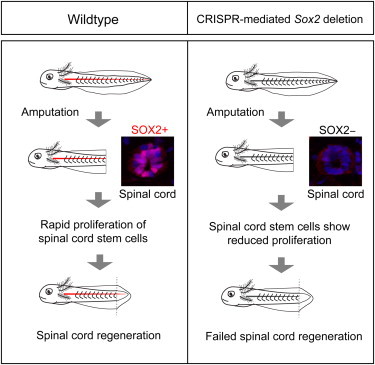
Summary
The salamander is the only tetrapod that functionally regenerates all cell types of the limb and spinal cord (SC) and thus represents an important regeneration model, but the lack of gene-knockout technology has limited molecular analysis. We compared transcriptional activator-like effector nucleases (TALENs) and clustered regularly interspaced short palindromic repeats (CRISPRs) in the knockout of three loci in the axolotl and find that CRISPRs show highly penetrant knockout with less toxic effects compared to TALENs. Deletion of Sox2 in up to 100% of cells yielded viable F0 larvae with normal SC organization and ependymoglial cell marker expression such as GFAP and ZO-1. However, upon tail amputation, neural stem cell proliferation was inhibited, resulting in spinal-cord-specific regeneration failure. In contrast, the mesodermal blastema formed normally. Sox3 expression during development, but not regeneration, most likely allowed embryonic survival and the regeneration-specific phenotype. This analysis represents the first tissue-specific regeneration phenotype from the genomic deletion of a gene in the axolotl.
Graphical Abstract
Highlights
-
•
CRISPRs yield more efficient and effective gene knockout than TALENs in axolotls
-
•
Sox2-CRISPR axolotl larvae have a normal spinal cord but show a regeneration phenotype
-
•
There is a lack of neural stem cell expansion during regeneration in Sox2-CRISPR axolotls
Tanaka and colleagues show that CRISPRs efficiently knock out target genes in the axolotl with less toxicity than TALENs. CRISPR-mediated Sox2 knockout resulted in normal spinal cord development, but spinal cord regeneration failure due to reduced proliferation of neural stem cells. This work represents the first tissue-specific regeneration phenotype upon genomic deletion of a gene in the axolotl.
Introduction
The molecular study of regeneration in animals such as the salamander serves as an important basis for understanding the limited regenerative abilities of other animals. The salamander is the only tetrapod where full cellular reconstitution of a lesioned spinal cord occurs and where the adult limb functionally regenerates all constituent tissues, and it therefore plays particular significance in understanding regenerative ability and how it may be improved (Kragl et al., 2008, 2009; Simon and Tanaka, 2013; Tanaka and Ferretti, 2009). The relevance of salamander regeneration to mammals was highlighted in an analysis of mouse digit tip regeneration, which showed that the molecular factors mediating tissue interactions between nerve and skin were similar to those first identified during salamander limb regeneration (Takeo et al., 2013). Furthermore, the extensive skin-regenerative capacities of the spiny mouse were shown to utilize an extracellular matrix (ECM) environment similar to those defined for salamander limb regeneration, suggesting that the implementation of certain ECM-expression programs is associated with deep regenerative ability (Seifert et al., 2012). Therefore, deepening our understanding of the molecular programs underlying the regenerative response in salamanders is an important endeavor.
Targeted overexpression of genes via electroporation, viral transduction, or transgenesis as well as knockdown of protein expression via electroporation morpholinos, respectively, has been an important means of molecularly analyzing regeneration in the axolotl (Echeverri and Tanaka, 2005; Kawakami et al., 2006; Khattak et al., 2013; Mercader et al., 2005; Roy et al., 2000; Whited et al., 2013). However, the lack of stable genomic knockout in the axolotl had limited molecular studies. Recently, exciting progress has been made on genome editing using zinc-finger nucleases, transcriptional activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeat (CRISPR) systems (for review, see Gaj et al., 2013). With these methods, highly efficient gene knockout or homologous gene knockin has been achieved in a variety of species including vertebrates such as Xenopus, zebrafish, and rodents. Studies implementing TALENs in Xenopus reported that targeted deletions reach efficiencies up to 100% of alleles (Lei et al., 2012; Suzuki et al., 2013). The effectiveness of TALENs was also recently demonstrated in the ribbed Iberian newt, Pleurodeles waltl (Hayashi et al., 2014). These results indicate that TALENs are a highly versatile system for targeting deletions and gene insertions into vertebrate genomes.
Recently, an alternative method of genome editing was described, based on nucleases of the bacterial antiphage “immune” system, CRISPR (for review, see Gaj et al., 2013; Mali et al., 2013a). Several groups engineered a two-component system for genome editing in which a single guide RNA (gRNA) was coexpressed with the Cas9 gene (Cong et al., 2013; Jinek et al., 2012; Mali et al., 2013b). This two-component system resulted in highly efficient generation of indels at the target site. In human embryonic stem cells, depending on the target, 1%–34% of expressing cells showed a deletion of at least one targeted allele. The method has also been used to demonstrate the possibility of generating genomic deletions in several model systems, including the axolotl (Blitz et al., 2013; Flowers et al., 2014; Hwang et al., 2013; Nakayama et al., 2013).
In most vertebrate models, mRNAs encoding TALENs and CRISPRs have been injected into the egg to generate genomic deletions from the beginning of development. Since regeneration is a late event, a question is whether this means of knocking out genes can be applied to studying specific cell types during regeneration or whether more sophisticated methods to induce knockouts later in development will be necessary. Here, we investigated the utility of the CRISPR system to knock out a key neural stem gene, Sox2, with the aim to study its role in the regenerating spinal cord. SOX2 is an SRY-related high-mobility group box transcription factor that supports embryonic pluripotency (Avilion et al., 2003), and it later plays a key role in the development, maintenance, and expansion of neural stem/progenitor cells (Bylund et al., 2003; Favaro et al., 2009; Takemoto et al., 2011). In addition, Sox2 is required to maintain the proliferative progenitor pool in various adult epithelia (Arnold et al., 2011; Que et al., 2009). In the axolotl, SOX2 is expressed in the mature spinal cord and in cultured spinal cord neurospheres that harbor multipotent neural stem cells that reconstitute the different spinal cord cell types (McHedlishvili et al., 2012; Tapia et al., 2012). The molecular factors that support the rapid expansion and self-renewal of neural stem cells during spinal cord regeneration after tail amputation were unknown. Here, we show by the CRISPR-mediated genomic deletion of Sox2 that axolotls harboring up to 100% Sox2 deleted cells survive embryonic development to generate larvae with a normal spinal cord cell complement and neuronal number. Morpholino knockdown of SOX2 in the embryo confirms lack of embryonic lethality. Upon tail amputation, however, the spinal cord cells do not increase their proliferation, resulting in defective expansion and an overall lack of spinal cord in the regenerating tail. Expression analysis of Sox3 shows overlapping expression with Sox2 during neural tube development, but not regeneration, rationalizing the spinal cord regeneration-specific phenotype. Despite severe defects in the regenerating spinal cord, the regenerative epidermis and the mesenchymal blastema form normally in Sox2-CRISPR animals.
Results
CRISPRs Mediate More Efficient, Less Toxic Gene Knockout Than TALENs in Axolotl
Germline-Transmitted Knockout of a GFP-Transgene
To determine if TALENs and CRISPRs can mediate efficient gene deletions, we initially chose to knock out a genomically inserted GFP-transgene in our germline-transgenic GFP-axolotl strain (Sobkow et al., 2006) to have an easy readout for successful gene knockout. Two pairs of GFP-TALENs and two pairs of CRISPR GFP-gRNAs in corresponding regions were designed according to published protocols (Cermak et al., 2011; Cong et al., 2013; Mali et al., 2013b) (Figures 1A–1A″; see the Experimental Procedures). We injected the TALEN mRNAs (mixture of left- and right-arm mRNAs) or CRISPR RNAs (mixture of Cas9 mRNA and GFP-gRNA) into freshly laid axolotl embryos with the aim of eliciting knockout of the genomic GFP as early as possible. In the egg injection experiments, we titrated the amount of injected TALEN (Table S1 available online) and CRISPR (Table S2) RNAs and found that the CRISPR RNAs were significantly less toxic to embryos (evaluated at 10 days postinjection) (Tables S1 and S2). We next assessed the efficiency of genomic modification. Injections of GFP-TALEN pair #1 or #3 mRNA, but not control TALENs (#1 left-arm plus #3 right-arm mRNAs), into embryos resulted in 24%–34% of the larvae showing a visible loss or reduction of GFP expression in at least some tissue/body cells in the resulting larvae (Figures 1B and C; Table S3). GFP expression was markedly reduced in the normally strongly expressing muscle, as confirmed on histological cross-sections (Figure 1C). The genomic generation of indels was confirmed by genomic PCR (Figure 1D). Animals modified by GFP-TALENs #1 and #3 were grown to adulthood, and both showed germline transmission of the GFP deletion with a representative transmission frequency of 56%.
Figure 1.
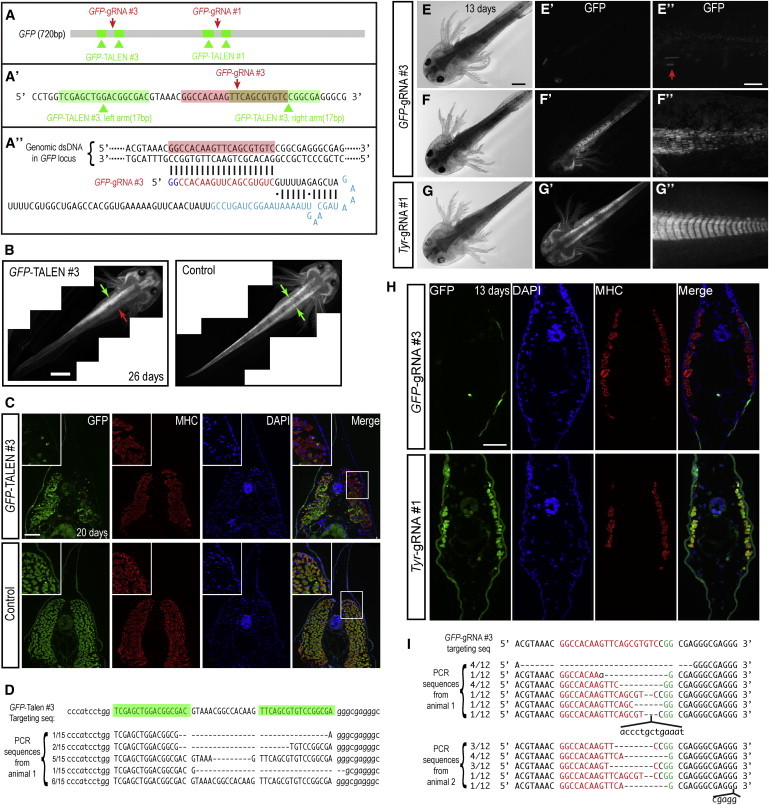
Deletions in a GFP-Transgene in the Axolotl Genome via TALEN or CRISPR Injections
(A–A″) TALENs (green) and CRISPR gRNAs (red) were designed to target the GFP sequence and injected into one-cell-stage eggs.
(B) Representative axolotl larvae that received GFP-TALEN #3 in which half of the body (red arrow compared to green arrow) has lost GFP expression. Control (GFP-TALEN #1 left-arm plus #3 right-arm mRNAs) injection showed bilaterally symmetric GFP expression (green arrows). Scale bar, 2 mm.
(C) Tail cross-sections from GFP-TALEN #3 knockdown versus control knockdown animals counterstained for DAPI and muscle-specific myosin heavy chain (MHC) immunostaining. GFP fluorescence signal is missing in muscle cells of the GFP-TALEN section, but not in a comparable region of the control animal. Region of interest is shown and zoomed by the white box. Scale bar, 200 μm.
(D) Analysis of genomic target site in two individuals injected with GFP-TALEN #3 by PCR amplification and sequencing of clones. A total of 9 out of 15 clones showed deletions or deletions plus insertions in the genomic sequence, reflecting a high rate of genome modification in the cells.
(E) Representative axolotl larva that received CRISPR GFP-gRNA#3 in which >50% of cells have lost GFP expression.
(E′) GFP expression is reduced throughout the body.
(E″) High-magnification view of tail tissue. Only a few remaining GFP cells are visible (red arrow).
(F–F″) Another representative axolotl larva with extensive knockout in 50% of the body.
(G–G″) Injection of CRISPR gRNAs targeting the axolotl Tyrosinase (Tyr) gene as a control shows no knockdown of the GFP-transgene. Scale bars represent 1 mm in (E) and 500 μm in (E″).
(H) Tail cross-sections from CRISPR GFP knockdown versus control Tyrosinase knockdown animals counterstained for DAPI and MHC immunostaining. GFP fluorescence signal is missing in most cells of the GFP-gRNA#3 section. Scale bars, 100 μm.
(I) Analysis of genomic target site in two individuals injected with GFP-gRNA#3 by PCR amplification and sequencing of clones. In both samples, all 12 clones showed deletions or deletions plus insertions in the genomic sequence, reflecting a high rate of genome modification in the cells.
See also Tables S1–S3.
In comparison to TALENs, injection of CRISPR GFP-gRNA #3 generated GFP deletions at a higher penetrance and efficiency. A total of 55% of injected animals showed at least some loss of GFP expression. Of the animals showing modification, 60% showed greater than 50% of cells lacking GFP expression (Table S3; Figures 1E–1F″). Control CRISPR Tyrosinase(Tyr)-gRNA did not yield knockdown of GFP expression (Figures 1G–1G″). Cryosections confirmed the loss of GFP expression in GFP-gRNA#3-injected, but not control Tyr-gRNA#1-injected, animals at the cellular level (Figure 1H). Finally, PCR cloning and sequencing of genomic DNA isolated from individual injected animals showed a large proportion of clones (cells) with deletions at the target GFP sequence (Figure 1I). Control-injected embryos showed no modification of the GFP locus. Germline transmission of CRISPRs is still to be determined. In summary, our results showed that GFP-TALENs and GFP-CRISPRs can successfully modify the GFP-transgene, with CRISPR mediating more efficient, less toxic effects.
Efficient Knockout of the Endogenous Tyrosinase Gene
To assess the effectiveness of TALENs and CRISPRs on an endogenous gene, we first chose the axolotl Tyrosinase (Tyr) gene as it is a nonessential gene whose knockout gives a clear, pigmentation defect. To knock out Tyr, two pairs of Tyr-TALENs (#3 and #4) and three CRISPR gRNAs (Figure 2A) were assessed by injection of RNAs into embryos and examined 13 days postinjection (Table S4). A total of 60% of animals injected with Tyr-TALEN #4 mRNA showed at least some loss of pigment in the body (Figures 2B and 2B′). In comparison, the most efficient CRISPR Tyr-gRNA#1 yielded 99% of animals showing loss in pigment, with 86% showing apparently complete loss of pigment (Table S4; Figures 2E and 2E′). Animals injected with control TALEN (left arm of Tyr-TALEN #4) and GFP-gRNA#3 showed no loss in pigmentation (Figures 2C, 2C′, 2F, and 2F′). Again, PCR amplification, cloning, and sequencing of the target Tyr sequence showed a higher rate of deletions from Tyr-CRISPR (Figure 2G) when compared to Tyr-TALEN (Figure 2D). Our results show that for two independent gene-knockout trials, CRISPRs showed a higher frequency and penetrance of gene knockdown with less animal lethality compared to TALENs.
Figure 2.
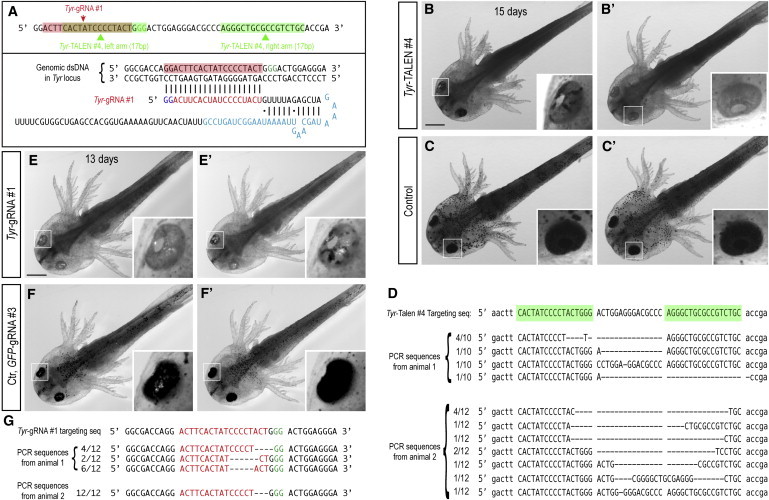
Deletions in the Tyr Gene via TALEN or CRISPR Injections
(A) TALEN and CRISPR gRNAs were designed to target the Tyr sequence and RNAs injected into one-cell-stage eggs.
(B and B′) Two representative axolotl larva that had been injected with Tyr-TALEN #4 mRNAs in which >50% of cells have lost pigmentation. Scale bar, 1 mm.
(C and C′) Two representative axolotl larvae injected with left-arm-only TALEN mRNAs show no loss of pigmentation.
(D) Analysis of the genomic target site in two individuals injected with Tyr-TALEN #4 by PCR amplification and sequencing of clones. In animal 1, seven out of ten clones showed deletions in the genomic sequence, while in animal 2, 11 out of 12 samples showed genomic modification, suggesting a high rate of modification in this animal.
(E and E′) Two representative axolotl larva that had been injected with Tyr-gRNA#1 in which nearly all (left panel) or >50% (right panel) of cells have lost pigmentation. Scale bar, 1 mm.
(F and F′) Two representative axolotl larvae injected with control GFP-gRNA#3 show no loss of pigmentation.
(G) Analysis of the genomic target site in two individuals injected with Tyr-gRNA#1 by PCR amplification and sequencing of clones. In sample 1, all (12 out of 12) clones showed deletions in the genomic sequence, while in animal 2, all (12 out of 12) samples showed the same genomic modification, suggesting that the deletion was made in the early egg/embryo.
See also Table S4.
Axolotls Harboring CRISPR-Mediated Deletions in Sox2 Show Normal Size and Organization of the Spinal Cord during Development
Given the higher success rate with CRISPRs, we sought to determine the role of Sox2 in axolotl spinal cord development and regeneration by the generation of CRISPR-mediated gene deletions. In axolotls, SOX2 expression was associated with neural stem cells that have the ability to clonally reconstitute spinal cord cell types during tail regeneration (McHedlishvili et al., 2012). In order to effectively disrupt the protein-coding sequence of Sox2, seven CRISPR targeting sites were selected as close as possible to the translational start codon (Figures 3A and 3B). Sox2-CRISPR RNAs were injected into fertilized eggs at the single-cell stage and analyzed 13 days postinjection. Of the seven Sox2-gRNAs designed, two of them, Sox2-gRNA#2 targeting the N-terminal region and Sox2-gRNA#4 targeting the middle of the sequence, gave identical, penetrant phenotypes described below, while a smaller proportion of Sox2-gRNA#5-injected animals gave a milder version of the phenotype and the other four Sox2-gRNAs gave no apparent phenotype. Of 487 eggs injected with Sox2-gRNA#2, 403 survived, hatched, and grew to normal sizes. Upon closer visual inspection of behavior, 274 (68%) showed a curved body (Figure 3C) when swimming, and many appeared to have excess blood in the olfactory bulb area (Figure 3C′, red arrow). The same phenotype was observed from Sox2-gRNA#4-injected animals (Figures 3E and 3E′). This phenotype was never observed in the GFP-gRNA#1-injected animals (Figures 3D, 3D′, 3F, and 3F′). Immunohistological examination of the olfactory bulb confirmed a severe reduction in SOX2-positive cells at the lumen of the olfactory bulb and a general loss of cell number (Figure S1A). The observation of identical phenotypes from two different Sox2-gRNAs, combined with the absence of the phenotype in all animals injected with GFP and Tyrosinase gRNAs, indicates that the phenotypes are specific to Sox2 deletion and not due to off-target effects.
Figure 3.
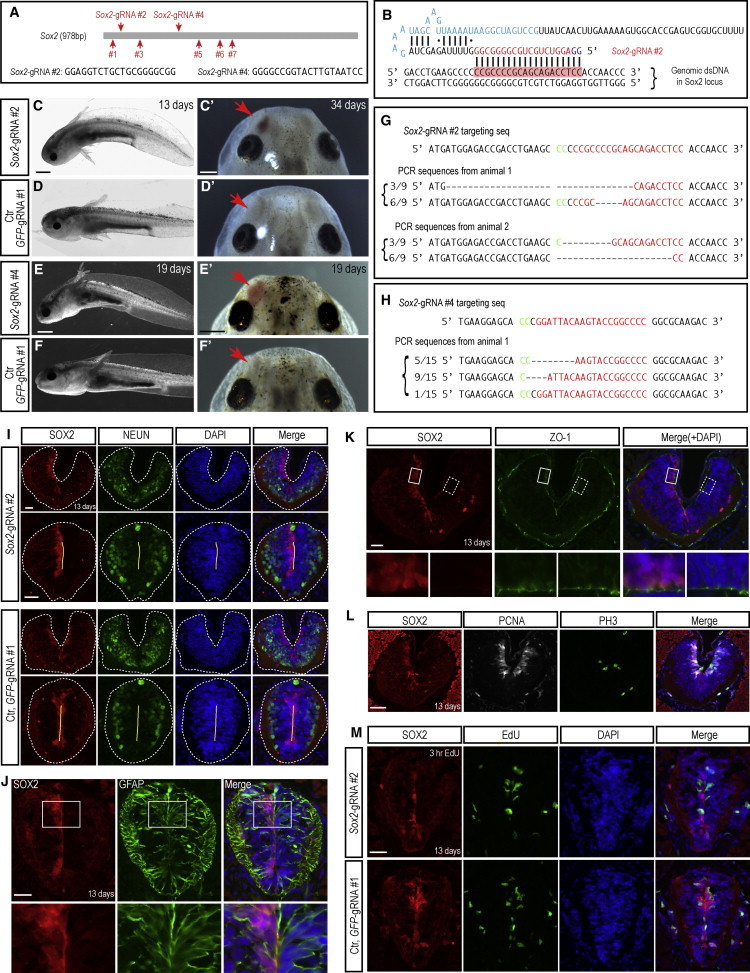
Normal Spinal Cord Organization in Animals with CRISPR-Mediated Deletions in the Sox2 Locus
(A) Schema depicting the locations of seven and sequence of two CRISPR gRNAs designed against the Sox2 coding sequence.
(B) Schema illustrating the binding between Sox2-gRNA#2 and the Sox2 target sequence.
(C–F″) Representative axolotl larvae that received Sox2-gRNA#2 (C and C′) and Sox2-gRNA#4 (E and E′) show curved body and/or olfactory bulb defects (C′ and E′, red arrows). Animals receiving control (Ctr) GFP-gRNA#1 show no such defects (D, D′, F, and F′). Scale bars represent 1 mm in (C) and (E) and 0.5 mm in (C′) and (E′).
(G and H) Analysis of the genomic target site in two (G) or one (H) individuals injected with Sox2-gRNA#2 or Sox2-gRNA#4, respectively, by PCR amplification and sequencing of clones. Note: these individuals show a very high proportion of clones with modification.
(I) Sox2-CRISPR animals maintain normal organization of neurons. Immunofluorescence staining for SOX2 (red) and NEUN (green) combined with DAPI (blue) on spinal cord cross-sections from animals injected with Sox2-gRNA#2 (upper panels) or control (Ctr) GFP-gRNA#1. Two individual spinal cord sections along the anterior-posterior axis are shown. A significant number of Sox2-gRNA#2-injected animals are lacking SOX2 protein expression in at least half of the spinal cord indicating biallelic loss in one-half of the body, suggesting modification at the two-cell stage. Both SOX2+ and SOX2− sides of the spinal cord have NEUN+ neurons at the outer circumference. Dashed lines indicate spinal cord area; yellow lines indicate apical surface. Scale bars, 50 μm.
(J and K) Sox2-deleted cells maintain GFAP and ZO-1 expression. Immunofluorescence for SOX2 (red) and GFAP (green, J) or ZO-1 (green, K) combined with DAPI (blue) from Sox2-gRNA#2-injected animals. Both the side with SOX2+ cells and the side without SOX2+ cells express GFAP. Square boxes denote insets shown at higher magnification below. Scale bars, 50 μm.
(L) Normal expression of proliferative markers in Sox2-deleted cells. Immunofluorescence for SOX2 (red), PCNA (white), and phosphohistone H3 (green) combined with DAPI (blue) on cross-sections. Scale bar, 50 μm.
(M) Incorporation of EdU in Sox2-deleted cells in the uninjured spinal cord. Immunofluorescence for SOX2 (red) combined with EdU (green) and DAPI (blue) on spinal cord cross-sections from animals injected with Sox2-gRNA#2, with 3 hr EdU labeling before harvest. Scale bar, 50 μm.
See also Figures S1 and S2.
The genetic knockout of Sox2 in mice results in preimplantation lethality (Avilion et al., 2003), and therefore, the survival through embryonic development was surprising, although knockdown of Sox2 by morpholino in amphibian or fish has not been reported. We therefore analyzed the penetrance of CRISPR-mediated Sox2 deletions within individual animals at two developmental stages: embryonic stage 15 (neural plate stage) and day 13 larvae. At stage 15, when there was no scorable phenotype, injected animals were randomly selected and genotyped by PCR. Among this cohort, 4 out of 11 animals yielded 100% of cloned inserts with deletions in Sox2, indicating that animals can survive to stage 15 with what appears to be complete penetrance of Sox2 deletion (Figure S1B). Day 13 larvae could be phenotypically sorted based on the curved-body phenotype (68% of total). Genotyping of the curved-body animals showed 8 out of 11 animals with 100% of clones containing deletions in Sox2 (Figures 3G and S1C). Analysis of animals injected with Sox2-gRNA#4 also showed a high-penetrance Sox2 genomic locus modification (Figure 3H). In contrast, genotyping of Sox2-gRNA#2-injected animals that did not show the curved-body/olfactory bulb phenotype showed only a very low frequency of gene modification (Figure S1D). These results show that animals harboring a very high penetrance of CRISPR-mediated Sox2 deletions in the genomes of their somatic cells can survive embryonic development to day 13 larvae.
To confirm our conclusions from the CRISPR-mediated deletions, we investigated the embryonic phenotype of knocking down SOX2 protein via injection of fluorescein isothiocyanate-coupled morpholinos against the Sox2 translational start site into one-cell-stage embryos. Sox2 morpholino or control morpholinos were injected at three different concentrations (Figure S2A) and yielded an indistinguishable proportion of viable embryos when screened at embryonic stage 37 (Figures S2A–S2C). Immunostaining for SOX2 on cross-sections of stage 16/17 animals confirmed downregulation of SOX2 expression in Sox2 morpholino-injected embryos (Figures S2D and S2E). The independent knockdown of SOX2 via morpholinos also revealed no detectable embryonic phenotype, confirming the results from CRISPR-mediated Sox2 deletion.
To initially characterize the effects of SOX2 deletion in the uninjured spinal cord by immunohistochemistry, we used animals that showed approximately half-body knockout of SOX2 to have an internal control. Based on our experience with GFP-CRISPR knockouts, animals can often show deletion of the gene in one-half of their body, presumably due to action of the CRISPR in one of the first two blastomeres during development. When we analyzed sections of the anterior spinal cord of such animals, at least one-half of the spinal cord was lacking SOX2 expression in the cells lining the spinal cord lumen (Figure 3I). Surprisingly, in such animals, the left and right side of the spinal cord were similar in size and organization, showing NEUN+ neuronal cells at the outer circumference (Figure 3I). We also observed normal expression of other markers associated with axolotl neural stem cells in the Sox2-deleted cells, including normal expression of GFAP and the tight junction protein ZO-1, as well as the proliferative markers proliferating cell nuclear antigen (PCNA) and phosphohistone H3 (PH3) (Figures 3J–3L). We further examined the profile of proliferation markers by injection of 5-ethynyl-2′-deoxyuridine (EdU). In the uninjured spinal cord, the incorporation of EdU was similar in the SOX2+ and the SOX2− side of the spinal cord (Figure 3M).
Defective Spinal Cord Regeneration due to Lack of Enhanced Proliferation after Tail Amputation in Sox2-CRISPR Animals
Since the uninjured spinal cord showed normal organization under conditions of Sox2 deletion, we went on to determine if there is a phenotype during tail regeneration. We amputated the tails of Sox2-CRISPR animals that showed the curved-body phenotype in order to select for animals that had a high penetrance of deletions (Figure 4A). We characterized the spinal cords in the removed portion of the tail next to the amputation plane to determine the penetrance of SOX2 deletion and the organization of the spinal cord at the starting point of the regeneration experiment. As expected, most animals showed massive or complete loss of the SOX2 spinal cord immunofluorescence signal at the amputation plane in the removed portion of the tail (Figures 4A, 4B, and S1E). Consistent with our analysis above, we found that the spinal cord size, organization, and PCNA expression was normal in spinal cords lacking SOX2+ cells (Figure 4B). PCNA expression in the uninjured ependymal cells of both control and Sox2-CRISPR animals showed differential intensity levels that reflect significant G1 and S phases of the slow cell cycle found in these neural stem cells under homeostatic conditions (A. Rodrigo-Albors, personal communication). We then examined whether the overall length of the regenerated spinal cord and tail was affected by following live animals at 6 and 10 days after tail amputation under oblique transillumination conditions. Animals injected with Sox2-gRNA#2 or with Sox2-gRNA#4 showed reduced or lack of spinal cord in the regenerated portion of the tail. (Figure 4C). In contrast, control GFP-gRNA#1-injected animals showed clear outgrowth of the spinal cord into the blastema (Figure 4C). We quantitated this phenotype by measuring the length of the overall tail regenerate versus the length of the visible spinal cord (Figure 4A). The overall tail regenerate length was unaffected in Sox2-CRISPR animals, while the spinal cord showed a significantly reduced length resulting in a shifted ratio of spinal cord/tail regenerate length (Figures 4D and 4E). We further correlated the extent of Sox2 deletion found in the removed portion of the tail (see above) with the spinal cord regeneration phenotype at 6 days and found that those specimens showing complete knockout of SOX2 immunofluorescence at the amputation plane displayed the strongest spinal cord growth inhibition (Figures S1E and S1F). Analysis of the live samples at 10 days postamputation showed a continued defect in spinal cord outgrowth into the blastema (Figures S3A–S3C). By this later time point, a mild effect on blastema length was observed in some Sox2-deleted animals.
Figure 4.
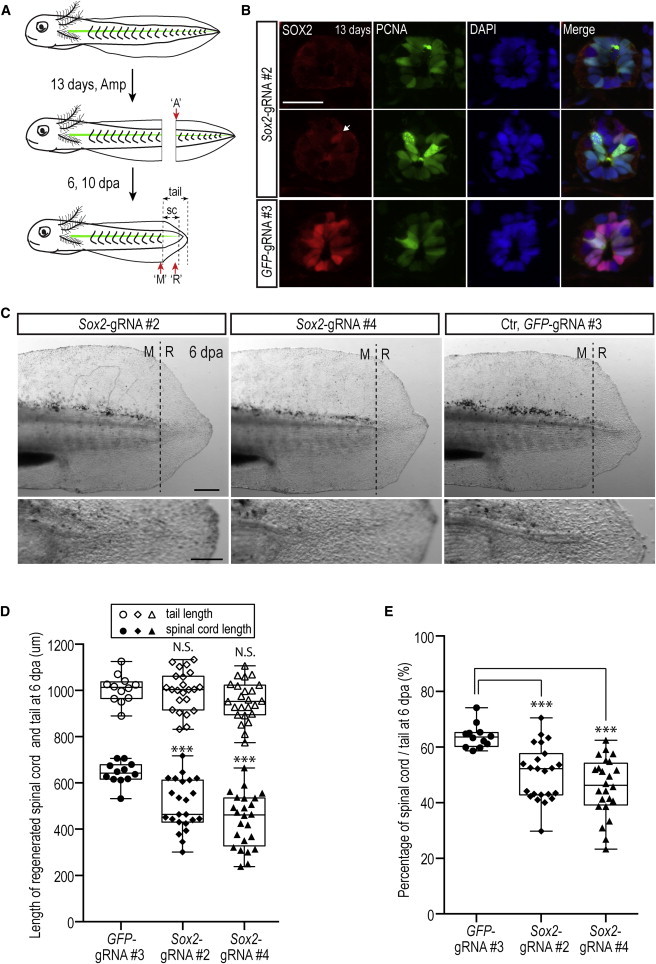
Deficient Spinal Cord Regeneration in Sox2-CRISPR Animals
(A) Cartoon illustrates regeneration experimental scheme. Amp, amputation; dpa, day post amputation; ‘A’, amputated spinal cord site; ‘M’, mature spinal cord area; ‘R’, regenerated area. Dashed line indicates the amputation plane, the tip of the spinal cord, and tip of the tail.
(B) Immunofluorescence for SOX2 (red) and PCNA (green) combined with DAPI (blue) on cross-sections from the removed piece of tail just at the amputation plane (‘A’ in A) from animals injected with Sox2-gRNA#2 or GFP-gRNA#3. Upper and middle panels are two adjacent sections from the same animal; one shows complete loss of the SOX2-positive signal, but the other shows the presence of a single Sox2-positive cell (arrow). Scale bar, 50 μm.
(C) Images of live, 6-day regenerates. Left: Sox2-gRNA#2; middle: Sox2-gRNA#4; right: control (Ctr), GFP-gRNA#3. Lower panels show larger-magnification images of regenerating spinal cord area. In animals injected with Sox2-gRNA#2 (left lower panels) and Sox2-gRNA#4 (middle lower panels), a clear spinal cord tube that extends into the blastema is not visible compared to the control sample (GFP-gRNA#3). ‘M’, mature spinal cord area; ‘R’, regenerated area. Dashed lines represent amputation planes. Scale bars represent 500 μm in the upper panel and 200 μm in the lower panel.
(D) Quantitation of spinal cord and tail length in CRISPR Sox2-gRNAs versus control GFP-gRNA-injected animals at 6 days of regeneration. Regenerated tail length was indistinguishable between the Sox2-gRNA#2- (n = 23), Sox2-gRNA#4- (n = 24), and GFP-gRNA#3-injected (n = 12) animals. In contrast, Sox2-gRNAs-injected animals showed a significantly shorter spinal cord outgrowth. Each bullet represents one individual animal. Error bars, SD; ∗∗∗p < 0.001.
(E) Spinal cord length to tail length ratio in 6-day regenerates. The spinal cord represents a smaller fraction of the total tail length in Sox2-gRNA-injected animals compared to controls. The same data set from (D) was used for data plotting. Each bullet represents one individual animal. Error bars, SD; ∗∗∗p < 0.001.
See also Figure S3.
To characterize the spinal cord regeneration defect in more detail, we examined immunohistological sections of the spinal cord at different distances behind the amputation plane and in the regenerate at 6 and 10 days postamputation. Behind the amputation plane, in the region of the spinal cord that acts as the source zone for the regenerate (McHedlishvili et al., 2007), the spinal cord showed comparable morphology and marker expression to control animals including robust GFAP expression as well as neuron-specific tubulin, TUJ1 expression at the outer circumference (Figures 5A–5D). However, in contrast to the Sox2-CRISPR spinal cords that prior to amputation showed normal cell numbers (Figure 5E), in the 6- and 10-day regenerates, the number of cells per cross-section in the source zone behind the amputation plane was reduced compared to control regenerates (M1 and M2; Figures 5F and S3D). This suggests that cells from the source zone contribute to the regenerate but are not replenished. The regenerating region of the spinal cord (R1–R5; Figures 5F and S3D) showed a starkly reduced number of GFAP+ spinal cord cells, especially in more distal regions of the regenerate, suggesting a defect in self-renewal and also a disorganization of TUJ1+ cells (Figures 5A, 5C, and S3E). Interestingly, despite the majority of the Sox2-gRNA#2-injected animals showing complete or nearly complete loss of SOX2 expression at the amputation plane (Figures 4B, S1C, and S1E), the regenerate routinely contained some cells that were immunopositive for SOX2. As described in Sox2 heterozygous knockout mice, we observed specific, cytoplasmic rather than nuclear staining in some of the regenerating samples in which the few remaining SOX2+ cells seem to have contributed to the regenerated spinal cord (Avilion et al., 2003) (Figures 5C and S3E).
Figure 5.
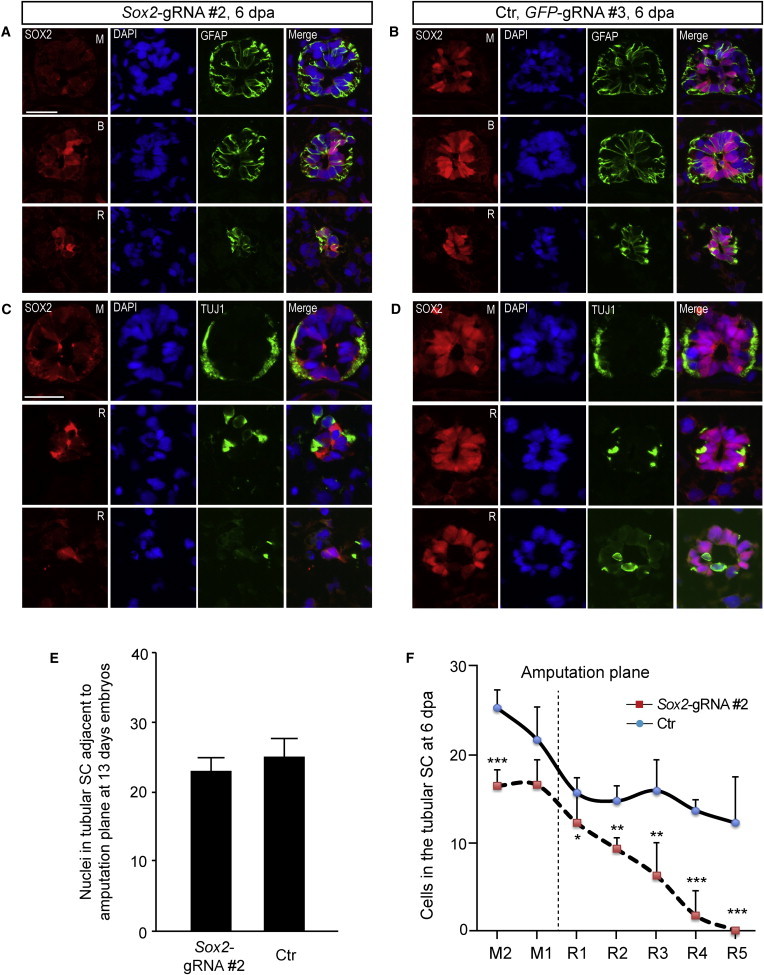
Cellular Analysis of Spinal Cord Regeneration defect in Sox2-CRISPR Animals
The defect in the regenerating spinal cord was investigated by immunofluorescence staining of cross-sections at different locations along the length of the regenerating spinal cord starting behind the amputation plane.
(A and B) Immunostaining for SOX2 (red) and GFAP (green) with DAPI shows that while the spinal cord in the mature region (M) shows similar morphology between the Sox2-gRNA#2- and GFP-gRNA#3-injected animals, in the middle region of the regeneration blastema (R), there are fewer cells in the spinal cord of the Sox2-gRNA#2-injected animals. Interestingly, some cells showing SOX2 staining are evident in the regenerating portion of the spinal cord. B, boundary between mature and regenerate. Scale bars, 50 μm.
(C and D) Immunostaining for SOX2 (red) and TUJ1 (green) with DAPI (blue) shows the comparable organization of neurons and axons in the mature portion of the Sox2-gRNA#2- and GFP-gRNA#3-injected animals. In contrast, the distribution of TUJ1+ cells is scattered and disorganized in the mid portion of the Sox2-gRNA#2-injected animals.
(E) Quantification of the number of nuclei in the intact spinal cord shows that Sox2-gRNA#2-injected animals have similar numbers of spinal cord cells to control (Ctr), GFP-gRNA#3-injected animals. Data are the mean value of determinants derived from four control and seven Sox2-gRNA#2 animals (each determinant is the average value of at least three adjacent sections close to amputation plane). Error bars, SD.
(F) Quantification of the number of nuclei in the regenerating spinal cord at different points along its length shows that cell number decreases markedly along the length of the regenerate in Sox2-gRNA#2-injected animals while it stays relatively constant in control GFP-gRNA#3-injected animals. Data are the mean value of determinants derived from four control and seven Sox2-gRNA#2 animals (each determinant is the average value of three sections, with 50 μm distance). Each bar on the x axis stands for 150 μm. Error bars, SD.
The above phenotype suggested that deletion of Sox2 in neural stem cells results in a defect in proliferative expansion of neural stem cells specifically after tail amputation. To gain further insight into these proliferative properties, we first examined PCNA expression in the 6-day regenerated spinal cords. While all cells in the regenerating spinal cord of Sox2-CRISPR animals were positive for PCNA (Figure 6A), they showed the varying signal intensities that were more similar to the slow-cycling, nonregenerating, control spinal cord cells (Figure 4B) compared to the more homogeneous staining typical of fast-cycling neural stem cells in the control regenerates (Figure 6A). This suggested that in Sox2-CRISPR animals, cells do not accelerate their cell cycle efficiently and possess a significant G1 phase. To corroborate this hypothesis, we performed pulse labeling with the S phase marker EdU on regeneration day 6 for 3 hr prior to harvesting the tail for immunohistochemical processing. In the Sox2-CRISPR animals, we observed a marked reduction in the percentage (from 75% to 31%) of TUJ1−NEUN− spinal cord cells that incorporated EdU (Figures 6B and 6C), indicating a decreased proliferative activity of the stem/progenitor cells. The timing or duration of the cell cycle in neural progenitors can be modulated at various phases of the cell cycle, including the Gap phases as well as S phase (Arai et al., 2011). We examined whether S phase may have slowed down in the Sox2-depleted cycling cells but found no evidence for a change in average EdU fluorescence intensity (Figure 6D and 6E), suggesting that the S phase properties were not altered but rather a higher percentage of cells remained in G1 or G2/M in the Sox2-CRISPR animals. These results indicate that lack of SOX2 results in a hampered proliferation and expansion of the neural stem/progenitor cell pool without changing a number of key markers of radial glial cell identity.
Figure 6.
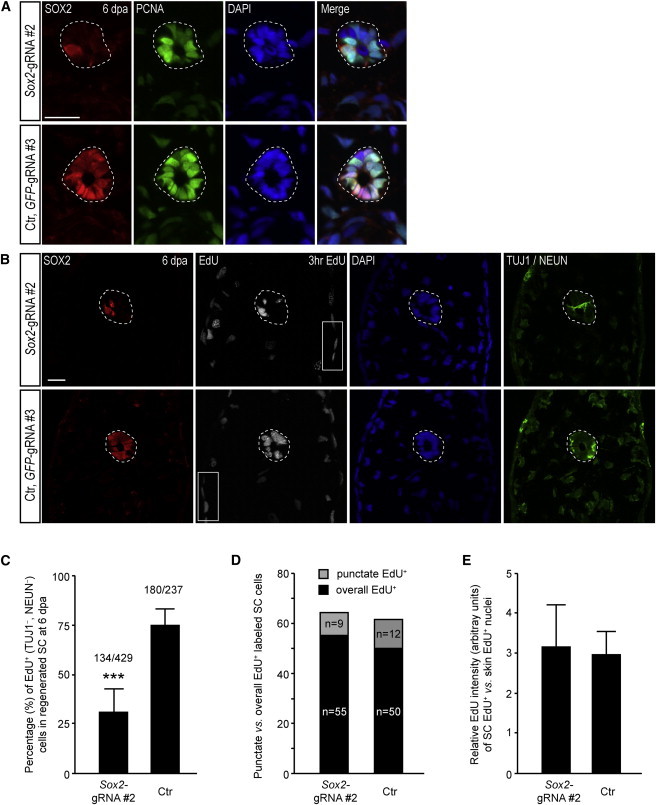
Reduced Proliferative Capacity of Regenerating Spinal Cord Cells in Sox2-CRISPR Animals
(A) Immunostaining for SOX2 (red) and PCNA (green) with DAPI (blue) shows that essentially all cells in the regenerating portion of Sox2-CRISPR animals at 6 dpa show PCNA staining (dotted lines). However, the heterogeneity in PCNA intensity levels more closely resembles the profile seen in an unamputated spinal cord (see Figure 4B) compared to the more uniform staining observed in a control regenerating spinal cord (Ctr). Scale bar, 50 μm.
(B) EdU incorporation was assayed to interrogate passage through the cell cycle. After a 3 hr EdU pulse, cells in the regenerating portion of the Sox2-gRNA#2-injected animals showed fewer EdU+ cells in the spinal cord (dotted lines) compared to GFP-gRNA#3-injected control (Ctr) animals. In contrast, the amount of EdU incorporation in epidermis (box) was similar between the two types of animals. Scale bar, 50 μm.
(C) Quantification of EdU+ (TUJ−, NEUN−) spinal cord cells as shown in (B) in the regenerated spinal cord within 300 μm of the amputation plane. Cells from Sox2-gRNA-injected animals showed a 31% incorporation rate compared to 75% in GFP-gRNA#3-injected control animals. In total, 429 and 237 spinal cord cells, from ten Sox2-gRNA#2- and four GFP-gRNA#3-injected control animals, respectively, were used for data plotting. Error bars, SD.
(D) Quantification of the distribution of EdU nuclear staining patterns shows that both types of animals have the same relative fraction “punctate” versus “overall” EdU label. Data derived from three Sox2-gRNA#2- and two GFP-gRNA#3-injected control animals were used for plotting.
(E) Quantification of the EdU intensity in spinal cord cells shows similar levels of incorporation, suggesting that in Sox2-gRNA#2-injected animals, S phase still progresses at a similar rate to controls, but more cells spend a longer time in G0, G1, or G2 phases. The “overall” EdU+ cells shown in (D), derived from three Sox2-gRNA#2- and two GFP-gRNA#3-injected control animals, were used for plotting. Error bars, SD.
Downregulation of Sox3 during Spinal Cord Regeneration
Considering the survival of Sox2-CRISPR animals through embryonic development but the strong regeneration-specific phenotype, we turned to Sox3 as a potential family member whose expression may rationalize the Sox2 dependence of regeneration, but not development. In frog and fish embryos, Sox3, whose overexpression caused expansion of the CNS and whose knockdown caused reduction of CNS tissue, was shown to control Sox2 expression, but not vice versa (Dee et al., 2008; Rogers et al., 2009). No phenotype for Sox2 knockdown in these models was described. Sox2 deletion in the mouse CNS showed a mild phenotype, with a compensatory upregulation of Sox3 (Miyagi et al., 2008). We therefore examined expression of the axolotl Sox3 gene during development and tail regeneration. Whole-mount in situ hybridization of axolotl embryos at stages 15, 17, 34, and 37 showed indistinguishable Sox2 and Sox3 expression in the CNS, including the spinal cord all along the body axis (Figures 7A and 7B). This finding most likely explains the lack of embryonic phenotype in animals with reduced SOX2 either by CRISPR- or morpholino-mediated knockdown. When we checked Sox3 expression during spinal cord regeneration by quantitative PCR and by in situ hybridization of longitudinal sections at day1 and day 6, we observed a specific downregulation of Sox3 in the regenerating portion of the spinal cord (Figures 7C–7E). These results would correspond to a heightened sensitivity of the regenerating spinal cord to Sox2 deletion compared to the developing spinal cord.
Figure 7.
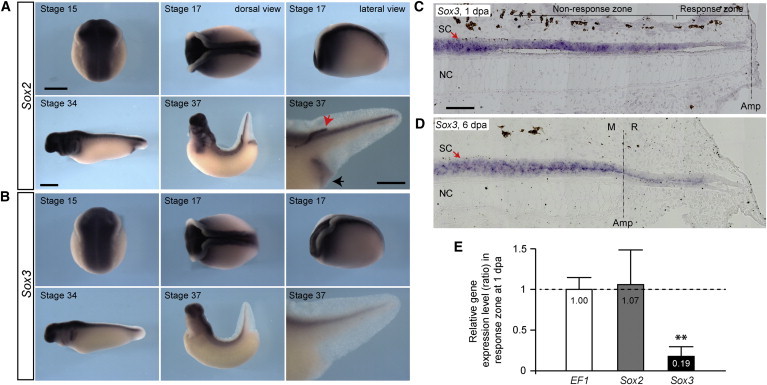
Sox3 Expression Overlaps with Sox2 during Development, but Not Regeneration
(A and B) Sox2 (A) and Sox3 (B) whole-mount in situ hybridization of axolotl embryos at stage 15, 17, 34, and 37. Note the indistinguishable Sox2 and Sox3 expression in the CNS, including the spinal cord, at all stages analyzed. Sox2, but not Sox3, is also expressed in the lateral line (red arrow) and cloaca (black arrow) at stage 37. Scale bars represent 1 mm in (A), left panels and 0.5 mm in (A), lower right panel.
(C) Sox3 in situ hybridization of a 1-day regenerating tail, longitudinal section. Sox3 expression is reduced in the 500 μm zone next to the amputation plane that will be the source of the regenerating spinal cord.
(D) Sox3 in situ hybridization of a 6-day regenerating tail, longitudinal section. Sox3 expression is low all along the regenerating portion of the spinal cord. SC, spinal cord; NC, nodal cord; M, mature; R, regenerate. Dashed lines represent amputation planes. Scale bar, 200 μm.
(E) Results of quantitative RT-PCR of Sox2 and Sox3 expression in the 500 μm response zone next to the amputation plane at 1 dpa as shown in (C) versus the 500 μm developing tail portion adjacent to the amputation plane at the day of amputation (0 dpa). Data are derived from three individual experiments and expressed as ratio of gene expression level in the response/nonresponse zone, with the ratio obtained in the case of control EF1 being set as1 and the ratios obtained in Sox2 and Sox3 being expressed relative to it. Error bars, SD; ∗∗p < 0.01.
Discussion
This work represents a number of important advances for the study of axolotl regeneration. First, we have shown for several genomic loci that TALENs and CRISPRs can induce targeted deletions in a large proportion of cells from injected animals. In all cases, CRISPRs yielded higher viability of injected animals and higher penetrance of the phenotype, reflecting deletion of both alleles within often a large majority of the cells. The Tyr and Sox2 experiments show strong cellular phenotypes associated with homozygous gene deletion. In addition, for the GFP-transgene, we have shown germline transmission of the deletion after TALEN-mediated gene deletion. We therefore conclude that CRISPRs will be an important mode of generating gene deletions and other genome modifications in the axolotl for regeneration studies.
Our work examining CRISPR-mediated Sox2 deletion showed mild phenotypes during development but a strong inhibition of spinal cord regeneration after tail amputation. The lack of a developmental phenotype due to SOX2 knockdown was corroborated by the injection of anti-Sox2 morpholinos that knocked down SOX2 immunostaining in the embryo but yielded the same frequency of normally developing embryos as control-injected cohorts. It is interesting that no knockdown experiments for Sox2 have been reported in amphibian and zebrafish embryos, raising the possibility that in these vertebrates, Sox2 is either not solely responsible for maintenance of pluripotent cells or that maternal stores allow survival through early stages. Indeed, when we examined Sox3, its distribution overlapped with Sox2 expression in the embryo, suggesting it can compensate for lack of Sox2, especially since Sox3 has been shown to be upstream of Sox2 in Xenopus embryos (Rogers et al., 2009). During regeneration, axolotl Sox3 was downregulated in the spinal cord, which would be consistent with an enhanced sensitivity of the regenerate to Sox2 knockdown.
Our analysis of tail regeneration in the Sox2-CRISPR animals showed that despite maintaining a number of radial glial cell markers, Sox2-deleted spinal cord shows defective proliferation of GFAP-positive cells and therefore, reduced outgrowth of the spinal cord at 6 and 10 days postamputation. In other work, we have defined the cell-cycle characteristics of SOX2+PCNA+-positive cells before and after regeneration and found that in the uninjured state, resident cells cycle slowly, on average once every 14 days. Upon tail amputation, the cell cycle accelerates to a 4-day cycle (A. Rodrigo-Albors, personal communication). Given that the SOX2−TUJ1−NEUN− cells at the amputation plane maintained expression of PCNA and normal levels of EdU incorporation before regeneration, we interpret this result to mean that the SOX2+ cells that would normally accelerate their cell cycle are, in the absence of SOX2, unable to accelerate their cell cycle in response to injury cues. The role of SOX2 in promoting proliferation of neural stem/progenitor cells has previously been documented in the brain and in CNS tumors (Favaro et al., 2009, 2014), and our work would support a role for SOX2 in injury mediated rapid cell cycles required to expand the pool of neural stem cells to reconstitute the missing spinal cord.
Experimental Procedures
For further details, see the Supplemental Experimental Procedures. All other methods were performed according to standard protocols.
CRISPR Design and RNA Synthesis
For better comparison, relevant TALENs and CRISPRs were designed against the identical sequence of each targeted gene locus. The design and RNA synthesis were carried out according to the published protocols (Bedell et al., 2012; Cermak et al., 2011; Hwang et al., 2013).
Axolotl Care and Egg Injection
Animal experiments were carried out according to German animal welfare legislation. Axolotl egg injection was performed according to previously published protocols (Khattak et al., 2014). Briefly, 125–500 pg TALEN mRNAs, 500–1,000 pg CRISPR RNAs, or 5–20 pmol of morpholinos was injected into freshly laid single-cell-stage embryos. For TALEN- and CRISPR-mediated GFP knockout experiments, eggs derived from the breeding of heterozygous GFP-transgenic and white axolotl animals were used for injection. For other experiments, eggs derived from either wild-type or white animals were used for injection. Axolotl larvae were kept individually in plastic cups with a change of fresh tap water every second day and fed Artemia daily. Axolotl larvae were anaesthetized within 0.01% ethyl-p-aminobenzoate (benzocaine; Sigma) prior to imaging or amputation.
DNA Extraction, Genotyping, and Sequencing
Genomic DNA extraction and PCR were carried out using REDExtract-N-Amp Tissue PCR Kit (Sigma) according to the manufacturer’s instructions. Each genomic locus was PCR amplified with gene-specific primer pairs (see the Supplemental Experimental Procedures). The resulting PCR products were cloned into pGEMT vector (Promega). Individual clones were sequenced with T7 primer.
Microscopy, Quantifications, and Statistics
Fluorescence images were acquired with a Zeiss Observer or confocal microscope. Bright-field or color images were acquired with an Olympus dissecting microscope. The length of the regenerated spinal cords and tails was measured with CellSens Standard software. Spinal cord cells were counted using Photoshop software. EdU+TUJ1−NEUN− cells, within 300 μm of amputation plane, in the regenerated spinal cord were used for quantification in Figure 4C. EdU fluorescence intensity (in Figure 4E) was measured with Software Image J, restricted to the spinal cells expressing overall EdU (TUJ1−, NEUN−) versus epidermal cells expressing overall EdU. The punctate EdU-labeled cells were most likely excluded because they were at either the beginning or the end of S phase during EdU pulse. After subtracting the background value, the relative EdU intensity was calculated by dividing the average EdU intensity value of each individual spinal cord cell with the average EdU intensity value, minimally derived from five epidermal cells of the same section. Data plotting was carried out using Microsoft Excel and Prism. Student’s t test was used for the p value calculation.
Author Contributions
J.F.F. designed, performed, and analyzed experiments and wrote the manuscript. M.S., A.T., Y.T., and K.R. performed experiments. E.M.T. provided funding, designed and analyzed experiments, and wrote the manuscript.
Acknowledgments
The authors are grateful to Beate Gruhl, Sabine Mögel, Mark Armisted, and Anja Wagner for outstanding animal care. The work was supported by an HFSP program grant, DFG 274/3-2, DFG-274/2-3/SFB655 from Cells into Tissues, and DFG 274/5-2/SPP1356 from Pluripotency, a CRTD seed grant on zinc finger nucleases, and from central funds from the DFG Research Center, CRTD.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Accession Numbers
The GenBank accession numbers for the Sox2 and Sox3 sequences reported in this paper are KJ999995 and KJ999996, respectively.
Supplemental Information
References
- Arai Y., Pulvers J.N., Haffner C., Schilling B., Nüsslein I., Calegari F., Huttner W.B. Neural stem and progenitor cells shorten S-phase on commitment to neuron production. Nat. Commun. 2011;2:154. doi: 10.1038/ncomms1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K., Sarkar A., Yram M.A., Polo J.M., Bronson R., Sengupta S., Seandel M., Geijsen N., Hochedlinger K. Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell. 2011;9:317–329. doi: 10.1016/j.stem.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilion A.A., Nicolis S.K., Pevny L.H., Perez L., Vivian N., Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedell V.M., Wang Y., Campbell J.M., Poshusta T.L., Starker C.G., Krug R.G., 2nd, Tan W., Penheiter S.G., Ma A.C., Leung A.Y. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitz I.L., Biesinger J., Xie X., Cho K.W. Biallelic genome modification in F(0) Xenopus tropicalis embryos using the CRISPR/Cas system. Genesis. 2013;51:827–834. doi: 10.1002/dvg.22719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylund M., Andersson E., Novitch B.G., Muhr J. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat. Neurosci. 2003;6:1162–1168. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- Cermak T., Doyle E.L., Christian M., Wang L., Zhang Y., Schmidt C., Baller J.A., Somia N.V., Bogdanove A.J., Voytas D.F. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dee C.T., Hirst C.S., Shih Y.H., Tripathi V.B., Patient R.K., Scotting P.J. Sox3 regulates both neural fate and differentiation in the zebrafish ectoderm. Dev. Biol. 2008;320:289–301. doi: 10.1016/j.ydbio.2008.05.542. [DOI] [PubMed] [Google Scholar]
- Echeverri K., Tanaka E.M. Proximodistal patterning during limb regeneration. Dev. Biol. 2005;279:391–401. doi: 10.1016/j.ydbio.2004.12.029. [DOI] [PubMed] [Google Scholar]
- Favaro R., Valotta M., Ferri A.L., Latorre E., Mariani J., Giachino C., Lancini C., Tosetti V., Ottolenghi S., Taylor V., Nicolis S.K. Hippocampal development and neural stem cell maintenance require Sox2-dependent regulation of Shh. Nat. Neurosci. 2009;12:1248–1256. doi: 10.1038/nn.2397. [DOI] [PubMed] [Google Scholar]
- Favaro R., Appolloni I., Pellegatta S., Sanga A.B., Pagella P., Gambini E., Pisati F., Ottolenghi S., Foti M., Finocchiaro G. Sox2 is required to maintain cancer stem cells in a mouse model of high-grade oligodendroglioma. Cancer Res. 2014;74:1833–1844. doi: 10.1158/0008-5472.CAN-13-1942. [DOI] [PubMed] [Google Scholar]
- Flowers G.P., Timberlake A.T., McLean K.C., Monaghan J.R., Crews C.M. Highly efficient targeted mutagenesis in axolotl using Cas9 RNA-guided nuclease. Development. 2014;141:2165–2171. doi: 10.1242/dev.105072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T., Gersbach C.A., Barbas C.F., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Sakamoto K., Sakuma T., Yokotani N., Inoue T., Kawaguchi E., Agata K., Yamamoto T., Takeuchi T. Transcription activator-like effector nucleases efficiently disrupt the target gene in Iberian ribbed newts (Pleurodeles waltl), an experimental model animal for regeneration. Dev. Growth Differ. 2014;56:115–121. doi: 10.1111/dgd.12103. [DOI] [PubMed] [Google Scholar]
- Hwang W.Y., Fu Y., Reyon D., Maeder M.L., Tsai S.Q., Sander J.D., Peterson R.T., Yeh J.R., Joung J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami Y., Rodriguez Esteban C., Raya M., Kawakami H., Martí M., Dubova I., Izpisúa Belmonte J.C. Wnt/beta-catenin signaling regulates vertebrate limb regeneration. Genes Dev. 2006;20:3232–3237. doi: 10.1101/gad.1475106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattak S., Schuez M., Richter T., Knapp D., Haigo S.L., Sandoval-Guzmán T., Hradlikova K., Duemmler A., Kerney R., Tanaka E.M. germline transgenic methods for tracking cells and testing gene function during regeneration in the axolotl. Stem Cell Rep. 2013;1:90–103. doi: 10.1016/j.stemcr.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattak S., Murawala P., Andreas H., Kappert V., Schuez M., Sandoval-Guzmán T., Crawford K., Tanaka E.M. Optimized axolotl (Ambystoma mexicanum) husbandry, breeding, metamorphosis, transgenesis and tamoxifen-mediated recombination. Nat. Protoc. 2014;9:529–540. doi: 10.1038/nprot.2014.040. [DOI] [PubMed] [Google Scholar]
- Kragl M., Knapp D., Nacu E., Khattak S., Schnapp E., Epperlein H.H., Tanaka E.M. Novel insights into the flexibility of cell and positional identity during urodele limb regeneration. Cold Spring Harb. Symp. Quant. Biol. 2008;73:583–592. doi: 10.1101/sqb.2008.73.034. [DOI] [PubMed] [Google Scholar]
- Kragl M., Knapp D., Nacu E., Khattak S., Maden M., Epperlein H.H., Tanaka E.M. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460:60–65. doi: 10.1038/nature08152. [DOI] [PubMed] [Google Scholar]
- Lei Y., Guo X., Liu Y., Cao Y., Deng Y., Chen X., Cheng C.H., Dawid I.B., Chen Y., Zhao H. Efficient targeted gene disruption in Xenopus embryos using engineered transcription activator-like effector nucleases (TALENs) Proc. Natl. Acad. Sci. USA. 2012;109:17484–17489. doi: 10.1073/pnas.1215421109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Esvelt K.M., Church G.M. Cas9 as a versatile tool for engineering biology. Nat. Methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHedlishvili L., Epperlein H.H., Telzerow A., Tanaka E.M. A clonal analysis of neural progenitors during axolotl spinal cord regeneration reveals evidence for both spatially restricted and multipotent progenitors. Development. 2007;134:2083–2093. doi: 10.1242/dev.02852. [DOI] [PubMed] [Google Scholar]
- McHedlishvili L., Mazurov V., Grassme K.S., Goehler K., Robl B., Tazaki A., Roensch K., Duemmler A., Tanaka E.M. Reconstitution of the central and peripheral nervous system during salamander tail regeneration. Proc. Natl. Acad. Sci. USA. 2012;109:E2258–E2266. doi: 10.1073/pnas.1116738109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercader N., Tanaka E.M., Torres M. Proximodistal identity during vertebrate limb regeneration is regulated by Meis homeodomain proteins. Development. 2005;132:4131–4142. doi: 10.1242/dev.01976. [DOI] [PubMed] [Google Scholar]
- Miyagi S., Masui S., Niwa H., Saito T., Shimazaki T., Okano H., Nishimoto M., Muramatsu M., Iwama A., Okuda A. Consequence of the loss of Sox2 in the developing brain of the mouse. FEBS Lett. 2008;582:2811–2815. doi: 10.1016/j.febslet.2008.07.011. [DOI] [PubMed] [Google Scholar]
- Nakayama T., Fish M.B., Fisher M., Oomen-Hajagos J., Thomsen G.H., Grainger R.M. Simple and efficient CRISPR/Cas9-mediated targeted mutagenesis in Xenopus tropicalis. Genesis. 2013;51:835–843. doi: 10.1002/dvg.22720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que J., Luo X., Schwartz R.J., Hogan B.L. Multiple roles for Sox2 in the developing and adult mouse trachea. Development. 2009;136:1899–1907. doi: 10.1242/dev.034629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers C.D., Harafuji N., Archer T., Cunningham D.D., Casey E.S. Xenopus Sox3 activates sox2 and geminin and indirectly represses Xvent2 expression to induce neural progenitor formation at the expense of non-neural ectodermal derivatives. Mech. Dev. 2009;126:42–55. doi: 10.1016/j.mod.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Gardiner D.M., Bryant S.V. Vaccinia as a tool for functional analysis in regenerating limbs: ectopic expression of Shh. Dev. Biol. 2000;218:199–205. doi: 10.1006/dbio.1999.9556. [DOI] [PubMed] [Google Scholar]
- Seifert A.W., Kiama S.G., Seifert M.G., Goheen J.R., Palmer T.M., Maden M. Skin shedding and tissue regeneration in African spiny mice (Acomys) Nature. 2012;489:561–565. doi: 10.1038/nature11499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon A., Tanaka E.M. Limb regeneration. Wiley Interdiscip. Rev. Dev. Biol. 2013;2:291–300. doi: 10.1002/wdev.73. [DOI] [PubMed] [Google Scholar]
- Sobkow L., Epperlein H.H., Herklotz S., Straube W.L., Tanaka E.M. A germline GFP transgenic axolotl and its use to track cell fate: dual origin of the fin mesenchyme during development and the fate of blood cells during regeneration. Dev. Biol. 2006;290:386–397. doi: 10.1016/j.ydbio.2005.11.037. [DOI] [PubMed] [Google Scholar]
- Suzuki K.T., Isoyama Y., Kashiwagi K., Sakuma T., Ochiai H., Sakamoto N., Furuno N., Kashiwagi A., Yamamoto T. High efficiency TALENs enable F0 functional analysis by targeted gene disruption in Xenopus laevis embryos. Biol. Open. 2013;2:448–452. doi: 10.1242/bio.20133855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto T., Uchikawa M., Yoshida M., Bell D.M., Lovell-Badge R., Papaioannou V.E., Kondoh H. Tbx6-dependent Sox2 regulation determines neural or mesodermal fate in axial stem cells. Nature. 2011;470:394–398. doi: 10.1038/nature09729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeo M., Chou W.C., Sun Q., Lee W., Rabbani P., Loomis C., Taketo M.M., Ito M. Wnt activation in nail epithelium couples nail growth to digit regeneration. Nature. 2013;499:228–232. doi: 10.1038/nature12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka E.M., Ferretti P. Considering the evolution of regeneration in the central nervous system. Nat. Rev. Neurosci. 2009;10:713–723. doi: 10.1038/nrn2707. [DOI] [PubMed] [Google Scholar]
- Tapia N., Reinhardt P., Duemmler A., Wu G., Araúzo-Bravo M.J., Esch D., Greber B., Cojocaru V., Rascon C.A., Tazaki A. Reprogramming to pluripotency is an ancient trait of vertebrate Oct4 and Pou2 proteins. Nat. Commun. 2012;3:1279. doi: 10.1038/ncomms2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whited J.L., Tsai S.L., Beier K.T., White J.N., Piekarski N., Hanken J., Cepko C.L., Tabin C.J. Pseudotyped retroviruses for infecting axolotl in vivo and in vitro. Development. 2013;140:1137–1146. doi: 10.1242/dev.087734. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


