Summary
Signaling factors including retinoic acid (RA) and thyroid hormone (T3) promote neuronal, oligodendrocyte, and astrocyte differentiation of cortical neural stem cells (NSCs). However, the functional specificity of transcriptional repressor checkpoints controlling these differentiation programs remains unclear. Here, we show by genome-wide analysis that histone deacetylase (HDAC)2 and HDAC3 show overlapping and distinct promoter occupancy at neuronal and oligodendrocyte-related genes in NSCs. The absence of HDAC3, but not HDAC2, initiated a neuronal differentiation pathway in NSCs. The ablation of the corepressor NCOR or HDAC2, in conjunction with T3 treatment, resulted in increased expression of oligodendrocyte genes, revealing a direct HDAC2-mediated repression of Sox8 and Sox10 expression. Interestingly, Sox10 was required also for maintaining the more differentiated state by repression of stem cell programming factors such as Sox2 and Sox9. Distinct and nonredundant actions of NCORs and HDACs are thus critical for control of lineage progression and differentiation programs in neural progenitors.
Graphical Abstract
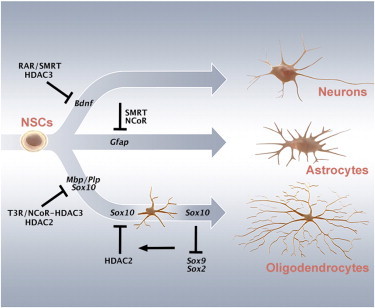
Highlights
-
•
ChIP-seq reveals distinct and overlapping occupancy of HDAC2 and HDAC3 in NSCs
-
•
Absence of NCOR promotes oligodendrocyte differentiation of NSCs
-
•
HDAC2 controls Sox10 expression in OL differentiation via a SOX2-occupied enhancer
-
•
Sox10 is required for maintaining the differentiated state in late OL precursors
ChIP-seq in neural stem cells revealed that HDAC2 and HDAC3 show overlapping and distinct promoter occupancy at neuronal and oligodendrocyte-related genes. HDAC3 knockdown initiated neuronal differentiation. Hermanson, Castelo-Branco, and colleagues show that ablation of Ncor or, in the presence of T3, Hdac2 resulted in increased expression of oligodendrocyte genes, unveiling an HDAC2-mediated repression of Sox10 expression. Sox10 was required for maintaining a more differentiated state by repression of stem cell factors including Sox2 and Sox9.
Introduction
Histone-deacetylase-associated (HDAC-associated) DNA-binding transcription factors, including REST (also known as neuron-restrictive silencer factor [NRSF]) and coregulators such as the nuclear receptor corepressor (NCOR/Ncor1) and silencing mediator of retinoic acid and thyroid hormone receptors (SMRT/Ncor2), have been shown to be essential regulators of neural proliferation and differentiation during brain development, as they actively repress lineage-characteristic gene expression and are required for self-renewal or progression of cell fate specification (Andreu-Agulló et al., 2009; Ballas et al., 2005; Hermanson et al., 2002b; Jepsen et al., 2007; Johnson et al., 2008; Lilja et al., 2013a; Miller and Gauthier, 2007; Wang et al., 2010). Mammalian HDACs can be divided into several classes, with the class I HDACs most strongly associated with NCOR and SMRT (Yang and Seto, 2008). NCOR and/or SMRT-HDAC complexes regulate histone modifications occurring at single lysines of histone tails that seem to display progenitor-specific signatures (Spivakov and Fisher, 2007). These modified lysines in turn function as molecular beacons recruiting specific complexes for regulation of gene expression (Perissi et al., 2010; Ruthenburg et al., 2007). Studies using inhibitors of HDACs, including valproic acid (VPA), have established that such compounds can affect the differentiation of embryonic and adult neural progenitors both negatively and positively and also exert toxic effects on neuroectodermal cells (Hsieh et al., 2004; Laeng et al., 2004; Marin-Husstege et al., 2002; Salminen et al., 1998). Genetic models of HDAC1 and HDAC2 ablation also support roles for class I HDACs in regulating forebrain neural progenitor characteristics, although the roles for individual HDACs remain unclear (Montgomery et al., 2009; Ye et al., 2009).
Despite the neurodevelopmental phenotypes and neural progenitor aberrations of mice harboring genetic deletions of Ncor versus Smrt being distinct (Hermanson et al., 2002b; Jepsen et al., 2000, 2007), functional and nonredundant specificity regarding the associated HDACs is not well understood (Perissi et al., 2010). HDAC activity has been shown to both be required and repressive for neurogenesis (Hsieh et al., 2004; Montgomery et al., 2009). Likewise, HDAC activity can be required for oligodendrocyte differentiation and proper myelination (Ye et al., 2009). However, HDAC inhibitors such as VPA have also been shown to have positive effects on oligodendrocyte generation and function (Liu et al., 2012), and the alleviation of transcription factors associated with HDAC complexes, such as REST (NRSF), results in increased expression of oligodendrocyte genes (Covey et al., 2012). Only a few studies have addressed the roles for individual HDACs and histone acetyl transferases (HATs) in oligodendrocyte differentiation of embryonic forebrain progenitors using genetic mouse models (Wang et al., 2010; Ye et al., 2009). The deletion of Hdac1 or Hdac2 individually in NSCs only had limited effects, while simultaneous deletion of the two factors resulted in a loss of markers of oligodendrocyte differentiation in cortical progenitors (Ye et al., 2009). Paradoxically, genetic and RNA knockdown of the HAT Cbp (also known as Crebbp) in the embryonic forebrain also resulted in decreased expression of oligodendrocyte markers in cortical NSCs, and Cbp haploinsufficiency leads to aberrant development of the corpus callosum (Wang et al., 2010).
To elucidate the roles for these factors in differentiation of cortical progenitors, we undertook an investigation of the functional roles for class I HDACs and nuclear receptor corepressors in the enforcement of NSC repression checkpoints that are subsequently released during differentiation to neurons and glia. By analysis of NSCs derived from rodents with gene deletions and/or specific RNA knockdown in wild-type primary-derived NSCs, we have revealed a series of distinct functional roles for the class I HDACs, HDAC2, and HDAC3, alone and in concert with NCOR or SMRT in the regulation of NSC differentiation into neurons and oligodendrocytes.
Results
HDAC2 and HDAC3 Show Both Unique and Overlapping Binding to Promoter Regions of Genes Associated with Neuronal and Oligodendrocyte Differentiation
To investigate specific roles for HDAC2 and HDAC3, we performed chromatin immunoprecipitation sequencing (ChIP-seq) (Gene Expression Omnibus [GEO] accession GSE57232; see Füllgrabe et al., 2013; Heldring et al., 2014) and subsequent single-gene ChIP analysis (see Lilja et al., 2013b) in neural stem cells (NSCs) derived from embryonic cortices of rats at embryonic day 15 (E15), which produce HDAC2 and HDAC3, but not HDAC1 (Figure 1C). HDAC2 and HDAC3 were determined to be present in the vicinity of a number of genes associated with transcriptional regulation of differentiation (Figure 1A). Several of the regions identified by ChIP-seq were confirmed by subsequent single-gene ChIP-quantitative PCR (ChIP-qPCR) experiments that demonstrated that HDAC2 and HDAC3 could bind both in an overlapping and distinct fashion near genes associated with development and differentiation, including Cebpb (C/EBPβ), Hoxd4, Ovol2, and Zfp7 (Figure 1A; data not shown). A more detailed analysis revealed differences in enrichment at certain genes critically involved in NSC differentiation. Whereas a significant enrichment of HDAC3 was found on the promoters of both Pax6 and Sox8, genes encoding transcription factors associated with neuronal and oligodendrocyte differentiation of NSCs, increased enrichment of HDAC2 was only found on the Sox8 promoter (Figure 1B). This observation was of particular interest due to a previous report demonstrating that class I HDACs, including HDAC2, are essential for proper progression of embryonic oligodendrocyte development (Ye et al., 2009).
Figure 1.
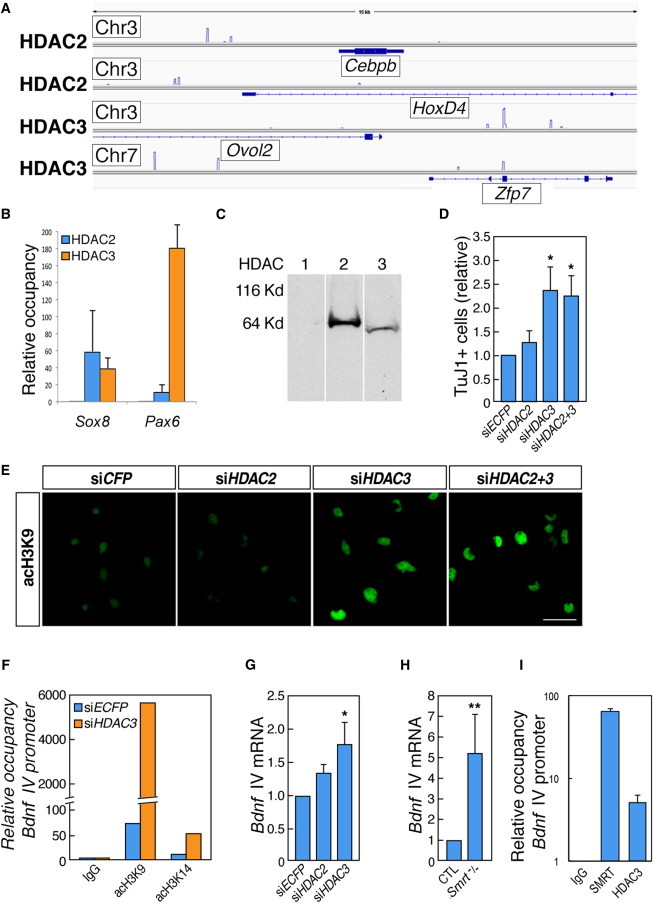
Single-Gene and Genome-wide ChIP Reveal Overlapping and Nonoverlapping Occupancies of HDAC2 and HDAC3 and HDAC3, but Not HDAC2, Knockdown Results in Increased Neuronal Differentiation of NSCs
(A) Genome browser images of normalized densities of HDAC2 and HDAC3 on regions of chromosomes 3 and 7 in the vicinity of genes encoding Cebpb, Ovol2, Hoxd4, and Zfp7 in cortically derived NSCs.
(B) qPCR analysis of regions in the vicinity of Sox8 and Pax6 after ChIP for HDAC2 and HDAC3 in NSCs.
(C) Immunoblotting of NSC protein lysates with antibodies to class I HDAC proteins. Molecular weights (approximately): HDAC1 and HDAC2, 60 kDa; HDAC3, 50 kDa.
(D) Quantification of the numbers of TuJ1-positive cells after treatment with control siRNA (siECFP), siHDAC2, siHDAC3, and siHDAC2+3.
(E) Immunofluorescence micrographs depicting NSCs stained for acetylated H3K9 after treatment with siECFP, siHDAC2, siHDAC3, and siHDAC2+3.
(F) qPCR of the Bdnf IV promoter following ChIP with anti-acetyl-H3K9 and anti-acetyl-H3K14 antibodies following transfection of control (siECFP) or Hdac3 siRNA.
(G) qRT-PCR analysis of Bdnf mRNA levels following transfection with siECFP, siHDAC2, or siHDAC3.
(H) qRT-PCR analysis of Bdnf mRNA levels in NSCs cultured from wild-type or Smrt−/− embryos.
(I) qPCR of the Bdnf IV promoter following ChIP with anti-SMRT and anti-HDAC3 antibodies in NSCs.
The scale bar represents 40 μm (E). All experiments were performed in n = 3–5 independent experiments, ∗p < 0.05, ∗∗p < 0.01 (t test). Error bars, SEM.
Similar to SMRT, HDAC3 Represses Neuronal Genes in Embryonic NSCs
While previous analyses of the functional roles for HDAC1 and HDAC2 by genetic mouse models have confirmed redundant roles for the two factors in embryonic development of the nervous system, our ChIP analysis suggested that there could be functional differences between HDAC2 and HDAC3 in embryonic NSCs. To examine the individual role(s) of these class I HDACs in NSCs, specific small interference RNA (siRNA) pools were used to rapidly and conditionally knockdown Hdac2 and Hdac3 mRNA in the cortically derived NSCs (see Figures S1A and S1B available online for efficiency of the siRNAs). NSCs transfected with siHDAC3 or a combination of siHDAC2 and siHDAC3 showed global hyperacetylation of H3K9 (Figure 1E) and a significant increase in TuJ1-positive cells (Figure 1D), as well as an increased H3K9 acetylation on the well-established HDAC target Bdnf IV promoter as compared to control siRNA (Figure 1F). In contrast, transfection of siHDAC2 had no significant effect on these parameters (Figures 1D–1F). Analysis of gene expression by quantitative RT-PCR (qRT-PCR) showed that siHDAC2 treatment had no significant effect on Bdnf mRNA levels in NSCs, whereas siHDAC3 delivery alone was sufficient to induce a significant increase in Bdnf gene expression (Figure 1G). As the gene encoding TUBB3, the protein detected by the antibody TuJ1, is a direct target for the HDAC-associated repressor REST, it is possible that the increase in TuJ1-positive cells could be due to a direct regulation and increased acetylation at the Tubb3 gene. We however also noted an increase in expression of the proneuronal gene Neurog2 6–24 hr after siHDAC3 delivery (data not shown), pointing to multiple putative mechanisms that could underlie increased neuronal differentiation and survival after siHDAC3 delivery. Together, these results demonstrate that HDAC3, but not HDAC2, is a nonredundant repressor of H3K9 acetylation, Bdnf gene expression, and neuronal differentiation in cortical NSCs.
To shed further light on the mechanisms underlying the functional specificity of HDAC2 and HDAC3, we turned to the closely associated nuclear receptors NCOR and SMRT. These HDAC-associated corepressors have in spite of their sequence homology previously been shown to play distinct functional roles in NSCs as spontaneous neuronal and astrocytic differentiation has been noted only in NSCs derived from Smrt-deficient mice, while only astrocytic and no neuronal differentiation has been reported in NSCs from Ncor-deficient or double-heterozygote mice (Hermanson et al., 2002b; Jepsen et al., 2007). ChIP analysis revealed that both SMRT and HDAC3 were enriched at the Bdnf IV promoter, and qRT-PCR investigation of Bdnf gene expression levels in Smrt−/− NSCs revealed a significant (>4-fold) increase compared to wild-type cells (Figures 1H and 1I). In contrast, Bdnf mRNA levels in Ncor−/− NSCs, which do not differentiate down the neuronal pathway, remained unchanged (data not shown; see further below). This result suggests that HDAC3, rather than HDAC2, is involved in at least a subset of the critical SMRT-mediated repressive events in NSCs.
Absence of the T3-Associated Corepressor NCOR Promotes Oligodendrocyte-Associated Gene Expression
NSCs derived from mice gene-deleted for either Ncor or Smrt, or from double-heterozygote animals, spontaneously differentiate into astrocytes, suggesting that regulation of this pathway depends on proper gene dosage of both Ncor and Smrt (Hermanson et al., 2002b; Jepsen et al., 2007). Interestingly, a substantial subset of the Ncor−/− NSCs display neither the astrocytic marker GFAP nor the stem cell marker nestin or neuronal marker TuJ1 (Hermanson et al., 2002b), suggesting the possibility that NCOR represses additional NSC fates. Indeed, gene expression profiling analysis by microarrays of Ncor−/− NSCs after 4 days in vitro (DIV) revealed a dramatic upregulation of several oligodendrocyte (OL)-associated genes, including myelin-basic protein (Mbp), myelin proteolipid protein (Plp), beta tubulin 4, and Nkx2.2, as well as early pan-glial markers such as S100β, whereas no significant upregulation of neuronal genes was detected (Figure 2B; Hermanson et al., 2002b). In contrast, gene expression profiling of Smrt−/− NSCs or NSCs treated with the HDAC inhibitor valproic acid (VPA) failed to detect any upregulation of OL-associated genes (Figure S2A; Figure S4 in Jepsen et al., 2007). Detailed analysis revealed that approximately 18% of Ncor−/− NSCs stained positive for the archetypical OL marker MBP and presented morphology typical for immature oligodendrocytes, whereas wild-type cells did not (Figures 2A and 2C). qRT-PCR experiments confirmed the upregulation of MBP expression in Ncor−/− as compared to wild-type (WT) NSCs at the mRNA level (Figure 2D). These results indicate that NCOR plays a critical role in a repression checkpoint of OL differentiation in embryonic multipotent NSCs and that absence of NCOR initiates specification into OL cell types.
Figure 2.
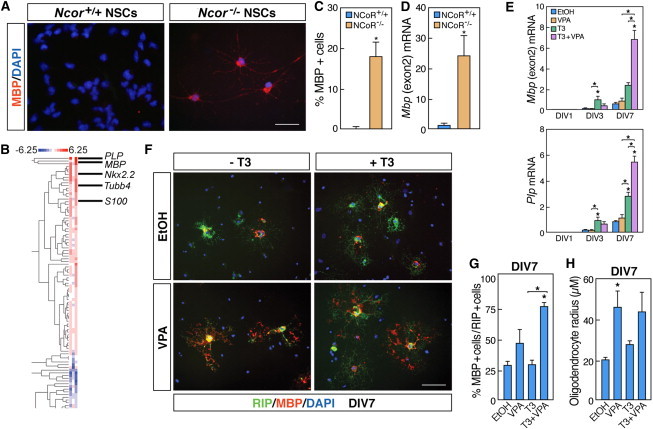
Ncor−/− NSCs Differentiate into Oligodendrocyte-like Cells Similar to T3 Treatment, and the HDAC Inhibitor VPA Enhances the T3-Mediated Differentiation
(A) Micrographs depicting NSCs isolated from wild-type (left panel) and Ncor−/− (right panel) embryos following immunocytochemistry with anti-MBP antibody (n = 3 independent experiments).
(B) RNA profiling data from Ncor−/− NSCs after 4 DIV, displaying the enrichment of increased expression of oligodendrocyte-associated genes (n = 3 independent samples).
(C) Quantification of MBP-positive cells in WT or Ncor−/− NSCs at 4 DIV (n = 3–5 independent experiments).
(D) qRT-PCR analysis of Mbp mRNA levels in wild-type or Ncor−/− NSCs (n = 3–5 independent experiments).
(E) qRT-PCR analysis of Mbp and Plp mRNA levels in NSCs treated with T3 and/or VPA. (n = 5 independent experiments, all treatments compared in each day).
(F and G) Micrographs, and corresponding quantification, depicting expression of RIP (green) and MBP (red) proteins in NSCs following treatment with T3 and/or VPA (n = 3 independent experiments). 20× objective.
(H) Quantification of oligodendrocyte radius following treatment of NSCs with T3 and/or VPA (n = 4 independent experiments, compared to EtOH treatment).
Scale bars represent 40 μm (A) and 80 μm (F). Statistical analysis: one-way ANOVA analysis of variance with Bonferroni’s multiple comparison test except (C) and (D) (t test); ∗p < 0.05. Error bars, SEM.
HDAC Inhibitors Promote OL Differentiation in Combination with T3 Stimulation, but Not Alone
NCOR was originally identified as interacting with the unliganded T3 receptor (T3R) to repress gene transcription. Indeed, it has been firmly established that T3 stimulation leads to transcriptional activation of its target genes by initiating removal of transcriptional corepressors such as NCOR from T3R, resulting in recruitment of coactivator proteins (Astapova et al., 2008; Hermanson et al., 2002a; Perissi et al., 2010). As T3 plays an essential role during OL development (Billon et al., 2002), and can lead to the specification of embryonic NSCs into the oligodendrocyte lineage (Johe et al., 1996), we reasoned that NCOR may repress OL differentiation through its role as a T3R corepressor. Treatment of wild-type embryonic NSCs with T3 induced OL differentiation in NSC cultures, as assessed by morphology and the upregulation of OL-characteristic genes, including Mbp and Plp, and early immunocytochemical markers, including RIP, at both the mRNA and protein level (Figures 2E and 2F), in accordance with previous reports (Johe et al., 1996).
While conventional models would associate NCOR-mediated repression with HDAC repressor function, alleviation of HDAC activity has paradoxically mostly been associated with inhibition of OL differentiation (Figure S2A) (Copray et al., 2009; Marin-Husstege et al., 2002; Ye et al., 2009), suggesting that HDACs are rather required for OL differentiation. To investigate whether T3 modulated the effects of HDAC inhibition on the OL lineage, embryonic NSCs were treated with T3 and VPA (0.5 mM), either alone or in combination. While T3 alone resulted in promotion of OL differentiation and gene expression, the expression of OL genes did not change in response to VPA alone (Figure 2E). In contrast, cotreatment of the NSCs with both T3 and VPA resulted in increased expression of Mbp and Plp after 7 DIV (Figure 2E). We also observed an increase in late markers of OL differentiation as compared to early markers in cells treated with both T3 and VPA at 7 DIV. Specifically, we found an increase in the number of cells displaying the late marker MBP double labeled with the early OL marker RIP, from 29% ± 3.2 in control conditions to 77% ± 3.4 in T3+VPA conditions (Figures 2F and 2G). In accordance with the increased MBP/RIP ratio and the possible increase in maturation, T3+VPA cotreatment resulted in morphological signs of late OL differentiation, including formation of myelin plaque-like structures (Figure 2F) and an increase in the average oligodendrocyte radius (Figure 2H). Notably, VPA treatment alone increased MBP immunoreactivity and the average oligodendrocyte radius in a very small subset of cells (Figure 2G and 2H), but it had no effect compared to the vehicle (EtOH) on the actual number of immature oligodendrocytes (Figures S2B and S2C).
HDAC2 Mediates Specific Control of Sox10 Expression in OL Differentiation
The ChIP-seq analysis revealed binding of HDAC2 to a subset of genes associated with oligodendrocyte differentiation, including Sox8, suggesting that HDAC2 might play a role in the oligodendrocyte lineage. Thus, we differentiated NSCs in the presence or absence of T3 for 3 days, lipofected the cells with siRNAs against Hdac2 and/or Hdac3, and then assessed the OL differentiation at 3 days after siRNA transfection (DAT) (see Figure S2D for scheme). In the absence of T3, no significant effects on the expression of genes associated with OL and glial development were seen after siRNA knockdown of Hdac2 or Hdac3 alone or in combination at 3 DAT (Figures 3A and 3B; data not shown). However, in the presence of T3, there was a significant upregulation of MBP and PLP expression following transfection with siHDAC2 or siHDAC3 (Figures 3A–3C), although we noted that the predominant effects on the expression of these genes were mediated by T3. Moreover, knockdown of Hdac2 but not Hdac3 in the oligodendrocyte cell lines Oli-neu (Jung et al., 1995) and CG4 (Louis et al., 1992) also induced an increased expression of oligodendrocyte markers as MBP, CNPase, and PLP (Figures 3D and 3E; Figures S3A, S3B, and S3H), which in the Oli-neu cells was accompanied by a dramatic change of morphology (Figure 3E; Figure S3A). These results suggest that the effects of VPA on OL differentiation by T3 are at least partially dependent on HDAC2 activity.
Figure 3.
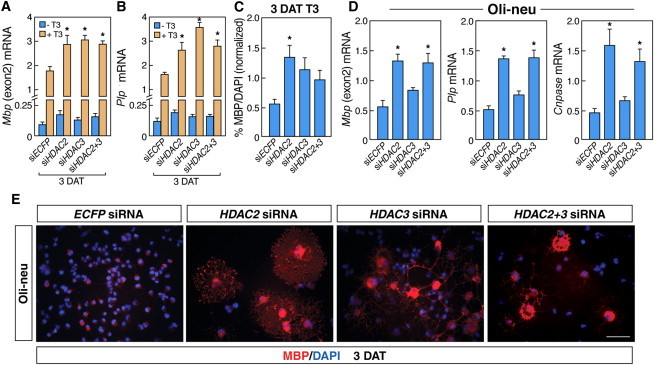
HDAC2 Regulates T3-Enhanced Oligodendrocyte Differentiation in NSCs
(A and B) qRT-PCR analysis demonstrating the effects of Hdac2 and/or Hdac3 siRNAs on Mbp and Plp mRNA levels in NSCs before and after T3 treatment (n = 4 independent experiments).
(C) Quantification of NSCs positive for MBP following transfection with control siRNA (ECFP) or siRNAs to Hdac2 and/or Hdac3 in the presence or absence of T3 relative to nuclear DAPI staining (n = 4 independent experiments).
(D) qRT-PCR results demonstrating the effects of Hdac2 and/or Hdac3 siRNAs on Mbp, Plp, and Cnpase mRNA levels in Oli-neu cells (n = 7, compared to ECFP siRNA).
(E) Immunocytochemistry of treated Oli-neu cells with MBP antibody (in red) and with DAPI (blue) counterstain.
All experiments: n = 3–5 independent experiments. Scale bars represent 80 μm (E). Statistical analysis: one-way ANOVA analysis of variance with Bonferroni's multiple comparison test; ∗p < 0.05. Error bars, SEM.
We next performed a functional screen to compare gene expression profiles of T3-treated NSCs to NSCs treated with VPA only or with VPA+T3, with particular attention to genes implicated in glial differentiation. While the expression of many OL-characteristic genes were influenced by T3 but not VPA, expression of the OL-associated transcription factor Sox10 was specifically increased 4-fold after VPA and VPA+T3 treatment, but unaffected by T3 alone (Figure 4A). Interestingly, Sox10 expression was increased by siHDAC2 upon T3 treatment (Figure 4B). In contrast, Sox10 mRNA levels were not affected by siHDAC3, by T3 treatment, or in Ncor−/− or Smrt−/− NSCs (Figures 2B, 4A, and 4B; Jepsen et al., 2007). In accordance with this result, Sox10 mRNA levels were also increased upon Hdac2, but not Hdac3 knockdown in Oli-neu cells (data not shown; see further below). A similar trend was seen for Sox8, detected as a direct HDAC2 target in the ChIP-seq experiments (Figure 1B). Transcriptional inhibitors of OL differentiation, such as Hes5, were not affected by treatment with VPA alone in the NSCs (Figure 4C), in contrast to what has been observed in postnatal oligodendrocyte precursor cells (OPCs) (Lyssiotis et al., 2007; Shen et al., 2008).
Figure 4.
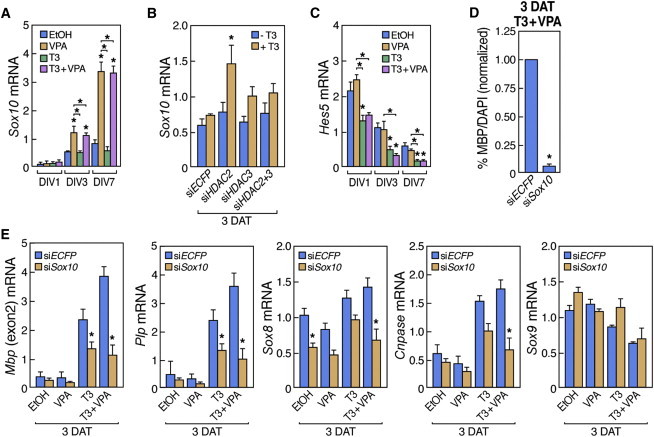
HDAC2 Regulates Sox10 Expression, and Sox10 Knockdown in NSCs and Oli-neu Cells Prevents Effects of T3 and VPA on Oligodendrocyte Differentiation
(A) qRT-PCR analysis of Sox10 mRNA levels in NSCs treated with T3 and/or VPA.
(B) qRT-PCR analysis of Sox10 mRNA levels in NSCs transfected with control siRNA (ECFP) or siRNAs to Hdac2 and/or Hdac3 in the presence or absence of T3.
(C) qRT-PCR analysis of Hes5 mRNA levels in NSCs treated with T3 and/or VPA.
(D) Quantification of NSCs expressing MBP following transfection with control siRNA (ECFP) or Sox10 siRNAs in the presence of T3 and VPA relative to nuclear DAPI staining.
(E) qRT-PCR demonstrating the effects of Sox10 siRNA on expression of Mbp, Plp, Sox8, Cnpase, and Sox9 following VPA and/or T3 treatment.
All experiments: n = 3–5 independent experiments. Statistical analysis: one-way ANOVA analysis of variance with Bonferroni's multiple comparison test; ∗p < 0.05. Error bars, SEM.
SOX10 is required for terminal OL differentiation as null mutant mice for this SoxE protein generate normal numbers of OL progenitors that fail to terminally differentiate (Stolt et al., 2002). To test whether SOX10 could mediate the additive effects of VPA in T3-mediated OL differentiation of embryonic NSCs, NSCs were differentiated in the presence of VPA and/or T3 and lipofected at 3 days with siRNA against Sox10 (Roh et al., 2006) (Figure S2D; Figure S1B for efficiency of the siRNA). Gene expression was then analyzed 3 DAT. Knockdown of Sox10 partially prevented the upregulation of MBP and PLP and blocked the terminal OL differentiation induced by the cotreatment of VPA and T3 at 3 DAT (Figures 4D and 4E). Sox10 knockdown also reduced the mRNA levels of Cnpase and Sox8 at 3 DAT, but importantly did not affect earlier markers, including Sox9, in differentiating NSCs (Figure 4E). We conclude that SOX10 is required for the proper OL differentiation effects observed when treating NSCs with VPA or siHDAC2 and T3.
To investigate specific roles for class I HDACs in the VPA-mediated potentiation of OL differentiation of NSCs, we next investigated the presence of HDAC2 and HDAC3 at previously characterized regulatory regions of Mbp (Farhadi et al., 2003) and Sox10 (Werner et al., 2007) in the embryonic NSCs (Figure 5A). ChIP-qPCR analysis indicated that while both HDAC2 and HDAC3 were present in the regulatory regions of the Mbp gene in NSCs and Oli-neu cells (Figure 5B; Figure S3D), HDAC2 was specifically enriched in the U2 enhancer of Sox10 (Figure 5B, left panel). This HDAC2 enrichment was observed both in comparison to other regions bound by HDAC2 as well as in comparison to the occupancy of HDAC3 at the same region (Figure 5B). Similar results were obtained in Oli-neu cells (Figures S3C and S3F). Indeed, an analysis of the acetylation state of the regulatory regions of Sox10 revealed relatively low acetylation levels of H3K14 and H4K16, but not H3K9, at the U2 enhancer in NSCs and to an extent also in the Oli-neu cells, strengthening the observation of increased HDAC2 occupancy at this specific enhancer (Figure 5C; Figure S3E). The U2 enhancer is an element that previously has been shown to regulate Sox10 expression specifically in the oligodendrocyte lineage starting at embryonic midgestation (Küspert et al., 2011; Werner et al., 2007). Together with our data, this suggests that HDAC2 may be specifically required for Sox10 gene regulation in early oligodendrocyte development. Class I HDACs and NCORs have been identified as binding partners of SOX2 (Engelen et al., 2011), and an in silico analysis revealed a putative binding site for SOX2 in the U2 enhancer of Sox10 (Figure S4). We therefore investigated whether this T3 and NCoR-independent HDAC2-mediated repression of Sox10 could be mediated in association with SOX2. Indeed, we found a significant enrichment of SOX2 occupancy on the U2, but not U1, enhancer compared to control regions (Figure 5D; data not shown). siRNA-mediated knockdown of Sox2, however, did not result in any significantly increased expression of Sox10 (data not shown), suggesting that other factors are sufficient to recruit HDAC2 to the U2 enhancer and mediate the essential repression.
Figure 5.
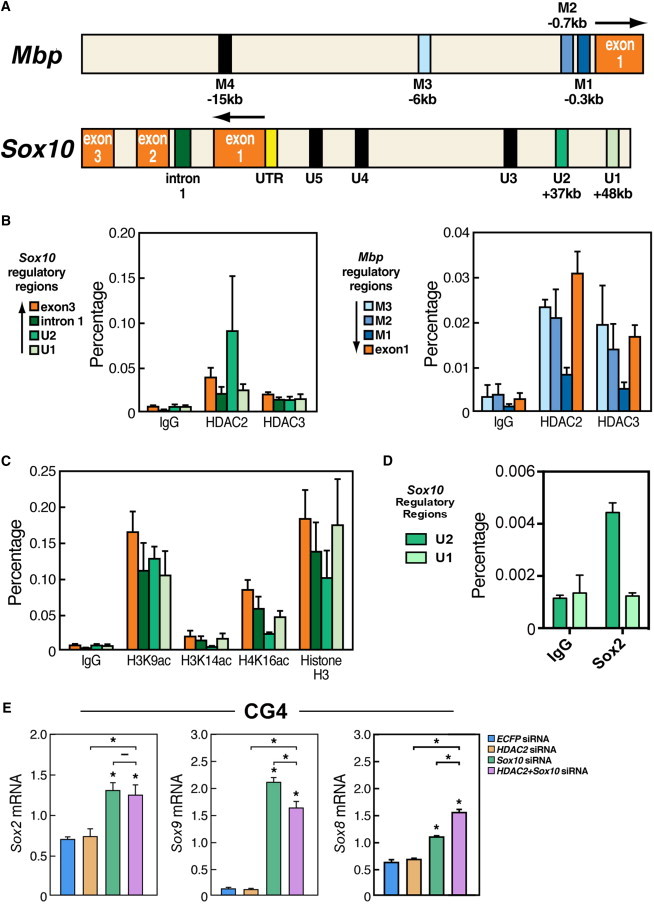
HDAC2 Occupies Developmentally Critical Regulatory Regions of Sox10 in NSCs, and Sox10 Controls a More Mature Signature of Postnatal Oligodendrocyte Precursors by Repression of Sox2 and Sox9
(A) Diagram depicting some of the regulatory regions of Mbp (M1-M4) and Sox10 (U1-U5) and their distance to the transcription start site.
(B) qPCR analysis of the indicated regions of Sox10 and Mbp following ChIP of HDAC2 or HDAC3. Percentage, percentage of input (1%).
(C) qPCR of the indicated regions of Sox10 following ChIP of acetylated H3K9, H3K14, or H4K16 from NSCs. Histone H3 is shown as a control for ChIP efficiency.
(D) qPCR of the indicated regions after ChIP of SOX2 revealed that SOX2 is bound to the U2, but not the U1, enhancer of the Sox10 gene.
(E) qRT-PCR demonstrating the effects of Sox10 siRNA and/or Hdac2 siRNA on the expression of Sox2, Sox8, and Sox9 in CG4 cells (postnatal OPCs) after Sox10 RNA knockdown.
All experiments: n = 2–3 independent experiments. Statistical analysis: one-way ANOVA analysis of variance with Bonferroni's multiple comparison test; ∗p < 0.05. Error bars, SEM.
Sox10, but Not HDACs, Is Required for Maintenance of the Differentiated State in Postnatal OL Progenitors
The differentiation of OPCs begins in early postnatal stages in the forebrain, although its peak occurs a few days later (Wegner, 2008). It is noteworthy that Hdac2 expression has been reported to decrease in cells of the oligodendrocyte lineage at these stages (Shen et al., 2005). Interestingly, we did not observe any morphological signs of premature differentiation of the proliferating postnatal CG4 upon siHDAC2 and/or siHDAC3 delivery (data not shown), in contrast to what we observed in embryonic Oli-neu cells or T3-treated NSCs (Figure 2; Figure S3A). Moreover, although moderate increases in gene expression levels of Mbp and Plp were observed following transfection with siHDAC2 in the CG4 cells (Figure S3H), these were found not to be associated with an siHDAC2-mediated Sox10 derepression as Sox10 levels were indeed rescued by siHDAC2, but Mbp and Plp expression levels were not in these cells (Figure S3H; data not shown). These results suggested that SOX10 could play alternative roles at this late stage of OL differentiation. Intriguingly, Sox10 siRNA delivery at this stage instead resulted in a dramatic increase in gene expression associated with the stem cell state (Pevny and Nicolis, 2010; Scott et al., 2010), including Sox2, Sox9, Sox8, and Hes5, without affecting markers of neuronal differentiation (Figure 5E; data not shown). This result stands in sharp contrast to the mild effects of Sox10 siRNA on the expression of these genes in NSCs and a reversible increase of Sox9 expression in Oli-neu cells (Figure 4E; data not shown). Taken together, our observations suggest that Sox10 expression is required for efficient progression of oligodendrocyte differentiation and expression of late OL characteristic genes and at a later stage serves to maintain the differentiated state and suppress the expression of genes associated with more undifferentiated progenitors and stem cells (Figure 6).
Figure 6.
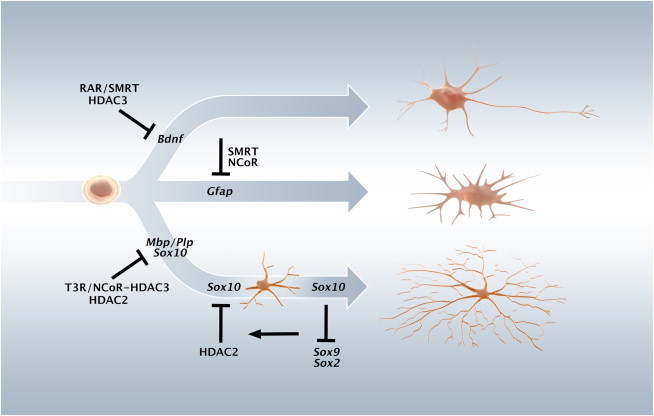
NCOR and HDACs Play Nonredundant Roles in the Regulation of Neural Stem Cell Differentiation
In multipotent embryonic NSCs, SMRT and HDAC3 are nonredundantly repressing neuronal differentiation, at least in part by repressing Bdnf expression required for the cell survival during the progression of neuronal differentiation. Astrocytic differentiation is repressed by both NCOR and SMRT. Oligodendrocyte differentiation is repressed by NCOR and HDAC2 in neural stem cells by repressing different OL-associated genes. In Oli-neu cells (prenatal oligodendrocyte progenitors), a repression of Sox10 is maintained by HDAC2 that occupies the regulatory U2 enhancer of Sox10. In CG4 cells (postnatal oligodendrocyte precursors), Sox10 is not only required for the progression of OL differentiation but also for retaining a more differentiated state by being required for efficient repression of Sox2 along with Sox9, Hes5, and other progenitor associated genes.
Discussion
Transcriptional corepressors such as NCOR and SMRT have distinct roles in repressing differentiation in NSCs, and we therefore hypothesized that associated HDACs also have specific functions albeit the reports so far have been unclear. Here we demonstrate that whereas HDAC3 and SMRT, and not HDAC2 or NCoR, are part of a repression checkpoint for neuronal differentiation in embryonic NSCs, NCoR and HDAC2 constitute specific repressor checkpoints in control of oligodendrocyte differentiation (Figure 6). The mechanism by which HDAC2 inhibits oligodendrocyte differentiation is at least in part, via repression of the key transcription factor SOX10, coinciding with binding of HDAC2 to the SOX2-occupied U2 enhancer of the Sox10 gene (Figure 6). Together, these data reveal nonredundant roles for class I HDACs and associated factors in NSC differentiation.
Our results suggest that specific HDACs play distinct roles in the differentiation of NSCs. Chemical or genetic ablation of class I HDAC activity have been reported to inhibit oligodendrocyte and Schwann cell differentiation and promote neuronal differentiation (Copray et al., 2009; Hsieh et al., 2004; Marin-Husstege et al., 2002). In these studies, HDACs are suggested to repress inhibitors of OL differentiation, thereby being required for proper oligodendrocyte differentiation. However, deletion of the HAT CREB-binding protein also results in decreased oligodendrocyte differentiation and corpus callosum deficits (Wang et al., 2010), and HDACs have also been shown to be required for neurogenesis (Montgomery et al., 2009), blurring a general consensus statement of the function of these factors in NSC differentiation. In the present study, loss of HDAC2 activity in the presence of T3 resulted in enhanced oligodendrocytic differentiation in embryonic cortical NSCs. Strikingly, HDAC2 does repress directly the key OL transcription factor Sox10, in contrast to the indirect effects of HDACs previously reported. Thus, the effects of HDAC inhibition on NSCs appear to be distinct depending on cell-type specificity, the temporal window of action, and the presence of additional factors. These cell-type and temporal specific effects of HDACs are in line with what has been reported for components of the NOTCH and WNT signaling, which can have both repressing and activating functions in oligodendrocyte development (Popko, 2003; Tawk et al., 2011).
Class B1 Sox proteins such as SOX2 are established repressors of differentiation and required to retain stem cell characteristics during development and in the adult (Arnold et al., 2011; Bylund et al., 2003; Engelen et al., 2011; Graham et al., 2003), but it has been shown that SOX2 can also act as a transcriptional activator in certain developmental contexts (Hoffmann et al., 2014). Our data on NCOR, HDAC2, and SOX10 suggest a transcriptional “transmission” mechanism where the appropriate “gear” controls the level of progenitor state and early and late progression of oligodendrocyte differentiation. In NSCs and embryonic OPCs, HDAC2 were found to bind to the critical U2 enhancer regulating Sox10 in association with SOX2, and alleviation of HDAC2 led to an increase in Sox10 expression. OLIG2 has recently been shown to activate the U2 enhancer (Küspert et al., 2011) and as such could also be a candidate for the upregulation of Sox10 expression. Nevertheless, in contrast to SOX2 (Figure 5D), OLIG2 is present not only at the U2 enhancer in immature oligodendrocytes but also at the U1, U3, U4, and U5 Sox10 enhancers (Yu et al., 2013), suggesting a more general role in the transcriptional regulation of SOX10. While OLIG2 is an important regulator of this system, we do not observe an upregulation of Olig2 upon VPA+T3 treatment or Hdac2 and/or Hdac3 siRNA in NSCs and Oli-neu cells, respectively (Figures S3G and S3I). It will be of immediate interest to elucidate additional factors responsible for HDAC2 recruitment to the U2 enhancer in NSCs and prenatal oligodendrocyte progenitors. Indeed, since Sox2 knockdown failed to induce a significant increase in Sox10 expression even in the presence of T3, it is plausible that other class B1 Sox factors such as SOX1/3 could be involved, and the role for class B2 factors binding similar sequences but with alternative actions, such as SOX14 or SOX21, should be further investigated.
In postnatal OPCs, increased levels of SOX10 seem to repress Sox2 expression and consequently promote late progression of oligodendrocyte differentiation. Intriguingly, forced Sox2 expression mediates dedifferentiation of maturely differentiated oligodendrocytes (Kondo and Raff, 2000, 2004) and SOX9 has been shown to promote neural stem cell state (Scott et al., 2010). Whereas there are few previous demonstrations of such key factors maintaining differentiated state in the central nervous system, the requirement of SOX10 for retaining a more differentiated state partly resembles the master role of PAX5 maintaining the differentiated state of mature B lymphocytes (Cobaleda et al., 2007). Our present observations that SOX10 is required for repression of Sox2 and Sox9 in late oligodendrocyte precursor cells, in addition to the distinct requirements for NCOR, SMRT, and HDAC-regulated repressor actions in the progression of neural stem and progenitor differentiation, suggest a distinct order of repressor and corepressor action to mediate the serial events that details the neuronal and oligodendrocyte lineages, with expression of Sox10 required to prevent reexpression of gene products promoting stem cell characteristics.
Experimental Procedures
Embryonic Neural Stem Cell Cultures
Rat embryonic neural stem cells were derived as previously described in detail elsewhere (Hermanson et al., 2002a; Johe et al., 1996; Joseph et al., 2009). In brief, rat cortical tissue from embryonic day 15.5 was dissociated and 800 × 103 cells were plated per 10 cm dish, previously coated with poly-L-ornithine and fibronectin (both from Sigma-Aldrich). Neural stem cells were then expanded in N2 media with 10 ng/ml of basic fibroblastic growth factor (R&D Systems) and passaged by light dissociation in the presence of HBSS, NaHCO3, and HEPES (from Invitrogen and Sigma-Aldrich). For differentiation assays, neural stem cells (passage 1) were plated at the following densities: 150 × 103 cells in 35 mm plates (Corning) or 300 × 103 cells in 60 mm plates (Corning). NSCs were differentiated for up to 7 DIV in the absence of FGF-2 (N2), in the presence of VPA (0.5 mM, a concentration at which it specifically inhibits class I HDACs, Sigma-Aldrich) (Göttlicher et al., 2001) or its vehicle 70% ethanol (EtOH), and with or without thyroid hormone (T3—50 ng/ml, Sigma-Aldrich). Factors were added daily and media was changed every second day. After 1, 3, or 7 DIV, cells were fixed with 10% formalin (Sigma-Aldrich) and analyzed by immunocytochemistry or qRT-PCR. For ChIP assays, NSCs were expanded after the first passage until confluency, without contact between adjacent colonies. Animals were treated in accordance with institutional and national guidelines, with all required ethical permits. In Figure S2, FGF2 (10 ng/ml) expanded NSCs were treated for 3 DIV with FGF2 (10 ng/ml), IGF-2 (5 ng/ml), and PDGF-BB (10 ng/ml) (OPC media).
OPC Cultures
The embryonic OPC cell line Oli-neu and the postnatal OPC cells were cultured as previously described (Jung et al., 1995; Louis et al., 1992); for details see Supplemental Information.
Transfection-Lipofection
siRNAs or pcDNAEF1-eGFP (Amaxa) were lipofected using Lipofectamine 2000 (Invitrogen), following the manufacturer’s instructions. Three micrograms of siRNA or 1 μg pcDNAEF1-eGFP was used per 35-mm plate. Opti-MEM I reduced serum medium was used to prepare the complexes. Cells were changed to Dulbecco's modified Eagle's medium (Invitrogen) and incubated with the complexes for 4 hr before returning to their original media. Cells were collected for immunocytochemistry or qRT-PCR analysis after 1 or 3 DIV. Lipofection of siRNA in differentiated conditions was performed after 3 DIV differentiation. The following target sequences for siRNAs (A4 synthesis, Dharmacon) were used.
ECFP Control
5′-GAAGAACGGCAUCAAGGCC-3′.
Sox10
5′- CUGCUGUUCCUUCUUGACC-3 (Roh et al., 2006).
Hdac2
(1) 5′-AAA UGU CGC UGA UCA UAA GAA-3′.
(2) 5′-AAG GUG UUC AAA UGC AAG CUA-3′.
Hdac3
(1) 5′-AAU AGC CUA GUC CUG CAU UAU-3′.
(2) 5′-AAC ACA GCU AAA CAA UAA GAU-3′
Optimal results for differentiation and gene expression in NSCs by HDAC2 and HDAC3 siRNAs were titrated to knockdowns in the range of 50%–75%. Higher efficiency resulted in increased cell death along with previous observations, and lower efficiency yielded less significant results.
Chromatin Immunoprecipitation and qPCR
ChIP-IT Express (Active Motif) was used according to the supplier’s recommendations. Cells were crosslinked using 1% formaldehyde for 10 min. Cells were rinsed twice with cold PBS, collected by scraping and pelleted at 2,000 rpm for 4 min at 4°C. Frozen pelleted cells were resuspended in lysis buffer and centrifuged for 10 min at 4°C, and the pelleted nuclei were resuspended in the shearing buffer. Chromatin was then sonicated using a Bioruptor 200 (Diagenode) at high frequency, 0.5 min/0.5min, for 12 min. Sonicated chromatin was analyzed in a 3% agarose gel to confirm efficient sonication. Input was collected for further analysis. Five micrograms of chromatin was incubated for 1 hr at 4°C with protein G magnetic beads and then immunoprecipitated overnight at 4°C with the following antibodies: α-H3K9ac (07-352; Upstate, 5 μl), α-H3K14ac (06-911; Upstate, 5 μl), α-H4K16ac (07-329; Upstate, 5 μl), α-HDAC2 (sc-7899x; Santa Cruz, 5 μg), α-HDAC3 (sc-11417x; Santa Cruz, 5 μg), α-SOX2 (sc-17320x; Santa Cruz, 1 μg), RNA polymerase II (Active Motif, positive control for ChIP-IT Express, 2 μg), Histone H3 (Abcam, ab1791, 2 μg), rabbit IgG (Active Motif, negative control, 2 μg; Santa Cruz, sc-2027, 1 μg), and goat IgG (Santa Cruz, sc-2028, 1 μg). After three washes with ChIP buffer 1 and two washes with ChIP buffer 2, the IP DNA was reverse crosslinked and resuspended in a final volume of 130 μl. Purified DNA and 1% input were analyzed by qPCR, using 5-fold dilutions of the input for standard curves and triplicates per sample. When primer dimers were detected, the relative quantity for the specific sample was considered to be zero. As the standard curve method was used, potential differences in primer efficiency were taken in account and occupancy in different regulatory regions could be compared for a specific antibody.
RNA Profiling
RNA profiling was performed as previously described (Jepsen et al., 2007); for details see Supplemental Information.
Statistical Analysis
Statistical analysis and graphs were performed using the software Prism 4 (Graph Pad). When comparing to a hypothetical control value (100%), one sample t test was used. When two groups were compared, statistical analysis was performed using two-tailed unpaired t test. When comparing three or more groups, one-way ANOVA analysis of variance with Bonferroni’s multiple comparison test was used. Comparisons were performed between all treatments, unless stated otherwise. The threshold value for statistical significance (α value) was set at 0.05 (∗p < 0.05). In all graphs, results are expressed as mean ± SEM. The “n” in the figure legends represents the number of independent experiments.
Acknowledgments
We would like to thank Dr. J.C. Louis (Amgen) for the CG4 cell line (Louis et al., 1992), Dr. J. Trotter (University of Mainz) for the Oli-neu cell line (Jung et al., 1995), J. Hightower for artwork, and Dr. M. Götz and members of the O.H. lab for valuable discussions. M.G.R. is a Howard Hughes Medical Institute Investigator. This study was supported by NIH grants (to M.G.R.), a Scientist Development Grant from the American Heart Association (to K.J.), and grants from the Swedish Brain Foundation (Hjärnfonden), David and Astrid Hagélen Foundation, SSMF (Svenska Sällskapet för Medicinsk Forskning), the Marie Curie Integration Grant, Seventh Framework Programme, European Union (to G.C.-B.), the Swedish Research Council (VR-MH), Karolinska Institutet Research Foundations, the Swedish Society of Medicine (SLS) (to G.C.-B. and O.H.), the Swedish Cancer Society (CF), the KI Cancer Network, the Swedish Foundation for Strategic Research (SSF), and the Swedish Childhood Cancer Foundation (BCF) (to O.H.).
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Contributor Information
Gonçalo Castelo-Branco, Email: goncalo.castelo-branco@ki.se.
Ola Hermanson, Email: ola.hermanson@ki.se.
Accession Numbers
The ChIP-seq data are freely accessible in the GEO database under accession number GSE57232. Additional descriptions of qRT-PCR, immunocytochemistry and ChIP-seq methods can be found in the Supplemental Information.
Supplemental Information
References
- Andreu-Agulló C., Morante-Redolat J.M., Delgado A.C., Fariñas I. Vascular niche factor PEDF modulates Notch-dependent stemness in the adult subependymal zone. Nat. Neurosci. 2009;12:1514–1523. doi: 10.1038/nn.2437. [DOI] [PubMed] [Google Scholar]
- Arnold K., Sarkar A., Yram M.A., Polo J.M., Bronson R., Sengupta S., Seandel M., Geijsen N., Hochedlinger K. Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell. 2011;9:317–329. doi: 10.1016/j.stem.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astapova I., Lee L.J., Morales C., Tauber S., Bilban M., Hollenberg A.N. The nuclear corepressor, NCoR, regulates thyroid hormone action in vivo. Proc. Natl. Acad. Sci. USA. 2008;105:19544–19549. doi: 10.1073/pnas.0804604105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N., Grunseich C., Lu D.D., Speh J.C., Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Billon N., Jolicoeur C., Tokumoto Y., Vennström B., Raff M. Normal timing of oligodendrocyte development depends on thyroid hormone receptor alpha 1 (TRalpha1) EMBO J. 2002;21:6452–6460. doi: 10.1093/emboj/cdf662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylund M., Andersson E., Novitch B.G., Muhr J. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat. Neurosci. 2003;6:1162–1168. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- Cobaleda C., Jochum W., Busslinger M. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature. 2007;449:473–477. doi: 10.1038/nature06159. [DOI] [PubMed] [Google Scholar]
- Copray S., Huynh J.L., Sher F., Casaccia-Bonnefil P., Boddeke E. Epigenetic mechanisms facilitating oligodendrocyte development, maturation, and aging. Glia. 2009;57:1579–1587. doi: 10.1002/glia.20881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covey M.V., Streb J.W., Spektor R., Ballas N. REST regulates the pool size of the different neural lineages by restricting the generation of neurons and oligodendrocytes from neural stem/progenitor cells. Development. 2012;139:2878–2890. doi: 10.1242/dev.074765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelen E., Akinci U., Bryne J.C., Hou J., Gontan C., Moen M., Szumska D., Kockx C., van Ijcken W., Dekkers D.H. Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes. Nat. Genet. 2011;43:607–611. doi: 10.1038/ng.825. [DOI] [PubMed] [Google Scholar]
- Farhadi H.F., Lepage P., Forghani R., Friedman H.C., Orfali W., Jasmin L., Miller W., Hudson T.J., Peterson A.C. A combinatorial network of evolutionarily conserved myelin basic protein regulatory sequences confers distinct glial-specific phenotypes. J. Neurosci. 2003;23:10214–10223. doi: 10.1523/JNEUROSCI.23-32-10214.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Füllgrabe J., Lynch-Day M.A., Heldring N., Li W., Struijk R.B., Ma Q., Hermanson O., Rosenfeld M.G., Klionsky D.J., Joseph B. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature. 2013;500:468–471. doi: 10.1038/nature12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göttlicher M., Minucci S., Zhu P., Krämer O.H., Schimpf A., Giavara S., Sleeman J.P., Lo Coco F., Nervi C., Pelicci P.G., Heinzel T. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 2001;20:6969–6978. doi: 10.1093/emboj/20.24.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham V., Khudyakov J., Ellis P., Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- Heldring N., Nyman U., Lönnerberg P., Onnestam S., Herland A., Holmberg J., Hermanson O. NCoR controls glioblastoma tumor cell characteristics. Neuro-oncol. 2014;16:241–249. doi: 10.1093/neuonc/not214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanson O., Glass C.K., Rosenfeld M.G. Nuclear receptor coregulators: multiple modes of modification. Trends Endocrinol. Metab. 2002;13:55–60. doi: 10.1016/s1043-2760(01)00527-6. [DOI] [PubMed] [Google Scholar]
- Hermanson O., Jepsen K., Rosenfeld M.G. N-CoR controls differentiation of neural stem cells into astrocytes. Nature. 2002;419:934–939. doi: 10.1038/nature01156. [DOI] [PubMed] [Google Scholar]
- Hoffmann S.A., Hos D., Küspert M., Lang R.A., Lovell-Badge R., Wegner M., Reiprich S. Stem cell factor Sox2 and its close relative Sox3 have differentiation functions in oligodendrocytes. Development. 2014;141:39–50. doi: 10.1242/dev.098418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J., Nakashima K., Kuwabara T., Mejia E., Gage F.H. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc. Natl. Acad. Sci. USA. 2004;101:16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepsen K., Hermanson O., Onami T.M., Gleiberman A.S., Lunyak V., McEvilly R.J., Kurokawa R., Kumar V., Liu F., Seto E. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell. 2000;102:753–763. doi: 10.1016/s0092-8674(00)00064-7. [DOI] [PubMed] [Google Scholar]
- Jepsen K., Solum D., Zhou T., McEvilly R.J., Kim H.-J., Glass C.K., Hermanson O., Rosenfeld M.G. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature. 2007;450:415–419. doi: 10.1038/nature06270. [DOI] [PubMed] [Google Scholar]
- Johe K.K., Hazel T.G., Muller T., Dugich-Djordjevic M.M., McKay R.D. Single factors direct the differentiation of stem cells from the fetal and adult central nervous system. Genes Dev. 1996;10:3129–3140. doi: 10.1101/gad.10.24.3129. [DOI] [PubMed] [Google Scholar]
- Johnson R., Teh C.H., Kunarso G., Wong K.Y., Srinivasan G., Cooper M.L., Volta M., Chan S.S., Lipovich L., Pollard S.M. REST regulates distinct transcriptional networks in embryonic and neural stem cells. PLoS Biol. 2008;6:e256. doi: 10.1371/journal.pbio.0060256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B., Andersson E.R., Vlachos P., Södersten E., Liu L., Teixeira A.I., Hermanson O. p57Kip2 is a repressor of Mash1 activity and neuronal differentiation in neural stem cells. Cell Death Differ. 2009;16:1256–1265. doi: 10.1038/cdd.2009.72. [DOI] [PubMed] [Google Scholar]
- Jung M., Krämer E., Grzenkowski M., Tang K., Blakemore W., Aguzzi A., Khazaie K., Chlichlia K., von Blankenfeld G., Kettenmann H. Lines of murine oligodendroglial precursor cells immortalized by an activated neu tyrosine kinase show distinct degrees of interaction with axons in vitro and in vivo. Eur. J. Neurosci. 1995;7:1245–1265. doi: 10.1111/j.1460-9568.1995.tb01115.x. [DOI] [PubMed] [Google Scholar]
- Kondo T., Raff M. Oligodendrocyte precursor cells reprogrammed to become multipotential CNS stem cells. Science. 2000;289:1754–1757. doi: 10.1126/science.289.5485.1754. [DOI] [PubMed] [Google Scholar]
- Kondo T., Raff M. Chromatin remodeling and histone modification in the conversion of oligodendrocyte precursors to neural stem cells. Genes Dev. 2004;18:2963–2972. doi: 10.1101/gad.309404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küspert M., Hammer A., Bösl M.R., Wegner M. Olig2 regulates Sox10 expression in oligodendrocyte precursors through an evolutionary conserved distal enhancer. Nucleic Acids Res. 2011;39:1280–1293. doi: 10.1093/nar/gkq951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laeng P., Pitts R.L., Lemire A.L., Drabik C.E., Weiner A., Tang H., Thyagarajan R., Mallon B.S., Altar C.A. The mood stabilizer valproic acid stimulates GABA neurogenesis from rat forebrain stem cells. J. Neurochem. 2004;91:238–251. doi: 10.1111/j.1471-4159.2004.02725.x. [DOI] [PubMed] [Google Scholar]
- Lilja T., Heldring N., Hermanson O. Like a rolling histone: epigenetic regulation of neural stem cells and brain development by factors controlling histone acetylation and methylation. Biochim. Biophys. Acta. 2013;1830:2354–2360. doi: 10.1016/j.bbagen.2012.08.011. [DOI] [PubMed] [Google Scholar]
- Lilja T., Wallenborg K., Björkman K., Albåge M., Eriksson M., Lagercrantz H., Rohdin M., Hermanson O. Novel alterations in the epigenetic signature of MeCP2-targeted promoters in lymphocytes of Rett syndrome patients. Epigenetics. 2013;8:246–251. doi: 10.4161/epi.23752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.S., Chopp M., Kassis H., Jia L.F., Hozeska-Solgot A., Zhang R.L., Chen C., Cui Y.S., Zhang Z.G. Valproic acid increases white matter repair and neurogenesis after stroke. Neuroscience. 2012;220:313–321. doi: 10.1016/j.neuroscience.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis J.C., Magal E., Muir D., Manthorpe M., Varon S. CG-4, a new bipotential glial cell line from rat brain, is capable of differentiating in vitro into either mature oligodendrocytes or type-2 astrocytes. J. Neurosci. Res. 1992;31:193–204. doi: 10.1002/jnr.490310125. [DOI] [PubMed] [Google Scholar]
- Lyssiotis C.A., Walker J., Wu C., Kondo T., Schultz P.G., Wu X. Inhibition of histone deacetylase activity induces developmental plasticity in oligodendrocyte precursor cells. Proc. Natl. Acad. Sci. USA. 2007;104:14982–14987. doi: 10.1073/pnas.0707044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Husstege M., Muggironi M., Liu A., Casaccia-Bonnefil P. Histone deacetylase activity is necessary for oligodendrocyte lineage progression. J. Neurosci. 2002;22:10333–10345. doi: 10.1523/JNEUROSCI.22-23-10333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller F.D., Gauthier A.S. Timing is everything: making neurons versus glia in the developing cortex. Neuron. 2007;54:357–369. doi: 10.1016/j.neuron.2007.04.019. [DOI] [PubMed] [Google Scholar]
- Montgomery R.L., Hsieh J., Barbosa A.C., Richardson J.A., Olson E.N. Histone deacetylases 1 and 2 control the progression of neural precursors to neurons during brain development. Proc. Natl. Acad. Sci. USA. 2009;106:7876–7881. doi: 10.1073/pnas.0902750106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V., Jepsen K., Glass C.K., Rosenfeld M.G. Deconstructing repression: evolving models of co-repressor action. Nat. Rev. Genet. 2010;11:109–123. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- Pevny L.H., Nicolis S.K. Sox2 roles in neural stem cells. Int. J. Biochem. Cell Biol. 2010;42:421–424. doi: 10.1016/j.biocel.2009.08.018. [DOI] [PubMed] [Google Scholar]
- Popko B. Notch signaling: a rheostat regulating oligodendrocyte differentiation? Dev. Cell. 2003;5:668–669. doi: 10.1016/s1534-5807(03)00331-9. [DOI] [PubMed] [Google Scholar]
- Roh J., Cho E.A., Seong I., Limb J.K., Lee S., Han S.J., Kim J. Down-regulation of Sox10 with specific small interfering RNA promotes transdifferentiation of Schwannoma cells into myofibroblasts. Differentiation. 2006;74:542–551. doi: 10.1111/j.1432-0436.2006.00084.x. [DOI] [PubMed] [Google Scholar]
- Ruthenburg A.J., Li H., Patel D.J., Allis C.D. Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen A., Tapiola T., Korhonen P., Suuronen T. Neuronal apoptosis induced by histone deacetylase inhibitors. Brain Res. Mol. Brain Res. 1998;61:203–206. doi: 10.1016/s0169-328x(98)00210-1. [DOI] [PubMed] [Google Scholar]
- Scott C.E., Wynn S.L., Sesay A., Cruz C., Cheung M., Gomez Gaviro M.V., Booth S., Gao B., Cheah K.S., Lovell-Badge R., Briscoe J. SOX9 induces and maintains neural stem cells. Nat. Neurosci. 2010;13:1181–1189. doi: 10.1038/nn.2646. [DOI] [PubMed] [Google Scholar]
- Shen S., Li J., Casaccia-Bonnefil P. Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. J. Cell Biol. 2005;169:577–589. doi: 10.1083/jcb.200412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S., Sandoval J., Swiss V.A., Li J., Dupree J., Franklin R.J., Casaccia-Bonnefil P. Age-dependent epigenetic control of differentiation inhibitors is critical for remyelination efficiency. Nat. Neurosci. 2008;11:1024–1034. doi: 10.1038/nn.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivakov M., Fisher A.G. Epigenetic signatures of stem-cell identity. Nat. Rev. Genet. 2007;8:263–271. doi: 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
- Stolt C.C., Rehberg S., Ader M., Lommes P., Riethmacher D., Schachner M., Bartsch U., Wegner M. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 2002;16:165–170. doi: 10.1101/gad.215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawk M., Makoukji J., Belle M., Fonte C., Trousson A., Hawkins T., Li H., Ghandour S., Schumacher M., Massaad C. Wnt/beta-catenin signaling is an essential and direct driver of myelin gene expression and myelinogenesis. J. Neurosci. 2011;31:3729–3742. doi: 10.1523/JNEUROSCI.4270-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Weaver I.C., Gauthier-Fisher A., Wang H., He L., Yeomans J., Wondisford F., Kaplan D.R., Miller F.D. CBP histone acetyltransferase activity regulates embryonic neural differentiation in the normal and Rubinstein-Taybi syndrome brain. Dev. Cell. 2010;18:114–125. doi: 10.1016/j.devcel.2009.10.023. [DOI] [PubMed] [Google Scholar]
- Wegner M. A matter of identity: transcriptional control in oligodendrocytes. J. Mol. Neurosci. 2008;35:3–12. doi: 10.1007/s12031-007-9008-8. [DOI] [PubMed] [Google Scholar]
- Werner T., Hammer A., Wahlbuhl M., Bösl M.R., Wegner M. Multiple conserved regulatory elements with overlapping functions determine Sox10 expression in mouse embryogenesis. Nucleic Acids Res. 2007;35:6526–6538. doi: 10.1093/nar/gkm727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.J., Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F., Chen Y., Hoang T., Montgomery R.L., Zhao X.H., Bu H., Hu T., Taketo M.M., van Es J.H., Clevers H. HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nat. Neurosci. 2009;12:829–838. doi: 10.1038/nn.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Chen Y., Kim B., Wang H., Zhao C., He X., Liu L., Liu W., Wu L.M., Mao M. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell. 2013;152:248–261. doi: 10.1016/j.cell.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


