Abstract
Strain AM-8B5T, isolated from Lake Sevan in Armenia, was characterized phenotypically, chemotaxonomically and phylogenetically. This chemo-organoheterotrophic, aerobic, facultatively anaerobic, catalase- and oxidase-positive, non-motile strain grew on NSY medium at NaCl concentrations of 0.0–0.2 % (w/v) and at 4–30 °C. Whole-cell fatty acids were dominated by summed feature 3 (including C16: 1ω7c and iso-C15: 0 2-OH), C16: 0 and C18: 1ω7c. C12: 0 2-OH and C16: 1 2-OH were the only hydroxylated fatty acids detected. Phylogenetic analysis as well as phenotypic and chemotaxonomic similarities indicated that the novel isolate was affiliated with the genus Polynucleobacter. 16S rRNA gene similarity values with the four previously described Polynucleobacter species ranged from 96.2 to 98.7 %. DNA–DNA hybridization experiments showed that the isolate did not belong to any of the previously described Polynucleobacter species. The isolate could be distinguished from all previously established Polynucleobacter species based on chemotaxonomic and phenotypic traits. The bacterium possessed a free-living lifestyle and represents a group of bacteria inhabiting the water column of many freshwater lakes. Based on the revealed phylogeny, and chemotaxonomic and phenotypic differences to previously described Polynucleobacter species, it is proposed that the isolate represents a novel species, Polynucleobacter difficilis sp. nov.; the type strain is AM-8B5T (=DSM 22349T=CIP 110078T).
K. Heckmann and H.-J. Schmidt established the genus Polynucleobacter for obligately endosymbiotic bacteria living in cells of several species of ciliates affiliated with the genus Euplotes (Hypotrichia) and they described the species Polynucleobacter necessarius to accommodate obligate endosymbionts of Euplotes aediculatus (Heckmann & Schmidt, 1987). None of these obligate endosymbionts could be cultivated as pure cultures (Heckmann & Schmidt, 1987; Vannini et al., 2007). However, Springer et al. (1996) cloned and sequenced the 16S rRNA gene of an endosymbiotic P. necessarius strain obligately associated with E. aediculatus. Their phylogenetic analyses of the obtained ribosomal sequence revealed the affiliation of the genus Polynucleobacter with the Betaproteobacteria. Investigations by cultivation-independent methods (e.g. Hiorns et al., 1997; Zwart et al., 2002; Burkert et al., 2003; Crump & Hobbie, 2005) and cultivation methods (Hahn, 2003; Watanabe et al., 2009) revealed a monophyletic genus-like group of bacteria including the previously characterized endosymbiotic P. necessarius. This large and diverse group of bacteria was subdivided in preliminary operational taxonomic units based on phylogenetic criteria (Hahn, 2003; Wu & Hahn, 2006a). The proposed operational taxonomic units were designated subclusters A (PnecA), B1 (PnecB1), B2 (PnecB2), C (PnecC) and D (PnecD). Furthermore, it was revealed that, in addition to obligate endosymbionts, this monophyletic group of bacteria also contains free-living strains (Hahn, 2003; Vannini et al., 2007) that thrive in the water column of freshwater habitats as planktonic bacteria (Hahn et al., 2005). Consequently, the descriptions of the genus Polynucleobacter and the species P. necessarius were emended by adding descriptions of free-living strains (Hahn et al., 2009). Strains closely related to the endosymbionts of E. aediculatus but differing in lifestyle were separated into the two subspecies P. necessarius subsp. necessarius (obligate endosymbionts) and P. necessarius subsp. asymbioticus (obligately free-living strains) (Hahn et al., 2009). Recently, strains affiliated with subclusters PnecD, PnecA and PnecB2 were described as the novel species P. cosmopolitanus (Hahn et al., 2010), P. rarus (Hahn et al., 2011a) and P. acidiphobus (Hahn et al., 2011b), respectively, whereas subcluster PnecC is represented by the previously established species P. necessarius (Heckmann & Schmidt, 1987; Hahn et al., 2009). Here, we have characterized isolate AM-8B5T, which represents the only strain cultivated so far that is affiliated with subcluster PnecB1. This strain has a free-living planktonic lifestyle, in common with all other Polynucleobacter strains that have been cultivated in pure culture, but cultivation and maintenance of this strain were more difficult than for any previously isolated Polynucleobacter strain.
Isolation and characterization
Strain AM-8B5T was isolated from the water column of eutrophic Lake Sevan located in Armenia. The lake possesses a surface area of 1416 km2 and a maximum depth of 98.7 m, and is located at an altitude of 1915 m (Hovhannissian, 1994). The geographical coordinates of the sampling site (station #4) are 40° 29′ 35.50″ N 45° 11′ 31.34″ E. The strain was isolated from a water sample taken from a depth of 60 m close to the bottom of the lake on 13 November 2006. At the time of sampling, the lake was still thermally stratified and the thermocline was located at a depth of 25 m. The sampled hypolimnic water was characterized by a pH of 8.7, conductivity of 110 μS cm−1, oxygen concentration of 7.17 mg l−1 (about 56 % saturation), and a temperature of 4.2 °C. The total number of bacteria in the water sample, determined by epifluorescent microscopy and direct counting, was 1.9×106 cells ml−1.
The strain was isolated and cultivated by using the filtration acclimatization method and NSY medium (Hahn et al., 2004). Strain AM-8B5T could be maintained on R2A (Remel) or NSY medium with a strength of 3 g l−1; however, better growth was observed on modified NSY medium enriched with 1 g l-cysteine l−1 and 1 g pyruvate l−1. However, even when using optimized medium, cultivation and maintenance of strain AM-8B5T remained delicate and more difficult than for any other of the >200 Polynucleobacter strain cultivated by us so far. Currently, it is not known which physiological trait of the strain makes cultivation more complicated than for the other Polynucleobacter strains. Growth at different temperatures and growth under anoxic conditions in an anaerobic chamber were examined on NSY agar or on NSY agar supplemented with nitrate (0.8 mM). NaCl tolerance was determined using NSY agar supplemented with various NaCl concentrations (0, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 1.0, 1.25, 1.5, 1.75 and 2.0 %, w/v). The temperature range supporting growth was tested on standard NSY agar plates exposed to different temperatures (4, 15, 20, 25, 30 and 35 °C). Utilization of various substrates was investigated in the same way as for previously described Polynucleobacter species (Hahn et al., 2009, 2010, 2011a, b). Briefly, growth enabled by utilization of a specific substrate was determined by comparison of OD at 575 nm established in liquid one-tenth-strength NSY medium (0.3 g l−1) with and without 0.5 g test substrate l−1. Differences of <10 %, 10–50 % and >50 % of the OD with and without test substrate were scored after 10 days of growth as no utilization (−), weak utilization (w) and good utilization (+), respectively.
Sequencing of the 16S rRNA gene was performed as described previously (Hahn, 2003; Hahn et al., 2005). Sequence similarity values were determined by using the software EzTaxon (Chun et al., 2007). Phylogenetic analyses with neighbour-joining (NJ), maximum-parsimony (MP) and maximum-likelihood (ML) algorithms were performed by using the software mega versions 4 and 5 (Tamura et al., 2007). For calculation of NJ trees, the Tamura–Nei substitution model and a gamma distribution with five categories were used. For ML tree calculation, the Tamura–Nei+G+I model was used as recommended by Model Test.
For analysis of the whole-cell fatty acid composition, the strain was grown on R2A agar plates for 7–9 days at 25 °C. During this time, the agar surface was kept moist with liquid R2A medium. The physiological age of the biomass harvested for fatty acid analysis was standardized as far as possible by observing growth development during incubation of the plates and choosing the moment of harvest according to growth state. Fatty acid methyl esters were obtained as described previously (Kämpfer & Kroppenstedt, 1996) and separated by GC (model 6890; Hewlett Packard). Peaks were computed automatically using the Microbial Identification systems MIDI 4.5 and 6.1 (Sasser, 1990). Chromatographic peaks were evaluated using the database TSBA 40. DNA for DNA–DNA reassociation experiments was obtained by disrupting cells by using a French pressure cell (Thermo Spectronic) and purifying DNA in the crude lysate by chromatography on hydroxyapatite as described by Cashion et al. (1977). DNA–DNA hybridization was carried out as described by De Ley et al. (1970) under consideration of the modifications described by Huß et al. (1983) using a model Cary 100 Bio UV/VIS-spectrophotometer equipped with a Peltier-thermostatted 6×6 multicell changer and a temperature controller with in situ temperature probe (Varian).
Phenotypic and chemotaxonomic traits
The results of the phenotypic and chemotaxonomic characterization of strain AM-8B5T are presented in Tables 1 and 2. The strain differed from P. necessarius subsp. asymbioticus, P. cosmopolitanus, P. rarus and P. acidiphobus strains in its inability to utilize acetate (Table 1).
Table 1.
Traits characterizing P. difficilis sp. nov. AM-8B5T and previously described Polynucleobacter species
Taxa: 1, AM-8B5T; 2, P. acidiphobus (Hahn et al., 2011b); 3, P. necessarius subsp. asymbioticus (Hahn et al., 2009); 4, P. cosmopolitanus (Hahn et al., 2010); 5, P. rarus (Hahn et al., 2011a). All taxa are non-motile. Note that sufficient phenotypic and chemotaxonomic characterizations are lacking for the obligately endosymbiotic strains of the subspecies P. necessarius subsp. necessarius. −, Negative; +, positive; W, weakly positive; +/−, some strains positive and some strains negative; +/W, some strains positive and some strains weakly positive; W/−, some strains weakly positive and some strains negative.
| Characteristic | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| Cell morphology | Short curved rods | Short curved rods | Straight or curved rods | Curved rods | Straight rods |
| Nucleoids visible (DAPI)* | Rarely | Rarely | Rarely | Rarely | Frequently |
| Cell length (μm) | 0.6–1.8 | 0.5–1.4 | 0.5–2.9 | 0.4–1.4 | 0.8–1.8 |
| Cell width (μm) | 0.4–0.5 | 0.4–0.5 | 0.3–0.5 | 0.3–0.5 | 0.6–0.8 |
| Catalase | W | + | + | + | + |
| Oxidase | W | + | + | + | + |
| Growth at: | |||||
| 5 °C | + | − | + | +/− | − |
| 35 °C | − | + | +/− | + | − |
| Maximum NaCl tolerance (%, w/v)† | 0.2 (W) | 0.3 | 0.3–0.5 | 0.3–0.5 | 0.3 (W) |
| Anaerobic growth on: | |||||
| NSY medium | − | + | +/− | +/− | − |
| NSY+0.8 mM nitrate | W | + | + | + | + |
| Growth in mineral medium with acetic acid and B12 | − | W | W/− | W/− | W |
| Assimilation of: | |||||
| Formic acid | W | − | W/− | − | − |
| Glyoxylic acid | − | − | W/− | W/− | + |
| Glycolic acid | − | − | − | − | W |
| Acetic acid | − | W | + | + | + |
| Oxalic acid | − | − | − | − | W |
| Propionic acid | − | − | +/− | +/W | − |
| Pyruvic acid | + | + | + | + | + |
| Malonic acid | − | W | +/− | +/− | − |
| Oxaloacetic acid | W | + | +/− | + | − |
| Malic acid | W | W | +/W | + | + |
| Succinic acid | + | W | + | + | + |
| Fumaric acid | W | − | +/W | + | + |
| Levulinic acid | − | − | W/− | W/− | + |
| Citric acid | − | − | − | +/− | − |
| d-Mannose | − | − | W/− | W/− | W |
| d-Glucose | − | W | W/− | W/− | − |
| d-Galacturonic acid | W | − | W | +/W | + |
| d-Galactose | − | − | W/− | W | − |
| d-Lyxose | W | − | W/− | − | W |
| d-Fructose | − | − | W/− | − | W |
| d-Fucose | W | − | W/− | − | W |
| d-Sorbitol | − | − | W/− | − | − |
| l-Glutamate | − | − | +/− | W/− | − |
| l-Aspartate | − | − | +/− | − | − |
| l-Cysteine | + | + | +/W | + | W |
| l-Alanine | − | − | W/− | +/W | − |
| l-Serine | − | − | − | − | − |
| l-Asparagine | − | − | W/− | − | − |
| Betaine | − | − | W/− | − | − |
| DNA G+C content (mol%) | 49.4 | 48.3 | 44–46 | 44.9 | 40.3 |
DAPI, 4′,6-diamidino-2-phenylindole.
Highest NaCl concentration (added to NSY medium) at which growth was observed.
Table 2.
Whole-cell fatty acid compositions of Polynucleobacter difficilis strain AM-8B5T and related strains
Taxa: 1, strain AM-8B5T; 2, P. acidiphobus MWH-PoolGreenA3T (Hahn et al., 2011a); 3, P. rarus MT-CBb6A5T (Hahn et al., 2011b); 4, P. cosmopolitanus MWH-MoIso2T (Hahn et al., 2010); 5, five P. cosmopolitanus strains (Hahn et al., 2010); 6, P. necessarius subsp. asymbioticus QLW-P1DMWA-1T (Hahn et al., 2009); 7, three P. necessarius subsp. asymbioticus strains (Hahn et al., 2009). Values are percentages of the summed fatty acids named in the peak library of the MIDI system (contents >0.2% are listed). Strains were grown on R2A agar plates for 7–9 days (strain AM-8B5T) or 3–5 days at 28 °C. tr, Trace, presence is variable; −, not detected.
| Fatty acid | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| Saturated | |||||||
| C12 : 0 | 3.6 | 3.8 | 3.0 | − | − | 3.4 | 3.4–5.5 |
| C14 : 0 | 3.0 | 0.9 | 0.4 | 0.7 | 0.6–2.3 | 0.9 | 0.3–1.2 |
| C15 : 0 | 1.4 | − | − | − | − | 0.3 | 0–0.3 |
| C16 : 0 | 16.4 | 24.2 | 19.8 | 15.4 | 11.0–15.4 | 22.2 | 15.5–29.6 |
| C17 : 0 | 0.8 | − | 0.3 | − | − | − | 0–0.5 |
| C18 : 0 | 0.8 | 0.5 | 0.5 | 0.8 | 0.5–1.1 | 1.2 | 0.5–2.4 |
| 10-Methyl-C19 : 0 | − | − | 0.4 | 0.7 | 0–0.7 | − | − |
| Unsaturated | |||||||
| C14 : 1ω5c | − | − | − | 0.6 | 0–1.1 | − | 0–0.6 |
| C15 : 1ω6c | tr | − | 0.3 | − | − | − | 0–0.6 |
| C16 : 1ω5c | 1.4 | 0.4 | 0.5 | 0.3 | 0–1.1 | 0.9 | 0–0.9 |
| C17 : 1ω6c | 1.1 | − | − | 0.5 | 0–0.7 | − | − |
| C18 : 1ω9c | tr | − | − | 0.3 | 0–2.0 | − | 0–0.4 |
| C18 : 1ω7c | 10.2 | 17.3 | 21.8 | 28.7 | 28.7–38.1 | 12.9 | 1.1–20.4 |
| 11-Methyl-C18 : 1ω7c | − | − | 7.5 | 3.7 | 0.4–3.7 | 3.1 | 1.1–8.1 |
| Hydroxylated | |||||||
| C12 : 0-2OH | 1.3 | − | − | − | − | 2.5 | 1.3–2.5 |
| C12 : 0-3OH | − | − | − | 11.1 | 7.1–11.2 | − | − |
| C14 : 0-2OH | − | 2.9 | − | − | − | − | − |
| C16 : 1-2OH | 0.7 | − | − | − | − | − | − |
| C16 : 0-3OH | − | 0.5 | 0.3 | − | − | − | − |
| Cyclic | |||||||
| C17 : 0 cyclo | − | 3.0 | − | − | − | − | − |
| Summed features* | |||||||
| 1 | 2.4 | − | 1.8 | − | − | 0.4 | 0.4–1.0 |
| 2 | 7.0 | 10.8 | 6.9 | 0.6 | 0.6–3.9 | 9.6 | 9.2–9.9 |
| 3 | 47.1 | 32.1 | 35.9 | 34.7 | 31.5–36.5 | 41.3 | 35.6–45.0 |
| 7 | 1.8 | 1.7 | − | 1.5 | 0–1.5 | 0.4 | 0.3–2.0 |
Summed features contain components which cannot be separated or identified by the MIDI system. The MIDI suggestion for feature 1 (ECL of 10.928) is C12 : 0 aldehyde; feature 2 comprises C14 : 0 3-OH and/or iso-C16 : 1; feature 3 comprises C16 : 1ω7c and/or iso-C15 : 0 2-OH; and feature 7 comprises C19 : 1ω6c and/or an as-yet undetermined component.
The whole-cell fatty acid pattern of strain AM-8B5T was typical for members of the genus Polynucleobacter isolated so far. The pattern was dominated by feature 3 of the MIDI system (C16: 1ω7c and/or iso-C15: 0 2-OH), C18: 1ω7c and C16: 0 (Table 2). As no other branched fatty acid components were detected, we concluded that the peak of feature 3 represented mainly C16: 1ω7c (Table 2). In comparison to other Polynucleobacter species, the novel isolate contained relatively high amounts of C15: 0 (1.4 %), summed features 1 (equivalent chain-length of 10.928; 2.4 %) and 7 (C19: 1ω6c and/or an as-yet undetermined compound; 1.8 %), and by the presence of C17: 0, C17: 1ω6c and C12: 0 2-OH, whereas other hydroxylated compounds were lacking in other Polynucleobacter species. A peculiar marker of the novel species was the presence of C16: 1 2-OH. The presence of relatively high amounts (2.7 %) of C14: 0, together with the presence of C17: 1ω6c and C16: 1 2-OH, seem to be the most reliable feature to distinguish P. difficilis from P. necessarius subsp. asymbioticus, which also contains C12: 0 2-OH.
Strain AM-8B5T, like several strains affiliated with the other four Polynucleobacter species described so far, possessed the trait that at least some in situ grown cells can pass through membrane filters with pore sizes of 0.2 μm (Hahn, 2004).
Phylogeny
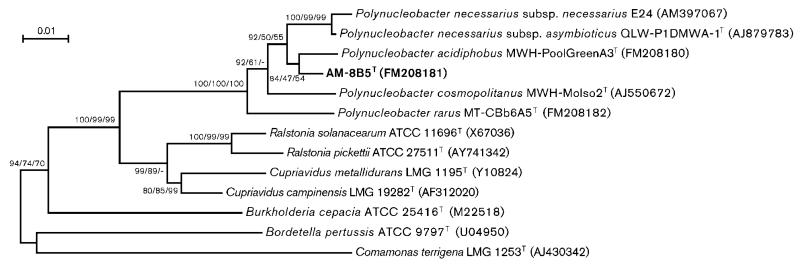
Phylogeny inference by NJ, MP and ML methods consistently resulted in clustering of the almost complete 16S rRNA gene sequence of strain AM-8B5T within the genus Polynucleobacter (Fig. 1). All three methods indicated that strain AM-8B5T represents a sibling taxon of P. acidiphobus. Analysis including 16S rRNA gene sequences of uncultivated bacteria (see below) revealed that strain AM-8B5T is affiliated with subcluster PnecB1 (Wu & Hahn, 2006a) of the genus Polynucleobacter. These phylogenetic results are further supported by previous phylogeny inference based on 16S–23S ITS sequences (Hahn et al., 2010). Furthermore, all three analyses of 16S rRNA sequences indicated that P. necessarius represents the closest relative of the subcluster (PnecB) represented by P. acidiphobus and AM-8B5T. In contrast, trees constructed by the ML method differed from trees calculated by the other two methods regarding the branching order of P. cosmopolitanus and P. rarus (Fig. 1).
Fig. 1.
Neighbour-joining tree based on almost complete 16S rRNA gene sequences reconstructing the phylogenetic position of Polynucleobacter difficilis sp. nov. strain AM-8B5T. Bootstrap percentages [1000 (NJ, MP) or 100 iterations (ML)] obtained by NJ/MP/ML methods are shown at nodes. Bar, 0.01 substitutions per nucleotide position.
Genotypic traits
Analysis of >400 sequences affiliated with the genus Polynucleobacter revealed a diagnostic oligonucleotide sequence specific for the 16S rRNA gene of bacteria affiliated with subcluster PnecB1. This sequence (5′-GCCGACTAGTTGTTGGGAATTTACATTCTCAG-3′; Escherichia coli positions 821–840) differs in a few base pairs from the sequences of strains affiliated with the other four Polynucleobacter subclusters. A blast search with this diagnostic sequence as query revealed 253 sequences with 100 % sequence identity. These sequences included many short partial 16S rRNA gene sequences. Therefore, a detailed phylogenetic analysis of these 253 sequences was omitted. Instead, a query with the reverse complement of the diagnostic sequence was performed by using the Probe Match service provided by the Ribosomal Database Project release 10 (Cole et al., 2009). This resulted in 250 hits with 100 % sequence identity. Almost all of these hits (241) were classified by the Ribosomal Database Project as Polynucleobacter bacteria; the remaining nine sequences were classified as ‘unclassified Burkholderiaceae’, ‘unclassified Burkholderiales’, ‘unclassified Comamonadaceae’ or ‘unclassified Betaproteobacteria’. blast searches with these nine sequences indicated that most of them are also affiliated with the genus Polynucleobacter, but two sequences seemed to represent uncultured bacteria not affiliated with this genus. In conclusion, the combined presence of this PnecB1-specific diagnostic sequence and a diagnostic sequence specific for the entire genus Polynucleobacter (5′-GAGCYGSTGTTTCTTCCC-3′; E. coli positions 445–463; Hahn et al., 2005) (Supplementary Table S1, available in IJSEM Online) seems to be suitable for reliable identification of members of the Polynucleobacter subcluster PnecB1. An overview of diagnostic sequences for identification of Polynucleobacter taxa is presented in Supplementary Table S1.
DNA–DNA reassociation experiments
The sequence similarities between 16S rRNA genes of strain AM-8B5T and the type strain of P. necessarius subsp. asymbioticus, a sequence representing the endosymbiotic P. necessarius subsp. necessarius ‘E24’, the type strain of P. cosmopolitanus, the type strain of P. rarus and the type strain of P. acidiphobus were 98.3, 97.9, 97.5, 96.2 and 98.7 %, respectively. Pairwise DNA–DNA reassociation experiments with strain AM-8B5T and the type strains of the three Polynucleobacter species sharing 16S rRNA sequence similarity values >97 % were performed in order to reveal whether the novel isolate belonged to a previously described species (Stackebrandt & Goebel, 1994). Note that experiments with DNA of a representative of P. necessarius subsp. necessarius could not be performed due to the lack of pure cultures (Heckmann & Schmidt, 1987; Vannini et al., 2007; Hahn et al., 2009). In all cases, the duplicate reassociation experiments resulted in DNA–DNA hybridization values ≤37.8 %. If the threshold value for delineation of prokaryotic species recommended by the ad hoc committee of Wayne et al. (1987) of 70 % DNA–DNA similarity is considered, strain AM-8B5T has to be viewed as a representative of a novel species that is distinct from all Polynucleobacter species described so far.
Proposal of a novel Polynucleobacter species
Results from the phylogenetic analysis and chemotaxonomic investigations demonstrated the affiliation of strain AM-8B5T with the genus Polynucleobacter (Tables 1 and 2, Fig. 1) but also revealed pronounced differences between this strain and strains affiliated with the four previously described Polynucleobacter species. DNA–DNA reassociation data indicated that the isolate does not belong to the previously described species P. necessarius, P. cosmopolitanus and P. acidiphobus and the 16S rRNA gene sequence similarity between strain AM-8B5T and the type strain of P. rarus of <97 % clearly indicated that the isolate also does not belong to the latter species (Stackebrandt & Goebel, 1994). Strain AM-8B5T can be discriminated from previously described Polynucleobacter species based on phenotypic, chemotaxonomic and genotypic traits. The strain differs from the P. acidiphobus type strain in its ability to utilize fumaric acid, d-galacturonic acid, and d-fucose, from the P. rarus type strain in its ability to utilize oxaloacetic acid, from all so far characterized P. cosmopolitanus strains in its ability to utilize d-lyxose and d-fucose, and from all so far characterized P. necessarius subsp. asymbioticus strains by the lack of acetate utilization (Table 2). The latter trait also discriminates strain AM-8B5T from all other Polynucleobacter species characterized so far. Regarding the fatty acid composition, strain AM-8B5T can be differentiated from each of the other Polynucleobacter species by differences in the relative occurrence of at least three fatty acid components (Table 2).
Based on these findings, we propose to establish the novel species, Polynucleobacter difficilis sp. nov., with strain AM-8B5T as the type strain. Furthermore, we propose to preliminarily include all strains phylogenetically affiliated with subcluster PnecB1 of the Polynucleobacter lineage (Wu & Hahn, 2006a) in this novel species. Further taxonomic investigations will be needed to clarify whether all strains affiliated with subcluster PnecB1 belong to P. difficilis sp. nov.
A consequence of proposing P. difficilis sp. nov. is a split of the taxon Polynucleobacter subcluster PnecB (Hahn, 2003), which is synonymous with tribe PnecB recently proposed in a review paper on phylogeny and ecology of freshwater bacterioplankton (Newton et al., 2011), into two separate species. Mainly for methodological reasons [i.e. the availability of a fluorescent in situ hybridization (FISH) probe; see Supplementary Table S1], this taxon was treated by ecologists as a taxonomic unit in several investigations on freshwater bacterioplankton (Wu & Hahn, 2006a, b; Salcher et al., 2008; Alonso et al., 2009; Newton et al., 2011). If the two species P. acidiphobus (PnecB2) and P. difficilis (PnecB1) possess distinct ecological characteristics, new molecular tools to distinguish the two taxa would be required for appropriate investigations of their ecology.
Ecology and biogeography of Polynucleobacter difficilis sp. nov.
blast searches (7 December 2010) with the 16S rRNA gene sequence of strain AM-8B5T resulted in 178 hits of almost full-length 16S rRNA genes with sequence similarities >99 %. Phylogenetic analyses of sequences of the top 250 blast hits and sequences representing the previously described Polynucleobacter species revealed that 182 sequences clustered together with AM-8B5T within subcluster PnecB1 (Wu & Hahn, 2006a). These sequences deposited in GenBank/EMBL/DDBJ were obtained from two estuaries (Chesapeake Bay and Delaware Bay) located in the USA (Shaw et al., 2008), an estuary (Weser) located in Europe (Selje et al., 2005), a lake in China (Z. H. Li, Y. Q. Ding and G. Y. Wang, unpublished data) and four high-altitude lakes located in Tibet (R. Zhang and W.-T. Liu, unpublished data). Furthermore, a large number of partial 16S rRNA sequences of about 500–600 bp lengths affiliated with subcluster PnecB1 were obtained from an experimental system strongly influenced by growth of phytoplankton (Horner-Devine et al., 2003) and deposited later in GenBank/EMBL/DDBJ (sequence accession numbers not mentioned by Horner-Devine et al., 2003). The majority of almost full-length sequences clustering within PnecB1 originated from the four Tibetan lakes. In contrast, despite the enormous number of deposited bacterial sequences from these intensively investigated lakes, sequences affiliated with subcluster PnecB2 (P. acidiphobus) were not obtained in this interesting study. Only two habitats are represented by sequences in both subclusters PnecB1 and PnecB2. These are the estuaries of Chesapeake Bay and Delaware Bay (Shaw et al., 2008), which potentially receive bacteria from ecologically distinct freshwater ponds and lakes drained by the large number of rivers and creeks discharging in these large bays. These differences in origin of sequences currently forming the two subclusters may indicate ecological differences between the two species representing the two subclusters. Therefore, generalization of results (Wu & Hahn, 2006a, b; Salcher et al., 2008; Alonso et al., 2009; Hahn et al., 2011b) obtained previously by using a PnecB-specific FISH probe (Wu & Hahn, 2006a) not distinguishing between the two subclusters PnecB1 and PnecB2 could be problematic. On the other hand, isolation of strain AM-8A5T from the water column of a lake and the exclusive origin of all other ribosomal sequences currently clustering within subcluster PnecB1 from the water columns of lakes and estuaries or experimental systems inoculated with water from lakes (Horner-Devine et al., 2003) indicates that PnecB1 bacteria share a planktonic lifestyle with all other free-living Polynucleobacter strains investigated so far. Whether the distribution of PnecB1 bacteria is restricted to freshwater habitats as with other free-living Polynucleobacter or populations also persist in saline systems (e.g. saline Tibetan lakes) remains to be revealed. Investigations by using the PnecB-specific FISH probe did not indicate that bacteria either affiliated with subcluster PnecB1 or PnecB2 thrive in saline lakes (Wu et al., 2006); however, the sequences deposited by R. Zhang and W.-T. Liu (unpublished data) in GenBank/EMBL/DDBJ seem to indicate that PnecB1 bacteria occur in some saline lakes.
Origination of PnecB1 sequences from habitats located in Asia, Europe and North America seems to indicate a cosmopolitan distribution of P. difficilis sp. nov. in freshwater habitats.
Description of Polynucleobacter difficilis sp. nov.
Polynucleobacter difficilis (dif.fi’ci.lis. L. masc. adj. difficilis difficult, troublesome, referring to difficulties in cultivating the type strain).
Curved, non-motile rods, 0.6–1.8 μm in length and 0.4–0.5 μm in width. Chemo-organotrophic, exhibits aerobic and anaerobic growth. Planktonic, free-living lifestyle. Inhabits various freshwater systems. Can be cultivated on NSY and R2A medium. Best growth is observed on modified NSY medium enriched with l-cysteine and pyruvate. Colonies grown on NSY agar are unpigmented, circular and convex with smooth surfaces. Mesophilic; growth occurs at 4 °C but not at 35 °C. Growth occurs without NaCl. Maximum NaCl concentration tolerated is 0.2 % (w/v). Oxidase- and catalase-positive. Utilizes formic acid, pyruvic acid, oxaloacetic acid, malic acid, succinic acid, fumaric acid, d-galacturonic acid, d-lyxose, d-fucose and l-cysteine when these substrates are provided in a medium containing low levels of NSY. Does not utilize glyoxylic acid, glycolic acid, acetic acid, oxalic acid, propionic acid, malonic acid, levulinic acid, citric acid, d-mannose, d-glucose, d-galactose, d-fructose, d-sorbitol, l-glutamate, l-aspartate, l-alanine, l-serine, l-asparagine or betaine. Whole-cell fatty acids are dominated by summed feature 3 (including C16: 1ω7c and iso-C15: 0 2-OH), C16: 0 and C18: 1ω7c. The components C12: 0, C15: 0, C17: 0, C17: 1ω6c, as well as C12: 0 2-OH and C16: 1 2-OH are present, but no other hydroxylated fatty acids. Tentatively, all strains affiliated with the genus Polynucleobacter and possessing the oligonucleotide sequence 5′-GCCGACTAGTTGTTGGGAATTTACATTCTCAG-3′ (E. coli positions 821–840) within the 16S rRNA gene shall be assigned to the proposed species.
The type strain is AM-8B5T (=DSM 22349T=CIP 110078T), isolated from Lake Sevan, Armenia. The DNA G+C content of the type strain is 49.4 mol%.
Supplementary Material
Acknowledgements
We are grateful to G. Pötter for performing the fatty acid analyses, to Bettina Sträubler for performing the DNA–DNA hybridization experiments, to P. Schumann and Birgit Grün for determination of the G+C value, and to P. Aumann for excellent technical assistance. We thank the Austrian Exchange Service (OeAD) for funding A. M.’s research stay at the Institute for Limnology. This study was supported by the Austrian Science Fund (project P19853 granted to M. W. H.) and the ESF project FREDI (FWF I 482-B09).
Abbreviations
- FISH
fluorescent in situ hybridization
- ML
maximum-likelihood
- MP
maximum-parsimony
- NJ
neighbour-joining
Footnotes
The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain AM-8B5T is FM208181.
A supplementary table is available with the online version of this paper.
References
- Alonso C, Zeder M, Piccini C, Conde D, Pernthaler J. Ecophysiological differences of betaproteobacterial populations in two hydrochemically distinct compartments of a subtropical lagoon. Environ Microbiol. 2009;11:867–876. doi: 10.1111/j.1462-2920.2008.01807.x. [DOI] [PubMed] [Google Scholar]
- Burkert U, Warnecke F, Babenzien D, Zwirnmann E, Pernthaler J. Members of a readily enriched beta-proteobacterial clade are common in surface waters of a humic lake. Appl Environ Microbiol. 2003;69:6550–6559. doi: 10.1128/AEM.69.11.6550-6559.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashion P, Holder-Franklin MA, McCully J, Franklin M. A rapid method for the base ratio determination of bacterial DNA. Anal Biochem. 1977;81:461–466. doi: 10.1016/0003-2697(77)90720-5. [DOI] [PubMed] [Google Scholar]
- Chun J, Lee J-H, Jung Y, Kim M, Kim S, Kim BK, Lim YW. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol. 2007;57:2259–2261. doi: 10.1099/ijs.0.64915-0. [DOI] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37(Suppl. 1):D141–D145. doi: 10.1093/nar/gkn879. other authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump BC, Hobbie JE. Synchrony and seasonality in bacterioplankton communities of two temperate rivers. Limnol Oceanogr. 2005;50:1718–1729. [Google Scholar]
- De Ley J, Cattoir H, Reynaerts A. The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem. 1970;12:133–142. doi: 10.1111/j.1432-1033.1970.tb00830.x. [DOI] [PubMed] [Google Scholar]
- Hahn MW. Isolation of strains belonging to the cosmopolitan Polynucleobacter necessarius cluster from freshwater habitats located in three climatic zones. Appl Environ Microbiol. 2003;69:5248–5254. doi: 10.1128/AEM.69.9.5248-5254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW. Broad diversity of viable bacteria in ‘sterile’ (0.2 micron) filtered water. Res Microbiol. 2004;155:688–691. doi: 10.1016/j.resmic.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Hahn MW, Stadler P, Wu QL, Pöckl M. The filtration-acclimatization method for isolation of an important fraction of the not readily cultivable bacteria. J Microbiol Methods. 2004;57:379–390. doi: 10.1016/j.mimet.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Hahn MW, Pöckl M, Wu QL. Low intraspecific diversity in a Polynucleobacter subcluster population numerically dominating bacterioplankton of a freshwater pond. Appl Environ Microbiol. 2005;71:4539–4547. doi: 10.1128/AEM.71.8.4539-4547.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Lang E, Brandt U, Wu QL, Scheuerl T. Emended description of the genus Polynucleobacter and the species Polynucleobacter necessarius and proposal of two subspecies, P. necessarius subsp. necessarius subsp. nov. and P. necessarius subsp. asymbioticus subsp. nov. Int J Syst Evol Microbiol. 2009;59:2002–2009. doi: 10.1099/ijs.0.005801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Lang E, Brandt U, Lünsdorf H, Wu QL, Stackebrandt E. Polynucleobacter cosmopolitanus sp. nov., free-living planktonic bacteria inhabiting freshwater lakes and rivers. Int J Syst Evol Microbiol. 2010;60:166–173. doi: 10.1099/ijs.0.010595-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Lang E, Tarao M, Brandt U. Polynucleobacter rarus sp. nov., a free-living planktonic bacterium isolated from an acidic lake. Int J Syst Evol Microbiol. 2011a;61:781–787. doi: 10.1099/ijs.0.017350-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Lang E, Brandt U, Spröer C. Polynucleobacter acidiphobus sp. nov., a representative of an abundant group of planktonic freshwater bacteria. Int J Syst Evol Microbiol. 2011b;61:788–794. doi: 10.1099/ijs.0.023929-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann K, Schmidt HJ. Polynucleobacter necessarius gen. nov., sp. nov., an obligately endosymbiotic bacterium living in the cytoplasm of Euplotes. Int J Syst Bacteriol. 1987;37:456–457. [Google Scholar]
- Hiorns WD, Methé BA, Nierzwicki-Bauer SA, Zehr JP. Bacterial diversity in Adirondack mountain lakes as revealed by 16S rRNA gene sequences. Appl Environ Microbiol. 1997;63:2957–2960. doi: 10.1128/aem.63.7.2957-2960.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner-Devine MC, Leibold MA, Smith V, Bohannan BJM. Bacterial diversity patterns along a gradient of primary productivity. Ecol Lett. 2003;6:613–622. [Google Scholar]
- Hovhannissian RH, Oganessian RO. Lake Sevan, yesterday, today. Erevan: Armenia National Academy of Science (in Russian, with extended summaries in English and Armenian) 1994 [Google Scholar]
- Huß VAR, Festl H, Schleifer KH. Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst Appl Microbiol. 1983;4:184–192. doi: 10.1016/S0723-2020(83)80048-4. [DOI] [PubMed] [Google Scholar]
- Kämpfer P, Kroppenstedt RM. Numerical analysis of fatty acid patterns of coryneform bacteria and related taxa. Can J Microbiol. 1996;42:989–1005. [Google Scholar]
- Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S. A guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol Rev. 2011;75:14–49. doi: 10.1128/MMBR.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcher MM, Pernthaler J, Zeder M, Psenner R, Posch T. Spatio-temporal niche separation of planktonic Betaproteobacteria in an oligo-mesotrophic lake. Environ Microbiol. 2008;10:2074–2086. doi: 10.1111/j.1462-2920.2008.01628.x. [DOI] [PubMed] [Google Scholar]
- Sasser M. Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Inc.; Newark, DE: 1990. MIDI Technical Note 101. [Google Scholar]
- Selje N, Brinkhoff T, Simon M. Detection of abundant bacteria in the Weser estuary using culture-dependent and culture independent approaches. Aquat Microb Ecol. 2005;39:17–34. [Google Scholar]
- Shaw AK, Halpern AL, Beeson K, Tran B, Venter JC, Martiny JB. It’s all relative: ranking the diversity of aquatic bacterial communities. Environ Microbiol. 2008;10:2200–2210. doi: 10.1111/j.1462-2920.2008.01626.x. [DOI] [PubMed] [Google Scholar]
- Springer N, Amann R, Ludwig W, Schleifer KH, Schmidt H. Polynucleobacter necessarius, an obligate bacterial endosymbiont of the hypotrichous ciliate Euplotes aediculatus, is a member of the beta-subclass of Proteobacteria. FEMS Microbiol Lett. 1996;135:333–336. doi: 10.1111/j.1574-6968.1996.tb08010.x. [DOI] [PubMed] [Google Scholar]
- Stackebrandt E, Goebel BM. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. mega4: Molecular Evolutionary Genetics Analysis (mega) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Vannini C, Pöckl M, Petroni G, Wu QL, Lang E, Stackebrandt E, Schrallhammer M, Richardson PM, Hahn MW. Endosymbiosis in statu nascendi: close phylogenetic relationship between obligately endosymbiotic and obligately free-living Polynucleobacter strains (Betaproteobacteria) Environ Microbiol. 2007;9:347–359. doi: 10.1111/j.1462-2920.2006.01144.x. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Komatsu N, Ishii Y, Negishi M. Effective isolation of bacterioplankton genus Polynucleobacter from freshwater environments grown on photochemically degraded dissolved organic matter. FEMS Microbiol Ecol. 2009;67:57–68. doi: 10.1111/j.1574-6941.2008.00606.x. [DOI] [PubMed] [Google Scholar]
- Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, et al. International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol. 1987;37:463–464. [Google Scholar]
- Wu QL, Hahn MW. Differences in structure and dynamics of Polynucleobacter communities in a temperate and a subtropical lake, revealed at three phylogenetic levels. FEMS Microbiol Ecol. 2006a;57:67–79. doi: 10.1111/j.1574-6941.2006.00105.x. [DOI] [PubMed] [Google Scholar]
- Wu QL, Hahn MW. High predictability of the seasonal dynamics of a species-like Polynucleobacter population in a freshwater lake. Environ Microbiol. 2006b;8:1660–1666. doi: 10.1111/j.1462-2920.2006.01049.x. [DOI] [PubMed] [Google Scholar]
- Wu QL, Zwart G, Schauer M, Kamst-van Agterveld MP, Hahn MW. Bacterioplankton community composition along a salinity gradient of sixteen high-mountain lakes located on the Tibetan Plateau, China. Appl Environ Microbiol. 2006;72:5478–5485. doi: 10.1128/AEM.00767-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart G, Crump BC, Kamst-van Agterveld MP, Hagen F, Han S-K. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat Microb Ecol. 2002;28:141–155. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.