Abstract
The laboratory mouse shares the majority of its protein-coding genes with humans, making it the premier model organism in biomedical research, yet the two mammals differ in significant ways. To gain greater insights into both shared and species-specific transcriptional and cellular regulatory programs in the mouse, the Mouse ENCODE Consortium has mapped transcription, DNase I hypersensitivity, transcription factor binding, chromatin modifications and replication domains throughout the mouse genome in diverse cell and tissue types. By comparing with the human genome, we not only confirm substantial conservation in the newly annotated potential functional sequences, but also find a large degree of divergence of sequences involved in transcriptional regulation, chromatin state and higher order chromatin organization. Our results illuminate the wide range of evolutionary forces acting on genes and their regulatory regions, and provide a general resource for research into mammalian biology and mechanisms of human diseases.
Supplementary information
The online version of this article (doi:10.1038/nature13992) contains supplementary material, which is available to authorized users.
Subject terms: Epigenomics
The Mouse ENCODE Consortium has mapped transcription, DNase I hypersensitivity, transcription factor binding, chromatin modifications and replication domains throughout the mouse genome in diverse cell and tissue types; these data were compared with those from human to confirm substantial conservation in the newly annotated potential functional sequences and to reveal pronounced divergence of other sequences involved in transcriptional regulation, chromatin state and higher order chromatin organization.
Supplementary information
The online version of this article (doi:10.1038/nature13992) contains supplementary material, which is available to authorized users.
Encyclopaedia of mouse epigenetic elements
The mouse is the premier model organism in biomedical research. To gain greater insights into the shared and species-specific transcriptional and cellular regulatory programs, the Mouse ENCODE Consortium has mapped transcription, DNase I hypersensitivity, transcription factor binding, chromatin modifications and replication domains throughout the mouse genome in diverse cell and tissue types. These finding are compared with the corresponding human data to confirm substantial conservation in the newly annotated potential functional sequences, and to reveal pronounced divergence of other sequences involved in transcriptional regulation, chromatin state and higher order chromatin organization. The data and their analyses provide a valuable resource for research into mammalian biology and mechanisms of human diseases.
Supplementary information
The online version of this article (doi:10.1038/nature13992) contains supplementary material, which is available to authorized users.
Main
Despite the widespread use of mouse models in biomedical research1, the genetic and genomic differences between mice and humans remain to be fully characterized. At the sequence level, the two species have diverged substantially: approximately one half of human genomic DNA can be aligned to mouse genomic DNA, and only a small fraction (3–8%) is estimated to be under purifying selection across mammals2. At the cellular level, a systematic comparison is still lacking. Recent studies have revealed divergent DNA binding patterns for a limited number of transcription factors across multiple related mammals3,4,5,6,7,8, suggesting potentially wide-ranging differences in cellular functions and regulatory mechanisms9,10. To fully understand how DNA sequences contribute to the unique molecular and cellular traits in mouse, it is crucial to have a comprehensive catalogue of the genes and non-coding functional sequences in the mouse genome.
Advances in DNA sequencing technologies have led to the development of RNA-seq (RNA sequencing), DNase-seq (DNase I hypersensitive sites sequencing), ChIP-seq (chromatin immunoprecipitation followed by DNA sequencing), and other methods that allow rapid and genome-wide analysis of transcription, replication, chromatin accessibility, chromatin modifications and transcription factor binding in cells11. Using these large-scale approaches, the ENCODE consortium has produced a catalogue of potential functional elements in the human genome12. Notably, 62% of the human genome is transcribed in one or more cell types13, and 20% of human DNA is associated with biochemical signatures typical of functional elements, including transcription factor binding, chromatin modification and DNase hypersensitivity. The results support the notion that nucleotides outside the mammalian-conserved genomic regions could contribute to species-specific traits6,12,14.
We have applied the same high-throughput approaches to over 100 mouse cell types and tissues15, producing a coordinated group of data sets for annotating the mouse genome. Integrative analyses of these data sets uncovered widespread transcriptional activities, dynamic gene expression and chromatin modification patterns, abundant cis-regulatory elements, and remarkably stable chromosome domains in the mouse genome. The generation of these data sets also allowed an unprecedented level of comparison of genomic features of mouse and human. Described in the current manuscript and companion works, these comparisons revealed both conserved sequence features and widespread divergence in transcription and regulation. Some of the key findings are:
Although much conservation exists, the expression profiles of many mouse genes involved in distinct biological pathways show considerable divergence from their human orthologues.
A large portion of the cis-regulatory landscape has diverged between mouse and human, although the magnitude of regulatory DNA divergence varies widely between different classes of elements active in different tissue contexts.
Mouse and human transcription factor networks are substantially more conserved than cis-regulatory DNA.
Species-specific candidate regulatory sequences are significantly enriched for particular classes of repetitive DNA elements.
Chromatin state landscape in a cell lineage is relatively stable in both human and mouse.
Chromatin domains, interrogated through genome-wide analysis of DNA replication timing, are developmentally stable and evolutionarily conserved.
Overview of data production and initial processing
To annotate potential functional sequences in the mouse genome, we used ChIP-seq, RNA-seq and DNase-seq to profile transcription factor binding, chromatin modification, transcriptome and chromatin accessibility in a collection of 123 mouse cell types and primary tissues (Fig. 1a, Supplementary Tables 1–3). Additionally, to interrogate large-scale chromatin organization across different cell types, we also used a microarray-based technique to generate replication-timing profiles in 18 mouse tissues and cell types (Supplementary Table 3)16. Altogether, we produced over 1,000 data sets. The list of the data sets and all the supporting material for this manuscript are also available at website http://mouseencode.org. Below we briefly outline the experimental approach and initial data processing for each class of sequence features.
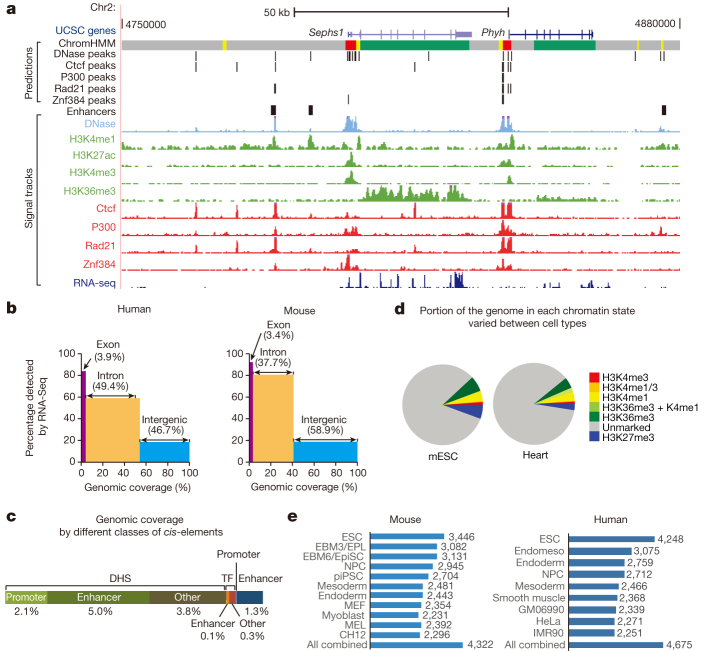
Figure 1. Overview of the mouse ENCODE data sets.
a, A genome browser snapshot shows the primary data and annotated sequence features in the mouse CH12 cells (Methods). b, Chart shows that much of the human and mouse genomes is transcribed in one or more cell and tissue samples. c, A bar chart shows the percentages of the mouse genome annotated as various types of cis-regulatory elements (Methods). DHS, DNase hypersensitive sites; TF, transcription factor. d, Pie charts show the fraction of the entire genome that is covered by each of the seven states in the mouse embryonic stem cells (mESC) and adult heart. e, Charts showing the number of replication timing (RT) boundaries in specific mouse and human cell types, and the total number of boundaries from all cell types combined. ESC, embryonic stem cell; endomeso, endomesoderm; NPC, neural precursor; GM06990, B lymphocyte; HeLa-S3, cervical carcinoma; IMR90, fetal lung fibroblast; EPL, early primitive ectoderm-like cell; EBM6/EpiSC, epiblast stem cell; piPSC, partially induced pluripotent stem cell; MEF, mouse embryonic fibroblast; MEL, murine erythroleukemia; CH12, B-cell lymphoma.
RNA transcriptome
To comprehensively identify the genic regions that produce transcripts in the mouse genome, we performed RNA-seq experiments in 69 different mouse tissues and cell types with two biological replicates each (Supplementary Table 3, Supplementary Information) and uncovered 436,410 contigs (Supplementary Table 4). Confirming previous reports13,17,18 and similar to the human genome, the mouse genome is pervasively transcribed (Fig. 1b), with 46% capable of producing polyadenylated messenger RNAs (mRNA). By comparison, 39% of the human genome is devoted to making mRNAs. In both species, the vast majority (87–93%) of exonic nucleotides were detected as transcribed, confirming the sensitivity of the approach. However, a higher percentage of intronic sequences were detected as transcribed in the mouse, and this might be owing to a greater sequencing depth and broader spectrum of biological samples analysed in mouse (Fig. 1b).
Candidate cis-regulatory sequences
To identify potential cis-regulatory regions in the mouse genome, we used three complementary approaches that involved mapping of chromatin accessibility, specific transcription factor occupancy sites and histone modification patterns. All of these approaches have previously been shown to uncover cis regulatory elements with high accuracy and sensitivity19,20.
By mapping DNase I hypersensitive sites (DHSs) in 55 mouse cell and tissue types21, we identified a combined total of ∼1.5 million distinct DHSs at a false discovery rate (FDR) of 1% (Supplementary Table 5)22. Genomic footprinting analysis in a subset (25) of these cell types further delineated 8.9 million distinct transcription factor footprints. De novo derivation of a cis-regulatory lexicon from mouse transcription factor footprints revealed a recognition repertoire nearly identical with that of the human, including both known and novel recognition motifs25.
We used ChIP-seq to determine the binding sites for a total of 37 transcription factors in various subsets of 33 cell/tissue types. Of these 37 transcription factors, 24 were also extensively mapped in the murine and human erythroid cell models (MEL and K562) and B-lymphoid cell lines (CH12 and GM12878)23. In total we defined 2,107,950 discrete ChIP-seq peaks, representing differential cell/tissue occupancy patterns of 280,396 distinct transcription factor binding sites (Supplementary Methods and Supplementary Table 6).
We also performed ChIP-seq for as many as nine histone H3 modifications (H3K4me1, H3K4me2, H3K4me3, H3K9me3, H3K27ac, H3K27me3, H3K36me3, H3K79me2 and H3K79me3) in up to 23 mouse tissues and cell types per mark. We applied a supervised machine learning technique, random-forest based enhancer prediction from chromatin state (RFECS), to three histone modifications (H3K4me1, H3K4me3 and H3K27ac)24, identifying a total of 82,853 candidate promoters and 291,200 candidate enhancers in the mouse genome (Supplementary Tables 7 and 8). To functionally validate the predictions, we randomly selected 76 candidate promoter elements (average size 1,000 bp, Supplementary Table 9) and 183 candidate enhancer elements (average size 1,000 bp, Supplementary Table 10). For candidate promoter elements, we cloned these previously unannotated sequences into reporter constructs, and performed luciferase reporter assays via transient transfection in pertinent mouse cell lines . For candidate enhancer elements, we performed functional validation assay using a high throughput method (see Supplementary Methods). Overall, 66/76 (87%) candidate promoters and 129/183 (70.5%) candidate enhancers showed significant activity in these assays, compared to 2/30 randomly selected negative controls (Supplementary Fig. 1c).
Collectively, our studies assigned potential regulatory function to 12.6% of the mouse genome (Fig. 1c).
Transcription factor networks
We explored the transcription factor networks and combinatorial transcription factor binding patterns in the mouse samples in two companion papers, and compared these networks to regulatory circuitry models generated for the human genome23,25. From genomic footprints, we constructed transcription-factor-to-transcription-factor cross-regulatory network in each of 25 cell/tissue types for a total of ∼500 transcription factors with known recognition sequences. Analyses of these networks revealed regulatory relationships between transcription factor genes that are strongly preserved in human and mouse, in spite of the extensive plasticity of the cis-regulatory landscape (detailed below). Whereas only 22% of transcription factor footprints are conserved, nearly 50% of cross-regulatory connections between mouse transcription factors are conserved in human through the innovation of novel binding sites. Moreover, analysis of network motifs shows that larger-scale architectural features of mouse and human transcription factor networks are strikingly similar25.
Chromatin states
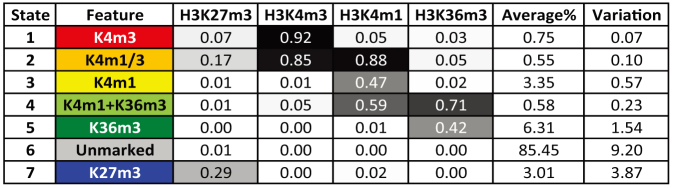
We produced integrative maps of chromatin states in 15 mouse tissue and cell types and six human cell lines (Supplementary Table 11), using a hidden Markov model (chromHMM)26,27 that allowed us to segment the genome in each cell type into seven distinct combination of chromatin modification marks (or chromatin states). One state is characterized by the absence of any chromatin marks, while every other state features either predominantly one modification or a combination of two modifications (Extended Data Table 1, Supplementary Information). The portion of the genome in each chromatin state varied with cell type (Fig. 1d, Supplementary Fig. 2). Similar proportions of the genome are found in the active states in each cell type, for both mouse and human. Interestingly, excluding the ‘unmarked’ state, the fraction of each genome that is in the H3K27me3-dominated, transcriptionally repressed state is the most variable, suggesting a profound role of transcriptional repression in shaping the cis-regulatory landscape during mammalian development.
Extended Data Table 1.
A seven-state chromHMM model learned from four histone modifications in 15 mouse cell types or lines and six human cell lines is shown
The numbers represent the emission probabilities of each histone modification (column) in each chromatin state (row). The enriched histone modifications in each state are summarized in the first column. The fraction of genome assigned in each state was calculated (Supplementary Fig. 2). The average and variation of these fraction values across all included cell types/tissues are listed in the last two columns.
Replication domains
Replication-timing, the temporal order in which megabase-sized genomic regions replicate during S-phase, is linked to the spatial organization of chromatin in the nucleus28,29,30,31, serving as a useful proxy for tracking differences in genome architecture between cell types32,33. Since different types of chromatin are assembled at different times during the S phase34, changes in replication timing during differentiation could elicit changes in chromatin structure across large domains. We obtained 36 mouse and 31 human replication-timing profiles covering 11 and 9 distinct stages of development, respectively (Supplementary Table 12). We defined ‘replication boundaries’ as the sites where replication profiles change slope from synchronously replicating segments (discussed later). A total of 64,535 and 50,194 boundaries identified across all mouse and human data sets, respectively, were mapped to 4,322 and 4,675 positions, with each cell type displaying replication-timing transitions at 50–80% of these positions (Fig. 1e).
Annotation of orthologous coding and non-coding genes
To facilitate a systematic comparison of the transcriptome, cis-regulatory elements and chromatin landscape between the human and mouse genomes, we built a high-quality set of human–mouse orthologues of protein coding and non-coding genes35. The list of protein-coding orthologues, based on phylogenetic reconstruction, contains a total of 15,736 one-to-one and a smaller set of one-to-many and many-to-many orthologue pairs (Supplementary Tables 13–15). We also inferred orthologous relationships among short non-coding RNA genes using a similar phylogenetic approach. We established one-to-one human–mouse orthologues for 151,257 internal exon pairs (Supplementary Table 16) and 204,887 intron pairs (Supplementary Table 17), and predicted 2,717 (3,446) novel human (respectively, mouse) exons (Supplementary Table 18). Additionally, we mapped the 17,547 human long non-coding RNA (lncRNA) transcripts annotated in Gencode v10 onto the mouse genome. We found 2,327 (13.26%) human lncRNA transcripts (corresponding to 1,679, or 15.48%, of the lncRNA genes) homologous to 5,067 putative mouse transcripts (corresponding to 3,887 putative genes) (Supplementary Fig. 3, Supplementary Table 19). Consistent with previous observations, only a small fraction of lncRNAs are constrained at the primary sequence level, with rapid evolutionary turnover36. Other comparisons of human and mouse transcriptomes, covering areas including pre-mRNA splicing, antisense and intergenic RNA transcription, are detailed in an associated paper37.
Divergent and conserved gene expression patterns
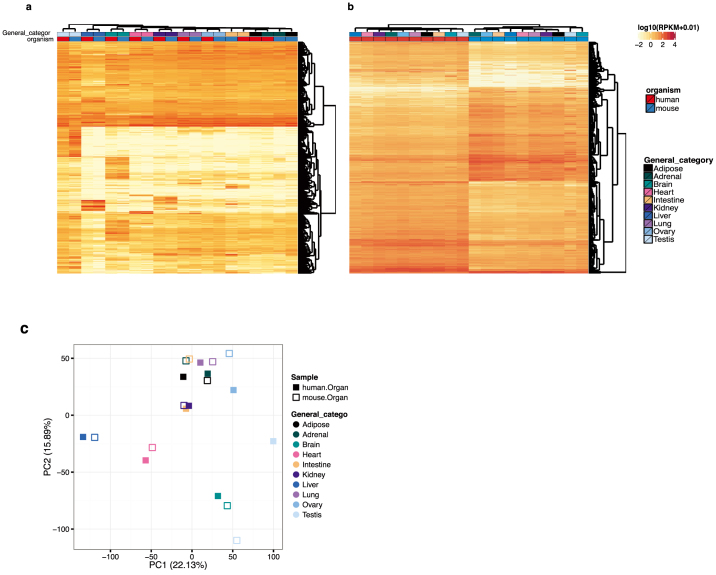
Previous studies have revealed remarkable examples of species-specific gene expression patterns that underlie phenotypic changes during evolution38,39,40,41,42. In these cases changes in expression of a single gene between closely related species led to adaptive changes. However, it is not clear how extensive the changes in expression patterns are between more distantly related species, such as mouse and human, with some studies emphasizing similarities in transcriptome patterns of orthologous tissues43,44,45 and others emphasizing substantial interspecies differences46. Our initial analyses revealed that gene expression patterns tended to cluster more by species rather than by tissue (Fig. 2a). To resolve the sets of genes contributing to different components in the clustering, we employed variance decomposition (see Methods) to estimate, for each orthologous human–mouse gene pair, the proportion of the variance in expression that is contributed by tissue and by species (Fig. 2b). This analysis revealed the sets of genes whose expression varies more across tissues than between species, and those whose expression varies more between species than across tissues. As expected, the clustering of the RNA-seq samples is dominated either by species or tissues, depending on the gene set employed (Extended Data Fig. 1a, b). Furthermore, removal of the ∼4,800 genes that drive the species-specific clustering (see ref. 47, Supplementary Fig. 1d therein) or normalization methods that reduce the species effects reveal tissue-specific patterns of expression in the same samples (Extended Data Fig. 1c). Categorizing orthologous gene pairs into these groups should enable more informative translation of research results between mouse and human. In particular, for gene pairs whose variance in expression is largest between tissues (and less between species), mouse should be a particularly informative model for human biology. In contrast, interpretation of studies involving genes whose variance in expression is larger between species needs to take into account the species variation. The relative contributions of species-specific and tissue-specific factors to each gene’s expression are further explored in two associated papers37,47.
Figure 2. Comparative analysis of the gene expression programs in human and mouse samples.
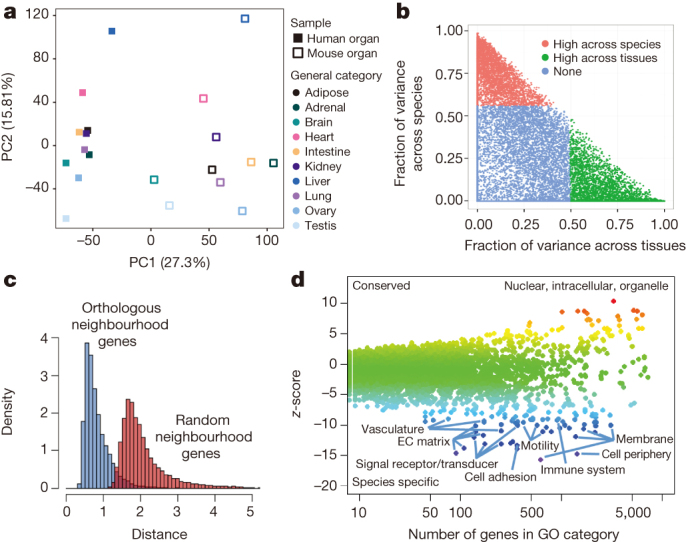
a, Principal component analysis (PCA) was performed for RNA-seq data for 10 human and mouse matching tissues. The expression values are normalized across the entire data set. Solid squares denote human tissues. Open squares denote mouse tissues. Each category of tissue is represented by a different colour. b, Gene expression variance decomposition (see Methods) estimates the relative contribution of tissue and species to the observed variance in gene expression for each orthologous human–mouse gene pair. Green dots indicate genes with higher between-tissue contribution and red dots genes with higher between-species contributions. c, Neighbourhood analysis of conserved co-expression (NACC) in human and mouse samples. The distribution of NACC scores for each gene is shown. d, A scatter plot shows the average of NACC score over the set of genes in each functional gene ontology category. Highlighted are those biological processes that tend to be more conserved between human and mouse and those processes that have been less conserved (see Supplementary Table 21 for list of genes).
Extended Data Figure 1. Clustering analysis of human and mouse tissue samples.
a, RNA-seq data from Ilumina Body Map (adipose, adrenal, brain, colon, heart, kidney, liver, lung, ovary and testis) were analysed together with that from the matched mouse samples using clustering analysis. Genes with high variance across tissues were used, resulting in cell samples clustering by tissues, not by species. b, Clustering employing genes with high variance between species shows clustering by species instead of tissues. c, Principal Component Analysis (PCA) was performed for RNA-seq data for 10 human and mouse matching tissues. The expression values are normalized within each species and we observed the clustering of samples by tissue types.
To further identify genes with conserved expression patterns and those that have diverged between humans and mice, we developed a novel method, referred to as neighbourhood analysis of conserved co-expression (NACC), to compare the transcriptional programs of orthologous genes in a way that did not require precisely matched cell lines, tissues or developmental stages, as long as a sufficiently diverse panel of samples is used in each species (Supplementary Methods). Observing that the orthologues of most sets of co-expressed genes in one species remained significantly correlated across samples in the other species, we use the mean of these small correlated sets of orthologous genes as a reference expression pattern in the other species. We compute Euclidean distance to the reference pattern in the multi-dimensional tissue/gene expression space as a relative measure of conservation of expression of each gene. Specifically, for each human gene (the test gene), we defined the most similarly expressed set of genes (n = 20) across all the human samples as that gene’s co-expression neighbourhood. We then quantify the average distance between the transcript levels of the mouse orthologue of the test gene and the transcript levels of each mouse orthologue of the neighbourhood genes across the mouse samples. We then invert the analysis, and choose a mouse test gene and define a similar gene co-expression neighbourhood in the mouse samples, and calculate the average distance between the expression of orthologues of the test gene and expression of neighbourhood genes across the human samples. The average change in the human-to-mouse and mouse-to-human distances, referred herein as a NACC score, is a symmetric measure of the degree of conservation of co-expression for each gene. The distribution of this quantity for each gene is shown in Fig. 2c, showing that genes in one species show a strong tendency to be co-expressed with orthologues of similarly expressed genes in the other species compared to random genes (also see Supplementary Information). We quantify the degree to which a specific biological process diverges between human and mouse as the average NACC scores of genes in each gene ontology category by calculating a z-score using random sampling of equal size sets of genes. Figure 2d shows that genes coding for proteins in the nuclear and intracellular organelle compartments, and involved in RNA processing, nucleic acid metabolic processes, chromatin organization and other intracellular metabolic processes, tend to exhibit more similar gene expression patterns between human and mouse. On the other hand, genes involved in extracellular matrix, cellular adhesion, signalling receptors, immune responses and other cell-membrane-related processes are more diverged (for a complete list of all GO categories and conservation analysis, see Supplementary Table 21). As a control, when we applied the NACC analysis to two different replicates of RNA-seq data sets from the same species, no difference in biological processes can be detected (Supplementary Fig. 5).
Several lines of evidence indicate that NACC is a sensitive and robust method to detect conserved as well as diverged gene expression patterns from a panel of imperfectly matched tissue samples. First, when we applied NACC to a set of simulated data sets, we found that NACC is robust for the diversity and conservation of the mouse–human sample panel (in Supplementary Fig. 6). Second, we randomly sampled subsets of the full panel of samples and demonstrated that the categories of human–mouse divergence shown in Fig. 2d are robust to the particular sets of samples we selected (Supplementary Fig. 7). Third, when we repeated NACC on a limited collection of more closely matched tissues and primary cell types (see Supplementary Methods), the biological processes detected as conserved and species-specific in the larger panel of mismatched human–mouse samples are largely recapitulated, although some pathways are detected with somewhat less significance, probably owing to the smaller number of data sets used (Supplementary Fig. 8). In summary, the NACC results support and extend the principal component analysis, showing that while large differences between mouse and human transcriptome profiles can be observed (revealed in PC1), genes involved in distinct cellular pathways or functional groups exhibit different degrees of conservation of expression patterns between human and mouse, with some strongly preserved and others changing markedly.
Prevalent species-specific regulatory sequences along with a core of conserved regulatory sequences
To better understand how divergence of cis-regulatory sequences is linked to the range of conservation patterns detected in comparisons of gene expression programs between species, we examined evolutionary patterns in our predicted regulatory sequences. Previous studies have identified a wide range of evolutionary patterns and rates for cis-regulatory regions in mammals5,8, but there are still questions regarding the overall degree of similarity and divergence between the cis-regulatory landscapes in the mouse and human. The variety of assays and breadth of tissue and cell-type coverage in the mouse ENCODE data therefore provide an opportunity to address this problem more comprehensively.
We first determined sequence homology of the predicted cis-elements in the mouse and human genomes. We established one-to-one and one-to-many mapping of human and mouse bases derived from reciprocal chained blastz alignments48 and identified conserved cis-regulatory sequences49. This analysis showed that 79.3% of chromatin-based enhancer predictions, 79.6% of chromatin-based promoter predictions, 67.1% of the DHS, and 66.7% of the transcription factor binding sites in the mouse genome have homologues in the human genome with at least 10% overlapping nucleotides, while by random chance one expects 51.2%, 52.3%, 44.3% and 39.3%, respectively (Fig. 3a, Supplementary Information for details). With a more stringent cutoff that requires 50% alignment of nucleotides, we found that 56.4% of the enhancer predictions, 62.4% of promoter predictions, 61.5% of DHS, and 53.3% of the transcription factor binding sites have homologues, compared with an expected frequency of 34%, 33.8%, 33.6% and 33.7% by random chance (Supplementary Fig. 9). The candidate mouse regulatory regions with human homologues are listed in Supplementary Tables 22–25. Thus, between half and two-thirds of candidate regulatory regions demonstrate a significant enrichment in sequence conservation between human and mouse. The remaining half to one-third have no identifiable orthologous sequence.
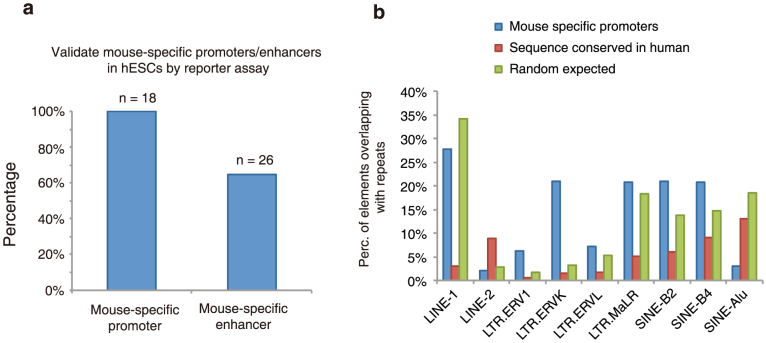
Figure 3. Comparative analysis of the cis-elements predicted in the human and mouse genome.
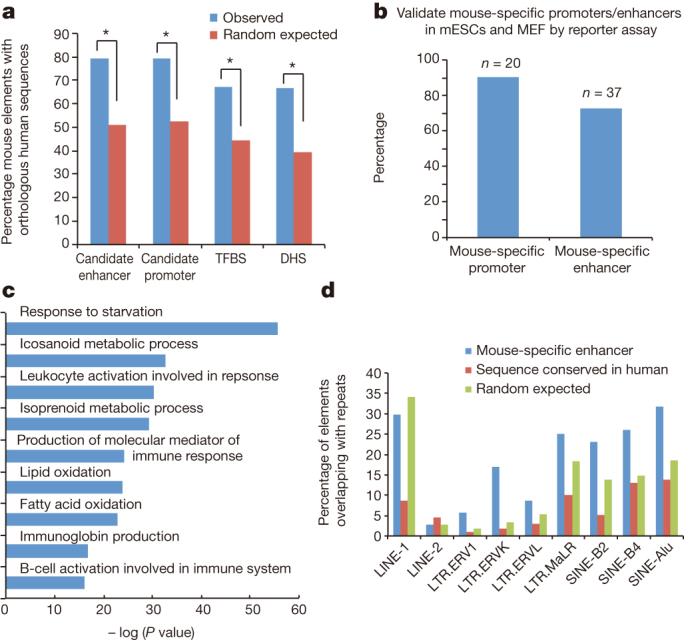
a, Chart shows the fractions of the predicted mouse cis-regulatory elements with homologous sequences in the human genome (Methods). TFBS, transcription factor binding site. b, A bar chart shows the fraction of the DNA fragments tested positive in the reporter assays performed either using mouse embryonic stem cells (mESCs) or mouse embryonic fibroblasts (MEF). c, A chart shows the gene ontology (GO) categories enriched near the predicted mouse-specific enhancers. d, A bar chart shows the percentage of the predicted mouse-specific enhancers containing various subclasses of LTR and SINE elements. As control, the predicted mouse cis elements with homologous sequences in the human genome or random genomic regions are included.
The candidate regulatory regions in mouse with no orthologue in human could arise either because they were generated by lineage-specific events, such as transposition, or because the orthologue in the other species was lost. Species-specific cis-regulatory sequences have been reported before3,14, but the fraction of regulatory sequences in this category remains debatable and may vary with different roles in regulation. We find that 15% (12,387 out of 82,853) of candidate mouse promoters and 16.6% (48,245 out of 291,200) of candidate enhancers (both predicted by patterns of histone modifications) have no sequence orthologue in humans (Supplementary Tables 26, 28, for details please refer to Supplementary Methods section). However, the question remains as to whether these species-specific elements are truly functional elements or simply correspond to false-positive predictions due to measurement errors or biological noise. Supporting the function of mouse-specific cis elements, 18 out of 20 randomly selected candidate mouse-specific promoters tested positive using reporter assays in mouse embryonic stem cells, where they were initially identified (Fig. 3b, Supplementary Table 27). Further, when these 18 mouse-specific promoters were tested using reporter assays in the human embryonic stem cells, all of them also exhibited significant promoter activities (Extended Data Fig. 2a, Supplementary Table 27), indicating that the majority of candidate mouse-specific promoters are indeed functional sequences, which are either gained in the mouse lineage or lost in the human lineage. Similarly, a majority of the candidate mouse-specific enhancers discovered in embryonic stem cells are also likely bona fide cis elements, as 70.2% (26 out of 37) candidate enhancers randomly selected from this group were found to exhibit enhancer activities in reporter assays (Fig. 3b, Supplementary Table 29). Like the candidate mouse-specific promoters, 61.5% (16 out of 26) of the candidate mouse-specific enhancers also show enhancer activities in human embryonic stem cells (Extended Data Fig. 2a).
Extended Data Figure 2. Comparative analysis of sequence conservation in the cis elements predicted in the human and mouse genome.
a, The predicted mouse-specific promoters and enhancers can function in human embryonic stem cells (hESCs). Percentages of predicted enhancers or promoters that test positive are shown in a bar chart. b, A bar chart shows the percentage of the predicted mouse-specific promoters containing various subclasses of LTR and SINE elements. As control, the predicted mouse cis elements with homologous sequences in the human genome or random genomic regions are included.
We next tested whether the rapidly diverged cis-regulatory elements would correspond to the same cellular pathways shown to be less conserved by the NACC analysis of gene expression programs. Indeed, gene ontology analysis revealed that the mouse-specific regulatory elements are significantly enriched near genes involved in immune function (Fig. 3c), in agreement with the divergent transcription patterns for these genes reported earlier and a previous report based on a smaller number of primate-specific candidate regulatory regions50. This suggests that regulation of genes involved in immune function tends to be species-specific50, just as the protein-coding sequences coding for immunity, pheromones and other environmental genes are frequent targets for adaptive selection in each species2,51. The target genes for mouse-specific transcription factor binding sites (Supplementary Table 30) are enriched in molecular functions such as histone acetyltransferase activity and high-density lipoprotein particle receptor activity, in addition to immune function (IgG binding).
We next investigated the mechanisms generating mouse-specific cis-regulatory sequences: loss in human, gain in mouse, or both. 89% (42,947 out of 48,245) of mouse-specific enhancers and 85% (10,535 out of 12,387) of mouse-specific promoters overlap with at least one class of repeat elements (compared to 78% by random chance). Confirming earlier reports52,53,54, we found that mouse-specific candidate promoters and enhancers are significantly enriched for repetitive DNA sequences, with several classes of repeat DNA highly represented (Fig. 3d and Extended Data Fig. 2b). Furthermore, mouse-specific transcription factor binding sites are highly enriched in mobile elements such as short interspersed elements (SINEs) and long terminal repeats (LTRs)55.
The 50% to 60% of candidate regulatory regions with sequences conserved between mouse and human are a mixture of (1) sequences whose function has been preserved via strong constraint since these species diverged, (2) sequences that have been co-opted (or exapted) to perform different functions in the other species, and (3) sequences whose orthologue in the other species no longer has a discernable function, but divergence by evolutionary drift has not been sufficient to prevent sequence alignment between mouse and human. Several companion papers delve deeply into these issues22,23,49. In particular, ref. 23 shows that the conservation of transcription factor binding at orthologous positions (falling in category (1)) is associated with pleiotropic roles of enhancers, as evidenced by activity in multiple tissues. References 22,49 describe the exaptation of conserved regulatory sequences for other functions.
We surveyed the conservation of function in the subset of mouse candidate cis elements that have sequence counterparts in the human genome. Of the 51,661 chromatin-based promoter predictions that have human orthologues, 44% (22,655) of them are still predicted as promoters in human on the basis of the same analysis of histone modifications (Supplementary Table 31, see Supplementary Methods for details). Of the 164,428 chromatin-based enhancer predictions that have human orthologues, 40% (64,962) of them are predicted as an enhancer in human (Supplementary Table 32). The remaining 56–60% of candidate mouse regulatory regions with a human orthologue fall into category (2) or (3) (see earlier), that is, the orthologous sequence in human either performs a different function or does not maintain a detectable function.
One caveat of the above observation is that the tissues or cell samples used in the survey were not perfectly matched. To better examine the conservation of biochemical activities among these predicted cis-regulatory elements with orthologues between mouse and human, we analysed the chromatin modifications at the promoter or enhancer predictions in a broad set of 23 mouse tissue and cell types with the neighbourhood co-expression association analysis (NACC) method described above. Instead of gene expression levels, we selected the histone modification H3K27ac as an indicator of promoter or enhancer activity as previously reported56. As shown in Fig. 4a, the promoter predictions (blue) show a significantly higher correlation in the level of H3K27ac in human and mouse than the random controls (red). Similarly, most chromatin-based enhancer predictions in the mouse genome exhibit conserved chromatin modification patterns in the human, albeit to a lesser degree than the promoters (Fig. 4b). NACC analysis on DNase-seq signal resulted in very similar distributions of conserved chromatin accessibility patterns at promoters (Fig. 4c) and enhancers (Fig. 4d). Thus many sequence-conserved candidate cis-regulatory elements appeared to have conserved patterns of activities in mice and humans.
Figure 4. Analysis of conservation in biochemical activities at the predicted mouse cis-regulatory sequences with human orthologues.
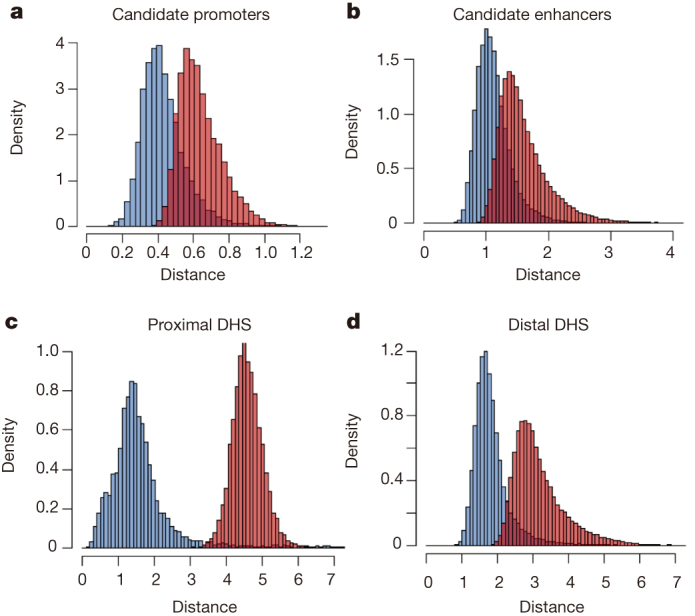
a, b, Histograms show the distribution of the NACC score for the chromatin modification H3K27ac signal at the predicted mouse promoters (a) or enhancers (b). c, d, Histograms show the distributions of NACC scores for DNase I signal at the promoter proximal (c) and distal (d) DNase I hypersensitive sites (DHS).
Taken together, these analyses show that the mammalian cis-regulatory landscapes in the human and mouse genomes are substantially different, driven primarily by gain or loss of sequence elements during evolution. These species-specific candidate regulatory elements are enriched near genes involved in stress response, immunity and certain metabolic processes, and contain elevated levels of repeated DNA elements. On the other hand, a core set of candidate regulatory sequences are conserved and display similar activity profiles in humans and mice.
Chromatin state landscape reflects tissue and cell identities
We examined gene-centred chromatin state maps in the mouse and human cell types (see Supplementary Methods) (Fig. 5a, Supplementary Fig. 10). In all cell types, the low-expressed genes were almost uniformly in chromatin states with the repressive H3K27me3 mark or in the state unmarked by these histone modifications. In contrast, expressed genes showed the canonical pattern of H3K4me3 at the transcription start site surrounded by H3K4me1, followed by H3K36me3-dominated states in the remainder of the transcription unit. A similar pattern was seen for all the active genes, regardless of the level of expression; the only exception was a tendency for the H3K4me3 to spread further into the transcription unit for the most highly expressed genes. The same binary relationship between chromatin state maps and expression levels of genes was observed in mouse and human cell types (Supplementary Fig. 10).
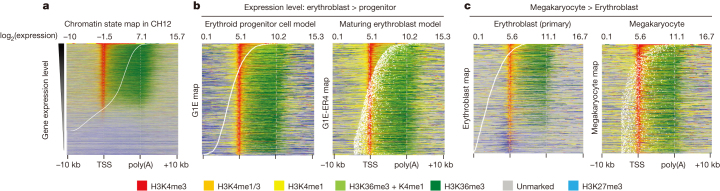
Figure 5. Chromatin landscape is stable within individual cell lineages.
a, Map displaying the distribution of chromatin states over the neighbourhoods of human–mouse one-to-one orthologue genes in CH12 cells. The gene neighbourhood intervals were sorted by the transcription level of each gene, shown by white dots. TSS, transcription start site. b, c, Distribution of chromatin states in human–mouse one-to-one orthologues that are differentially expressed genes between erythroid progenitor and erythroblasts models (b) and between erythroblast and megakaryocyte (c).
For both mouse and human cells, the majority of the genome was in the unmarked state in each cell type, consistent with previous observations in Drosophila57 and human cell lines12 (Supplementary Fig. 2). About 55% of the mouse genome was in an unmarked state in all the 15 cell types examined, while 65% is unmarked in all six human cell types. For genes that were in the unmarked state in mouse, their orthologues in human also tended to be in the unmarked state, and vice versa, leading to a positive correlation for the amount of gene neighbourhoods in unmarked states (Supplementary Fig. 11). Strong correlations were also observed in profiles of other chromatin marks averaged over cell lines and tissues37. The genes in the unmarked zones were depleted of transcribed nucleotides relative to the number expected based on fraction of the genome included, and the levels of the transcripts mapped there were lower than those seen in the active chromatin states (Supplementary Fig. 12).
Previous studies revealed limited changes of the chromatin states in lineage-restricted cells as they undergo large-scale changes in gene expression during maturation58,59,60. The chromatin state maps recapitulated this result, showing very similar patterns of chromatin modification in a cell line model for proliferating erythroid progenitor cells (G1E) and in maturing erythroblasts (G1E-ER4 cells treated with oestradiol) across genes whose expression level changed significantly during maturation (Fig. 5b, Supplementary Fig. 10b). This limited change raised the possibility that the chromatin landscape, once established during lineage commitment, dictates a permissive (or restrictive) environment for the gene regulatory programs in each cell lineage60, and that the chromatin states may differ between cell lineages. We tested this by examining the chromatin state maps for genes that were differentially expressed between haematopoietic cell lineages (erythroblasts versus megakaryocytes), and we found marked differences between the two cell types (Fig. 5c and Supplementary Fig. 10b). Genes expressed at a higher level in megakaryocytes than in erythroblasts were all in active chromatin states in megakaryocytes, but many were in inactive chromatin states in erythroblasts (Fig. 5c). In the converse situation, genes expressed at a higher level in erythroblasts than in megakaryocytes showed more inactive states in the cells in which they were repressed (Supplementary Fig. 10b). These greater differences in chromatin states correlating with differential expression of genes between, but not within, cell lineages support the model that chromatin states are established during the process of lineage commitment. The clustering of cell types together by lineage based on chromatin state maps (Supplementary Fig. 10c) also supports the model that the landscape of active and repressed chromatin is established no later than lineage commitment, and that this landscape is a defining feature of each cell type. Greater differences in chromatin states correlating with differences in gene expression were also observed when comparing average chromatin profiles in human and mouse37.
Mouse chromatin states inform interpretation of human disease-associated sequence variants
To investigate whether the mouse chromatin states were informative on sequence variants linked to human diseases by genome-wide association studies (GWAS), we combined the chromatin state segmentations of the fifteen mouse samples into a refined segmentation, which we used to train a self-organizing map (SOM)61 on four histone modification ChIP-seq data sets (H3K4me3, H3K4me1, H3K36me3 and H3K27me3) for each mouse sample. We mapped 4,265 single nucleotide polymorphisms (SNPs) from the human GWAS studies uniquely onto the mouse genome and scored these SNPs onto the trained SOM to determine whether SNP subsets were enriched in specific areas of the map. As shown in Fig. 6a, the highest enriched H3K4me1 unit in the kidney contains five GWAS hits (P value < 3.95 × 10−14) on different chromosomes related to blood characteristics such as platelet counts (Fig. 6a, Extended Data Table 2a). Similarly, the second highest enriched unit in liver H3K36me3 contained six GWAS hits (P value < 7.54 × 10−31) related to cholesterol and alcohol dependence out of twelve in that unit (Fig. 6b, Extended Data Table 2b). In contrast, one of the highest units in brain H3K27me3 has five GWAS hits (P value < 4.93 × 10−33) on different chromosomes associated with brain disorders/response to addictive substances (Fig. 6c, Extended Data Table 2c). This unit is different from the other examples in that it is enriched for H3K27me3 signal in multiple tissues, with brain being the highest. 801 out of the 1,350 units of the map showed statistical enrichment of SNPs of 0.05 after Holm–Bonferroni correction for multiple hypothesis testing, 55% of which (accounting for 1,750 GWAS hits) had signal for at least one histone mark that ranked within the top 100 units on the map (Fig. 6d). The best histone marks for enriched GWAS units were primarily H3K4me1 (23%), H3K36me3 (18%) and H3K27me3 (12%), with H3K4me3 accounting for less than 2% of the remainder. Together these results suggest that the chromatin state maps can be used to identify potential sites for functional characterization in mouse for human GWAS hits. Indeed, ref. 23 shows that conserved DNA segments bound by orthologous transcription factors in human and mouse are enriched for trait-associated SNPs mapped by GWAS.
Figure 6. Human GWAS hits when mapped onto mouse genome are associated with specific chromatin states.
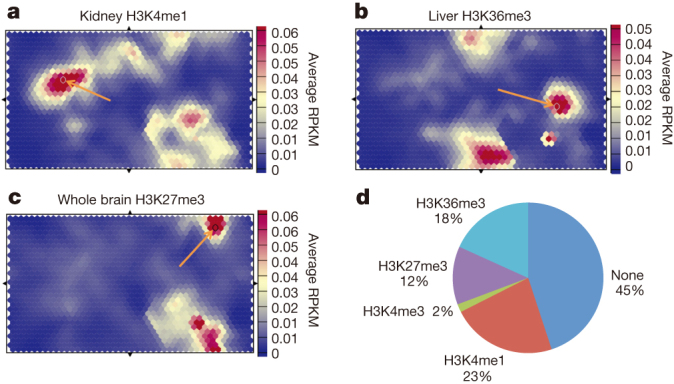
a, A self-organization map of histone modification H3K4me1 shows association between kidney H3K4me1 state and specific GWAS hits associated with urate levels (Methods). b, Liver-specific H3K36me3 unit shows enrichment in GWAS hits related to cholesterol, alcohol dependence and triglyceride levels. c, Brain-specific H3K27me3 high unit shows enrichment in GWAS SNPs associated with neurological disorders. d, Characterization of every unit with statistically significant GWAS enrichments in terms of highest histone modification signal in at least one sample. Units with no signal in top 100 map units for every histone modification are listed as none. RPKM, reads per kilobase per million reads mapped.
Extended Data Table 2.
Self-organizing map of histone modifications shows enrichment of human GWAS SNPs when mapped onto mouse
a, Kidney-specific H3K4me1 that shows enrichment of specific GWAS hits associated with urate levels and metabolites. b, Liver-specific H3K36me3 unit shows enrichment in GWAS hits related to cholesterol, alcohol dependence and triglyceride levels. c, Brain-specific H3K27me3 signals show enrichment in GWAS SNPs associated with neurological disorders.
Large-scale chromatin domains are developmentally stable and evolutionarily conserved
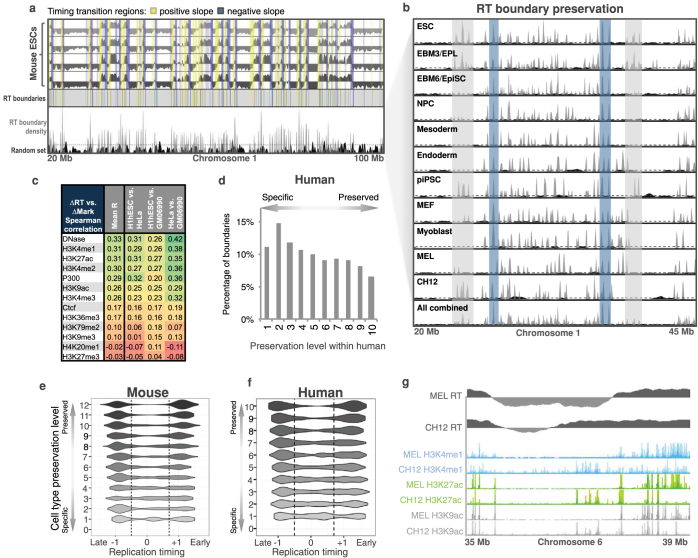
We mapped the positions of early and late replication timing boundaries in each of 36 mouse and 31 human profiles (Fig. 7a). Significantly clustered boundary positions (above the 95th percentile of re-sampled positions) were identified and peaks in boundary density were aligned between cell types using a common heuristic (Extended Data Fig. 3a, b, Supplementary Fig. 13). After alignment, consensus boundaries were further classified by orientation and amount of replication timing separation, resulting in a more stringent filtering of boundaries (Supplementary Figs 14, 15). Overall, we found that 88% of boundary positions (versus 20% expected for random alignment; Fisher exact test P < 2 × 10−16) aligned position and orientation between two or more cell types in both mouse and human (that is, 12% were cell-type-specific, Fig. 7b, Extended Data Fig. 3). Pair-wise comparisons of boundaries were consistent with developmental similarity between cell types (Supplementary Fig. 16). The earliest and latest replicating boundaries were most well preserved between cell types, while those of mid-S replicating boundaries were highly variable (Extended Data Fig. 3e, f).
Figure 7. Replication timing boundaries preserved among tissues are conserved in mice and humans.
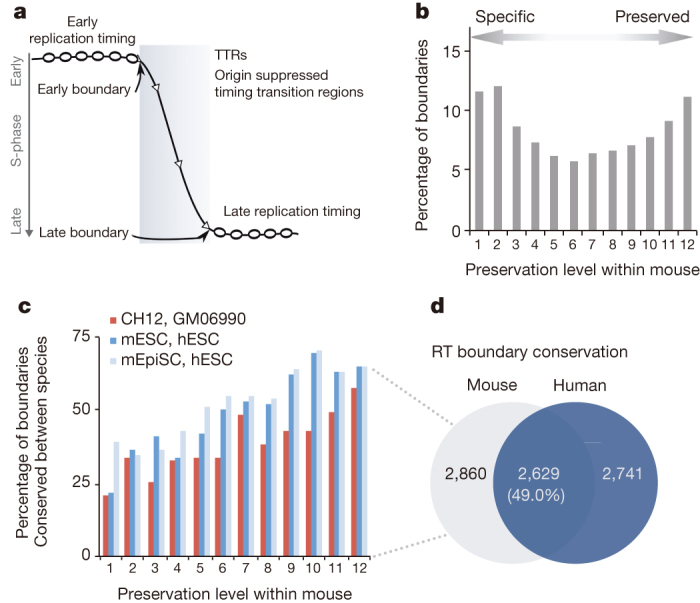
a, Depiction of a timing transition region (TTR) between the early and late replication domains. Early and late boundaries are defined as slope changes at either end of TTRs. b, Boundaries conserved between species for matched mouse and human cell types as a function of preservation among mouse cell types. c, Percentage of boundaries conserved between species (bar graph) and overall conservation of boundaries between comparable mouse and human cell types (CH12 versus GM06990, mESC versus hESC, mouse epiblast stem cells (mEpiSC) versus hESC) as a function of preservation among mouse cell types. d, A Venn diagram compares the replication timing boundaries identified in the mouse and human genome.
Extended Data Figure 3. Replication timing boundaries preserved among tissues are conserved during evolution.
a, Heat map of TTR overlap with positive (yellow) or negative (blue) slope. Replication timing (RT) boundaries were identified as clustered TTR endpoints (grey) above the 95th percentile (dashed line) of randomly resampled positions (black). b, Examples of constitutive boundaries (blue regions) and regulated boundaries (grey regions) highlighted. c, Spearman correlations between differences in chromatin feature enrichment and differences in RT in non-overlapping 200-kb windows. d, Percentage of boundaries preserved between the indicated number of human cell types. e, f, Distribution of boundary replication timing in mouse (e) and human (f) as a function of preservation level between cell types. g, Comparison of changes in replication timing versus various histone marks across a segment of mouse chromosome 6.
Interestingly, the greatest number of boundaries was detected in embryonic stem cells in both species, with significant reduction in boundary numbers during differentiation (Supplementary Fig. 16), consistent with consolidation of domains and by proxy large-scale chromatin organization into larger ‘constant timing regions’ during differentiation62. Given that over half of the mouse and human genomes exhibit significant replication timing changes during development16,63, these observations support the model that developmental plasticity in replication timing is derived from differential regulation of replication timing within constant timing regions whose boundaries are preserved during development.
Although conservation of replication timing between mouse and human has been reported29,30, the conservation of replicating timing boundaries has not been examined. We converted boundary coordinates ± 100 kb across boundary positions between species, revealing significant overlap (Fig. 7c, d; P < 2.2 × 10−16 by Fisher’s exact test relative to a randomized boundary list). The level of conservation of the positions of boundaries improved from a median of 27% for cell-type-specific boundaries to 70% for boundaries preserved in nine or more cell types (Fig. 7c), demonstrating that boundaries most highly preserved during development were the most conserved across species. This was consistent with results for transcription (Fig. 2), as well as the previous observation that suggests that an increased plasticity of replication timing during development is associated with increased plasticity of replication timing during evolution64. Together, these findings identify evolutionarily labile versus constrained domains of the mammalian genome at the megabase scale.
Given the link between replication and chromatin assembly, we compared replication timing and levels of other chromatin properties in 200-kb windows across the genome (Supplementary Fig. 17). Features associated with active enhancers (H3K4me1, H3K27ac, DNase I sensitivity) were more closely correlated to replication timing than features associated with active transcription (RNA polymerase II, H3K4me3, H3K36me3, H3K79me2). By contrast, the correlation of replication timing to repressive features, such as H3K9me3, was poor and cell-type-specific, consistent with prior results. A more stringent comparison of differences in chromatin to differences in replication timing between cell types (Extended Data Fig. 3c, g, Supplementary Fig. 17) again revealed that marks of enhancers, including p300, H3K4me1 and H3K27ac, and DNase I sensitivity were more strongly correlated to replication timing than marks of active transcription.
Conclusion
By comparing the transcriptional activities, chromatin accessibilities, transcription factor binding, chromatin landscapes and replication timing throughout the mouse genome in a wide spectrum of tissues and cell types, we have made significant progress towards a comprehensive catalogue of potential functional elements in the mouse genome. The catalogue described in the current study should provide a valuable reference to guide researchers to formulate new hypotheses and develop new mouse models, in the same way as the recent human ENCODE studies have impacted the research community12.
We provide multiple lines of evidence that gene expression and their underlying regulatory programs have substantially diverged between the human and mouse lineages although a subset of core regulatory programs are largely conserved. The divergence of regulatory programs between mouse and human is manifested not only in the gain or loss of cis-regulatory sequences in the mouse genome, but also in the lack of conservation in regulatory activities across different tissues and cell types. This finding is in line with previous observations of rapidly evolving transcription factor binding in mammals, flies and yeasts, and highlights the dynamic nature of gene regulatory programs in different species3,4,7,65. Furthermore, by comprehensively delineating the potential cis-regulatory elements we demonstrated that specific groups of genes and regulatory elements have undergone more rapid evolution than others. Of particular interest is the finding that cis-regulatory sequences next to immune-system-related genes are more divergent. The finding of species-specific cis-elements near genes involved in immune function suggests rapid evolution of regulatory mechanisms related to the immune system. Indeed, previous studies have uncovered extensive differences in the immune systems among different mouse strains and between humans and mice66, ranging from relative makeup of the innate immune and adaptive immune cells66, to gene expression patterns in various immune cell types67, and transcriptional responses to acute inflammatory insults68,69. At least some of these differences may be attributed to distinct regulatory mechanisms67, and our finding that many predicted mouse cis elements near genes with immune function lack sequence conservation supports the model that evolution of cis-regulatory sequences contributes to differences in the immune systems between humans and mice. More generally, our findings are consistent with the view that changes in transcriptional regulatory sequences are a source for phenotypic differences in species evolution.
How can species-specific gains or loss of cis-regulatory elements during evolution be compatible with their putative regulatory function? The finding of different rates of divergence associated with regulatory programs of distinct biological pathways suggests complex forces driving the evolution of the cis-regulatory landscape in mammals. We discovered that specific classes of endogenous retroviral elements are enriched at the species-specific putative cis-regulatory elements, implicating transposition of DNA as a potential mechanism leading to divergence of gene regulatory programs during evolution. Previous studies have shown that endogenous retroviral elements can be transcribed in a tissue-specific manner70,71, with a fraction of them derived from enhancers and necessary for transcription of genes involved in pluripotency72,73. Future studies will be necessary to determine whether retroviral elements at or near enhancers are generally involved in driving tissue-specific gene expression programs in different mammalian species.
Despite the divergence of the regulatory landscape between mouse and human, the pattern of chromatin states (defined by histone modifications) and the large-scale chromatin domains are highly similar between the two species. Half of the genome is well conserved in replication timing (and by proxy, chromatin interaction compartment) with the other half highly plastic both between cell types and between species. It will be interesting to investigate the significance of these conserved and divergent classes of DNA elements at different scales, both with regard to the forces driving evolution and for implications of the use of the laboratory mouse as a model for human disease.
Supplementary information
This file contains Supplementary Methods and Materials, Supplementary Figures 1-22, Supplementary Tables 1-3, 9, 10, 14, 15, 20, 27, 29, 33 and Supplementary References. Supplementary Tables 4-8, 11-13, 16-19, 21-26, 28, 30-32 are available at http://mouse.encodedcc.org. (PDF 28647 kb)
Acknowledgements
This work is funded by grants R01HG003991 (B.R.), 1U54HG007004 (T.R.G.), 3RC2HG005602 (M.P.S.), GM083337 and GM085354 (D.M.G.), F31CA165863 (B.D.P.), RC2HG005573 and R01DK065806 (R.C.H.) from the National Institutes of Health, and BIO2011-26205 from the Spanish Plan Nacional and ERC 294653 (to R.G.). J.V. is supported by a National Science Foundation Graduate Research Fellowship under grant no. DGE-071824. K.B., M.P., J.H. and P.F. acknowledge the Wellcome Trust (grant number 095908), the NHGRI (grant number U01HG004695) and the European Molecular Biology Laboratory. We thank G. Hon for helping the analysis of high-throughput enhancer validation. L.S. is supported by R01HD043997-09. S.L. was supported by grants F32HL110473 and K99HL119617.
Extended data figures and tables
PowerPoint slides
Author Contributions
F.Y., Y.C., A.B., J.V., W.W., T.R., M.A.Beer, R.C.H., J.A.S., M.P.S., R.G., T.R.G., D.M.G. and B.R. led the data analysis effort, R.Sandstrom, Z.M., C.D., B.D.P., Y.S., R.C.H., J.A.S., M.P.S., R.G., T.R.G., D.M.G. and B.R. led the data production. F.Y., M.A.Beer, L.E., Y.C., P.C., A.B., A.K., S.L., Y.L., J.V., R.Sandstrom, R.E.T., E.R., E.H., A.P.R., S.N., R.H., W.W., T.M., R.S.H., C.J., A.M., B.D.P., T.R., T.K., D.Lee, O.D., J.T., C.Z., A.D., D.D.P., S.D., P.P., J.Lagarde, G.B., A.T., K.B., M.P., P.F. and J.H. analysed data. Y.S., D.M., L.P., Z.Y., S.K., Z.M., T.K., G.E., J.Lian, S.M.W., R.K., M.A.Bender, S.L., Y.L., M.Z., R.B., M.T.G., A.J., S.V., K.L., D.B., F.N., M.D., T.C., R.S.H., P.J.S., M.S.W., T.A.R., E.G., A.S., T.K., E.H., D.D., M.D.B., L.S., A.R., S.J., R.Samstein, E.E.E., S.H.O., D.Levasseur, T.P., K.-H.C., A.S., C.D., P.T., W.W., C.A.K., C.S.M., T.M., D.J., N.D., B.D.P., T.R., C.D., L.-H.S., M.F., J.D. produced data. F.Y., Y.C., W.W., T.R., B.D.P., S.L., Y.L., C.J., C.D., A.D., A.B., D.D.P., S.D., C.N., A.M., J.A.S., M.P.S., R.G., T.R.G., D.M.G., R.C.H., M.A.Beer., B.R. wrote the manuscript. The role of the NHGRI Project Management Group (P.J.G., R.F.L., L.B.A., X.-Q.Z., M.J.P., E.A.F.) in the preparation of this paper was limited to coordination and scientific management of the Mouse ENCODE consortium.
Competing interests
The authors declare no competing financial interests.
Footnotes
Lists of participants and their affiliations appear in the Supplementary Information.
Feng Yue, Yong Cheng, Alessandra Breschi, Jeff Vierstra, Weisheng Wu, Tyrone Ryba, Richard Sandstrom, Zhihai Ma, Carrie Davis, Benjamin D. Pope, Yin Shen and Michael A. Beer: These authors contributed equally to this work.
Contributor Information
John A. Stamatoyannopoulos, Email: jstam@u.washington.edu
Michael P. Snyder, Email: mpsnyder@stanford.edu
Roderic Guigo, Email: roderic.guigo@crg.cat.
Thomas R. Gingeras, Email: gingeras@cshl.edu
David M. Gilbert, Email: gilbert@bio.fsu.edu
Ross C. Hardison, Email: ross@bx.psu.edu
Michael A. Beer, Email: mbeer@jhu.edu
Bing Ren, Email: biren@ucsd.edu.
References
- 1.Paigen K. One hundred years of mouse genetics: an intellectual history. I. The classical period (1902–1980) Genetics. 2003;163:1–7. doi: 10.1093/genetics/163.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chinwalla AT, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 3.Odom DT, et al. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nature Genet. 2007;39:730–732. doi: 10.1038/ng2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt D, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stefflova K, et al. Cooperativity and rapid evolution of cobound transcription factors in closely related mammals. Cell. 2013;154:530–540. doi: 10.1016/j.cell.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson MD, Odom DT. Evolution of transcriptional control in mammals. Curr. Opin. Genet. Dev. 2009;19:579–585. doi: 10.1016/j.gde.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Borneman AR, et al. Divergence of transcription factor binding sites across related yeast species. Science. 2007;317:815–819. doi: 10.1126/science.1140748. [DOI] [PubMed] [Google Scholar]
- 8.Zheng W, Gianoulis TA, Karczewski KJ, Zhao H, Snyder M. Regulatory variation within and between species. Annu. Rev. Genomics Hum. Genet. 2011;12:327–346. doi: 10.1146/annurev-genom-082908-150139. [DOI] [PubMed] [Google Scholar]
- 9.Wray GA. The evolutionary significance of cis-regulatory mutations. Nature Rev. Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- 10.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 11.Hawkins RD, Hon GC, Ren B. Next-generation genomics: an integrative approach. Nature Rev. Genet. 2010;11:476–486. doi: 10.1038/nrg2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature489, 57–74 (2012) [DOI] [PMC free article] [PubMed]
- 13.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature447, 799–816 (2007) [DOI] [PMC free article] [PubMed]
- 15.Stamatoyannopoulos JA, et al. An encyclopedia of mouse DNA elements (Mouse ENCODE) Genome Biol. 2012;13:418. doi: 10.1186/gb-2012-13-8-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiratani I, et al. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res. 2010;20:155–169. doi: 10.1101/gr.099796.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nature Rev. Genet. 2009;10:833–844. doi: 10.1038/nrg2683. [DOI] [PubMed] [Google Scholar]
- 18.Xu Z, et al. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maston GA, Landt SG, Snyder M, Green MR. Characterization of enhancer function from genome-wide analyses. Annu. Rev. Genomics Hum. Genet. 2012;13:29–57. doi: 10.1146/annurev-genom-090711-163723. [DOI] [PubMed] [Google Scholar]
- 20.Hardison RC, Taylor J. Genomic approaches towards finding cis-regulatory modules in animals. Nature Rev. Genet. 2012;13:469–483. doi: 10.1038/nrg3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vierstra, J. et al. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science10.1126/science.1246426 (in the press) [DOI] [PMC free article] [PubMed]
- 23.Cheng, Y. et al. Principles of regulatory information conservation between mouse and human. Nature10.1038/nature13985 (this issue) [DOI] [PMC free article] [PubMed]
- 24.Rajagopal N, et al. RFECS: a random-forest based algorithm for enhancer identification from chromatin state. PLoS Comput. Biol. 2013;9:e1002968. doi: 10.1371/journal.pcbi.1002968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stergachis, A. B. et al. Conservation of trans-acting circuitry during mammalian regulatory evolution. Nature10.1038/nature13972 (this issue) [DOI] [PMC free article] [PubMed]
- 26.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nature Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffman MM, et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 2013;41:827–841. doi: 10.1093/nar/gks1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryba T, et al. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 2010;20:761–770. doi: 10.1101/gr.099655.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yaffe E, et al. Comparative analysis of DNA replication timing reveals conserved large-scale chromosomal architecture. PLoS Genet. 2010;6:e1001011. doi: 10.1371/journal.pgen.1001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baker A, et al. Replication fork polarity gradients revealed by megabase-sized U-shaped replication timing domains in human cell lines. PLoS Comput. Biol. 2012;8:e1002443. doi: 10.1371/journal.pcbi.1002443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moindrot B, et al. 3D chromatin conformation correlates with replication timing and is conserved in resting cells. Nucleic Acids Res. 2012;40:9470–9481. doi: 10.1093/nar/gks736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takebayashi S-i, Dileep V, Ryba T, Dennis JH, Gilbert DM. Chromatin-interaction compartment switch at developmentally regulated chromosomal domains reveals an unusual principle of chromatin folding. Proc. Natl Acad. Sci. USA. 2012;109:12574–12579. doi: 10.1073/pnas.1207185109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lande-Diner L, Zhang J, Cedar H. Shifts in replication timing actively affect histone acetylation during nucleosome reassembly. Mol. Cell. 2009;34:767–774. doi: 10.1016/j.molcel.2009.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu, Y.-C., Bansa, l. S., Rasmussen, M. D., Herrero, J. & Kellis, M. Phylogenetic identification and functional validation of orthologs and paralogs across human, mouse, fly, and worm. bioRxiv10.1101/005736 (31 May 2014)
- 36.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pervouchine, D. et al. Enhanced transcriptome maps from multiple mouse tissues reveal evolutionary constraint in gene expression for thousands of genes. bioRxiv10.1101/010884 (30 October 2014) [DOI] [PMC free article] [PubMed]
- 38.McLean CY, et al. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature. 2011;471:216–219. doi: 10.1038/nature09774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shubin N, Tabin C, Carroll S. Deep homology and the origins of evolutionary novelty. Nature. 2009;457:818–823. doi: 10.1038/nature07891. [DOI] [PubMed] [Google Scholar]
- 40.Jones FC, et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484:55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grossman SR, et al. Identifying recent adaptations in large-scale genomic data. Cell. 2013;152:703–713. doi: 10.1016/j.cell.2013.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fraser HB. Gene expression drives local adaptation in humans. Genome Res. 2013;23:1089–1096. doi: 10.1101/gr.152710.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brawand D, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- 44.Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science. 2012;338:1593–1599. doi: 10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barbosa-Morais NL, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- 46.Sabeti PC, et al. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 47.Lin, S. et al. Comparison of the transcriptional landscapes between human and mouse tissues. Proc. Natl Acad. Sci. USA (in the press) [DOI] [PMC free article] [PubMed]
- 48.Schwartz S, et al. Human–mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Denas, O. et al. Genome-wide comparative analysis reveals human-mouse regulatory landscape and evolution. bioRxiv10.1101/010926 (30 October 2014) [DOI] [PMC free article] [PubMed]
- 50.King DC, et al. Finding cis-regulatory elements using comparative genomics: some lessons from ENCODE data. Genome Res. 2007;17:775–786. doi: 10.1101/gr.5592107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ponting CP. The functional repertoires of metazoan genomes. Nature Rev. Genet. 2008;9:689–698. doi: 10.1038/nrg2413. [DOI] [PubMed] [Google Scholar]
- 52.Bourque G, et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18:1752–1762. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kunarso G, et al. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nature Genet. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- 54.Jacques P-É, Jeyakani J, Bourque G. The majority of primate-specific regulatory sequences are derived from transposable elements. PLoS Genet. 2013;9:e1003504. doi: 10.1371/journal.pgen.1003504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sundaram, V. et al. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 10.1101/gr.168872.113 (15 October 2014) [DOI] [PMC free article] [PubMed]
- 56.Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol. Cell. 2013;49:825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Filion GJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.John S, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nature Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jin F, Li Y, Ren B, Natarajan R. PU.1 and C/EBPα synergistically program distinct response to NF-κB activation through establishing monocyte specific enhancers. Proc. Natl Acad. Sci. USA. 2011;108:5290–5295. doi: 10.1073/pnas.1017214108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu W, et al. Dynamics of the epigenetic landscape during erythroid differentiation after GATA1 restoration. Genome Res. 2011;21:1659–1671. doi: 10.1101/gr.125088.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mortazavi A, et al. Integrating and mining the chromatin landscape of cell-type specificity using self-organizing maps. Genome Res. 2013;23:2136–2148. doi: 10.1101/gr.158261.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hiratani I, et al. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 2008;6:e245. doi: 10.1371/journal.pbio.0060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hansen RS, et al. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc. Natl Acad. Sci. USA. 2010;107:139–144. doi: 10.1073/pnas.0912402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ryba T, et al. Replication timing: a fingerprint for cell identity and pluripotency. PLoS Comput. Biol. 2011;7:e1002225. doi: 10.1371/journal.pcbi.1002225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moses AM, et al. Large-scale turnover of functional transcription factor binding sites in Drosophila. PLoS Comput. Biol. 2006;2:e130. doi: 10.1371/journal.pcbi.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mestas J, Hughes CCW. Of mice and not men: differences between mouse and human immunology. J. Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 67.Shay T, et al. Conservation and divergence in the transcriptional programs of the human and mouse immune systems. Proc. Natl Acad. Sci. USA. 2013;110:2946–2951. doi: 10.1073/pnas.1222738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seok J, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl Acad. Sci. USA. 2013;110:3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wells CA, et al. Genetic control of the innate immune response. BMC Immunol. 2003;4:5. doi: 10.1186/1471-2172-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Faulkner GJ, et al. The regulated retrotransposon transcriptome of mammalian cells. Nature Genet. 2009;41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 71.Xie W, et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153:1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu X, et al. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nature Struct. Mol. Biol. 2014;21:423–425. doi: 10.1038/nsmb.2799. [DOI] [PubMed] [Google Scholar]
- 73.Fort A, et al. Deep transcriptome profiling of mammalian stem cells supports a regulatory role for retrotransposons in pluripotency maintenance. Nature Genet. 2014;46:558–566. doi: 10.1038/ng.2965. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file contains Supplementary Methods and Materials, Supplementary Figures 1-22, Supplementary Tables 1-3, 9, 10, 14, 15, 20, 27, 29, 33 and Supplementary References. Supplementary Tables 4-8, 11-13, 16-19, 21-26, 28, 30-32 are available at http://mouse.encodedcc.org. (PDF 28647 kb)