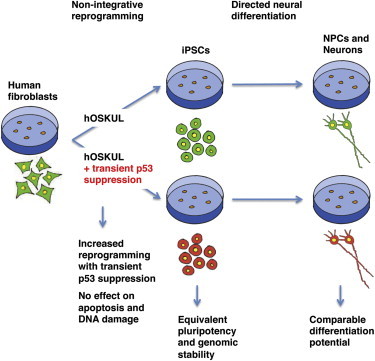
Summary
The discovery of human-induced pluripotent stem cells (iPSCs) has sparked great interest in the potential treatment of patients with their own in vitro differentiated cells. Recently, knockout of the Tumor Protein 53 (p53) gene was reported to facilitate reprogramming but unfortunately also led to genomic instability. Here, we report that transient suppression of p53 during nonintegrative reprogramming of human fibroblasts leads to a significant increase in expression of pluripotency markers and overall number of iPSC colonies, due to downstream suppression of p21, without affecting apoptosis and DNA damage. Stable iPSC lines generated with or without p53 suppression showed comparable expression of pluripotency markers and methylation patterns, displayed normal karyotypes, contained between 0 and 5 genomic copy number variations and produced functional neurons in vitro. In conclusion, transient p53 suppression increases reprogramming efficiency without affecting genomic stability, rendering the method suitable for in vitro mechanistic studies with the possibility for future clinical translation.
Graphical Abstract
Highlights
-
•
Transient p53 suppression increases reprogramming efficiency through p21 inhibition
-
•
No adverse effect on DNA damage and apoptosis is observed during reprogramming
-
•
Stable iPSC lines display normal karyotypes and expression of pluripotency markers
-
•
The iPSC lines retain their differentiation potential and form functional neurons
In this article, Clausen and colleagues show that transient p53 suppression increases reprogramming efficiency in defined conditions without affecting apoptosis and DNA damage. Moreover, iPSC lines generated with or without transient p53 suppression are identical with respect to their pluripotent phenotype and their mutational load and can give rise to functional neurons in vitro.
Introduction
Dr. Wilmut and colleague’s discovery in 1996 that mammalian somatic cells can be reprogrammed into a totipotent state of development by fusion with an enucleated oocyte (Campbell et al., 1996) has paved the way for autologous stem cell therapy by exploiting a patient’s own in vitro differentiated cells. Ten years later, with the revolutionizing finding that induced pluripotent stem cells (iPSCs) can be established from somatic cells by overexpression of merely four transcription factors (Takahashi and Yamanaka, 2006), a major step toward a clinical translation was taken. However, new coding mutations were recently reported to arise during the reprogramming process (Gore et al., 2011; Laurent et al., 2011; Martins-Taylor and Xu, 2012; Ng et al., 2011), rendering the process too hazardous for regenerative use in humans.
Several nonintegrative reprogramming strategies including Sendai virus (Fusaki et al., 2009) and episomal plasmids (Okita et al., 2011, 2013; Yu et al., 2009) have been tested to produce safe, transplantable iPSCs. Yet, recent whole-genome sequencing studies have failed to demonstrate a reduction in the mutational load in iPSC lines derived by integration-free reprogramming methods (Gore et al., 2011).
The cell-cycle regulator p53 acts as an important safeguard mechanism, by preventing cells from undergoing uncontrolled proliferation in response to DNA damage (Hong et al., 2009). The downstream DNA damage response (DDR) involves a series of events that lead to either cell-cycle arrest induced by p21 or apoptosis induced by PUMA. Importantly, p53 has also been shown to act as a critical barrier to the reprogramming process, and knockout of the TP53 gene in mouse and human fibroblasts was shown to produce significantly more iPSC colonies (Hong et al., 2009; Kawamura et al., 2009; Marión et al., 2009). Although the inhibition of p53 is clearly advantageous for the reprogramming efficiency, knocking out TP53 may possess critical safety issues because it was found to cause genomic instability (Chen et al., 2012; Lake et al., 2012; Marión et al., 2009; Menendez et al., 2010).
Recently, transient suppression of p53 with nonintegrative plasmids was shown to improve the reprogramming efficiency of human fibroblasts (Okita et al., 2011) and blood cells (Okita et al., 2013) by use of nonintegrative plasmids. Yet it remains unknown whether transient suppression of p53 will also give rise to genomic instability, which may in turn have detrimental effects on the gene expression, the epigenetic status, and the differentiation capacity of the resulting iPSC lines.
In this study, we set out to establish an optimized nonintegrative reprogramming approach under defined conditions in order to study the functional effects of transient p53 suppression in normal human dermal fibroblasts (NHDFs) during and after reprogramming.
Results
Increased Reprogramming Efficiency with Transient p53 Suppression
To study the effect of transient p53 suppression during reprogramming of human fibroblasts, a nonintegrative reprogramming system was established in defined conditions (Figure 1A and Figure S1 available online). Seven days after reprogramming with or without (w/wo) transient expression of a short hairpin to TP53 (shp53), a subpopulation of small, highly proliferative cells was observed in both conditions (Figures 1B and 1F). At days 14–28, iPSC colonies were observed (Figures 1C, 1D, 1G, and 1H), which stained positive for TRA-1-81 (Figures 1E and 1I). The growth rate was, in general, higher during reprogramming with transient p53 suppression (0.12 cell doublings per day) compared to without (0.06 cell doublings per day), although not to a significant degree (Figure 1J). In contrast, a fibroblast line, which stably express shp53 (PLK.O; Godar et al., 2008) displayed a significantly higher growth rate (0.42 cell doublings per day), yet, this line proved resistant to reprogramming (data not shown). Alkaline phosphatase staining and counting on day 28 showed a significant (7.5-fold) increase in the number of iPSC colonies with transient p53 suppression (0.12%) compared to without (0.016%) (Figure 1K), which was confirmed in several independent experiments comprising NHDFs from different individuals.
Figure 1.
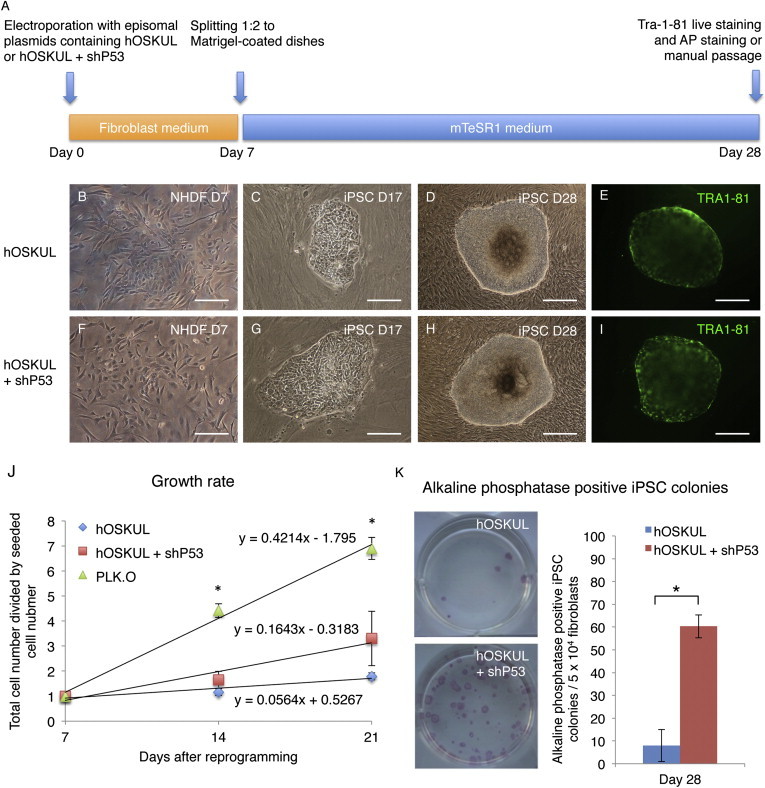
Increased Reprogramming with Transient p53 Suppression in Defined Conditions
(A) Timeline showing the reprogramming in defined conditions with the episomal plasmids hOCT4, hSOX2, hKLF4, hL-MYC, and hLIN28 (hOSKUL) with or without (w/wo) a short hairpin to p53 (shp53).
(B–D and F–H) Morphology of normal human dermal fibroblasts (NHDFs) and induced pluripotent stem cells (iPSCs) at days 7, 17, and 28 after reprogramming w/wo shp53.
(E and I) Tra-1-81 staining of primary iPSC colonies 28 days after reprogramming w/wo shp53.
(J) Growth rate of cells electroporated with hOSKUL, hOSKUL + shp53, or a fibroblast line stably expressing shp53 (PLK.O) at days 7, 21, and 28 after reprogramming.
(K) Alkaline phosphatase staining and counting of primary iPSC colonies at day 28 after reprogramming w/wo shp53.
Scale bars, 200 μm (B, C, F, and G) and 400 μm (D, E, H, and I). Y error bars depict SD of three independent experiments. ∗p < 0.05. See also Figure S1.
Transient p53 Suppression Increases Reprogramming Efficiency through Inhibition of p21 without Affecting Apoptosis and DNA Damage
To investigate the underlying effect of the increased reprogramming efficiency with transient p53 suppression, temporal changes in pluripotency markers (SSEA4, TRA1-60), tumor suppression (p53), cell-cycle regulation (p21), apoptosis (PUMA), and DNA damage (H2A.X) were studied by flow cytometry. Although the PLK.O line stably expressing shp53 showed a 43.6% reduction in p53-positive cells compared to NHDFs, the pluripotency markers SSEA4 and TRA1-60 remained low during the entire reprogramming experiment (data not shown). In contrast, the analyses revealed a significant increase in SSEA4 and TRA1-60 double-positive cells with transient p53 suppression on day 21 (10.0% versus 3.1%) and on day 28 (16.6% versus 8.3%) after reprogramming (Figure 2A). A significant decrease in p53-positive cells was also observed with transient p53 suppression at days 7 (82.8% versus 90%), 14 (79% versus 92.7%), and 21 (63.8% versus 89.4%) (Figure 2B), which correlated with a significant decrease in p21-positive cells at days 7 (92.6% versus 96.3%), 14 (87.9% versus 95.6%), and 21 (68.1% versus 93.4%) (Figure 2C). In contrast, no significant effect of transient p53 suppression on apoptosis was observed, as evaluated by the proapoptotic marker PUMA (Figure 2D), a mitochondrial membrane assay (Figure S2A), and Annexin V in TRA1-60-positive cells (Figure S2B). Moreover, analysis with H2A.X, which is associated with replication-induced DNA damage (Garcia-Canton et al., 2012), detected no significant effect of transient p53 suppression on DNA damage, neither in the total cell population (Figure 2E) nor in TRA1-60-positive cells (Figures S2C and S2D).
Figure 2.
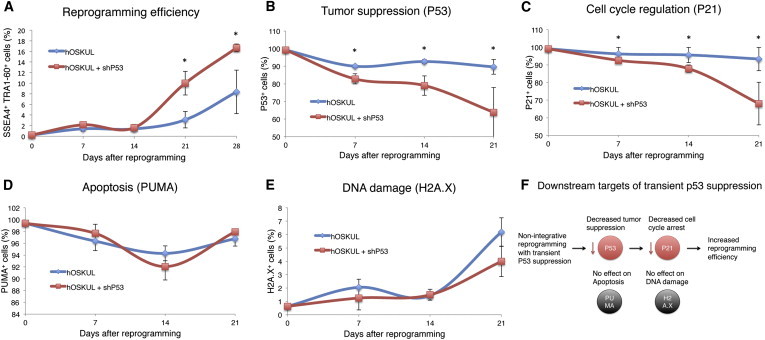
Transient p53 Suppression Increases Reprogramming Efficiency without Affecting Apoptosis and DNA Damage
Flow cytometry of normal human dermal fibroblasts (NHDFs) on days 0, 7, 14, 21, and 28 after reprogramming with the episomal plasmids hOCT4, hSOX2, hKLF4, hL-MYC, and hLIN28 (hOSKUL) with or without (w/wo) a short hairpin to p53 (shp53).
(A–E) Flow cytometry was performed with (A) the pluripotency markers SSEA4 and TRA1-60, (B) the tumor suppressor p53, (C) the cell-cycle regulator p21, (D) the proapoptotic marker PUMA, and (E) the double-stranded DNA damage marker H2A.X.
(F) Overview depicting the mechanisms involved in transient p53 suppression during reprogramming. Y error bars depict SD of three independent experiments. ∗p < 0.05. See also Figure S2.
Next, gene expression analysis of TP53 and its downstream DDR targets P21 and PUMA, as well as the pluripotency marker NANOG was performed. The silencing effect of shp53 resulted in a significant decrease in TP53 expression in unsorted cells 7 days after reprogramming (Figure S2E), and in P21 expression 14 days after reprogramming (Figure S2F), whereas expression of PUMA was unaffected (Figure S2G). In addition, no significant effect of shp53 suppression in TRA1-60-sorted cells was observed (Figures S2I–S2K). In contrast, the pluripotency marker NANOG was significantly upregulated in both unsorted and sorted cells 21 and 14 days after reprogramming with shp53, respectively (Figures S2H and S2L).
iPSCs Generated by Transient p53 Suppression Display Normal In Vitro Characteristics
To examine whether transient p53 suppression confers a negative effect on the long-term stability, six iPSC lines established w/wo transient p53 suppression were characterized in detail. Overall, there were no differences in morphology (Figures 3A and 3E). The pluripotency markers OCT4, NANOG, SSEA3, SSEA4, TRA1-60, and TRA1-81 were all detected by immunocytochemical staining (Figures 3B–3D and 3F–3H) and quantitative real-time PCR analyses showed a comparable upregulation of POU5F1 (OCT4), SOX2, NANOG, ZFP42, DNMT3B, and TP53 and downregulation of P21 compared with NHDFs (Figure 3I). Moreover, integration analyses confirmed lack of expression from exogenous genes (Figure S3A). A comparable hypomethylation of the pluripotency-associated genes OCT4, NANOG, SALL4, and RAB25 and hypermethylation of the fibroblast-associated gene UBE1L was also observed (Figure 3J). Genome-wide transcriptome profiling showed no significant up- or downregulated genes in iPSC lines generated w/wo transient p53 suppression and analysis with PluriTest showed that all iPSC lines scored within the predefined pluripotency and novelty scores (Figure 3K). Finally, in vitro differentiation confirmed by the ability to generate embryoid bodies (Figures 3L, 3M, 3P, and 3Q), which stained positive for Beta-III-tubulin (TUJI) (Figures 3N and 3R), smooth muscle actin (SMA), and alpha-fetoprotein (AFP) (Figures 3O and 3S).
Figure 3.
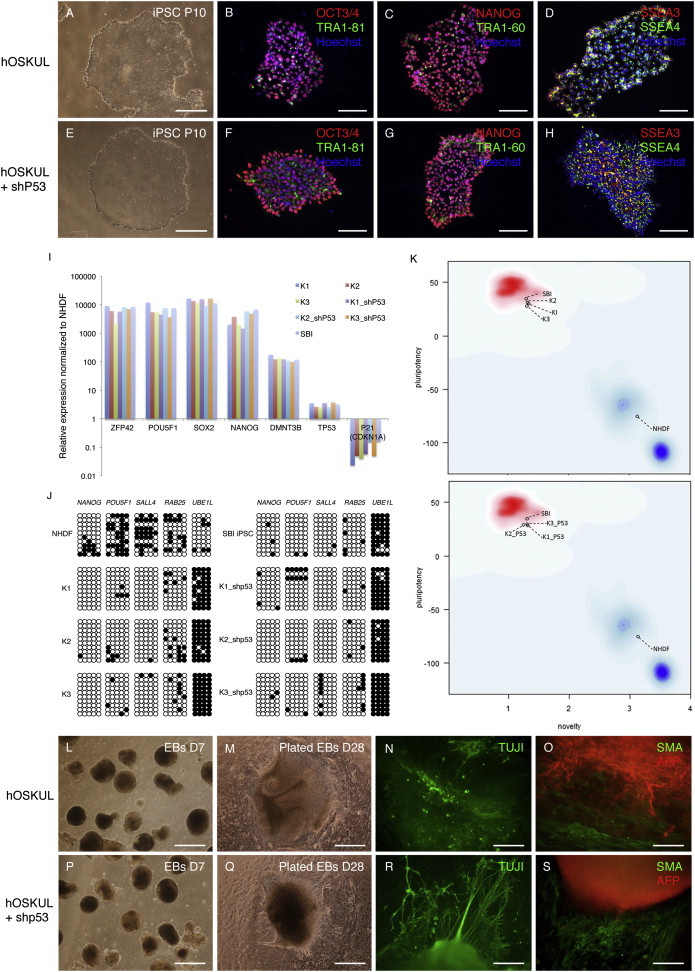
iPSCs Generated by Transient p53 Suppression Display Normal In Vitro Characteristics
(A–H) (A and E) Phase contrast morphology and (B–H) immunocytochemical staining of induced pluripotent stem cells (iPSCs) generated with or without (w/wo) a short hairpin to p53 (shp53) with the pluripotency markers OCT3/4, TRA1-81, and Hoechst (B and F), NANOG, TRA1-60, and Hoechst (C and G), and SSEA3, SSEA4, and Hoechst (D and H).
(I) Quantitative real-time PCR with the pluripotency markers ZFP42, POU5F1 (OCT4), SOX2, NANOG, DNMT3B, and TP53 and P21. Relative expression is shown as the fold change (2−ΔΔCt) with normal human dermal fibroblasts (NHDFs) as reference. A commercial iPSC line (SBI) was included.
(J) Schematic view of the methylation state of selected CpG sites in POU5F1 (OCT4), SAL4, RAB25, and UBE1L. Each row represents a separate clone and each circle represents a CpG site. Open circles represent unmethylated sites and black circles represent methylated sites.
(K) PluriTest algorithm results of transcriptome profiles including a reference iPSC line (SBI) and NHDFs. The PluriTest results are plotted in a density distribution for previously referenced pluripotent stem cells (red cloud) and somatic cells (blue cloud).
(L–S) (L, M, P, and Q) Morphology of embryoid bodies (EBs) on day 7 (L and P), plated EBs on day 28 (M and Q), and immunocytochemical staining of plated EBs on day 28 with TUJI (N and R), smooth muscle actin (SMA), and alpha-fetoprotein (AFP) (O and S).
Scale bars, 400 μm (A, E, L, M, P, and Q), 100 μm (B–D and F–H), or 200 μm (N, O, R, and S). See also Figure S3 and Tables S1–S3.
iPSCs Generated by Transient p53 Suppression Show Genomic Stability
The genomic stability of fibroblasts and iPSC lines generated w/wo transient p53 suppression was analyzed in detail by chromosome studies and copy number variation (CNV) analyses. All the lines presented normal karyotype and did not contain any microscopically visible structural or numerical abnormalities (46, XY) (Figure S3B). CNV analyses showed no significant differences between the numbers of CNVs in iPSC lines generated w/wo transient p53 suppression, displaying an average of 1.66 genomic CNVs (four gains and six losses), which were not present in the donor NHDFs (Table S1). Three of the CNVs were detected in iPSC lines without transient p53 suppression, with one line containing two CNVs (K1), one line containing a single CNV (K3), and one line containing no CNVs (K2). In contrast, seven of the CNVs were detected in iPSC lines with transient p53 suppression, with one line (K1_shp53) containing five CNVs, whereas the other two lines (K2_shp53 and K3_shp53) had only a single CNV each, the latter in a noncoding region.
iPSCs Generated by Transient p53 Suppression Can Differentiate to Functional Neurons In Vitro
To evaluate the differentiation potential of iPSC lines established w/wo transient p53 suppression, directed neural differentiation was performed (Figure 4A). At day 12, all the iPSC lines had formed a uniform monolayer of tightly packed neuroepithelial cells (Figures 4B and 4F), and at days 21, a population of neural progenitor cells (NPCs) was formed, which stained positive for the NPC markers SOX2, NESTIN (Figures 4J and 4N), and VIMENTIN (Figures 4K and 4O), whereas OCT4 was negative. After subsequent culture and passage for 35 days, a network of neural structures was observed (Figures 4D and 4H), containing neurons with a bi- or multipolar morphology with long axonal connections (Figures 4E and 4I). At this stage, most of the cells stained positive for TUJI (Figures 4L and 4P) and the glutamatergic marker vGLUT (Figures 4M and 4Q). Functional characterization on day 80 showed that the iPSC lines responded to glutamate/glycine (Figures 4R and 4T), GABA, and acetylcholine (Figures 4S and 4U), indicating the presence of receptors for these neurotransmitters.
Figure 4.
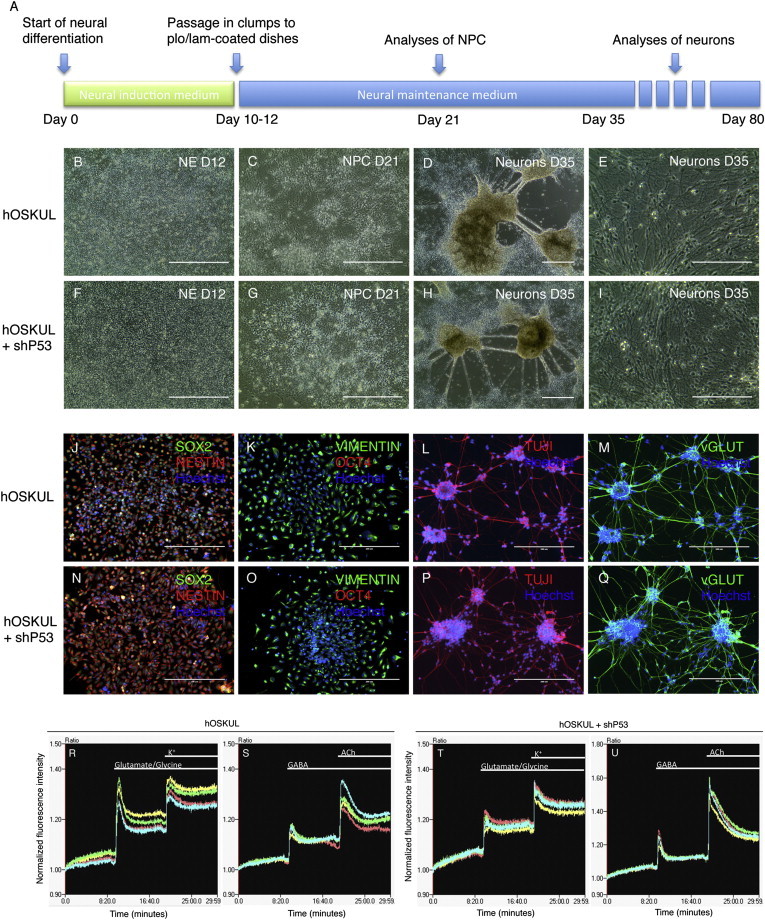
iPSCs Generated by Transient p53 Suppression Can Differentiate to Functional Neurons In Vitro
(A) Timeline showing the directed neural differentiation of induced pluripotent stem cell (iPSC) lines generated with or without (w/wo) a short hairpin to p53 (shp53).
(B–I) Phase contrast morphology of neuroepithelium (NE) at day 12 (B and F), neural progenitor cells (NPC) at day 21 (C and G), and neurons at day 35 (D, E, H, and I).
(J–Q) Immunocytochemistry of NPCs at day 21 with SOX2, NESTIN, and Hoechst (J and N) and VIMENTIN, OCT4, and Hoechst (K and O) and of neurons at day 35 with TUJI and Hoechst (L–P) and vGLUT and Hoechst (M and Q).
(R–U) Intracellular calcium kinetics in iPSC-derived neurons generated without (R and S) or with shp53 (T and U). Baseline fluorescence was recorded for 10 min before application of 300 μM glutamate/10 μM glycine, 25 mM K+, 100 μM GABA and 300 μM acetylcholine. The fluorescence was normalized to the first data point of each of the traces.
Scale bars, 200 μm (B–D, F–H, and J–Q) and 100 μm (E and I).
Discussion
In this study, we report the establishment of a nonintegrative reprogramming approach in defined conditions, based on transient suppression of p53. Using this method, we were able to consistently generate AP and TRA1-81-positive iPSC colonies from individuals with different genders and ages. Compared to the results of Okita et al. (2011), who reported an average of 30 iPSC colonies per 1 × 105 human fibroblasts (0.03%), we observed a 3-fold increase (0.11%) at defined conditions. In contrast, a fibroblast line stably expressing shp53 did not produce any iPSC colonies, and it is likely that the uncontrolled growth of this line has made it resistant to reprogramming due to accumulation of DNA damage or severe shortening of telomeres.
To study temporal changes during reprogramming with transient p53 suppression, flow cytometry with the pluripotency markers SSEA4/TRA1-60, which were previously used for isolation of fully reprogrammed iPSCs (Kahler et al., 2013), was performed. A significant increase in the amount of SSEA4/TRA1-60 double-positive cells was observed on day 21 (3-fold) and on day 28 (2-fold) after reprogramming with shp53. Moreover, transient p53 suppression also accelerated the temporal appearance of fully reprogrammed iPSCs, as the cells became earlier positive for SSEA4/TRA1-60.
As expected from the silencing effect of shp53, p53 was significantly suppressed during reprogramming with transient p53 suppression, which correlated with suppression of p21, whereas expression of the proapoptotic marker PUMA remained unchanged. These results imply that transient p53 suppression increases reprogramming by activating cell proliferation through p21 suppression, rather than by decreasing apoptosis (Figure 2F). Moreover, the effect of transient p53 suppression appears to occur in fibroblasts prior to reprogramming, because no significant differences in TP53, P21, and PUMA were observed in TRA1-60-sorted cells (Figures S2I–S2L). The latter is in agreement with a recent study, which demonstrated that embryonic stem cells (ESCs) possess a nonfunctional p53-p21 axis, in which p53 activates specific microRNAs, which inhibit p21 expression, thereby affecting cell-cycle regulation (Dolezalova et al., 2012). In contrast, NANOG was upregulated with transient p53 suppression in both unsorted and TRA1-60-sorted cells; thus, NANOG could be a potential downstream target of p53. In addition, knockout of tp53 in mouse fibroblasts was recently shown to increase reprogramming through actions of p21 by promoting a mesenchymal-to-epithelial (MET) transition (Brosh et al., 2013), which could also apply in human.
Marión and colleagues previously reported that knockout of TP53 in human and mouse fibroblasts allows for efficient reprogramming at the expense of increased DNA damage, which was attributed to a decrease in apoptosis of DNA damaged cells (Marión et al., 2009). In contrast, we found that transient p53 suppression did not induce DNA damage or result in a decrease in apoptosis during reprogramming. The underlying mechanism is likely to constitute a combination of nonintegrative plasmids, which prevents excessive DNA damage, and a low background expression of TP53, which may be sufficient to sustain the apoptotic pathway.
To examine whether transient suppression of p53 had a negative effect on the long-term stability of iPSCs, we performed detailed characterization of iPSC lines established w/wo transient p53 suppression. All iPSC lines were highly similar with respect to expression of pluripotency markers and showed a comparable upregulation of TP53 and downregulation of P21 compared to NHDFs. Methylation analyses showed a comparable hypomethylation of the pluripotency-associated genes OCT4, NANOG, SALL4, and RAB25 and hypermethylation of the fibroblast-specific gene UBE1L, as previously reported for iPSCs and ESCs (Nishino et al., 2011). Furthermore, genome-wide transcriptome analyses showed no significant up- or downregulated genes and analysis with PluriTest, which is a fast and ethically superior alternative to teratoma assays in mice (Buta et al., 2013), showed that all the iPSC lines clustered within the predefined pluripotency and novelty scores. Finally, in vitro differentiation confirmed the potential of the iPSC lines to generate cells of the three germ layers. To date, some of the iPSC lines (K1 and K1_shp53) have been cultured for more than 35 passages, without showing signs of reduced proliferation.
iPSCs present an ideal system for studies on genomic stability and confer the ability to determine whether a given alteration is new, because the parental cells can also be analyzed (Laurent et al., 2011). The iPSC lines generated w/wo shp53 displayed normal karyotypes, and structural CNV analyses showed that they contained between zero and five CNVs (1.66 CNVs in average), which were not present in the parental NHDFs. These numbers are comparable with recent reports using single-nucleotide variants (SNVs) analyses that reported an average of five coding mutations, of which around half was preexisting in the parental fibroblasts (Cheng et al., 2012; Gore et al., 2011). Moreover, no significant difference between the overall numbers of CNVs in iPSC lines generated w/wo transient p53 suppression was found, thus corroborating recent studies that demonstrated that episomal reprogramming is not inherently mutagenic (Cheng et al., 2012; Gore et al., 2011).
Lin and colleagues reported that tp53 knockout ESC showed impaired differentiation and remained in a pluripotent state (Lin et al., 2005). In contrast, transient p53 suppression did not affect the neural differentiation potential, and all the iPSC lines successfully formed NPCs and functional neurons. Recently, transplantation of NPCs from plasmid-derived iPSCs (Yu et al., 2009) was reported to increase the neurovascular coupling and behavioral recovery in a mouse model of ischemic stroke without any signs of tumor formation 1 year after transplantation (Mohamad et al., 2013). In accordance, transplantation of NPCs from the iPSC lines established by transient p53 suppression in relevant animal models would be highly useful for evaluation of safety and efficacy in regenerative stem cell therapy.
The present study demonstrates that transient p53 suppression increases reprogramming efficiency without affecting apoptosis and DNA damage. Furthermore, iPSC lines generated w/wo transient p53 suppression are identical with respect to their pluripotent phenotype, their mutational load, and their differentiation capacity, rendering the method suitable for in vitro studies on patient-specific disease pathology with a potential for clinical translation.
Experimental Procedures
A complete description of experimental procedures can be found in Supplemental Experimental Procedures.
Reprogramming
Normal human dermal fibroblasts (NHDFs; Lonza) were electroporated with plasmids encoding hOCT4 or hOCT4 with a short hairpin to TP53 (shp53) in combination with hSOX2, hKLF4, hL-MYC, and hLIN28 (Addgene plasmids 27076, 27077, 27078 and 27080), abbreviated hOSKUL or hOSKUL + shp53, respectively (Okita et al., 2011, 2013) and cultured in mTeSR1 medium (STEMCELL Technologies) and Matrigel-coated dishes (BD Biosciences) from day 7 to day 28.
Flow Cytometry and Apoptotic Measurements
Flow cytometry was performed with monoclonal antibodies against SSEA4 and TRA1-60 (BD Biosciences), p53, p21, H2A.X (Cell Signaling Technology), and PUMA (Novus Biologicals) on a FACSarray Bioanalyzer or a BD Accuri C6 (BD Biosciences). Unlabeled and isotype-labeled NHDFs were used as controls for gating.
Characterization of iPSC Lines
Six iPSC lines were established with hOSKUL (K1, K2, and K3) or hOSKUL + shp53 (K1_shp53, K2_shp53, and K3_shp53) and characterized at passage 10 according to established pluripotency criteria (Martí et al., 2013). Genome-wide transcriptome analysis was performed using the HT12 v4 BeadChip microarray (Illumina). DNA methylation analyses were performed with the Cells-to-CpG Bisulfite Conversion Kit (Life Technologies), and PCR-amplified sequences from ten clones per chromosomal region were examined. Primers are specified in Table S2.
Chromosome Studies and Copy Number Variation Analyses
Metaphase chromosomes were investigated with G-banding using standard procedures. Copy number variation (CNV) analysis was performed using the high resolution CytoScan HD chromosome microarray platform (Affymetrix), and data were processed using Affymetrix Chromosome Analysis Suite (ChAS) and manually corrected for false-positive hits.
Directed Neural Differentiation
Directed neural differentiation was carried out according to Shi et al. (2012). Primary antibodies are listed in Table S3.
Statistical Analyses
Statistical analyses comprised a two-tailed Student’s t test with ∗p < 0.05 considered significant.
Acknowledgments
We would like to thank Dr. Keisuke Okita and Prof. Shinya Yamanaka for providing the plasmids. Furthermore, we would like to thank Ida Jørring, Bente Smith Thorup, Ulla Bekker Poulsen, and Tina Christoffersen for excellent technical assistance. We thank the following for financial support: The Danish National Advanced Technology Foundation (patient-specific stem cell-derived models for Alzheimer’s disease), The Danish Council for Independent Research (COGNITO), and the People Programme (Marie Curie Actions) of the European Union's Seventh Framework Programme FP7/2007-2013 under REA grant agreement PIAPP-GA-2012-324451.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Accession Numbers
The GEO (http://www.ncbi.nlm.nih.gov/geo/) accession number for the microarray data reported in this paper is GSE48665. CNV raw data are available upon request.
Supplemental Information
References
- Brosh R., Assia-Alroy Y., Molchadsky A., Bornstein C., Dekel E., Madar S., Shetzer Y., Rivlin N., Goldfinger N., Sarig R., Rotter V. p53 counteracts reprogramming by inhibiting mesenchymal-to-epithelial transition. Cell Death Differ. 2013;20:312–320. doi: 10.1038/cdd.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buta C., David R., Dressel R., Emgård M., Fuchs C., Gross U., Healy L., Hescheler J., Kolar R., Martin U. Reconsidering pluripotency tests: do we still need teratoma assays? Stem Cell Res. (Amst.) 2013;11:552–562. doi: 10.1016/j.scr.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell K.H., McWhir J., Ritchie W.A., Wilmut I. Sheep cloned by nuclear transfer from a cultured cell line. Nature. 1996;380:64–66. doi: 10.1038/380064a0. [DOI] [PubMed] [Google Scholar]
- Chen Z., Zhao T., Xu Y. The genomic stability of induced pluripotent stem cells. Protein and cell. 2012;3:271–277. doi: 10.1007/s13238-012-2922-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Hansen N.F., Zhao L., Du Y., Zou C., Donovan F.X., Chou B.-K., Zhou G., Li S., Dowey S.N., NISC Comparative Sequencing Program Low incidence of DNA sequence variation in human induced pluripotent stem cells generated by nonintegrating plasmid expression. Cell Stem Cell. 2012;10:337–344. doi: 10.1016/j.stem.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezalova D., Mraz M., Barta T., Plevova K., Vinarsky V., Holubcova Z., Jaros J., Dvorak P., Pospisilova S., Hampl A. MicroRNAs regulate p21(Waf1/Cip1) protein expression and the DNA damage response in human embryonic stem cells. Stem Cells. 2012;30:1362–1372. doi: 10.1002/stem.1108. [DOI] [PubMed] [Google Scholar]
- Fusaki N., Ban H., Nishiyama A., Saeki K., Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc. Jpn. Acad., Ser. B, Phys. Biol. Sci. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Canton C., Anadón A., Meredith C. γH2AX as a novel endpoint to detect DNA damage: applications for the assessment of the in vitro genotoxicity of cigarette smoke. Toxicol. In Vitro. 2012;26:1075–1086. doi: 10.1016/j.tiv.2012.06.006. [DOI] [PubMed] [Google Scholar]
- Godar S., Ince T.A., Bell G.W., Feldser D., Donaher J.L., Bergh J., Liu A., Miu K., Watnick R.S., Reinhardt F. Growth-inhibitory and tumor-suppressive functions of p53 depend on its repression of CD44 expression. Cell. 2008;134:62–73. doi: 10.1016/j.cell.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore A., Li Z., Fung H.-L., Young J.E., Agarwal S., Antosiewicz-Bourget J., Canto I., Giorgetti A., Israel M.A., Kiskinis E. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H., Takahashi K., Ichisaka T., Aoi T., Kanagawa O., Nakagawa M., Okita K., Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahler D.J., Ahmad F.S., Ritz A., Hua H., Moroziewicz D.N., Sproul A.A., Dusenberry C.R., Shang L., Paull D., Zimmer M. Improved methods for reprogramming human dermal fibroblasts using fluorescence activated cell sorting. PLoS ONE. 2013;8:e59867. doi: 10.1371/journal.pone.0059867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T., Suzuki J., Wang Y.V., Menendez S., Morera L.B., Raya A., Wahl G.M., Izpisúa Belmonte J.C. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake B.B., Fink J., Klemetsaune L., Fu X., Jeffers J.R., Zambetti G.P., Xu Y. Context-dependent enhancement of induced pluripotent stem cell reprogramming by silencing Puma. Stem Cells. 2012;30:888–897. doi: 10.1002/stem.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent L.C., Ulitsky I., Slavin I., Tran H., Schork A., Morey R., Lynch C., Harness J.V., Lee S., Barrero M.J. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8:106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T., Chao C., Saito S., Mazur S.J., Murphy M.E., Appella E., Xu Y. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat. Cell Biol. 2005;7:165–171. doi: 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
- Marión R.M., Strati K., Li H., Murga M., Blanco R., Ortega S., Fernandez-Capetillo O., Serrano M., Blasco M.A. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martí M., Mulero L., Pardo C., Morera C., Carrió M., Laricchia-Robbio L., Esteban C.R., Izpisua Belmonte J.C. Characterization of pluripotent stem cells. Nat. Protoc. 2013;8:223–253. doi: 10.1038/nprot.2012.154. [DOI] [PubMed] [Google Scholar]
- Martins-Taylor K., Xu R.-H. Concise review: Genomic stability of human induced pluripotent stem cells. Stem Cells. 2012;30:22–27. doi: 10.1002/stem.705. [DOI] [PubMed] [Google Scholar]
- Menendez S., Camus S., Izpisua Belmonte J.C. p53: guardian of reprogramming. Cell Cycle. 2010;9:3887–3891. doi: 10.4161/cc.9.19.13301. [DOI] [PubMed] [Google Scholar]
- Mohamad O., Drury-Stewart D., Song M., Faulkner B., Chen D., Yu S.P., Wei L. Vector-free and transgene-free human iPS cells differentiate into functional neurons and enhance functional recovery after ischemic stroke in mice. PLoS ONE. 2013;8:e64160. doi: 10.1371/journal.pone.0064160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S., Bazett-Jones D.P., Alitalo K., Lahesmaa R., Nagy A., Otonkoski T. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- Nishino K., Toyoda M., Yamazaki-Inoue M., Fukawatase Y., Chikazawa E., Sakaguchi H., Akutsu H., Umezawa A. DNA methylation dynamics in human induced pluripotent stem cells over time. PLoS Genet. 2011;7:e1002085. doi: 10.1371/journal.pgen.1002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K., Matsumura Y., Sato Y., Okada A., Morizane A., Okamoto S., Hong H., Nakagawa M., Tanabe K., Tezuka K. A more efficient method to generate integration-free human iPS cells. Nat. Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- Okita K., Yamakawa T., Matsumura Y., Sato Y., Amano N., Watanabe A., Goshima N., Yamanaka S. An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells. 2013;31:458–466. doi: 10.1002/stem.1293. [DOI] [PubMed] [Google Scholar]
- Shi Y., Kirwan P., Livesey F.J. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat. Protoc. 2012;7:1836–1846. doi: 10.1038/nprot.2012.116. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Yu J., Hu K., Smuga-Otto K., Tian S., Stewart R., Slukvin I.I., Thomson J.A. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


