Abstract
Background
pfmdr1 and its variants are molecular marker which are responsible for antibiotics resistance in Plasmodium falciparum, a parasitic carrier for malaria disease. A novel strategy to treat malaria disease is by disrupting parasite lactate dehydrogenase (pLDH), a crucial enzyme for Plasmodium survival during their erythrocytic stages. This research was aimed to investigate and characterize the pfmdr1 and pldh genes of P. falciparum isolated from Nusa Tenggara Indonesia.
Methods
Genomic DNA of P.falciparum was isolated from malaria patients in Nusa Tenggara Indonesia. pfmdr1 was amplified using nested PCR and genotyped using Restriction Fragment Length Polymorphism (RFLP). pldh was amplified, sequenced, and analyzed using NCBI public domain databases and alignment using Clustal W ver. 1.83.
Results
Genotyping of the pfmdr1 revealed that sequence diversity was extremely high among isolates. However, a sequence analysis of pldh indicated that open reading frame of 316 amino acids of the gene showing 100% homology to the P. falciparum 3D7 reference pldh (GeneBank: XM_001349953.1).
Conclusion
This is the first report which confirms the heterologous of pfmdr1 and the homologous sequences of P.falciparum pldh isolated from Nusa Tenggara Islands of Indonesia, indicating that the chloroquine could not be used effectively as antimalarial target in the region and the pLDH-targeted antimalarial compound would have higher chance to be successful than using chloroquine for curbing malaria worldwide.
Keywords: Malaria, Plasmodium falciparum, Drug resistance gene, pfmdr1, pldh, Indonesia
Introduction
Malaria, also known as “King of Diseases” is a major infectious disease and has caused enormous problems in tropical and subtropical regions(1-3). According to WHO (2008), 3.3 billion people were reported at risky condition from 109 countries of which 881,000 was deaths (4). In Indonesia, 73.6% of municipalities/cities are endemic area of malaria (5). Recently, the malaria cases in Indonesia are concentrated in the eastern regions, contributing to more than 80% of the country’s population (6). The latest survey conducted in 2011 showed that 45,000 cases were confirmed to be malaria (7).
Even though intensive prevention and eradication programs were performed, malaria is still becoming a main health problem worldwide (7, 8), and the emergence of P. falciparum resistance isolates particularly to choloquine makes this problem even worse (9). The resistance to the chloroquine is resulted from point mutation in multi-drug resistance-1 (PfMDR-1) gene, which causes the diversity in the genes (10-12). Detection of molecular marker of anti-malarial drug resistance is the latest method to monitor anti-malarial drug resistance in Plamodium (13).
The increasing resistance of malaria strains to conventional anti-malarial drug has stimulated the need for the development of new compounds with novel modes of action. Paracite lactate dehidrogenase (pLDH), a crucial enzyme for Plasmodium survival during their erythrocytic stages, has also been identified to be a novel target for antimalarials (14, 15). Compunds that inhibit the enzyme function can represent therapeutic agents to target the disease. Therefore, study of sequence homologous of the enzyme is necessary to predict the effectiveness of the compounds.
This research was carried out to detect molecular markers of antimalarial drug resistance based on P. falciparum multidrug resistance 1 (PfMDR-1) gene and the sequence of P. falciparum lactate dehydrogenase (PfLDH) gene.
Materials and Methods
Blood samples
Blood samples were collected in 2010 from patients with fever by finger prick in several islands of Nusa Tenggara Indonesia (Lombok, Sumbawa, Alor, Kupang). Thick and thin blood smears were made and stained with Giemsa. The slides were examined for the presence of malaria parasite by light microscopy. After they were confirmed microscopy, the infected blood (approximately 1-5 ml) was drawn from the venous blood of infected patients. The drawn blood was washed with RPMI medium to get rid of the white blood cells on the buffy coat layer (16). Then, these samples were used for genomic DNA isolation of parasites.
Isolation of genomic DNA of parasites
DNA was isolated from blood sample using standard method (17). The isolated DNAs was then diluted in TE buffer and used for pfmdr1 and pldh amplification. The integrity of DNA samples isolated was monitored by agarose gel electrophoresis.
Nested PCR for Plasmodium Identification and pfmdr1 Amplification
Identification of Plasmodium was performed using nested PCR (primers provided in Table 1) as previously described (18). Afterward, the target region of pfmdr1 was amplified by PCR using primers in Table 1 and checked for polymorphisms in two codons (86 and 1034) of the pfmdr1 using PCR-RFLP methodology (19, 20). The amplicon was analyzed on 1% agarose gel containing 0.5 ug/ml of ethidium bromide and the band was visualized under UV light.
Table 1.
List of primers used in this study
| Primers | Sequence (5’ to 3’) |
|---|---|
| rPLU5 | TTAAAATTGTTGCAGTTAAAACG |
| rPLU6 | CCTGTTGTTGCCTTAAACTTC |
| rFAL1 | TTAAACTGGTTTGGGAAAACCAAATATATT |
| rFAL2 | ACACAATGAACTCAATCATGACTACCCGTC |
| rVIV1 | CGCTTCTAGCTTATTCCACATAACTGATAC |
| rVIV2 | ACTTCCAAGCCGAAGCAAAGAAAGTCCTTA |
| fpmdr1 86 | |
| MDR-A | TTGAACAAAAAAGAGTACCGCTG |
| MDR-B | TCGTACCAATTCCTGAACTCAC |
| fpmdr1 1034 | |
| 1034F | TATGTCAAGCGGAGTTTTTGC |
| 1034R | TCTGAATCTCCTTTTAAGGAC |
| pLDH S Kpn | AGAGAGGGTACCGCACCAAAAGCA |
| pLDH AS Eco | CACACAGATTCTTAAGCTTAACATTCTC |
Molecular analyses of pfmdr1
RFLP analysis of pfmdr1 codon 86 and 1034 were conducted by digestion of the PCR product with AflIII and DdeI (New England Biolabs, Beverly, MA), respectivelly, at 37oC for 1 h. For each locus, RFLP products were electrophoresed on 1% agarose gels and visualized by UV transillumination.
Amplification of pldh
Oligonucleotide primers pLDH S Kpn and pLDH AS Eco corresponding to pfldh open reading frame (ORF) were constructed based on pfldh sequence (K1 strain) (Table 1). PCR was performed in 25 μl reaction volume containing 10 pmol of each primer, 1.25 mM MgCl2, 200 μM of dNTPs, 100 ng of P. falciparum genomic DNA and 2.5 U of Pfu Polymerase. The temperature gradients (55-65oC) were used to determine the optimum annealing te-mperature. The thermal cycling programs for PCR consisted of initial denaturation at 94 °C for 5 min, followed by 35 cycles at 94 °C for 60 s, at 60 °C for 45 s, at 72 °C for 60 s and final extension at 72 °C for 10 min. The amplicon was analyzed on 1% agarose gel conaining 0.5 ug/ml of ethidium bromide and the band was visualized under UV light.
Cloning, sequencing, and sequence analysisof pfldh
The PCR products were purified from gel by gel extraction kit (Qiagen, USA). The purified pLDH PCR product was ligated into EcoRV site of pBlueScript II KS+ vector at 16oC for 1 h. Ligation was performed at 16oC for 30 min using reaction mixtures as follow: 5x dilution of phosphorilated pldh (1.5 μl), 1.5 μl of dephosphorilated pBluescript II KS+ vector, and 2x Mix ligation kit (2.5 μl). By using heat shock transformation technique, the resulted recombinant plasmid (pBluS-pLDH) was transformed into E.coli top 10 competent cells and plated on LB-ampicillin/IPTG/X-gal plates followed by incubation at 37oC overnight. Since, the pBluescript II KS+ vector has β-galactosidase gene, LB medium containing X-gal was degraded perfectly by the E. coli bearing the plamid. The indicator of the degradation is blue colony for non-recombinant plasmid-bearing colony and white colony for recombinant plasmid-bearing colony. Then, a single colony (white colony) carrying the insert was screened by colony PCR using the gene specific oligonucleotide primers to detect the insert. The DNA plasmid was purified using QIAGEN Miniprep kit and the presence of insert was verified by EcoR1 and XhoI restriction digestion of purified recombinant plasmid. Several clones were selected to be sequenced. The sequences of cloned fragment were analyzed using public domain database of NCBI (http://blas-t.ncbi.nlm.nih.gov/). The sequences were aligned using Clustal W ver. 1.83 (http://ww-w.genebee.-msu.su/clu-stal/clustal.php) (21).
Results
A positive reaction of nested PCR indicated that 250-bp amplification product was generated with P.falciparum-specific primer and 120-bp amplification product from P. vivax-specific primer. Mixed infection of both P. falciparum and P. vivax was showed by the appearence of two band (120-bp and 250-bp). Results of nested PCR were shown in Fig. 1.
Fig. 1.
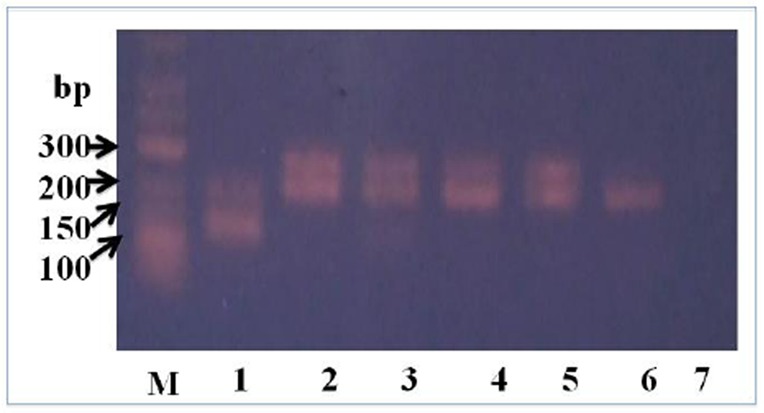
An example of results from nested PCR examined in this study. M = 1 kb DNA marker, Lane 1 = P. vivax, 2-5 = P. falciparum, 6 = positive control, 7 = negative control
Out of 311 malaria samples based on microscopy results, 155 (50%) samples were confirmed to have a Plasmodium spp infection by PCR. The results of nested PCR indicated that the positive samples consist of 131 (85%) for P. falciparum, 22 (14%) for P.vivax, and 2 (1%) for mixture of both species of malaria.
Then, the pfmdr1 was amplified from P. falciparum genome and used for restriction fragment length polymorphism analysis. The results of nested PCR indicated that more than 90% of the genes were successfully amplified. AflIII restriction enzyme was used to analyse and detect mutation point at codon 86 (N86Y) and DdeI for mutation point at codon 1034 (S1034). Amplification of pfmdr1 generated 372-bp PCR product. Restriction of the fragment using AflIII produced 248-bp and 124-bp in the mutation of N to Y at possition 86. Whereas, the amplification of pfmdr1 using 1034-F and 1034-R (Table 1) generated 189-bp PCR product.
The samples inspected in Lombok, Sumb-awa, and Kupang islands had 100% mutation in the pfmdr1, especially in N86Y and S1034C. Eventhough 33.3% of samples isolated from Alor had mutation in N86Y, S1034C were 100% mutated. The diversity in the pfmdr1 indicating that the chloroquine could not be effectively used as antimalarial target in the region.
Gene coding for LDH was amplified using P. falciparum genome isolated from Nusa Tenggara regions of Indonesia as template. Since, discrepancy of annealing temperature between sense and antisense primers, the use of 60oC and 67oC annealing temperature showed maximum amplification of pldh about 951 bp. The amplified product was obtained and then ligated with pBlueScript II KS+. Electrophoresis results of pldh amplification and pBlu-eScript II KS restriction were shownin Fig. 2A.
Fig. 2.
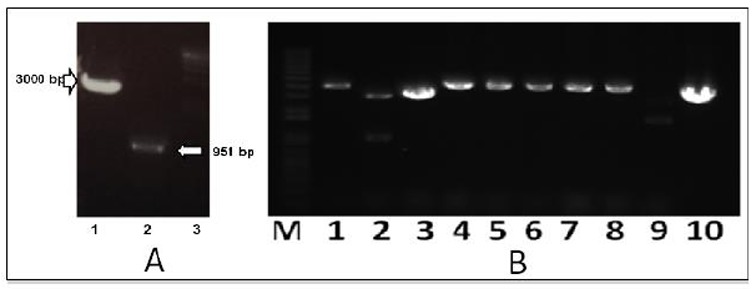
PCR amplification product of P. Falciparum ldh (A) and ligation result confirmation (B). Lane A1: digested pBlueScript II KS+, A2: pldh, A3: 1 kb DNA Marker, Lane BM: 1 kb DNA marker, lane 1-9: single digestion of recombinant plasmid comparing to empty plasmid (pBlueScript II KS+) (B10)
To obtain adequate amount of PCR product for ligation, concentration of PCR product was quantified in gel agarose by comparing the PCR product band density with the 1 kb and HindIII-λ DNA marker density.pldh PCR products was subsequently ligated with linearized pBlueScript II KS after phosphorilation using T4 Polynucleotide kinase.
Ligation results were confirmed not only using PCR colony, but also using size comparison between recombinant plasmid and empty plasmid (pBlueScript II KS). Therefore, plasmid isolation was carried out from colonies which have the right size of PCR product and then digested using EcoR1 restriction enzyme. Electrophoresis results of the digestion were shown in Fig. 2B. The figure showed that the size of recombinant plasmid was higher than the size of empty plasmid, indicating that the insert was perfectly ligated with pBluescript II KS+ vector.
Once the correct recombinant plasmid was detected, the colony carrying the plasmid was cultured in LB medium containg ampicilin and grown overnight at 37oC in a shaker. The obtained DNA was sequenced from both directions using sequencing primers given in Table 1.
The sequencing of pBlueScript-PfLDH revealed that the complete ORF comprised of 951 base pairs initiated with an ATG start codon and ending with a TTA codon. The P.falciparum ldh encoding putative protein of 316 amino acids contains no intron in the whole sequences.
Discussion
Molecular diagnostic method, such as PCR, has become widely used for the detection of malaria parasites in mixed and low level infection. However, the success of the method depends on a several factor, especially quality of DNA isolated from blood sample. In this research, sensitivity of PCR method was 55% indicating that Plasmodium DNA was not detected in other 45% samples which had microscopically detected parasites. A possible explanation for the dramatic difference between the microscopy and PCR method is the low quality of Plasmodium DNA obtained from blood sample. It is well known that degraded DNA, a high content of human DNA or hemoglobin, the use of heparin or inadequate condition of blood collecting, storage and amplification of samples can inhibit the PCR method (22).
As a gold standard treatment and as the first-line antimalarial drug for malaria, chloroquine has been used extensivelly to halt plasmodium pandemic worldwide because of its cheaper, less drawbacks, and easy to get. However, the use of the compound was banned in several regions because of increasing chloroquine resistance of parasites. Thus, detection of chloroquine resistance molecular marker of Plasmodium in Indonesia is critical path to design a novel antimalarial drug to overcome the disease.
P.falciparum genomic DNA was isolated from malaria patients in Indonesia (Lombok, Sumbawa, Kupang, and Alor) and subsequently used for the pfmdr1 amplification. Genotyping of the pfmdr1 amplified fragments using Restriction Fragment Length Polymorphism (RFLP) showed that high diversity sequences were observed among isolates. The sequence diversity of parasitemia genomes in the pfmdr1 (mutation of pfmdr1 N86Y and S1034C) was detected in Lombok and Sumbawa Islands (West Nusa Tenggara), Alor and Kupang (East Nusa Tenggara). Mutation at codon 86 of pfmdr1 (N86Y) was detected by AflIII restriction enzyme in which asparagin (N) was substituted by tyrosin (Y) in the position. Moreover, DdeI restriction enzyme was used successfully to detect the substitution of serine (S) with cystein (C) at codon 1034 (S1034S). Point mutations in pfmdr1 mainly N86Y, S1034, N1042D, and D1246Y have shown to modulate chloroquine resistance (23). Therefore, detection of point mutation in the positions suggested that the chloroquine could not be used as antimalaria in the regions.
Study of drug resistance gene in other region of Indonesia, Madagascar and Angola reported an association of pfmdr1 Y86 mutant alleles with chloroquine clinical failures in P. falciparum malaria (24-26). In addition, pmdr1 mutations in P. falciparum can confer resistance to high levels of chloroquine, and that these pfmdr1 mutations has an important role in the resistance of P. falciparum to other drugs (23).
Gene of a novel antimalarial target, parasite lactate dehidrogenase (pLDH) as a important enzyme for ATP production of parasite during anaerobic glucose metabolism in their erythrocytic stages, has also been amplified, sequenced, and compared with the P. falciparum 3D7 reference pldh. Sequencing results showed that pldh isolated from several islands in Nusa Tenggara Indonesia contains no intron and is present in a single copy on chromosome 13. The same characteristic of pldh of P.falciparum was also found in the previous research (27).
Sequence analysis of pldh was performed using NCBI public domain database and aligned using Clustal W ver. 1.83. Alignment to P. falciparum 3D7 reference pldh (GeneBank: XM_001349953.1) indicated that the open reading frame of 316 amino acids of the gene showing 100% homology. The sequences of the pldh showed that there was no variation between the P.falciparum pldh obtained from Nusa Tenggara regions of Indonesia and the pldh sequence from BankGene XM_001-349953.1.
Translation of the obtained sequence indicated that the key catalytic residues in the amino acids (Arg109, Asp168, Arg171, His19-5) (28) are conserved in all P.falciparum LDH. Moreover, the characteristic of five-amino acid insert, DKEWN, in the substrate specific loop (in front of the catalytic residue R-109) of malaria parasite LDH, that was conserved in all plasmodial LDH (F.vivax, P.malariae, P.ovale, P.knowlesi, P.falciparum), was also found to be present in the P.falciparum isolated from Indonesia. The five amino acid residues adjacent to the active site are likely to provide a good target for the rational design of new antimalarial compounds (29).
Cofactor binding in the pLDH, which is characterized by two main conserved interactions of Leu163 and Gly164, was also available in the obtained P. Falciparum pldh. The Leu163 perform acceptor of proton in the hydrogen bond formed with nitrogen of carboxyamidase side chain of nicotinamide. The Gly164 amino acids forms hydrogen bond with a water molecule to cofactor that acts as bridge between the pLDH enzyme and cofactor. In addition, several conserve residues (Ala98, Val26, Phe52, Asp53, and Ile54) which have pivotal role to bind with the adenosin of NADH are also available in the P. falciparumpldh isolated from several islands in Indonesia.
Conclusion
The DNA sequences of P.falciparum pldh isolated from Indonesia are the same with the 3D7 reference pLDH gene, indicating that the pLDH-targeted antimalarial compound would be potentially used to design of new antimalarial agents instead of the chloroquine to control malaria worldwide.
Acknowledgments
We are thankful to Prof Mulyanto (Medical Faculty of Mataram University) for providing the malaria patients’ blood samples and Susilayati S.Si for P. falciparum genomic DNA isolation. This work was supported by grant for International Research Collaboration and Scientific Publication from Directorate General of Higher Education, Ministry of Education and Culture, Indonesia.
Footnotes
The authors declare that there is no conflict of interest.
References
- 1.Snow RW, Guerra CA, Noor AM, Mynt HY, Hay SI. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434(7030):214–7. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McMichael AJ. Global environmental change and human population health: a conceptual and scientific challenge for epidemiology. Int J Epidemiol. 1993;22(1):1–8. doi: 10.1093/ije/22.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Cox J, Hay SI, Abeku TA, Checchi F, Snow RW. The uncertain burden of Plasmodium falciparum epidemics in Africa. Trends Parasitol. 2007;23(4):142–148. doi: 10.1016/j.pt.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO. Malaria Rapid Diagnostic Tests Performance: Results of WHO Product Testing of Malaria RDTs Round 1. France. 2008 [Google Scholar]
- 5.Setiawan B. Current malaria management: guideline 2009. Acta Med Indones-Indones J Intern Med. 2010;42(4):258–261. [PubMed] [Google Scholar]
- 6.WHO. Malaria in the Greater Mekong Subregion: Regional and Country Profiles. India. 2010 [Google Scholar]
- 7.WHO. Malaria. 2012 Available from http://www.who.int/mediacentre/factshee-ts/fs094/en/
- 8.Kain KC, Harrington MA, Tennyson S, Keystone JS. Imported malaria: prospective analysis of problems in diagnosis and management. Clin Infec Dis. 1998;27(1):142–9. doi: 10.1086/514616. [DOI] [PubMed] [Google Scholar]
- 9.Awad-El-Kariem FM, Miles MA, Warhurst DC. Chloroquine resistant Plasmodium falciparum isolates from the Sudan lack two mutations in the pfmdr1 gene though to be associated with chloroqione resistance. Trans R Soc Trop Med Hyg. 1992;86(6):587–9. doi: 10.1016/0035-9203(92)90140-8. [DOI] [PubMed] [Google Scholar]
- 10.Djimde A, Doumbo PK, Cortese JF, Kayentao K, Doumbo S, Diourte Y, Coulibaly D, Dicko A, Su XZ, Nomura T, Fidock DA, Wellems TE, Plowe CV. A molecular marker for chloroquine-resistant falciparum malaria. N Eng J Med. 2001;344(4):257–63. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- 11.Sidhu ABS, Valderramos SG, Fidock DA. Pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol Microbiol. 2005;57(4):913–26. doi: 10.1111/j.1365-2958.2005.04729.x. [DOI] [PubMed] [Google Scholar]
- 12.Anderson T. Mapping the spread of malaria drug resistance. PloS Medicine. 2009;6(4):1–2. doi: 10.1371/journal.pmed.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wongsrichanalai C, Pickard AL, Wernsdorfer WH, Meshnick SR. Epidemiology of drug-resistant malaria. Lancet Infect Dis. 2002;2(4):209–18. doi: 10.1016/s1473-3099(02)00239-6. [DOI] [PubMed] [Google Scholar]
- 14.Shoemark DK, Cliff MJ, Session RB, Clarke AR. Enzymatic properties of lactate dehidrogenase enzyme for Plasmodium falciparum. FEBS J. 2007;274(4):2738–2748. doi: 10.1111/j.1742-4658.2007.05808.x. [DOI] [PubMed] [Google Scholar]
- 15.Penna-Coutinho J, Cortopassi WA, Oliveira AA, Franca TC, Krettli AU. Antimalarial activity of potential inhibitors of Plasmodium falciparum lactate dehidrogenase enzyme selected by docking studies. PloS ONE. 2011;6(7):e21237. doi: 10.1371/journal.pone.0021237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson TJ, Su XZ, Bockarie M, Lagog M, Day KP. Twelve microsatelite markers for characerization of Plasmodium falciparum from finger-prick blood samples. Parasitol. 1999;119(Pt 2):113–25. doi: 10.1017/s0031182099004552. [DOI] [PubMed] [Google Scholar]
- 17.Warhurst DC, William JE. Laboratory diagnosis of malaria. J Clin Pathol. 1996;49(7):533–38. doi: 10.1136/jcp.49.7.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boonma P, Christensen PR, Suwanarusk R, Price RN, Russell B, Le-Uthai U. Comparison of three molecular methods for the detection and specification of P. vivax and P. falciparum. Malar J. 2007;6:124. doi: 10.1186/1475-2875-6-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopes D, Rungsihirunrat K, Nogueira F, Seugorn A, Gil JP, do Rosario VE, Cravo P. Molecular characterization of drug-resistant Plasmodium falciparum from Thailand. Malar J. 2002;1(12):1–11. doi: 10.1186/1475-2875-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatabu T, Iwagami M, Kawazu S, Taguchi N, Escueta AD, Villacorte EA, Rivera PT, Kano S. Association of molecular markers in Plasmodium falciparum crt and mdr1 with in vitro chloroquine resistance: A Philippine study. Parasitol Intern. 2009;58(2):166–70. doi: 10.1016/j.parint.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 21.Singh V, Kaushal DC, Rathaur S, Kumar N, Kaushal NA. Cloning, overexpression purification, and characterization of Plasmodium knowlesi lactate dehidrogenase. Protein Exp Purif. 2012;84(2):195–203. doi: 10.1016/j.pep.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 22.Scopel KKG, Fontes CJF, Nunes AC, Horta MdF, Braga EM. Low sensitivity of nested PCR using Plasmodium DNA extracted from stained thick blood smears: an epidemiological retrospective study among subjects with low parasitemia in an endemic area of the Brazilian Amazon region. Malar J. 2004;3:8. doi: 10.1186/1475-2875-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wellem T, Plowe C. Chloroquine-resistant malaria. J Infect Dis. 2001;184(6):770–6. doi: 10.1086/322858. [DOI] [PubMed] [Google Scholar]
- 24.Syafruddin D, Asih PBS, Anggarwal SL, Shankar AH. Frequency distribution of antimalarial drug-resistant alleles among isolates of Plasmodium falciparum in Purworejo District, Central Java Province, Indonesia. Am J Trop Med Hyg. 2003;69(6):614–20. [PubMed] [Google Scholar]
- 25.Andriantsoanirina V, Ratsimbasoa A, Bouchier C, Tichit M, Jahevitra M, Rabearimanana S, Raherinafy R, Mercereau-Puijalon O, Durand R, Menard D. Chloroquine clinical failures in Plasmodium falciparum malaria are associated with mutant pfmdr-1, not pfcrt in Madagascar. PLoS One. 2010;5:e13281. doi: 10.1371/journal.pone.0013281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Figueiredo P, Benchimol C, Lopez D, Bernardino L, Rosario VED, Nogueira Varandas L. Prevalence of pfmdr1, pfcrt, pfdhfr and pfdhps mutations associated with drug resistance, in Luanda, Angola. Malar J. 2008;7:236. doi: 10.1186/1475-2875-7-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bzik DJ, Fox BA, Gonyer K. Expression of Plasmodium falciparum lactate dehydrogenase in Escherichia coli. Mol Biochem Parasitol. 1993;59(1):155–66. doi: 10.1016/0166-6851(93)90016-q. [DOI] [PubMed] [Google Scholar]
- 28.Brown WM, Yowell CA, Hoard A, Jagt TAV, Hunsaker LA, Deck LM, Royer RE, Piper RC, Dame JB, Makler MT, Jagt DLV. Comparative structural analysis and kinetic properties of lactate dehydrogenases from the four species of human malaria parasites. Biochemistry. 2004;43(20):6219–29. doi: 10.1021/bi049892w. [DOI] [PubMed] [Google Scholar]
- 29.Balik DT, Holbrook JJ. Determination of the DNA and amino acid sequences of the lactate dehydrogenase gene from Plasmodium falciparum strain K1 and PF FCBR: a route to the design of new antimalarials. Turk J Biol. 2001;25(5):241–50. [Google Scholar]


