., Tesar P., Schöler H.R. Conserved and divergent roles of FGF signaling in mouse epiblast stem cells and human embryonic stem cells. Cell Stem Cell. 2010;6:215–226. doi: 10.1016/j.stem.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Griffin C.T., Brennan J., Magnuson T. The chromatin-remodeling enzyme BRG1 plays an essential role in primitive erythropoiesis and vascular development. Development. 2008;135:493–500. doi: 10.1242/dev.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang C.T., Yang J., Han P., Cheng H.L., Shang C., Ashley E., Zhou B., Chang C.P. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature. 2010;466:62–67. doi: 10.1038/nature09130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L., Jothi R., Ronan J.L., Cui K., Zhao K., Crabtree G.R. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc. Natl. Acad. Sci. USA. 2009;106:5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L., Ronan J.L., Wu J., Staahl B.T., Chen L., Kuo A., Lessard J., Nesvizhskii A.I., Ranish J., Crabtree G.R. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc. Natl. Acad. Sci. USA. 2009;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L., Miller E.L., Ronan J.L., Ho W.Q., Jothi R., Crabtree G.R. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat. Cell Biol. 2011;13:903–913. doi: 10.1038/ncb2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z.Q., Li J., Sachs L.M., Cole P.A., Wong J. A role for cofactor-cofactor and cofactor-histone interactions in targeting p300, SWI/SNF and Mediator for transcription. EMBO J. 2003;22:2146–2155. doi: 10.1093/emboj/cdg219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C., Hargreaves D.C., Hodges C., Elias L., Ho L., Ranish J., Crabtree G.R. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeser M.D., Aslanian A., Dong M.Q., Yates J.R., 3rd, Emerson B.M. BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. J. Biol. Chem. 2008;283:32254–32263. doi: 10.1074/jbc.M806061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidder B.L., Palmer S., Knott J.G. SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells. 2009;27:317–328. doi: 10.1634/stemcells.2008-0710. [DOI] [PubMed] [Google Scholar]
- Kim J.K., Huh S.O., Choi H., Lee K.S., Shin D., Lee C., Nam J.S., Kim H., Chung H., Lee H.W. Srg3, a mouse homolog of yeast SWI3, is essential for early embryogenesis and involved in brain development. Mol. Cell. Biol. 2001;21:7787–7795. doi: 10.1128/MCB.21.22.7787-7795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Fedoriw A.M., Magnuson T. An essential role for a mammalian SWI/SNF chromatin-remodeling complex during male meiosis. Development. 2012;139:1133–1140. doi: 10.1242/dev.073478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klochendler-Yeivin A., Fiette L., Barra J., Muchardt C., Babinet C., Yaniv M. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 2000;1:500–506. doi: 10.1093/embo-reports/kvd129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., Wan M., Zhang Y., Gu P., Xin H., Jung S.Y., Qin J., Wong J., Cooney A.J., Liu D., Songyang Z. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat. Cell Biol. 2008;10:731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- Lickert H., Takeuchi J.K., Von Both I., Walls J.R., McAuliffe F., Adamson S.L., Henkelman R.M., Wrana J.L., Rossant J., Bruneau B.G. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- Meshorer E., Yellajoshula D., George E., Scambler P.J., Brown D.T., Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neganova I., Zhang X., Atkinson S., Lako M. Expression and functional analysis of G1 to S regulatory components reveals an important role for CDK2 in cell cycle regulation in human embryonic stem cells. Oncogene. 2009;28:20–30. doi: 10.1038/onc.2008.358. [DOI] [PubMed] [Google Scholar]
- Pal S., Yun R., Datta A., Lacomis L., Erdjument-Bromage H., Kumar J., Tempst P., Sif S. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol. Cell. Biol. 2003;23:7475–7487. doi: 10.1128/MCB.23.21.7475-7487.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V., Jepsen K., Glass C.K., Rosenfeld M.G. Deconstructing repression: evolving models of co-repressor action. Nat. Rev. Genet. 2010;11:109–123. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S.A., Flynn R.A., Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaniel C., Ang Y.S., Ratnakumar K., Cormier C., James T., Bernstein E., Lemischka I.R., Paddison P.J. Smarcc1/Baf155 couples self-renewal gene repression with changes in chromatin structure in mouse embryonic stem cells. Stem Cells. 2009;27:2979–2991. doi: 10.1002/stem.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead E., White J., Faast R., Conn S., Goldstone S., Rathjen J., Dhingra U., Rathjen P., Walker D., Dalton S. Pluripotent cell division cycles are driven by ectopic Cdk2, cyclin A/E and E2F activities. Oncogene. 2002;21:8320–8333. doi: 10.1038/sj.onc.1206015. [DOI] [PubMed] [Google Scholar]
- Surani M.A., Hayashi K., Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Tay Y., Zhang J., Thomson A.M., Lim B., Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- Tesar P.J., Chenoweth J.G., Brook F.A., Davies T.J., Evans E.P., Mack D.L., Gardner R.L., McKay R.D. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Trotter K.W., Archer T.K. The BRG1 transcriptional coregulator. Nucl. Recept. Signal. 2008;6:e004. doi: 10.1621/nrs.06004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill C., Qutob M.S., Yee S.P., Torchia J. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J. Biol. Chem. 2000;275:40463–40470. doi: 10.1074/jbc.M007864200. [DOI] [PubMed] [Google Scholar]
- Wang W., Xue Y., Zhou S., Kuo A., Cairns B.R., Crabtree G.R. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- Wang Y., Yates F., Naveiras O., Ernst P., Daley G.Q. Embryonic stem cell-derived hematopoietic stem cells. Proc. Natl. Acad. Sci. USA. 2005;102:19081–19086. doi: 10.1073/pnas.0506127102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Yabuuchi A., McKinney-Freeman S., Ducharme D.M., Ray M.K., Chawengsaksophak K., Archer T.K., Daley G.Q. Cdx gene deficiency compromises embryonic hematopoiesis in the mouse. Proc. Natl. Acad. Sci. USA. 2008;105:7756–7761. doi: 10.1073/pnas.0708951105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Gu H., Lin H., Chi T. Essential roles of the chromatin remodeling factor BRG1 in spermatogenesis in mice. Biol. Reprod. 2012;86:186. doi: 10.1095/biolreprod.111.097097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.I., Lessard J., Crabtree G.R. Understanding the words of chromatin regulation. Cell. 2009;136:200–206. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Wang Z., Sharova L., Sharov A.A., Ling C., Piao Y., Aiba K., Matoba R., Wang W., Ko M.S. BAF250B-associated SWI/SNF chromatin-remodeling complex is required to maintain undifferentiated mouse embryonic stem cells. Stem Cells. 2008;26:1155–1165. doi: 10.1634/stemcells.2007-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo A.S., Crabtree G.R. ATP-dependent chromatin remodeling in neural development. Curr. Opin. Neurobiol. 2009;19:120–126. doi: 10.1016/j.conb.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang C., Schones D.E., Zeng C., Cui K., Zhao K., Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


