Summary
Cell therapies treating pathological muscle atrophy or damage requires an adequate quantity of muscle progenitor cells (MPCs) not currently attainable from adult donors. Here, we generate cultures of approximately 90% skeletal myogenic cells by treating human embryonic stem cells (ESCs) with the GSK3 inhibitor CHIR99021 followed by FGF2 and N2 supplements. Gene expression analysis identified progressive expression of mesoderm, somite, dermomyotome, and myotome markers, following patterns of embryonic myogenesis. CHIR99021 enhanced transcript levels of the pan-mesoderm gene T and paraxial-mesoderm genes MSGN1 and TBX6; immunofluorescence confirmed that 91% ± 6% of cells expressed T immediately following treatment. By 7 weeks, 47% ± 3% of cells were MYH+ve myocytes/myotubes surrounded by a 43% ± 4% population of PAX7+ve MPCs, indicating 90% of cells had achieved myogenic identity without any cell sorting. Treatment of mouse ESCs with these factors resulted in similar enhancements of myogenesis. These studies establish a foundation for serum-free and chemically defined monolayer skeletal myogenesis of ESCs.
Highlights
-
•
CHIR99021 treatment induces 90% of hESCs into mesoderm in vitro without FACS
-
•
43% and 47% of treated hESCs become PAX7+ve MPCs and skeletal myocytes, respectively
-
•
CHIR99021/FGF2/N2-derived myotubes twitch and contract in vitro
-
•
Treatment of mESCs similarly enhances skeletal myogenesis
In this article, Skerjanc and colleagues show a monolayer-based protocol for the in vitro differentiation of human embryonic stem cells (ESCs) into the skeletal muscle lineage. Ninety-one percent ± 6% of hESCs stain positive for T following CHIR99021-based mesoderm induction. After 50 days of culture, 47% ± 3% of cells are MYH+ve myocytes/myotubes surrounded by a 43% ± 4% population of PAX7+ve muscle progenitor cells (MPCs).
Introduction
Cell therapies to reverse muscle atrophy and to strengthen skeletal muscle would greatly enhance and extend the lives of patients with muscle wasting conditions due to diseases and/or aging. Embryonic stem cells (ESCs) have unlimited proliferation potential, and no need for locating a suitable immunotype-matched donor as with adult-derived stem cells (Araki et al., 2013). However, a major obstacle in the development of ESC-based therapies targeting muscle has been the generation of a homogeneous myogenic population from in vitro differentiation, thus requiring optimization to enrich for muscle lineage cells.
Several studies have validated the potential of mouse and human ESCs (mESCs and hESCs, respectively) and induced pluripotent stem cells (iPSCs), in skeletal muscle therapy (Barberi et al., 2007; Chang et al., 2009; Darabi et al., 2008, 2011a, 2011b, 2012; Sakurai et al., 2008). Cells were differentiated into paraxial mesoderm-like muscle progenitors, either by a standard serum-based embryoid body (EB) differentiation protocol (Chang et al., 2009; Sakurai et al., 2008) or by transient expression of PAX3 or PAX7 (Darabi et al., 2008, 2011a, 2012). These in vitro derived progenitors were able to engraft into adult myofibers of mice, replenish the muscle stem cell (satellite cell) niche, and enhance muscle contractile function (Chang et al., 2009; Darabi et al., 2008, 2011a, 2012; Sakurai et al., 2008). Despite promising results, these protocols are not appropriate for the generation of muscle progenitor cells (MPC) for clinical applications due to the inefficiency of differentiation and the use of viral vectors and potential insertional mutations (Thomas et al., 2003).
Previous studies from our lab have used a serum-containing EB-induced differentiation supplemented with low levels of retinoic acid (RA) to enhance myogenesis from mouse (Kennedy et al., 2009) and human (Ryan et al., 2012) ESCs. However, serum-containing EB-differentiation of hESCs produced relatively low yields of skeletal muscle (<5%) and is undefined (Al Madhoun et al., 2011; Kennedy et al., 2009; Ryan et al., 2012). In contrast, directed differentiation uses knowledge of embryogenesis to recreate embryonic conditions in vitro using combinations of signaling molecules, to support the differentiation into one lineage (Murry and Keller, 2008). Applying the serum-free directed differentiation approach should greatly improve the efficiency of hESC-derived myogenesis for molecular analysis and for future use in cell therapies.
Wnt signaling is critically important for the development of the primitive streak and paraxial mesoderm (Liu et al., 1999), marked by the T and MSGN1 or TBX6 genes, respectively, and in the formation of posterior somites and the tail bud (Takada et al., 1994), marked by the transcription factors PAX3, MEOX1, and PAX7. In the canonical pathway (reviewed in Clevers, 2006), Wnt binds to Frizzled cell-surface receptors, initiating a signaling cascade that inhibits GSK3B, preventing B-CATENNIN (CTNNB1) degradation, and allowing CTNNB1 to accumulate and translocate into the nucleus. Nuclear CTNNB1 enhances transcription by interaction with T cell factors or lymphocyte enhancer factors (Clevers, 2006).
It has previously been shown that the GSK3 inhibitor CHIR99021 (CHIR) can augment mesoderm induction (Tan et al., 2013), leading to cardiomyogenesis in ESCs (Lian et al., 2012). Recombinant proteins BMP4 and ACTIVIN-A (INHBA) have similarly been used to induce mesoderm and cardiac muscle from ESCs (Kattman et al., 2011; Murry and Keller, 2008). These studies implicate BMP4/INHBA or CHIR treatment as a potential method for generating skeletal muscle. Furthermore, we have shown that overexpression of WNT3A or CTNNB1 enhances the formation of premyogenic mesoderm in P19 embryonal carcinoma cells, resulting in increased myogenesis (Petropoulos and Skerjanc, 2002). The loss of CTNNB1 function via dominant-negative mutation or knockdown results in the loss of MPC formation and myogenesis, supporting the use of CHIR to induce myogenesis.
The PAX3/7 population that is present in the central dermomyotome appears to represent an MPC pool that is maintained throughout embryogenesis and is responsible for almost all skeletal muscle (Buckingham, 2007; Kuang et al., 2007). FGF2 prevents expression of the myoblast commitment transcription factors MYF5, MYOD1, and MYOG—collectively known as the myogenic regulatory factors (MRF)—during satellite cell activation and thus can be utilized to enhance proliferation of PAX3/7 expressing MPCs in vitro (Fedorov et al., 1998; Hall et al., 2010). Satellite cells are more efficient in reconstituting the satellite cell niche during transplantation into muscle if they do not yet upregulate the MRFs (Kuang et al., 2007; Montarras et al., 2005). We have previously shown that N2 supplemented media can enhance the terminal differentiation of myoblasts and myocytes, which would ensure MPCs are capable of complete myogenesis in vitro (Al Madhoun et al., 2011; Ryan et al., 2012).
In this report, we describe the robust skeletal myogenesis of hESCs and mESCs using CHIR to induce mesoderm, FGF2 treatment to expand the MPC population, and N2-mediated terminal differentiation. This chemically defined, serum- and transgenic-free protocol yields a nearly homogeneous myogenic population from hESCs, consisting of 43% ± 4% PAX7+ve MPCs and 47% ± 3% Myosin Heavy Chain+ve (MYH) skeletal muscle.
Results
GSK3 Inhibition Enhanced Premyogenic Mesoderm Formation from Human ESC
Our objective was to devise a robust, serum-free directed differentiation method to obtain skeletal muscle from hESC in vitro. We adapted a recently developed chemically defined, feeder-free culture system to both expand pluripotent cells and induce mesoderm differentiation (Chen et al., 2011). E8 maintenance media and Matrigel extracellular matrix were observed to support pluripotency based on OCT4 expression, and hESCs retained a normal karyotype after prolonged collagenase passaging (data not shown). Removal of FGF2 and TGFB1 from E8 media—referred to as E6 media—was observed to support cell expansion, but not pluripotency based on the loss of OCT4, SOX2, and NANOG gene and protein expression. Therefore, E6 medium provided a basal medium to which myogenic-inductive signaling molecules could be added. To control seeding density during differentiation procedures, hESCs were seeded as single cells in E8 media supplemented with ROCK inhibitor (Y27632) (Gauthaman et al., 2010). After overnight seeding, E8 media and ROCK inhibitor were removed and replaced with mesendoderm inductive conditions (R.L.C., S-P.D., and W.L.S., unpublished data).
Because previous studies have shown that CHIR or BMP4/INHBA treatment enhanced mesoderm induction in hESCs (Cao et al., 2013; Kattman et al., 2011; Lian et al., 2012), we initially sought to determine an optimal dose of CHIR and/or BMP4/INHBA to induce premyogenic mesoderm. For optimization of CHIR treatment, concentrations of 2.5 and 10 μM were chosen to encompass a range of concentrations previously used to induce cardiac tissue-fated mesoderm (Cao et al., 2013; Lian et al., 2012). Cells were plated on matrigel-coated dishes overnight in the presence of Y-27632 before differentiation was induced (Gauthaman et al., 2010). Factors maintaining pluripotency were removed from the media and treated with CHIR or vehicle for 2, 4, and 8 days in length: time points that encompass expression of T and early somite markers PAX3 and MEOX1 in serum-EB induced skeletal myogenesis (Ryan et al., 2012). Analysis by quantitative PCR (qPCR) showed significant upregulation of T and MSGN1 after 2 days of treatment with 10 μM CHIR, leading to significant upregulation of PAX3 on day 8 (Figure 1A). Although 2.5 μM CHIR treatment was able to upregulate T and MSGN1 expression after 2 days of treatment, it did not upregulate PAX3 at day 8 for any treatment length. Longer application of 10 μM CHIR, for 4 or 8 days, did not enhance PAX3 expression as effectively and appeared to have toxicity toward the cells (data not shown). MEOX1 gene expression appeared similar between both CHIR concentrations. Thus, the application of 10 μM CHIR for the first 2 days of differentiation was determined optimal for enhancing the formation of PAX3+ve premyogenic mesoderm.
Figure 1.
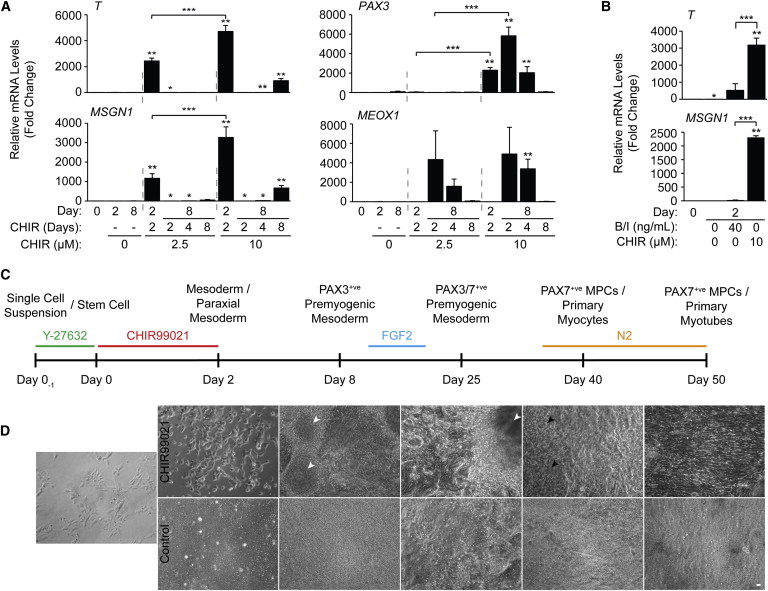
Ten Micromolar CHIR Applied for 2 Days Produces Optimal Levels of Paraxial and Premyogenic Mesoderm Gene Expression
(A) hESCs were treated in monolayer with or without 2.5 or 10 μM CHIR for 2, 4, or 8 days. RNA was harvested at days 0, 2, and 8 to be analyzed by qPCR for markers of early and premyogenic mesoderm. Results are expressed as fold change over day 0 (n = 3 independent experiments, ∗p ≤ 0.05 versus day 0, ∗∗p ≤ 0.05 versus day 0 and control, ∗∗∗p ≤ 0.05 2.5 versus 10 μM).
(B) CHIR-differentiated cells were compared to BMP4/INHBA treatment by qPCR for markers of early mesoderm. (n = 3 independent experiments, ∗p ≤ 0.05 versus day 0, ∗∗p ≤ 0.05 versus day 0 and control, ∗∗∗p ≤ 0.05 BMP4/INHBA versus CHIR).
(C) Schematic overview of the CHIR directed differentiation of hESCs on a nonlinear scale timeline, along with highlighting key supplemental factors applied during the differentiation, and the predicted developmental stages of skeletal myogenesis.
(D) Representative phase contrast live-cell images demonstrating the progressive changes in hESC morphology with and without CHIR treatment. 3D cell clusters (white arrowheads) and the presence of skeletal myocytes (black arrowheads) are indicated at specific time points (scale bar, 20 μm).
Results are shown ±SEM.
We titrated levels of BMP4/INHBA to optimize T expression and found by qPCR that 40 ng/ml of BMP4/INHBA enhanced T transcript levels, although not as efficiently as CHIR treatment (Figure 1B). Furthermore, BMP4/INHBA treatment did not significantly enhance MSGN1 expression, suggesting a lack of skeletal muscle-fated mesoderm. Thus, CHIR treatment was better than BMP4/INHBA treatment to induce premyogenic mesoderm in hESCs.
CHIR Treatment Resulted in up to 90% of hESCs Entering the Myogenic Lineage
hESCs were differentiated using the 2 day CHIR treatment protocol devised in Figure 1A and cultured until day 50 as summarized in Figure 1C, outlining the stages of myogenesis and the chief chemical and recombinant factor additions. Live-cell imaging was performed, showing the progressive morphological changes from stem cells to myotubes (Figure 1D). CHIR-treated cells appeared morphologically distinct from control cells by day 2 of differentiation, and proliferation was reduced as judged by lower confluency compared to controls.
By day 8, CHIR-treated cells developed distinct 3D cell clusters (Figure 1D, white arrowheads) (Rohwedel et al., 1994). FGF2 was applied from days 12 to 20, which corresponds to the time frame that we previously detected somite or MPC markers PAX3, MEOX1, and PAX7 along with MYF5 (Ryan et al., 2012). Therefore, this time frame was an ideal target for FGF2 treatment to preferentially expand MPCs while suppressing early potential expression of the MRFs. N2 supplemented media was applied at day 35—coinciding with later detection of all MRFs previously (Ryan et al., 2012)—to promote maturation of the myogenic cultures. Skeletal myocytes were observed by day 40, following 5 days of culture in N2 medium (Figure 1D, black arrowheads). The myocytes were, for the most part, randomly arranged among other nonbipolar cells. By day 50, cultures revealed large areas of aligned myocytes and fused myotubes, organized radially outward from the 3D clusters (Figure S1 available online). No appreciable skeletal myogenesis was observed in control cultures that were given DMSO vehicle in place of CHIR from days 0 to 2 and treated identically to CHIR-treated cells thereafter. Thus, CHIR treatment resulted in the formation of abundant mesoderm that could be developed into easily visualized myocytes and myotubes by day 50 of differentiation.
CHIR treatment resulted in detection of T protein in an average of 91% ± 6% of cells on day 2, compared to no T present in the control-treated cells (Figures 2A and 2E). At day 8, an average of 58% ± 10% of cells expressed PAX3 after CHIR treatment compared to 1% ± 1% in control cells (Figures 2B and 2F). Skeletal myocytes were prominent by day 40 following 5 days of growth in N2 medium, with an average of 14% ± 3% of CHIR-treated hESCs expressing MYH and were surrounded by a 37% ± 2% population of PAX7+ve cells (Figures 2C and 2G). In addition to myocytes, some myotubes were present (Figure S2A, white arrowheads), and skeletal muscle contractions could be observed by light microscopy (Movie S1). When the cultures were left in N2 media from day 35 until day 50, 43% ± 4% of cells remained PAX7 positive, whereas 47% ± 3% were MYH positive (Figures 2D and 2G). A macroscopic view of the culture dishes suggests that the 3D clusters function as myogenic foci, with high concentrations of PAX7+ve cells delaminating from their edges and myocytes/myotubes emanating outward (Figures S2B and S2C). Thus, CHIR/FGF2/N2 treatment can induce around 90% of hESCs to enter the myogenic lineage.
Figure 2.
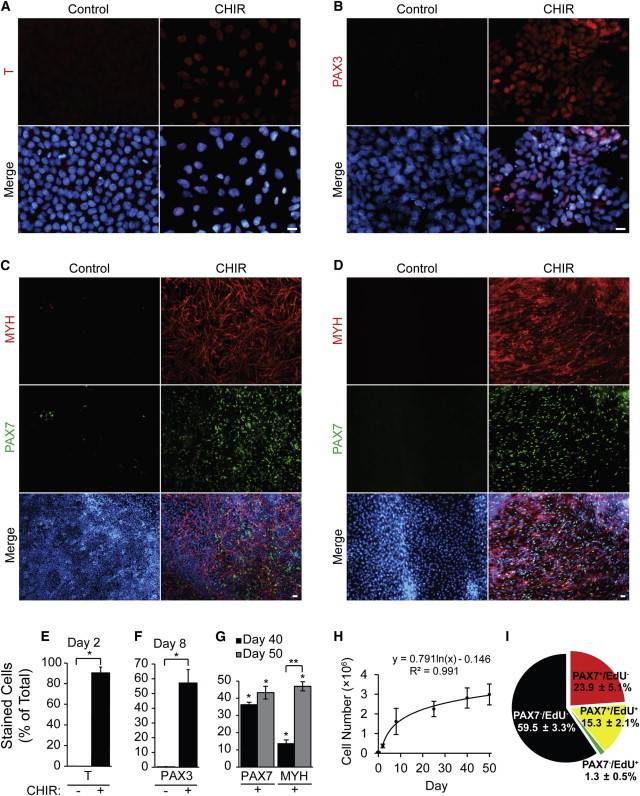
CHIR Directed Differentiation of hESCs Induces up to a 90% Myogenic Population
(A–G) hESCs were differentiated as described in Figure 1. Myogenic induction was quantified by staining with antibodies against (A) and (E) T at day 2, (B) and (F) PAX3 at day 8, (C) and (G) MYH and PAX7 at day 40, and (D) and (G) MYH and PAX7 at day 50. Target populations are expressed as a percentage of total number of cells, which was quantified by Hoechst staining (n = 3 independent experiments, ∗p ≤ 0.05 versus control, ∗∗p ≤ 0.05 day 40 versus day 50, scale bar, 20 μm).
(H) Total number of cells was obtained by trypsinizing and counting one well of a 12-well culture dish at various time points of the differentiation protocol (n = 3 independent experiments).
(I) Day 50 CHIR-treated cells were pulsed with EdU for 4 hr and fixed and stained additionally with PAX7. EdU and PAX7 single or double-positive cells were quantified and expressed as percentage of total number of cells (n = 3 independent experiments).
Results are shown ±SEM.
We also quantified total cell number at various time points to investigate the scalability of hESC-derived myogenic cells with our protocol. The total cell number followed a logarithmic growth curve over the course of the differentiation protocol, resulting in roughly 20-fold more cells at day 50 compared to the initial number present at day 0 (Figure 2H). We also characterized the proliferation potential of our MPCs at day 50 by ethynyl-deoxyuridine (EdU) incorporation (Figures 2I and S2D). During a 4 hr labeling pulse, 42% ±10% of PAX7+ve MPCs incorporated EdU, demonstrating that our differentiation protocol yields a persistent population of actively proliferating MPCs at the final time point assayed in these studies.
We examined the temporal pattern of expression of myogenic factors by qPCR during CHIR-directed differentiation of hESCs to confirm the immunofluorescence results. Day 2 of CHIR-treated hESCs showed significant upregulation of T transcripts, that was orders of magnitude higher than our previous results using a serum/EB method (Figure 3A) (Ryan et al., 2012). Significant upregulation of the paraxial mesoderm markers MSGN1 and TBX6 accompanied T at day 2, and all were downregulated by day 8, which marked peaks of the premyogenic mesoderm genes PAX3 and MEOX1 (Figure 3B), in agreement with our previously published results (Ryan et al., 2012). Two control patient-derived iPSC lines showed similar mesoderm gene elevation in response to our CHIR-based protocol (data not shown).
Figure 3.
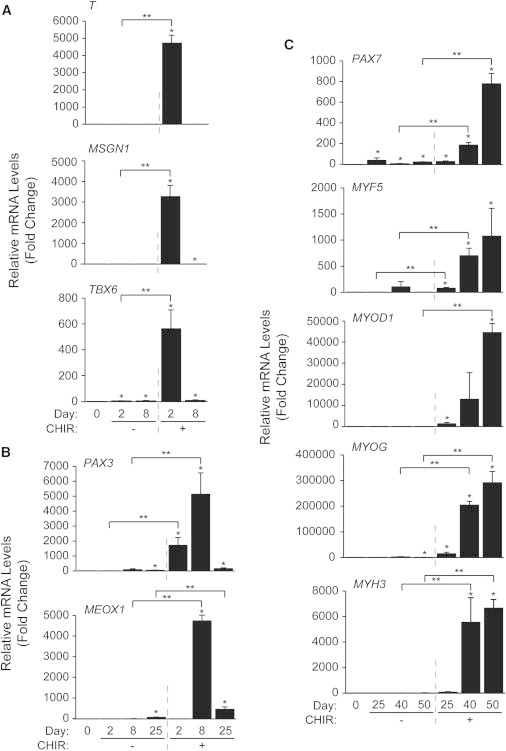
qPCR Profiling of CHIR-Treated hESCs Highlights a Clear Progression through Specified Mesodermal Subtypes, Muscle Progenitor Stages, and Committed Myogenic Cultures
hESCs were differentiated as described in Figure 1. RNA was harvested at days 0, 2, 8, 25, 40, and 50 to be analyzed by qPCR for markers several of (A) early and paraxial mesoderm, (B) premyogenic mesoderm, and (C) committed or differentiated skeletal muscle. Results are expressed as fold change over day 0 (n = 3 independent experiments, ∗p ≤ 0.05 versus day 0, ∗∗p ≤ 0.05 versus control).
Results are shown ±SEM.
Expression of PAX3 and MEOX1 was downregulated by day 25, although significant levels were still detected until day 50 (data not shown). Although PAX7, MYF5, MYOD1, and MYOG transcripts were significantly upregulated by day 25, their expression continued to increase until day 50 (Figure 3C). Expression of MYH3 transcripts also increased at days 40 and 50, suggesting an enduring MPC population coexisting with terminally differentiated skeletal myocytes/myotubes in agreement with our immunofluorescence data (Figures 2D, 2G, 2I, and S2D). Control cells showed no significant expression of MYF5, MYOD1, or MYH3 transcripts. There was, however, very low but statistically significant upregulated expression of PAX7 and MYOG mRNA in control cells. Overall, CHIR-treated cells exhibited waves of expression of mesoderm, presomitic mesoderm, muscle progenitor, myoblast, and muscle structural genes.
Because Wnt signaling is important for the development of a multitude of tissues and not simply premyogenic mesoderm (Clevers, 2006), we screened our cultures for markers of other lineages. Therefore, we assayed for other cell types that may be present in the 9% T−ve cells early in differentiation, and later in the 10% of PAX7−ve/MYH−ve cells. CHIR treatment led to a significant drop in the pluripotency marker SOX2 (Figure 4A) by day 2, indicating an efficient loss of pluripotency. SOX2 is also expressed in ectoderm lineages pertaining to the neural tube and future spinal cord (D’Amour and Gage, 2003; Wood and Episkopou, 1999). Thus, levels of these tissues in our day 8 CHIR cultures should be low for SOX2 expression. FOXA2—a marker of notochord, floor plate, and future endoderm (Monaghan et al., 1993; Sasaki and Hogan, 1993)—was reduced in both CHIR-treated and control cells. Significant upregulation of NOG and SOX10 was detected at day 8 of CHIR-mediated differentiation, which conventionally mark the roof plate and neural crest during embryonic development (Pusch et al., 1998; Valenzuela et al., 1995). Although neural crest cells may have been present early in differentiation, there was no significant upregulation in the neuronal marker NEUROG1 at later time points (Figure 4B), and immunofluorescent staining for NEFL—a neurofilament protein expressed in central and peripheral nervous system axons—was also negative (data not shown). Significant expression of traditional cardiac muscle lineage genes NKX2-5 and MYH6 (Akazawa and Komuro, 2005; Lyons et al., 1990) was detected with CHIR treatment at day 50, although cells exhibiting a cardiac muscle phenotype were not readily observed in these cultures (Figure S2B). As NKX2-5 and MYH6 have previously been detected in developing tongue skeletal muscle (Diez-Roux et al., 2011), we also assessed additional markers that are expressed exclusively in cardiomyocytes, NPPA and MYL2. No expression of these markers was detected in our day 50 differentiated cultures (Figure 4C). Therefore, neither mature neuronal tissue nor cardiac muscle were present in terminally differentiated cultures.
Figure 4.

Low Levels of Neural Crest Transcripts Were Present in CHIR-Treated Cultures
hESCs were differentiated, harvested, and analyzed as described above for markers of (A) pluripotency and neural ectoderm markers, (B) differentiated neural and traditional cardiac markers, (C) specific markers of mature cardiomyocytes, and (D) patterned dermomyotome genes (n = 3 independent experiments, ∗p ≤ 0.05 versus day 0, ∗∗p ≤ 0.05 versus control). In (C) RNA from serum-differentiated hESCs and human heart (HH) cardiomyocyte-positive controls were obtained (n = 1). All hESC fold changes (black bars) were plotted on the primary y axis, and HH fold change (gray bar) was plotted on the secondary y axis.
Results are shown ±SEM.
To determine the patterning of the dermomyotome, we performed qPCR for the regional dermomyotome markers EN1, SIM1, LBX1, and MET (Figure 4D). The dermomyotome is patterned into the dorsomedial half—expressing EN1 and giving rise to epaxial muscle, forming the deep back muscles—and the ventrolateral half, expressing SIM1 and giving rise to hypaxial muscle, forming the ventral and limb muscle (Cheng et al., 2004; Ordahl and Le Douarin, 1992; Pourquié et al., 1996). Proper migration of the hypaxial MPCs into the limb requires the expression of LBX1 and MET (Vasyutina and Birchmeier, 2006). Therefore, presence of LBX1 and MET expression would support the existence of MPCs similar to those found in migrating limb muscle. We found a significant increase of the three regional markers of the dermomyotome—EN1, SIM1, LBX1—and a trend toward elevated MET with CHIR. However, MET was expressed at high levels throughout the differentiation, shown by low Ct values, making changes as a result of CHIR-treatment difficult to identify (data not shown). Thus, day 25–50 cultures containing muscle progenitors expressed markers of epaxial, hypaxial, and migratory dermomyotome.
CHIR Treatment Enhanced Skeletal Myogenesis in Mouse ESCs
A similar CHIR-based differentiation protocol was performed in mouse embryonic stem cells (mESCs) and compared with BMP4/INHBA-mediated differentiation. Pax3/7+ve cells, presumably representing MPCs, were present in both BMP4/INHBA- and CHIR-treated mESCs at day 15, although PAX3/7+ve cells were visibly more abundant with CHIR (Figure 5A). Interestingly, MYH staining primarily revealed cardiomyocytes in BMP4/INHBA-treated cultures despite the presence of PAX3/7+ve cells (Figure 5B). CHIR, conversely, promoted the differentiation of primarily skeletal and not cardiac myocytes. Control cultures showed negligible or no staining with either PAX3/7 or MYH antibodies. Thus, CHIR treatment upregulated PAX3/7 expression, leading to enhanced skeletal myogenesis in mESCs.
Figure 5.
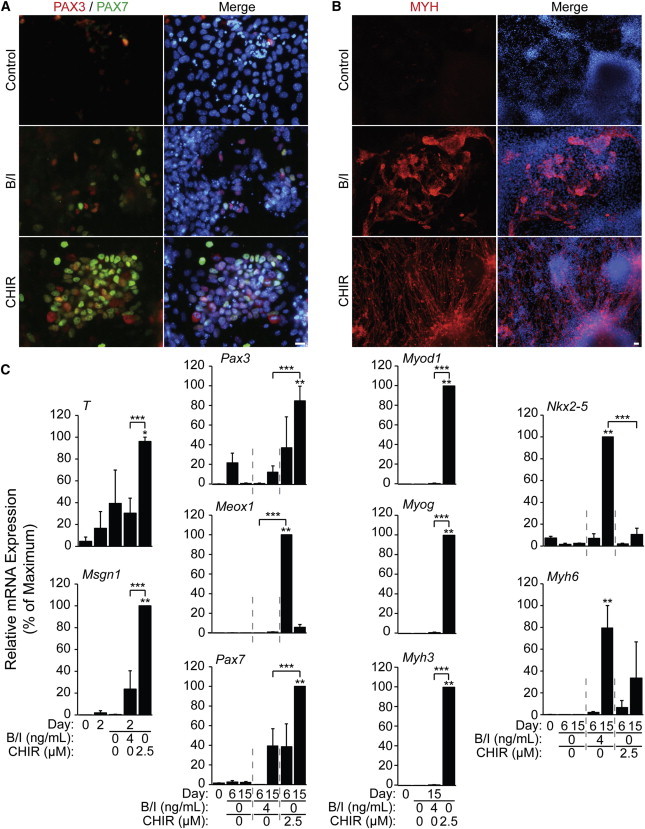
CHIR Enhances Skeletal Myogenesis in a 15 Day Differentiation of mESCs
mESCs were aggregated in differentiation medium for 2 days prior to treatment. CHIR, BMP4/INHBA, or vehicle control was applied from days 2 to 4.
(A and B) Immunofluorescent staining was carried out at day 15 with antibodies against (A) PAX3 and PAX7 or (B) MYH (scale bar, 20 μm).
(C) RNA was harvested at days 0 and 2 (prior to CHIR treatment) and days 4, 6, and 15 (following treatment). Samples were analyzed by qPCR for select markers of skeletal and cardiomyogenesis.
Results are expressed relative to the maximum fold change over day 0 (n = 3 independent experiments, ∗p ≤ 0.05 versus day 0, ∗∗p ≤ 0.05 versus day 0 and control, ∗∗∗p ≤ 0.05 BMP4/INHBA versus CHIR). Results are shown ±SEM.
Established markers of skeletal myogenesis were examined by qPCR and demonstrated corroborating trends between CHIR-induced differentiation in mESCs and hESCs (Figure 5C). Indeed, CHIR treatment significantly enhanced the expression of key paraxial mesoderm, premyogenic mesoderm, and MRF genes, including T, Msgn1, Pax3, Meox1, Myod1, Myog, and Myh3. BMP4/INHBA-treated cells exhibited some upregulation of the same mesodermal genes. However, BMP4/INHBA treatment did not upregulate expression of the MRFs but did upregulate high levels of the cardiomyocyte markers Nkx2-5 and Myh6. Thus, mESCs respond to CHIR treatment by upregulating a similar set of muscle lineage genes as identified in hESCs, albeit in a much-reduced time frame.
Discussion
Our studies define a method whereby human and mouse ESCs are differentiated in vitro into highly enriched populations of skeletal muscle progenitor cells and myocytes/myotubes by activating Wnt signaling via the GSK3 inhibitor CHIR99021. A mere 2 day exposure to the compound causes immediate changes in growth and morphology toward mesoderm, and sets in motion downstream cascades of myogenic transcription factors across weeks of culture. In due course, we achieve approximately 90% purity of skeletal muscle lineage cells from hESCs after 50 days in culture.
Although forced expression of PAX3/7 or MYOD in hESCs have resulted in enriched myogenic populations (Albini et al., 2013; Darabi et al., 2012; Goudenege et al., 2012; Rao et al., 2012), we sought to develop a viral- and transgenic- free method of in vitro differentiation. A chemically defined protocol should be more readily adaptable to a broader number of cell lines and should allow for the examination of all of the stages of differentiation, some of which may be bypassed by transgenic overexpression (Albini et al., 2013).
Our study contrasts with previously published methods of transgene-free hESC myogenesis that were dependent upon serum, showing low overall myogenic induction and/or myogenic progenitor formation (Awaya et al., 2012; Hwang et al., 2013; Ryan et al., 2012). For example, we previously obtained a 4% level of MYH+ve skeletal myocytes and 7% PAX3+ve MPCs with RA treatment (Ryan et al., 2012). Our current CHIR-directed protocol improves on the serum/EB methods by directing a larger proportion of the cultures into mesoderm, leading to enriched myogenic lineage populations.
Several studies have shown that Wnt signaling modulation by CHIR can maintain ESC pluripotency (Wu et al., 2013), and also can cause differentiation into tissues of all three germ layers (Denham et al., 2012; Lian et al., 2012; Tahamtani et al., 2013). However, supporting factors were required immediately following CHIR treatment for these approaches. Cell seeding density may also determine how GSK3 inhibition can generate skeletal muscle specific mesoderm versus other cell fates. Seeding density was critical first and foremost in determining how hESCs responded to external differentiation cues in our system (R.L.C., S-P.D., and W.L.S., unpublished data). At lower seeding densities (<1 × 105 cells/ml), hESC survival was reduced even in the presence of ROCK inhibitor, suggesting that cell-cell contacts are important for the initial seeding process. Additionally, sparse populations of cells responded to BMP4/INHBA treatment with increased cell death compared to higher densities. Furthermore, at higher seeding densities (>2 × 105 cells/ml), overconfluence was observed after 48 hr, again leading to compromised cell viability. Other monolayer approaches have used GSK3 to drive cardiomyogenesis; however, these protocols induce GSK3 inhibition once cells reach confluence or with supplemental BMP4 inhibition (Cao et al., 2013; Lian et al., 2012). These protocols also emphasize the critical importance of insulin depletion after GSK3 inhibition, or the need for Wnt inhibition following mesoderm induction lest cardiomyogenesis will not occur in vitro (Kattman et al., 2011; Lian et al., 2013). This is supported by the role of Dkk-mediated Wnt inhibition in specifying embryonic mesoderm to the primary heart field (Ueno et al., 2007).
Of particular relevance to the results presented in our study was the differentiation of mesoderm from hESCs via 5 days of 5 μM CHIR treatment (Tan et al., 2013). Intermediate to the desired cell types, CHIR treatment caused a marked upregulation of T mRNA and protein levels, similar to our findings. Though the authors followed CHIR treatment with BMP4—which typically effects lateral plate mesoderm—to develop hematopoietic and endothelial cell types and not skeletal muscle (Pick et al., 2007; Rufaihah et al., 2011). They also found INHBA following CHIR treatment was necessary for pronounced endoderm commitment (Tan et al., 2013).
Although CHIR can coax differentiation of hESCs into all three germ layers, we obtained surprisingly 43% ± 4% PAX7+ve and 47% ± 3% MYH+ve cell populations at the experimental endpoint. However, trace expression of other ectoderm and mesoderm lineages can be detected early in the protocol. It is possible that other tissues, such as neural crest present early in our protocol, aid in the myogenic differentiation pathway; attempts to reach 100% T+ve cultures could demand additional signaling molecules to replace factors from these tissues (Nitzan and Kalcheim, 2013; Rios et al., 2011). It is likely that our current protocol can still be improved by the inclusion of additional signaling molecules at the correct time in the pathway, such as RA and Notch signaling, and/or by the use of fluorescence-activated cell sorting (FACS) or microdissection to further enrich the MPCs.
Indeed, we attempted to improve our protocol according to a recent study published while our manuscript was in review that generated high proportions of skeletal myocytes from aggregated human iPSCs using combinatorial treatment with CHIR, FGF2, and the cAMP signaling activator Forskolin (Xu et al., 2013). Hence, we included Forskolin in our culture system to determine whether activation of cAMP signaling with could improve skeletal myocyte yield in the context of our protocol (Figure S3). qPCR analysis showed that, in the context of our monolayer differentiation protocol with 10 μM CHIR alone, Forskolin does not provide additional benefit to myogenesis as it does with aggregate based differentiation. By day 7, however, conditions that included Forskolin were observably denser than CHIR alone treated cultures, suggesting that Forskolin increases cell proliferation (data not shown). One major difference between our protocol and the published work by Zon and colleagues is that the myogenic cultures generated with Forskolin treatment appear to terminally differentiate beyond the MPC stage to over 90% MYOG+ve (Xu et al., 2013), whereas our system maintained a significant population of proliferating PAX7+ve MPCs (Figures 2D, 2G, 2I, and S2D), suggesting that our culture system will provide replicative MPCs that could maintain homeostasis in the transplanted muscle.
During the revisions of this manuscript, two additional manuscripts were published that generated transgene-free human ESC myocytes. Barberi and colleagues utilized a lower concentration of CHIR and longer treatment duration to direct skeletal myogenesis in embryonic stem cells (Borchin et al., 2013). We initially determined that similar CHIR conditions used by Borchin et al. (2013) induced less skeletal muscle-specific mesoderm than the optimal 10 μM for 2 days (Figure 1A). Indeed, our protocol generated twice as many PAX7+ve MPCs at comparable time points (Figures 2C and 2G). This recent publication, however, introduced a rigorous HNK−/ACHR−/CXCR4+/MET+ FACS profile for isolating MPCs from bulk cultures. Therefore, future studies in embryonic-derived MPCs may benefit from implementing our differentiation protocol to generate greater numbers of starting cells, which could be enriched via the HNK−/ACHR−/CXCR4+/MET+ profile.
In addition, Suzuki and colleagues present a viable alternative to monolayer directed skeletal myogenesis (Hosoyama et al., 2014). The authors’ “EZ Sphere” suspension-culture system uses high concentrations of FGF2 and EFGF to yield remarkably similar proportions of PAX7+ve populations at the experimental endpoint as those presented here (Figures 2D, 2G, 2I, and S2D). One caveat to the EZ Sphere method, however, is potentially upward of 30% of cells are neural progenitors (Hosoyama et al., 2014).
We show that CHIR can mediate efficient skeletal myogenesis in mESCs using a 2 day CHIR treatment similar to that used for hESCs, illustrating the effectiveness of our protocol across these species. Our current findings are consistent with our previous results showing that Wnt signaling via CTNNB1 is important for murine P19 embryonal carcinoma cell differentiation, using gain- and loss-of-function as well as a dominant-negative approach (Petropoulos and Skerjanc, 2002; Wong et al., 2013). In addition, the greater experimental variability with mouse compared to human ESC differentiation into muscle could be due to the EB-based nature of the mESC serum-free protocol. Therefore, we are adapting the more consistent monolayer approach used with our hESC method to mESCs. Although hESCs are more therapeutically relevant, mESCs continue to play an integral role in our ability to study cellular differentiation, primarily due to their substantially shorter differentiation time, creating a simpler high-throughput model system for detailed molecular analysis.
Overall, the results presented here have provided a method to differentiate mouse and human ESCs into highly enriched skeletal muscle lineage cultures. Most importantly, we show a stable PAX7+ve population of MPCs even after 7 weeks of culture. Ultimately, these studies provide the foundation for in vitro study of ESC skeletal myogenesis, which could contribute to future drug testing or stem cell therapies, leading to the repair of muscle in patients with muscular wasting disorders.
Experimental Procedures
Human Cell Culture
All hESC media and components are formulated in Table S1. H9 hESCs were maintained feeder free on BD Matrigel (BD Bioscience) -coated dishes in E8 media prepared in lab (Chang et al., 2013; Chen et al., 2011; Thomson et al., 1998).
Cells were prepared for differentiation by plating 1.5 × 105 cells per well on BD Matrigel coated 12-well dishes in E8 supplemented with 10 μM Y-27632 (Tocris Bioscience) overnight. The medium was then replaced with E6 medium (Thomson et al., 1998), supplemented with CHIR99021 (Tocris Bioscience) or BMP4 and INHBA (R&D Systems) for 2 days. DMSO was used as the vehicle control at a final concentration of 0.1%. CHIR99021 or BMP4 and INHBA were washed out at day 2, and cells were grown in unsupplemented E6 until day 12. From days 12 to 20, the medium was replaced with StemPro-34 media. Cells were returned to E6 media from days 20 to 35. The medium was then replaced with N2 medium until the endpoint of the experiment. All media were changed daily. When indicated (Figure S3), 20 μM Forskolin (Tocris Bioscience) and 10 ng/ml FGF2 was used from days 0 to 7 in conjunction with 0.5 μM CHIR99021, or from days 2 to 7 following 2 days of 10 μM CHIR99021 treatment.
Mouse Cell Culture
All mESC media and components are formulated in Table S2. R1 mESCs were maintained feeder free and passaged every other day in Maintenance Media, prepared in lab.
Cells were prepared for differentiation by plating 5 × 105 cells on 0.1% gelatin coated 10 cm dishes overnight in Serum Free Maintenance Media to deplete serum (Kattman et al., 2011). The cells were then trypsinized and aggregated into embryoid bodies at 1 × 105 cells/ml on Petri dishes in Serum Free Differentiation Medium for 2 days. At day 2, EBs were trypsinized and reaggregated at 1 × 105 cells/ml on Petri dishes in Serum Free Differentiation Media—supplemented with 5 ng/ml VEGF (R&D Systems), and either CHIR99021 or BMP4/INHBA—from days 2 to 4 of differentiation. DMSO was used as the vehicle control at a final concentration of 0.1%. From days 4 to 10, the medium was replaced with StemPro-34 Media. EBs were plated on BD Matrigel coated dishes on day 6. The medium was replaced at day 10 with N2 medium until the endpoint of the experiment. All media were changed every other day.
Gene Expression Analysis
RNA was isolated from cells using either the RNeasy kit (QIAGEN) or Total RNA Mini Kit (FroggaBio). RNA ranging from 0.2 to 1 μg of each sample was reverse transcribed using the Quantitect Reverse Transcription kit (QIAGEN). qPCR was carried out using a Mastercycler Realplex and analyzed with Realplex software (Eppendorf). For real-time detection of mRNA expression, 1/40 of the total first strand synthesis product was used as a template for PCR amplification using either GoTaq qPCR Master Mix (Promega) or Kapa SYBR Fast qPCR kit (Kapa Biosystems). Primer pairs (Table S3) were selected from PrimerBank (Wang and Seed, 2003) or generated by Primer-BLAST (Altschul et al., 1990) and tested for equivalent efficiency. Each reaction was carried out in duplicate, and fold changes were calculated using the comparative Ct method as described earlier (Livak and Schmittgen, 2001). The resulting Ct values were normalized to either GAPDH in hESCs or Actb in mESCs. Results are shown ±SE of the mean of three independent experiments, unless otherwise stated.
Immunofluorescence
Cells were fixed for immunofluorescence with 4% formaldehyde for 15 min and permeabilized with PBS containing 0.5% Triton X-100 for 10 min. Cells were then blocked for 1 hr with PBS containing 0.1% bovine serum albumin, 0.1% Triton X-100, and 10% goat or donkey serum. Primary antibodies against T (Abcam), PAX3 (R&D Systems), PAX7 and MF20 (Developmental Studies Hybridoma Bank), or NEFL (Sigma-Aldrich) were incubated overnight at 4°C. Cy3-conjugated donkey anti-rabbit, goat anti-mouse IgG2a, goat anti-mouse IgG2b, Alexa-488-conjugated donkey anti-goat, and DyLight488-conjugated goat anti-mouse IgG1 secondary antibodies (Jackson ImmunoResearch) were used for detection as appropriate for 1 hr at room temperature. Cells were mounted in sPBS:glycerol containing Hoechst dye to identify cell nuclei. EdU labeling was performed using the Click-iT EdU Alexa Fluor488 Imaging Kit (Life Technologies). Cells we pulsed with EdU for 4 hr before fixing and staining as per the manufacturer’s protocol.
Cells were visualized with a Leica DMI6000 B microscope (Leica Microsystems), and pictures were acquired using a Micropublisher 3.3 RTV camera (Q Imaging). Staining was quantified by performing either manual or automated cell counts using the Volocity software (PerkinElmer) and represented as a proportion of total nuclei. On average, 2.5 × 104 cells were quantified from each experiment. Results are shown ±SE of the mean of three independent experiments.
Statistical Analysis
Statistical differences between means were calculated using the Student’s t test. p values ≤ 0.05 were considered significant.
Acknowledgments
The authors would like to acknowledge Nicolas Tremblay for his assistance in the quantification of immunofluorescent images. This work was funded by an award from the Muscular Dystrophy Association to I.S.S. (218371), and by grants from the Natural Sciences and Engineering Research Council (RGPIN 293170-11) and Canadian Institutes of Health Research (MOP-89910) to W.L.S. M.S. was supported in part by the Queen Elizabeth II Graduate Scholarship in Science and Technology. R.L.C. was funded, in part, by the Government of Ontario Ministry of Economic Development and Innovation for the Ontario Research Fund supporting the International Regulome Consortium. S.-P.D. was partially supported by a Fonds de la Recherche en Santé du Québec postdoctoral fellowship. W.L.S. was supported by the Canada Research Chair in Integrative Stem Cell Biology.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Contributor Information
William L. Stanford, Email: wstanford@ohri.ca.
Ilona S. Skerjanc, Email: iskerjan@uottawa.ca.
Supplemental Information
References
- Akazawa H., Komuro I. Cardiac transcription factor Csx/Nkx2-5: Its role in cardiac development and diseases. Pharmacol. Ther. 2005;107:252–268. doi: 10.1016/j.pharmthera.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Al Madhoun A.S., Mehta V., Li G., Figeys D., Wiper-Bergeron N., Skerjanc I.S. Skeletal myosin light chain kinase regulates skeletal myogenesis by phosphorylation of MEF2C. EMBO J. 2011;30:2477–2489. doi: 10.1038/emboj.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albini S., Coutinho P., Malecova B., Giordani L., Savchenko A., Forcales S.V., Puri P.L. Epigenetic reprogramming of human embryonic stem cells into skeletal muscle cells and generation of contractile myospheres. Cell Reports. 2013;3:661–670. doi: 10.1016/j.celrep.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Araki R., Uda M., Hoki Y., Sunayama M., Nakamura M., Ando S., Sugiura M., Ideno H., Shimada A., Nifuji A., Abe M. Negligible immunogenicity of terminally differentiated cells derived from induced pluripotent or embryonic stem cells. Nature. 2013;494:100–104. doi: 10.1038/nature11807. [DOI] [PubMed] [Google Scholar]
- Awaya T., Kato T., Mizuno Y., Chang H., Niwa A., Umeda K., Nakahata T., Heike T. Selective development of myogenic mesenchymal cells from human embryonic and induced pluripotent stem cells. PLoS ONE. 2012;7:e51638. doi: 10.1371/journal.pone.0051638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberi T., Bradbury M., Dincer Z., Panagiotakos G., Socci N.D., Studer L. Derivation of engraftable skeletal myoblasts from human embryonic stem cells. Nat. Med. 2007;13:642–648. doi: 10.1038/nm1533. [DOI] [PubMed] [Google Scholar]
- Borchin B., Chen J., Barberi T. Derivation and FACS-Mediated Purification of PAX3+/PAX7+ Skeletal Muscle Precursors from Human Pluripotent Stem Cells. Stem Cell Rev. 2013;1:620–631. doi: 10.1016/j.stemcr.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham M. Skeletal muscle progenitor cells and the role of Pax genes. C. R. Biol. 2007;330:530–533. doi: 10.1016/j.crvi.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Cao N., Liang H., Huang J., Wang J., Chen Y., Chen Z., Yang H.-T. Highly efficient induction and long-term maintenance of multipotent cardiovascular progenitors from human pluripotent stem cells under defined conditions. Cell Res. 2013;23:1119–1132. doi: 10.1038/cr.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H., Yoshimoto M., Umeda K., Iwasa T., Mizuno Y., Fukada S., Yamamoto H., Motohashi N., Miyagoe-Suzuki Y., Takeda S. Generation of transplantable, functional satellite-like cells from mouse embryonic stem cells. FASEB J. 2009;23:1907–1919. doi: 10.1096/fj.08-123661. [DOI] [PubMed] [Google Scholar]
- Chang W.Y., Lavoie J.R., Kwon S.Y., Chen Z., Manias J.L., Behbahani J., Ling V., Kandel R.A., Stewart D.J., Stanford W.L. Feeder-independent derivation of induced-pluripotent stem cells from peripheral blood endothelial progenitor cells. Stem Cell Res. (Amst.) 2013;10:195–202. doi: 10.1016/j.scr.2012.11.006. [DOI] [PubMed] [Google Scholar]
- Chen G., Gulbranson D.R., Hou Z., Bolin J.M., Ruotti V., Probasco M.D., Smuga-Otto K., Howden S.E., Diol N.R., Propson N.E. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Alvares L.E., Ahmed M.U., El-Hanfy A.S., Dietrich S. The epaxial-hypaxial subdivision of the avian somite. Dev. Biol. 2004;274:348–369. doi: 10.1016/j.ydbio.2004.07.020. [DOI] [PubMed] [Google Scholar]
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- D’Amour K.A., Gage F.H. Genetic and functional differences between multipotent neural and pluripotent embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2003;100(Suppl 1):11866–11872. doi: 10.1073/pnas.1834200100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darabi R., Gehlbach K., Bachoo R.M., Kamath S., Osawa M., Kamm K.E., Kyba M., Perlingeiro R.C.R. Functional skeletal muscle regeneration from differentiating embryonic stem cells. Nat. Med. 2008;14:134–143. doi: 10.1038/nm1705. [DOI] [PubMed] [Google Scholar]
- Darabi R., Santos F.N.C., Filareto A., Pan W., Koene R., Rudnicki M.A., Kyba M., Perlingeiro R.C. Assessment of the myogenic stem cell compartment following transplantation of Pax3/Pax7-induced embryonic stem cell-derived progenitors. Stem Cells. 2011;29:777–790. doi: 10.1002/stem.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darabi R., Pan W., Bosnakovski D., Baik J., Kyba M., Perlingeiro R.C.R. Functional myogenic engraftment from mouse iPS cells. Stem Cell Rev. 2011;7:948–957. doi: 10.1007/s12015-011-9258-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darabi R., Arpke R.W., Irion S., Dimos J.T., Grskovic M., Kyba M., Perlingeiro R.C.R. Human ES- and iPS-derived myogenic progenitors restore DYSTROPHIN and improve contractility upon transplantation in dystrophic mice. Cell Stem Cell. 2012;10:610–619. doi: 10.1016/j.stem.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denham M., Bye C., Leung J., Conley B.J., Thompson L.H., Dottori M. Glycogen synthase kinase 3β and activin/nodal inhibition in human embryonic stem cells induces a pre-neuroepithelial state that is required for specification to a floor plate cell lineage. Stem Cells. 2012;30:2400–2411. doi: 10.1002/stem.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Roux G., Banfi S., Sultan M., Geffers L., Anand S., Rozado D., Magen A., Canidio E., Pagani M., Peluso I. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011;9:e1000582. doi: 10.1371/journal.pbio.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov Y.V., Jones N.C., Olwin B.B. Regulation of myogenesis by fibroblast growth factors requires beta-gamma subunits of pertussis toxin-sensitive G proteins. Mol. Cell. Biol. 1998;18:5780–5787. doi: 10.1128/mcb.18.10.5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthaman K., Fong C.-Y., Bongso A. Effect of ROCK inhibitor Y-27632 on normal and variant human embryonic stem cells (hESCs) in vitro: its benefits in hESC expansion. Stem Cell Rev. 2010;6:86–95. doi: 10.1007/s12015-009-9107-8. [DOI] [PubMed] [Google Scholar]
- Goudenege S., Lebel C., Huot N.B., Dufour C., Fujii I., Gekas J., Rousseau J., Tremblay J.P. Myoblasts derived from normal hESCs and dystrophic hiPSCs efficiently fuse with existing muscle fibers following transplantation. Mol. Ther. 2012;20:2153–2167. doi: 10.1038/mt.2012.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J.K., Banks G.B., Chamberlain J.S., Olwin B.B. Prevention of muscle aging by myofiber-associated satellite cell transplantation. Sci. Transl. Med. 2010;2:57ra83. doi: 10.1126/scitranslmed.3001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoyama T., McGivern J.V., Van Dyke J.M., Ebert A.D., Suzuki M. Derivation of myogenic progenitors directly from human pluripotent stem cells using a sphere-based culture. Stem Cells Transl. Med. 2014;3:564–574. doi: 10.5966/sctm.2013-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang Y., Suk S., Lin S., Tierney M., Du B., Seo T., Mitchell A., Sacco A., Varghese S. Directed in vitro myogenesis of human embryonic stem cells and their in vivo engraftment. PLoS ONE. 2013;8:e72023. doi: 10.1371/journal.pone.0072023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattman S.J., Witty A.D., Gagliardi M., Dubois N.C., Niapour M., Hotta A., Ellis J., Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Kennedy K.A., Porter T., Mehta V., Ryan S.D., Price F., Peshdary V., Karamboulas C., Savage J., Drysdale T.A., Li S.C. Retinoic acid enhances skeletal muscle progenitor formation and bypasses inhibition by bone morphogenetic protein 4 but not dominant negative beta-catenin. BMC Biol. 2009;7:67. doi: 10.1186/1741-7007-7-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S., Kuroda K., Le Grand F., Rudnicki M.A. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X., Hsiao C., Wilson G., Zhu K., Hazeltine L.B., Azarin S.M., Raval K.K., Zhang J., Kamp T.J., Palecek S.P. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. USA. 2012;109:E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X., Zhang J., Zhu K., Kamp T.J., Palecek S.P. Insulin inhibits cardiac mesoderm, not mesendoderm, formation during cardiac differentiation of human pluripotent stem cells and modulation of canonical Wnt signaling can rescue this inhibition. Stem Cells. 2013;31:447–457. doi: 10.1002/stem.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Wakamiya M., Shea M.J., Albrecht U., Behringer R.R., Bradley A. Requirement for Wnt3 in vertebrate axis formation. Nat. Genet. 1999;22:361–365. doi: 10.1038/11932. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lyons G.E., Schiaffino S., Sassoon D., Barton P., Buckingham M. Developmental regulation of myosin gene expression in mouse cardiac muscle. J. Cell Biol. 1990;111:2427–2436. doi: 10.1083/jcb.111.6.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan A.P., Kaestner K.H., Grau E., Schütz G. Postimplantation expression patterns indicate a role for the mouse forkhead/HNF-3 alpha, beta and gamma genes in determination of the definitive endoderm, chordamesoderm and neuroectoderm. Development. 1993;119:567–578. doi: 10.1242/dev.119.3.567. [DOI] [PubMed] [Google Scholar]
- Montarras D., Morgan J., Collins C., Relaix F., Zaffran S., Cumano A., Partridge T., Buckingham M. Direct isolation of satellite cells for skeletal muscle regeneration. Science. 2005;309:2064–2067. doi: 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- Murry C.E., Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Nitzan E., Kalcheim C. Neural crest and somitic mesoderm as paradigms to investigate cell fate decisions during development. Dev. Growth Differ. 2013;55:60–78. doi: 10.1111/dgd.12004. [DOI] [PubMed] [Google Scholar]
- Ordahl C.P., Le Douarin N.M. Two myogenic lineages within the developing somite. Development. 1992;114:339–353. doi: 10.1242/dev.114.2.339. [DOI] [PubMed] [Google Scholar]
- Petropoulos H., Skerjanc I.S. Beta-catenin is essential and sufficient for skeletal myogenesis in P19 cells. J. Biol. Chem. 2002;277:15393–15399. doi: 10.1074/jbc.M112141200. [DOI] [PubMed] [Google Scholar]
- Pick M., Azzola L., Mossman A., Stanley E.G., Elefanty A.G. Differentiation of human embryonic stem cells in serum-free medium reveals distinct roles for bone morphogenetic protein 4, vascular endothelial growth factor, stem cell factor, and fibroblast growth factor 2 in hematopoiesis. Stem Cells. 2007;25:2206–2214. doi: 10.1634/stemcells.2006-0713. [DOI] [PubMed] [Google Scholar]
- Pourquié O., Fan C.M., Coltey M., Hirsinger E., Watanabe Y., Bréant C., Francis-West P., Brickell P., Tessier-Lavigne M., Le Douarin N.M. Lateral and axial signals involved in avian somite patterning: a role for BMP4. Cell. 1996;84:461–471. doi: 10.1016/s0092-8674(00)81291-x. [DOI] [PubMed] [Google Scholar]
- Pusch C., Hustert E., Pfeifer D., Südbeck P., Kist R., Roe B., Wang Z., Balling R., Blin N., Scherer G. The SOX10/Sox10 gene from human and mouse: sequence, expression, and transactivation by the encoded HMG domain transcription factor. Hum. Genet. 1998;103:115–123. doi: 10.1007/s004390050793. [DOI] [PubMed] [Google Scholar]
- Rao L., Tang W., Wei Y., Bao L., Chen J., Chen H., He L., Lu P., Ren J., Wu L. Highly efficient derivation of skeletal myotubes from human embryonic stem cells. Stem Cell Rev. 2012;8:1109–1119. doi: 10.1007/s12015-012-9413-4. [DOI] [PubMed] [Google Scholar]
- Rios A.C., Serralbo O., Salgado D., Marcelle C. Neural crest regulates myogenesis through the transient activation of NOTCH. Nature. 2011;473:532–535. doi: 10.1038/nature09970. [DOI] [PubMed] [Google Scholar]
- Rohwedel J., Maltsev V., Bober E., Arnold H.H., Hescheler J., Wobus A.M. Muscle cell differentiation of embryonic stem cells reflects myogenesis in vivo: developmentally regulated expression of myogenic determination genes and functional expression of ionic currents. Dev. Biol. 1994;164:87–101. doi: 10.1006/dbio.1994.1182. [DOI] [PubMed] [Google Scholar]
- Rufaihah A.J., Huang N.F., Jamé S., Lee J.C., Nguyen H.N., Byers B., De A., Okogbaa J., Rollins M., Reijo-Pera R. Endothelial cells derived from human iPSCS increase capillary density and improve perfusion in a mouse model of peripheral arterial disease. Arterioscler. Thromb. Vasc. Biol. 2011;31:e72–e79. doi: 10.1161/ATVBAHA.111.230938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan T., Liu J., Chu A., Wang L., Blais A., Skerjanc I.S. Retinoic acid enhances skeletal myogenesis in human embryonic stem cells by expanding the premyogenic progenitor population. Stem Cell Rev. 2012;8:482–493. doi: 10.1007/s12015-011-9284-0. [DOI] [PubMed] [Google Scholar]
- Sakurai H., Okawa Y., Inami Y., Nishio N., Isobe K. Paraxial mesodermal progenitors derived from mouse embryonic stem cells contribute to muscle regeneration via differentiation into muscle satellite cells. Stem Cells. 2008;26:1865–1873. doi: 10.1634/stemcells.2008-0173. [DOI] [PubMed] [Google Scholar]
- Sasaki H., Hogan B.L. Differential expression of multiple fork head related genes during gastrulation and axial pattern formation in the mouse embryo. Development. 1993;118:47–59. doi: 10.1242/dev.118.1.47. [DOI] [PubMed] [Google Scholar]
- Tahamtani Y., Azarnia M., Farrokhi A., Sharifi-Zarchi A., Aghdami N., Baharvand H. Treatment of human embryonic stem cells with different combinations of priming and inducing factors toward definitive endoderm. Stem Cells Dev. 2013;22:1419–1432. doi: 10.1089/scd.2012.0453. [DOI] [PubMed] [Google Scholar]
- Takada S., Stark K.L., Shea M.J., Vassileva G., McMahon J.A., McMahon A.P. Wnt-3a regulates somite and tailbud formation in the mouse embryo. Genes Dev. 1994;8:174–189. doi: 10.1101/gad.8.2.174. [DOI] [PubMed] [Google Scholar]
- Tan J.Y., Sriram G., Rufaihah A.J., Neoh K.G., Cao T. Efficient derivation of lateral plate and paraxial mesoderm subtypes from human embryonic stem cells through GSKi-mediated differentiation. Stem Cells Dev. 2013;22:1893–1906. doi: 10.1089/scd.2012.0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C.E., Ehrhardt A., Kay M.A. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 2003;4:346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Ueno S., Weidinger G., Osugi T., Kohn A.D., Golob J.L., Pabon L., Reinecke H., Moon R.T., Murry C.E. Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2007;104:9685–9690. doi: 10.1073/pnas.0702859104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela D.M., Economides A.N., Rojas E., Lamb T.M., Nuñez L., Jones P., Lp N.Y., Espinosa R., 3rd, Brannan C.I., Gilbert D.J. Identification of mammalian noggin and its expression in the adult nervous system. J. Neurosci. 1995;15:6077–6084. doi: 10.1523/JNEUROSCI.15-09-06077.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasyutina E., Birchmeier C. The development of migrating muscle precursor cells. Anat. Embryol. (Berl.) 2006;211(Suppl 1):37–41. doi: 10.1007/s00429-006-0118-9. [DOI] [PubMed] [Google Scholar]
- Wang X., Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 2003;31:e154. doi: 10.1093/nar/gng154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J., Mehta V., Voronova A., Coutu J., Ryan T., Shelton M., Skerjanc I.S. β-catenin is essential for efficient in vitro premyogenic mesoderm formation but can be partially compensated by retinoic acid signalling. PLoS ONE. 2013;8:e57501. doi: 10.1371/journal.pone.0057501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood H.B., Episkopou V. Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Mech. Dev. 1999;86:197–201. doi: 10.1016/s0925-4773(99)00116-1. [DOI] [PubMed] [Google Scholar]
- Wu Y., Ai Z., Yao K., Cao L., Du J., Shi X., Guo Z., Zhang Y. CHIR99021 promotes self-renewal of mouse embryonic stem cells by modulation of protein-encoding gene and long intergenic non-coding RNA expression. Exp. Cell Res. 2013;319:2684–2699. doi: 10.1016/j.yexcr.2013.08.027. [DOI] [PubMed] [Google Scholar]
- Xu C., Tabebordbar M., Iovino S., Ciarlo C., Liu J., Castiglioni A., Price E., Liu M., Barton E.R., Kahn C.R. A zebrafish embryo culture system defines factors that promote vertebrate myogenesis across species. Cell. 2013;155:909–921. doi: 10.1016/j.cell.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


