Abstract
There is an ongoing debate about the clinical significance of Sphingomonas paucimobilis as a virulent bacterial pathogen. In the present study, we investigated the presence of different virulence factors and genes in Sphingomonas bacteria. We utilized phylogenetic, comparative genomics and bioinformatics analysis to investigate the potentiality of Sphingomonas bacteria as virulent pathogenic bacteria. The 16S ribosomal RNA gene (16S rDNA) phylogenetic tree showed that the closest bacterial taxon to Sphingomonas is Brucella with a bootstrap value of 87 followed by Helicobacter, Campylobacter, Pseudomonas, and then Legionella. Sphingomonas shared no virulence factors with Helicobacter or Campylobacter, despite their close phylogenic relationship. In spite of the phylogenetic divergence between Sphingomonas and Pseudomonas, they shared many major virulence factors, such as adherence, antiphagocytosis, iron uptake, proteases, and quorum sensing. In conclusion, Sphingomonas spp. contains several major virulence factors resembling Pseudomonas sp., Legionella sp., Brucella sp., and Bordetella sp. virulence factors. Similarity of virulence factors did not match phylogenetic relationships. These findings suggest horizontal gene transfer of virulence factors rather than sharing a common pathogenic ancestor. Sphingomonas spp. is potential virulent bacterial pathogen.
Keywords: Sphingomonas spp, virulence factors, phylogenetics, comparative genomics, bioinformatics, Pseudomonas sp
Introduction
Sphingomonas is a group of Gram-negative, rod-shaped, non-spore-forming, chemoheterotrophic, strictly aerobic bacterium that produces yellow or off-white pigmented colonies. The distinctiveness of Sphingomonas lies in its possession of ubiquinone 10 as its major respiratory quinone, presence of glycosphingolipids (GDLs) in their cell envelopes, and its metabolic versatility.1 The genome of Sphingomonas is approximately 3,948 kB and contains 70 structural RNAs. It encodes for approximately 3,914 proteins.2 Sphingomonas utilize glucose as its primary source of carbon. However, it can also utilize a wide variety of other sugars such as arabinose, fucose, glactose, lactose, mannose, melibiose, sucrose, trehalose, and xylose. Moreover, Sphingomonas can also degrade polysaccharides, and many of the strains were also able to utilize one or more of the contaminants as their source of carbon.2 The capability of Sphingomonas to utilize a wide range of organic compounds, and to grow and survive under low-nutrient conditions has resulted in its widespread distribution in various environments, including drinking water, soil, air, sinks, shower curtains, and corroding copper pipes. In addition, Sphingomonas paucimobilis has also been found in several clinical specimens, which include hospital water supplies; temperature probe respirators; stocked distilled water; blood; removes; hospital dialysis equipment; patients with meningitis, septicemia, bacteremia, and peritonitis; and wound infections.3 Reported cases of nosocomial infections caused by S. paucimobilis are rarely serious and could be effectively treated with antibiotics. On the contrary, some other reports have concluded that S. paucimobilis nosocomial infections have the ability to severely threaten immune-compromised or ill patients causing health problems with consistent exposure to the source of infection such as shower curtains.4 For example, in 2007, reports concluded that S. paucimobilis was the cause of bacteremia outbreak in the hemato/oncology units in Gülhane Military Hospital in Ankara, Turkey. The reports also showed that the clinical isolates were traced back neither to the health care workers nor to the environmental isolates.5 Moreover, it was strongly documented that S. paucimobilis created significant problems in various clinical settings, being the most widespread cause of nosocomial infections including bacteremia/septicemia caused by contaminated solutions such as distilled water, hemodialysis fluid, and sterile drug solutions. Cases of pseudo-bacteremia have been recorded in association with S. paucimobilis, as have many cases of unusual infections both invasive and severe, eg, septic arthritis and osteomyelitis.6 Moreover, S. paucimobilis caused bloodstream infection in a patient with Down syndrome. It was thereby concluded that S. paucimobilis should be recognized as a nosocomial infectious agent in patients with Down syndrome and immunosuppressive disorders.7 In addition, it was also reported that S. paucimobilis has the ability to cause infections in both previously healthy and immune-compromised children8 and can act as a causal agent of osteomyelitis in an immune-competent patient.9 Frequent S. paucimobilis infections were observed among our diabetic foot ulcer patients (23% of observed Gram-negative infections). S. paucimobilis general infection rate was 9.5%, falling just behind Staphylococcus aureus (unpublished data). There is still an ongoing debate about the clinical virulence of S. paucimobilis with a possible conclusion that its clinical importance cannot be neglected. Henceforth, this study employs comparative genomics and bioinformatics techniques in order to investigate the pathogenic potentials of Sphingomonas spp.
Materials and Methods
Phylogenetic relationships reconstruction
Partial 16S rDNA sequences of selected pathogenic bacteria, namely, S. paucimobilis (D16144.1), Bacillus anthracis (GQ280034.1), Bartonella bacilliformis (AF442955.1), Brucella sp. (DQ413258.1), Burkholderia sp. (AB379686.1), Campylobacter jejuni (AY621112.1), Clostridium difficile (HM245939.1), Corynebacterium sp. (D83375.1), Escherichia coli (KC504012.1), Haemophilus sp. (AB004027.1), Helicobacter sp. (AY034821.1), Legionella pneumophila (NR_041742.1), Neisseria meningitidis (AF059671.1), Pseudomonas sp. (AB379690.1), Salmonella typhimurium (DQ153191.1), Shigella spp. (JN626189.1), Vibrio cholerae (Z21856.1), Yersinia pestis (AJ232235.1), were all acquired from the GenBank. These sequences were then aligned using the Bioedit built-in clustal W program (gap opening penalty = 10, gap extension penalty = 5, delay divergent sequences = 40%). The resulting alignments were compared, and the final alignments were improved manually and prepared in FASTA and MEGA formats using format converter tool v2.2.5 available online at http://www.hiv.lanl.gov/content/sequence/FORMAT_CONVERSION/form.html.
In order to establish the phylogenetic relationships among taxa, tree was constructed using the maximum likelihood (ML) method based on the Tamura–Nei model.10 The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search was(were) obtained automatically by applying Neighbor-Joining and BIONJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach, and then the topology was selected with superior log-likelihood value. The tree was drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 18 nucleotide sequences. Codon positions included were first + second + third + noncoding. All positions containing gaps and missing data were eliminated. There were a total of 253 positions in the final dataset. Evolutionary analyses were conducted in MEGA6.11
Comparative genomics and bioinformatics analysis
Virulence genes sequences and functions, corresponding to different major bacterial virulence factors of selected pathogens, were collected from GenBank and validated in virulence factors of pathogenic bacteria database at http://www.mgc.ac.cn/VFs/. Supplementary Table 1 shows the tested major pathogenic virulence factors. Selected gene sequences were tested against available Sphingomonas gene information using Sphingomonas nucleotide BLAST tool available at http://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch&PROG_DEF=blastn&BLAST_PROG_DEF=megaBlast&BLAST_SPEC=MicrobialGenomes_13687&DB_GROUP=AllMG. The search set all Sphingomonas complete genomes, and selected organism was Sphingomonas (tax Id: 13687). The program selected for the search was blastn, optimizes for fairly similar sequences because of evolutionary divergence of the tested and query taxa.
Results and Discussion
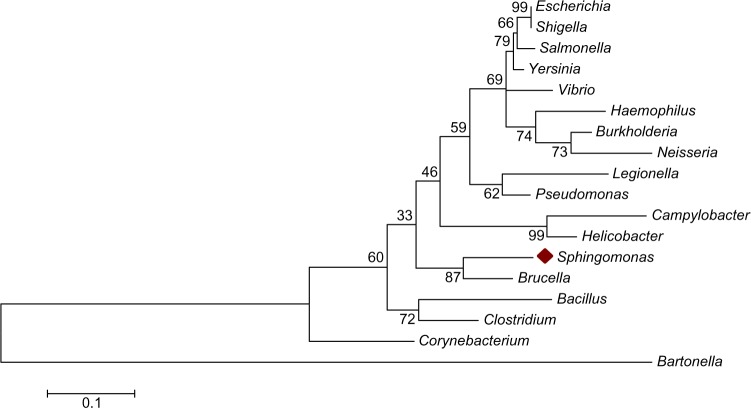
In this study, the presence of the major known bacterial virulence factors in Sphingomonas spp. was examined. In order to decide on the accurate common pathogenic bacterial species for comparison with Sphingomonas spp., a phylogenetic tree using the ML method was constructed using partial 16S rDNA sequences of selected pathogenic bacteria as mentioned above (Fig. 1). The phylogenetic tree showed that the selected bacterial species are divided into two major clades (groups), namely, Gram-positive bacteria and Gram-negative bacteria. The Gram-positive bacterial group contained Clostridium spp., Corynebacterium sp., and Bacillus spp., whereas the Gram-negative bacterial group contained the remaining bacterial species. These results also agree with the known taxonomic arrangement of the tested bacterial species with the exception of Bartonella spp. that accrued in an independent clad outside the Gram-negative bacteria. More importantly, the 16S rDNA phylogenetic tree showed Brucella sp. to be the closest bacterial taxon to Sphingomonas with a bootstrap value of 87 followed by Helicobacter spp., Campylobacter sp., Pseudomonas sp., and then Legionella sp. Based on the suggested phylogenetic relationships, the following bacterial species which include Brucella sp., Helicobacter spp., Campylobacter sp., Pseudomonas sp., and Legionella sp. were selected for further comparative genomic and bioinformatics analyses. Table 1 presents the five selected bacterial genera with its corresponding species, the selected hosts, and the diseases caused by these pathogens. The selected pathogens were mainly human pathogens having also the ability to infect mammals, protozoa (Legionella sp.), and plants (Pseudomonas syringae). All the virulent factors acquired by these pathogens (Table 2) were tested for their presence in Sphingomonas genomic information. The major categories of bacterial virulence factors such as adherence, endotoxin, mobility, secretion systems, quorum sensing, and many others are shown in (Table 2).
Figure 1.
Partial 16SrDNA-based Maximum Likelihood (ML) phylogenetic tree for a major pathogenic bacterial taxa utilizing base substitution Tamura–Nei model.
Table 1.
Major pathogenic taxa used in the comparative analysis against Sphingomonas spp.
| GENUS | SPECIES | HOST | DISEASE |
|---|---|---|---|
| Brucella | B. abortus | Human and cattle | Brucellosis, Osteoarthritis, endocarditis and several neurological disorders. |
| B. canis | Human and dogs | ||
| B. melitensis | Human goats and sheep | ||
| B. ovis | Sheep | ||
| B. suis | Human and pigs | ||
| Helicobacter | H. acinonychis | Humans and other mammals | Bacterial carcinogen, Gastroduodenal diseases |
| H. hepaticus | |||
| H. pylori | |||
| Campylobacter | C. fetus | Humans | Bacterial gastroenteritis Guillain-Barre syndrome (GBS) |
| C. jejuni | |||
| Legionella | L. pneumophila | Humans and protozoa | Legionnaires’ disease |
| Pseudomonas | P. aeruginosa | Human | Eye, burn and wound infections |
| P. syringae | Plant | Bacterial speck and bacterial blight |
Table 2.
Major pathogenic virulence factors and its corresponding genes and functions used in the comparative analysis against Sphingomonas spp.
| GENUS | MAJOR TESTED VIRULENCE FACTORS | CORRESPONDING TESTED GENES OR FUNCTION |
|---|---|---|
| Brucella | Immune-evasion | Btp1/TcpB (Toll-interleukin-1 receptor (TIR) domain) |
| Intracellular survival | Cyclic β-1,2-glucan synthase (Cgs), gmd; manA; manC; per; pgm; pmm/manB; wbkA; wbkB; wbkC; wzm; wzt; and RicA (Rab2 interacting conserved protein A) | |
| Regulation | BvrR-BvrS two-component system | |
| Secretion system | BMEII0025; BMEII0026; BMEII0027; BMEII0028; BMEII0029; BMEII0030; BMEII0031; BMEII0032; BMEII0033; BMEII0034; BMEII0035; type IV secretion system | |
| Helicobacter | Adherence | babA; babB; hopZ; sabA; |
| Endotoxin | gluE; gluP; kdtB; lpxB; rfaC; rfaJ; rfbD; rfbM; wbcJ; wbpB; | |
| Enzyme | ureA; ureB; ureE; ureF; ureG; ureH; ureI; | |
| Molecular mimicry | fucT; fucU; neuA; neuB; | |
| Motility | flaA; flaB; flgG | |
| Proinflammatory effect | napA; oipA; | |
| Secretion system | cag1; cag10; cag11; cag12; cag13; cag14; cag15; cag16; cag17; cag18/cagL; cag19; cag2; cag20; cag21; cag22; cag23 (cagE/picB); cag24; cag25; cag3; cag4; cag5 (virD4); cag6; cag7; cag8; cag9; virB11; type IV secretion system. | |
| Toxin | vacA | |
| Type IV secretory protein | cagA | |
| Pathogenicity islands | cag-PAI | |
| Campylobacter | Adherence | cadF; Cj1415c; Cj1416c; Cj1421c; Cj1422c; Cj1423c; Cj1425c; Cj1426c; Cj1427c; Cj1429c; Cj1430c; Cj1431c; Cj1432c; Cj1433c; Cj1434c; Cj1435c; Cj1436c; Cj1437c; Cj1438c; Cj1440c; Cj1442c; fcl; glf; gmhA2; kfiD; kpsC; kpsD; kpsE; kpsF; kpsM; kpsS; kpsT; Cj0983; Cj1135; Cj1136; Cj1137c; Cj1138; Cj1139c; Cj1140; Cj1144c; Cj1145c; gmhA; htrB; neuA1; neuB1; neuC1; waaC; waaD; waaE; waaF; waaV; porA; peb1 A. |
| Invasion | ciaB, ciaC (invasion antigens) | |
| Motility | Cj0371; Cj1312; Cj1313; flaA; flaB; flaC; flaD; flaG; flgB; flgC; flgD; flgE; flgE2; flgG; flgG2; flgH; flgI; flgK; flhA; flhB; flhF; fliA; fliD; fliE; fliF; fliG; fliH; fliI; fliL; fliM; fliN; fliP; fliQ; fliR; fliS; fliY; motA; motB; pflA; ptmA; ptmB. | |
| Secretion system | Cjp54; virB10; virB11; virB4; virB8; virB9; virD4; type IV secretion system. | |
| Toxin | cdtA; cdtB; cdtC; | |
| Legionella | Adherence | htpB; omp28; pilB; pilC; pilD; |
| Enzyme | mip; | |
| Iron uptake | ccmC; iraaB; frgA; feoA; feoB; | |
| Motility | flaA; flgA; flgB; flgC; flgD; flgE; flgF; flgG; flgH; flgI; flgK; flgL; flhA; flhB; flhF; fliA; fliD; fliE; fliF; fliG; fliH; fliJ; fliM; fliN; fliO; fliP; fliQ; fliR; fliS; motA; motB; | |
| Nutrient acquisition | phtA; | |
| Regulation | csrA; letA; letS; relA; rpoS; | |
| Secretion system | lspD; lspE; lspF; lspG; lspH; lspI; lspJ; lspK; lspL; lspM; pilD; (Type II secretion system). dotA; dotB; dotC; dotD; icmB; icmC; icmD; icmE; icmF; icmG; icmH; icmJ; icmK; icmL; icmM; icmN; icmO; icmP; icmQ; icmR; icmS; icmT; icmV; icmW; icmX; lepA; lepB; lidA; lvgA; ralF; sdeA/laiA; sdeB; sdeC; sdeD; sidA; sidB; sidC; sidE; sidF; sidG; sidH; vipA; vipD; vipE; vipF; wipA; wipB; wipC; ylfA; ylfB (type IV secretion system) | |
| Stress protein | gspA (global stress gene) katA; katB; sodB; sodC; | |
| Toxin | RtxA | |
| Unclassified | enhA; enhB; enhC; ligA; | |
| Pseudomonas | Adherence | fleN; fleQ; fleR; flgC; flgD; flgE; flgF; flgG; flgH; flgI; flgJ; flgK; flgL; flhA; flhB; flhF; fliC; fliD; fliE; fliF; fliG; fliH; fliI; fliJ; fliM; fliN; fliO; fliP; fliQ; fliR; waaA; waaC; waaF; waaG; waaP; wzy; wzz; chpA; chpB; chpC; chpD; chpE; fimT; fimU; fimV; pilA; pilB; pilC; pilD; pilE; pilF; pilG; pilH; pilI; pilJ; pilK; pilM; pilN; pilO; pilP; pilQ; pilR; pilS; pilT; pilU; pilV; pilW; pilX; pilY1; pilY2; |
| Antiphagocytosis | Alg44; Alg8; algA; algB; algD; algE; algF; algG; algI; algJ; algK; algL; algP; algQ; algR; algU; algX; algZ; mucA; mucB; mucC; | |
| Biosurfactant | rhlA; rhlB; | |
| Iron uptake | fptA; pchA; pchB; pchC; pchD; pchE; pchF; pchG; pchH; pchI; pchR; fpvA; pvdA; pvdD; pvdE; pvdS; | |
| Pigment | phzM; phzS (Pyocyanin) | |
| Protease | aprA; lasA; lasB. | |
| Regulation | lasI; lasR; rhlL; rhlR; | |
| Secretion system | xcpP; xcpQ; xcpR; xcpS; xcpT; xcpU; xcpV; xcpW; xcpX; xcpY; xcpZ (Type II secretion system) | |
| Toxin | toxA; exoS; exoT; exoU; exoY; plcH; |
Table 3 and Figure 2 present the shared virulence factors among Sphingomonas and the selected five bacterial pathogens. Results in Table 3 showed that Sphingomonas spp. shares the genes responsible for intracellular survival ability (Cgs, manC, and pgm) with Brucella sp. with e-values ranging from 0 to 3.00E − 09.12–14 In addition, Sphingomonas spp. shares the genes encoding for Type IV secretion system such as BMEII0026 with Brucella sp. with e-value of 6.00E − 04 and identity similarity of 90%. On the contrary, Sphingomonas spp. shared no virulence factors with Helicobacter spp. or Campylobacter sp., despite their close phylogenic relationship when compared to Pseudomonas sp. and Legionella sp. Sphingomonas spp. shared Legionella sp. genes responsible for adherence and motility, namely, htpB and flip. Moreover, they also shared the gene responsible for stress tolerance and sodB. The sodB encodes for superoxide dismutase, which is a cytoplasmic iron superoxide dismutase important for intracellular survival and transmission.15
Table 3.
Comparative analysis against Sphingomonas spp. against major bacterial virulence factors and functions from different pathogenic bacteria.
| BACTERIAL TAXA | MAJOR BACTERIAL VIRULENCE FACTORS (VFS) | SUB VFS | RELATED GENE | E VALUE | IDENT | Sphingomonas GENBANK ACCESSION |
|---|---|---|---|---|---|---|
| Brucella | Intracellular survival and Immuno-modulatory activity | CβG (cyclic β-1,2 glucan) | cgs | 6.00E-59 | 66% | CP006644.1 |
| Mannose-1-phosphate guanylyltransferase | manC | 3.00E-09 | 77% | NC_009511.1 | ||
| Phosphoglucomutase | pgm | 0 | 74% | NC_009511.1 | ||
| Secretion system | VirB type IV secretion system | BMEII 0025 | 4.00E-04 | 83% | CP006644.1 | |
| VirB type IV secretion system | BMEII 0026 | 6.00E-04 | 90% | NC_020561.1 | ||
| VirB type IV secretion system | BMEII 0035 | 4.00E-06 | 72% | NC_020561.1 | ||
| Legionella | Adherence | Hsp60 | htpB | 2.00E-25 | 64% | NC_020561.1 |
| Motility | Flagella | fliP | 4.00E-11 | 68% | NC_020561.1 | |
| Stress protein | SodB | sodB | 3.00E-05 | 82% | NC_020561.1 | |
| Pseudomonas | Adherence | Flagella | fleQ | 4.00E-65 | 72% | NC_009511.1 |
| flgE | 5.00E-19 | 67% | NC_009511.1 | |||
| flgF | 7.00E-14 | 71% | NC_009511.1 | |||
| flgG | 4.00E-55 | 68% | NC_020561.1 | |||
| flgH | 4.00E-29 | 72% | NC_020561.1 | |||
| flgI | 6.00E-93 | 68% | NC_020561.1 | |||
| flgJ | 6.00E-05 | 89% | NC_020561.1 | |||
| flgK | 1.00E-04 | 94% | NC_009511. | |||
| flhA | 2.00E-141 | 69% | NC_020561.1 | |||
| flhB | 1.00E-18 | 68% | NC_020561.1 | |||
| flhF | 6.00E-05 | 89% | NC_020561.1 | |||
| fliC | 1.00E-32 | 72% | NC_009511.1 | |||
| fliE | 6.00E-04 | 74% | NC_020561.1 | |||
| fliF | 3.00E-04 | 66% | NC_020561.1 | |||
| fliG | 8.00E-15 | 69% | NC_020561.1 | |||
| fliH | 4.00E-05 | 85% | CP006644.1 | |||
| fliI | 1.00E-115 | 71% | NC_009511. | |||
| fliN | 3.00E-16 | 70% | NC_009511.1 | |||
| fliP | 9.00E-102 | 75% | NC_020561. | |||
| fliQ | 2.00E-08 | 84% | NC_020561.1 | |||
| fliR | 7.00E-08 | 72% | NC_020561.1 | |||
| GENUS | MAJOR VFS | SUB VFS | RELATED GENE | E VALUE | IDENT | ACCESSION |
| Pseudomonas | Adherence | LPS (lipopolysaccharide) | waaA | 2.00E-04 | 89% | CP006644.1 |
| waaG | 4.00E-07 | 76% | NC_020561.1 | |||
| waaP | 1.00E-04 | 83% | NC_020561.1 | |||
| Type IV pili | chpA | 7.00E-26 | 67% | NC_020561.1 | ||
| chpC | 8.00E-05 | 82% | NC_020561.1 | |||
| chpE | 3.00E-05 | 86% | CP006644.1 | |||
| fimU | 3.00E-04 | 93% | NC_009511.1 | |||
| pilB | 3.00E-04 | 93% | NC_009511.1 | |||
| pilD | 4.00E-05 | 76% | CP006644.1 | |||
| pilF | 4.00E-05 | 97% | CP006644.1 | |||
| pilG | 2.00E-04 | 80% | NC_020561.1 | |||
| pilH | 2.00E-04 | 77% | NC_009511.1 | |||
| pilI | 7.00E-06 | 84% | CP006644.1 | |||
| pilJ | 5.00E-21 | 76% | NC_020561.1 | |||
| pilK | 3.00E-06 | 83% | NC_020561.1 | |||
| pilN | 8.00E-04 | 82% | CP006644.1 | |||
| pilQ | 1.00E-09 | 73% | NC_009511.1 | |||
| pilR | 9.00E-54 | 70% | NC_009511.1 | |||
| pilS | 6.00E-06 | 74% | NC_020561.1 | |||
| pilT | 1.00E-07 | 83% | NC_009511.1 | |||
| pilU | 1.00E-12 | 75% | NC_009511.1 | |||
| pilV | 2.00E-06 | 72% | CP006644.1 | |||
| pilW | 1.00E-05 | 88% | NC_009511.1 | |||
| Antiphagocytosis | Alginate | Alg 44 | 7.00E-04 | 85% | NC_020561.1 | |
| algA | 1.00E-21 | 65% | NC_009511.1 | |||
| algB | 2.00E-36 | 69% | NC_009511.1 | |||
| algD | 2.00E-23 | 72% | CP006644.1 | |||
| algF | 3.00E-05 | 91% | CP006644.1 | |||
| algG | 8.00E-05 | 85% | CP006644. | |||
| algI | 1.00E-59 | 67% | NC_009511.1 | |||
| algJ | 7.00E-04 | 94% | NC_009511.1 | |||
| algP | 1.00E-07 | 80% | CP006644.1 | |||
| algZ | 5.00E-05 | 91% | CP006644.1 | |||
| Pseudomonas | Biosurfactant | Rhamnolipid | rhlA | 5.00E-04 | 89% | NC_009511.1 |
| Iron uptake | Pyochelin | fptA | 6.00E-08 | 86% | NC_009511.1 | |
| pchA | 8.00E-04 | 93% | NC_009511.1 | |||
| pchB | 6.00E-04 | 67% | NC_020561.1 | |||
| pchC | 1.00E-04 | 80% | NC_009511.1 | |||
| pchD | 1.00E-09 | 67% | NC_009511.1 | |||
| pchE | 5.00E-12 | 94% | CP006644.1 | |||
| pchF | 9.00E-17 | 79% | CP006644.1 | |||
| pchG | 1.00E-05 | 79% | NC_009511.1 | |||
| pchH | 2.00E-12 | 75% | CP006644.1 | |||
| pchI | 3.00E-16 | 74% | NC_020561.1 | |||
| Pyoverdine | fpvA | 3.00E-12 | 68% | NC_020561.1 | ||
| pvdA | 6.00E-05 | 85% | NC_020561.1 | |||
| pvdD | 6.00E-33 | 67% | CP006644.1 | |||
| pvdE | 7.00E-06 | 82% | NC_009511.1 | |||
| Pigment | Pyocyanin | phzM | 5.00E-05 | 77% | NC_009511.1 | |
| Pyocyanin | phzS | 6.00E-05 | 68% | NC_009511.1 | ||
| Protease | Alkaline protease | aprA | 2.00E-12 | 79% | CP006644.1 | |
| LasA | lasA | 2.00E-04 | 86% | NC_020561.1 | ||
| LasB (Elastase) | lasB | 9.00E-04 | 86% | CP006644.1 | ||
| Regulation | Quorum sensing | rhlL | 3.00E-04 | 93% | NC_009511.1 | |
| rhlR | 5.00E-09 | 72% | NC_009511.1 | |||
| Secretion system | xcp secretion system | xcpQ | 2.00E-46 | 70% | NC_009511.1 | |
| xcpR | 3.00E-174 | 72% | NC_009511.1 | |||
| xcpS | 1.00E-44 | 66% | NC_009511.1 | |||
| xcpT | 3.00E-23 | 69% | NC_009511.1 | |||
| xcpU | 2.00E-07 | 78% | NC_020561.1 | |||
| xcpW | 4.00E-04 | 73% | NC_009511.1 | |||
| xcpX | 1.00E-05 | 88% | NC_020561.1 | |||
| Toxin | PLC (Phospholipase C) | plcH | 6.00E-33 | 68% | CP006644.1 |
Figure 2.
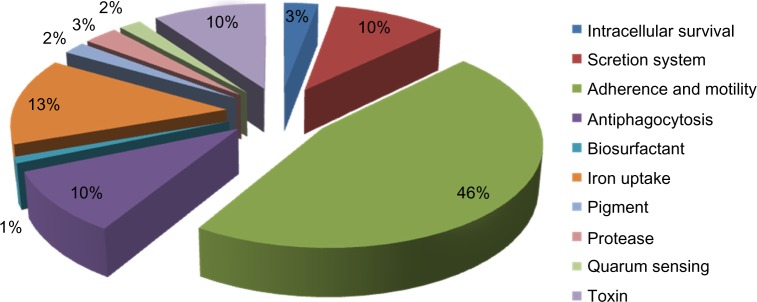
Percentage of different virulence factors associated with Sphingomonas spp.
Regardless of the phylogenetic divergence between Sphingomonas spp. and Pseudomonas sp., it was observed from our results that they shared several major virulence factors such as adherence, antiphagocytosis, iron uptake, proteases, quorum sensing, and others. With regard to adherence, they shared 20 genes, including flgK with e-value of 1.00E − 04 and identity similarity of 94%, and flgJ with e-value of 6.00E − 05 and identity similarity of 89%. Flagella plays an important role as a virulence factor such as in swimming motility toward the infection site and a role in biofilm formation and other pathogenic adaptations.16–20 Type IV pili play an important role in adherence by assisting the pathogens to attach with their host cells and causing a twitching motility that allows the bacteria to move along the cell surface, and in biofilm formation.17,21–25 Both Sphingomonas spp. and Pseudomonas sp. shared many genes implicated in Type IV pili biogenesis and mechanical function of pili, such as pilF with e-value of 4.00E − 05 and identity similarity of 97%, and pilB with e-value of 3.00E − 04 and identity similarity of 93%.
Furthermore, Sphingomonas spp. and Pseudomonas sp. shared many genes implicated in antiphagocytosis through the production of alginate. They shared 10 alginate genes, including algJ with e-value of 7.00E − 04 and identity similarity of 94%, and algZ with e-value of 5.00E − 05 and identity similarity of 91%. The production of alginate permits pathogenic bacteria to form biofilm and contributes to the persistence of bacteria in the lung by acting as an adhesin, which prevents the bacteria from being expelled from the infection site, and the alginate slime layer makes it more difficult for phagocytes to ingest and kill the bacteria.26–30 Another important bacterial virulence factor shared between Sphingomonas spp. and Pseudomonas sp. is quorum sensing. Sphingomonas spp. showed acquiring of both rhlL and rhlR with e-values of 3.00E − 4 and 5.00E − 9, respectively. These results show that Sphingomonas spp. possesses only rhl system of quorum sensing. While in Pseudomonas sp., quorum sensing consists of two separate but interrelated systems, namely, las and rhl, which are found to regulate the production of multiple virulence factors and are also crucial for proper biofilm formation.31–33
It is also worth mentioning that both Sphingomonas spp. and Pseudomonas sp. share seven genes encoding for xcp secretion system (Type II secretion system), including xcpX and xcpR with e-values of 1.00E − 5 and 3.00E − 174, respectively. The xcp secretion system is found to be responsible for secretion of toxins and enzymes into the extracellular fluid.34,35 It was also observed that both Sphingomonas spp. and Pseudomonas sp. shared many genes involved in iron uptake using both pyochelin (10 genes) and pyoverdine (4 genes). The pyochelin is effective at promoting iron uptake in Pseudomonas aeruginosa, catalyzes the formation of tissue-damaging free radicals, and also binds other transition metals (eg, Mo(IV), Co(II)) with appreciable affinity, and is also implicated in the delivery of both Co(II) and Mo(IV) to P. aeruginosa cells.36,37 The pyoverdine is effective at acquiring iron from transferrin and lactoferrin. Moreover, pyoverdine is cytotoxic because of its ability to stimulate the production of reactive oxygen species.38,39 Interestingly, Sphingomonas spp. and Pseudomonas sp. shared only one gene responsible for toxin production, namely, plcH that is responsible for degrading the phospholipids surfactant, which functions to reduce the surface tension so that the alveoli do not collapse completely when air leaves them during breathing. It is noteworthy that sphingomyelin (PlcH is a multifunctional enzyme) has been isolated from Pseudomonas aeruginosa in 2003.40,41
Our comparative analysis was further extended to search for other bacterial toxins that Sphingomonas spp. may acquire. Table 4 shows other toxins that were found to be shared between Sphingomonas spp. and Bordetella pertussis, which is a Gram-negative species and strictly aerobic coccobacilli. B. pertussis is a strict human pathogen causing whooping cough, a highly contagious respiratory disease marked by severe, spasmodic coughing episodes.42 It was also observed that Sphingomonas spp. contains genes for invasive adenylate cyclase/hemolysin, cyclolysin secretion protein, which is a bifunctional toxin harboring both adenylate cyclase and hemolytic activities, and functions primarily as an anti-inflammatory factor.43–45 Moreover, Sphingomonas spp. contains genes responsible for pertussis toxin and its secretion system, which assists in the attachment of B. pertussis to ciliated respiratory cells, important immunogen and activate cyclic adenosine phosphate (cAMP), histamine sensitizing factor (HSF), lymphocytosis promoting factor (LPF), islet-activating protein (IAP); interferes with leucocyte function; and is hemolytic.46,47
Table 4.
Suggested Sphingomonas spp. toxin information in relation to Bordetella pertussis toxins.
| BORDETELLA TOXIN | RELATED GENE | PRODUCT NAME | BORDETELLA PERTUSSIS TOHAMA I | Sphingomonas SPP. | ||
|---|---|---|---|---|---|---|
| PROTIEN GENBANK ID | E VALUE | IDENT | ACCESSION | |||
| Cya (Invasive Adenylate cyclase/haemolysin) | cyaA | Bifunctional hemolysin-adenylate cyclase precursor | 33591934 | 1.00E-21 | 79% | CP006644.1 |
| cyaD | Cyclolysin secretion protein | 33591936 | 8.00E-04 | 81% | NC_009511.1 | |
| cyaE | Cyclolysin secretion protein | 33591937 | 2.00E-04 | 89% | NC_009511.1 | |
| Ptx (Pertussis toxin) | ptlA | Pertussis toxin transport protein | 33594643 | 6.00E-04 | 84% | CP006644.1 |
| ptlC | Putative bacterial secretion system protein | 33594645 | 3.00E-06 | 88% | NC_009511.1 | |
| ptlD | Putative membrane protein | 33594646 | 8.00E-04 | 78% | CP006644. | |
| ptlF | Putative bacterial secretion system protein | 33594649 | 5.00E-04 | 76% | NC_009511.1 | |
| ptlH | Putative bacterial secretion system protein | 33594651 | 6.00E-10 | 71% | CP006644.1 | |
| ptxA | Pertussis toxin subunit 1 precursor | 33594638 | 1.00E-04 | 83% | CP006644.1 | |
Conclusion
Results of this study showed that Sphingomonas spp. contains several major virulence factors, mainly resembling those of Pseudomonas sp. Other virulence factors from Legionella sp., Brucella sp., and Bordetella sp. have also been observed. Moreover, the similarity of virulence factors did not correspond to the phylogenetic relationships. These findings suggest horizontal gene transfer of virulence factors rather than sharing a common pathogenic ancestor.
Highlights.
We selected Sphingomonas spp. to test its potentiality for being potential virulent pathogen.
We tested the phylogenetic relationship of Sphingomonas spp. against known virulent pathogens.
We screened the presence of various virulent factors from different virulent pathogens in Sphingomonas spp.
Sphingomonas spp. contains several major virulence factors, mainly resembling those of Pseudomonas sp.
Supplementary Material
Supplementary Table 1. This table shows the tested major pathogenic virulence factors.
Acknowledgments
We like to thank all members of the Information Technology Department in Strategist Center for Diabetes Research, College of Medicine, King Saud University for facilitating the conduction of the data analysis.
Glossary
Abbreviations
- GDLs
glycosphingolipids
- 16S rDNA
16S ribosomal RNA gene
- NJ
Neighbor-Joining
- MCL
maximum composite likelihood
- tax ID
taxon identity
- cAMP
cyclic adenosine phosphate
- HSF
histamine sensitizing factor
- LPF
lymphocytosis promoting factor
- IAP
islet-activating protein
Footnotes
ACADEMIC EDITOR: Jike Cui, Associate Editor
FUNDING: Authors disclose no funding sources.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review by minimum of two reviewers. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Designed the study methodology, collected information, performed phylogenetic and bioinformatics analyses, and prepared the manuscript: ATMS. Assisted in collecting information, performed phylogenetic and bioinformatics analyses, and prepared the manuscript: SKD, HAB. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Arellano-Reynoso B, Lapaque N, Salcedo S, et al. Cyclic β-1,2-glucan is a brucella virulence factor required for intracellular survival. Nat Immunol. 2005;6(6):618–25. doi: 10.1038/ni1202. [DOI] [PubMed] [Google Scholar]
- 2.Busse H-J, Denner EBM, Buczolits S, Salkinoja-Salonen M, Bennasar A, Kämpfer P. Sphingomonas aurantiaca sp. nov., Sphingomonas aerolata sp. nov. and Sphingomonas faeni sp. nov., air- and dustborne and Antarctic, orange-pigmented, psychrotolerant bacteria, and emended description of the genus Sphingomonas. Int J Syst Evol Microbiol. 2003;53(5):1253–60. doi: 10.1099/ijs.0.02461-0. [DOI] [PubMed] [Google Scholar]
- 3.White DC, Sutton SD, Ringelberg DB. The genus Sphingomonas: physiology and ecology. Curr Opin Biotechnol. 1996;7(3):301–6. doi: 10.1016/s0958-1669(96)80034-6. [DOI] [PubMed] [Google Scholar]
- 4.Kelley ST, Theisen U, Angenent LT, St, Amand A, Pace NR. Molecular analysis of shower curtain biofilm microbes. Appl Environ Microbiol. 2004;70(7):4187–92. doi: 10.1128/AEM.70.7.4187-4192.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kilic A, Senses Z, Kurekci AE, et al. Nosocomial outbreak of Sphingomonas paucimobilis bacteremia in a hemato/oncology unit. Jpn J Infect Dis. 2007;60(6):394–6. [PubMed] [Google Scholar]
- 6.Ryan MP, Pembroke JT, Adley CC. Ralstonia pickettii: a persistent gram-negative nosocomial infectious organism. J Hosp Infect. 2006;62(3):278–84. doi: 10.1016/j.jhin.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 7.Özdemir M, Pekcan S, Demircili ME, et al. A rare cause of bacteremia in a pediatric patient with down syndrome: Sphingomonas paucimobilis. Int J Med Sci. 2011;8(7):537–9. doi: 10.7150/ijms.8.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bayram N, Devrim I, Apa H, Gulfidan G, Turkyılmaz HN, Gunay I. Sphingomonas paucimobilis infections in children: 24 case reports. Mediterr J Hematol Infect Dis. 2013;5(1):e2013040. doi: 10.4084/MJHID.2013.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pascale R, Russo E, Esposito I, Leone S, Esposito S. Sphingomonas paucimobilis osteomyelitis in an immunocompetent patient. A rare case report and literature review. New Microbiol. 2013;36(4):423–26. [PubMed] [Google Scholar]
- 10.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10(3):512–26. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 11.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guidolin LS, Ciocchini AE, IñóndeIannino N, Ugalde RA. Functional mapping of Brucella abortus cyclic β-1,2-glucan synthase: identification of the protein domain required for cyclization. J Bacteriol. 2009;191(4):1230–8. doi: 10.1128/JB.01108-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomes Cardoso P, Costa Macedo G, Azevedo V, Costa Oliveira S. Brucella spp noncanonical LPS: structure, biosynthesis, and interaction with host immune system. Microb Cell Fact. 2006;5(1):1–11. doi: 10.1186/1475-2859-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haag AF, Myka KK, Arnold MFF, Caro-Hernandez P, Ferguson GP. Importance of lipopolysaccharide and cyclic β-1,2-glucans in Brucella-mammalian infections. Int J Microbiol. 2010;2010:124509. doi: 10.1155/2010/124509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sadosky AB, Wilson JW, Steinman HM, Shuman HA. The iron superoxide dismutase of Legionella pneumophila is essential for viability. J Bacteriol. 1994;176(12):3790–9. doi: 10.1128/jb.176.12.3790-3799.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feldman M, Bryan R, Rajan S, et al. Role of flagella in pathogenesis of pseudomonas aeruginosa pulmonary infection. Infect Immun. 1998;66(1):43–51. doi: 10.1128/iai.66.1.43-51.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Toole GA Kolter R. Flagellar and twitching motility are necessary for pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30(2):295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 18.Ciacci-Woolwine F, McDermott PF, Mizel SB. Induction of cytokine synthesis by flagella from gram-negative bacteria may be dependent on the activation or differentiation state of human monocytes. Infect Immun. 1999;67(10):5176–85. doi: 10.1128/iai.67.10.5176-5185.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adamo R, Sokol S, Soong G, Gomez MI, Prince A. Pseudomonas aeruginosa flagella activate airway epithelial cells through asialoGM1 and toll-like receptor 2 as well as toll-like receptor 5. Am J Respir Cell Mol Biol. 2004;30(5):627–34. doi: 10.1165/rcmb.2003-0260OC. [DOI] [PubMed] [Google Scholar]
- 20.Dasgupta N, Wolfgang MC, Goodman AL, et al. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol Microbiol. 2003;50(3):809–24. doi: 10.1046/j.1365-2958.2003.03740.x. [DOI] [PubMed] [Google Scholar]
- 21.Hahn HP. The type-4 pilus is the major virulence-associated adhesin of Pseudomonas aeruginosa – a review. Gene. 1997;192(1):99–108. doi: 10.1016/s0378-1119(97)00116-9. [DOI] [PubMed] [Google Scholar]
- 22.Keizer DW, Slupsky CM, Kalisiak M, et al. Structure of a pilin monomer from Pseudomonas aeruginosa: implications for the assembly of pili. J Biol Chem. 2001;276(26):24186–93. doi: 10.1074/jbc.M100659200. [DOI] [PubMed] [Google Scholar]
- 23.Skerker JM, Berg HC. Direct observation of extension and retraction of type IV pili. Proc Natl Acad Sci U S A. 2001;98(12):6901–4. doi: 10.1073/pnas.121171698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mattick JS. Type IV pili and twitching motility. Annu Rev Microbiol. 2002;56(1):289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 25.Whitchurch CB, Leech AJ, Young MD, et al. Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol Microbiol. 2004;52(3):873–93. doi: 10.1111/j.1365-2958.2004.04026.x. [DOI] [PubMed] [Google Scholar]
- 26.Pier GB, Coleman F, Grout M, Franklin M, Ohman DE. Role of alginate O acetylation in resistance of mucoid Pseudomonas aeruginosa to opsonic phagocytosis. Infect Immun. 2001;69(3):1895–901. doi: 10.1128/IAI.69.3.1895-1901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nivens DE, Ohman DE, Williams J, Franklin MJ. Role of alginate and its O acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J Bacteriol. 2001;183(3):1047–57. doi: 10.1128/JB.183.3.1047-1057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song Z, Wu H, Ciofu O, et al. Pseudomonas aeruginosa alginate is refractory to Th1 immune response and impedes host immune clearance in a mouse model of acute lung infection. J Med Microbiol. 2003;52(9):731–40. doi: 10.1099/jmm.0.05122-0. [DOI] [PubMed] [Google Scholar]
- 29.Stapper AP, Narasimhan G, Ohman DE, et al. Alginate production affects Pseudomonas aeruginosa biofilm development and architecture, but is not essential for biofilm formation. J Med Microbiol. 2004;53(7):679–90. doi: 10.1099/jmm.0.45539-0. [DOI] [PubMed] [Google Scholar]
- 30.Franklin MJ, Douthit SA, McClure MA. Evidence that the algI/algJ gene cassette, required for O acetylation of Pseudomonas aeruginosa alginate, evolved by lateral gene transfer. J Bacteriol. 2004;186(14):4759–73. doi: 10.1128/JB.186.14.4759-4773.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55(1):165–99. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 32.Erickson DL, Endersby R, Kirkham A, et al. Pseudomonas aeruginosa quorum-sensing systems may control virulence factor expression in the lungs of patients with cystic fibrosis. Infect Immun. 2002;70(4):1783–90. doi: 10.1128/IAI.70.4.1783-1790.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith RS, Iglewski BH. P. aeruginosa quorum-sensing systems and virulence. Curr Opin Microbiol. 2003;6(1):56–60. doi: 10.1016/s1369-5274(03)00008-0. [DOI] [PubMed] [Google Scholar]
- 34.Chapon-Hervé V, Akrim M, Latifi A, Williams P, Lazdunski A, Bally M. Regulation of the xcp secretion pathway by multiple quorum-sensing modulons in Pseudomonas aeruginosa. Mol Microbiol. 1997;24(6):1169–78. doi: 10.1046/j.1365-2958.1997.4271794.x. [DOI] [PubMed] [Google Scholar]
- 35.Filloux A, Michel G, Bally M. GSP-dependent protein secretion in gram-negative bacteria: the Xcp system of Pseudomonas aeruginosa. FEMS Microbiol Rev. 1998;22(3):177–98. doi: 10.1111/j.1574-6976.1998.tb00366.x. [DOI] [PubMed] [Google Scholar]
- 36.Ankenbauer RG, Quan HN. FptA, the Fe(III)-pyochelin receptor of Pseudomonas aeruginosa: a phenolate siderophore receptor homologous to hydroxamate siderophore receptors. J Bacteriol. 1994;176(2):307–19. doi: 10.1128/jb.176.2.307-319.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poole K, McKay GA. Iron acquisition and its control in Pseudomonas aeruginosa: many roads lead to Rome. Front Biosci. 2003;8:d661–86. doi: 10.2741/1051. [DOI] [PubMed] [Google Scholar]
- 38.Meyer JM, Neely A, Stintzi A, Georges C, Holder IA. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect Immun. 1996;64(2):518–23. doi: 10.1128/iai.64.2.518-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao R, Kisaalita WS. Iron acquisition from transferrin and lactoferrin by Pseudomonas aeruginosa pyoverdin. Microbiology. 1997;143(7):2509–15. doi: 10.1099/00221287-143-7-2509. [DOI] [PubMed] [Google Scholar]
- 40.Terada LS, Johansen KA, Nowbar S, Vasil AI, Vasil ML. Pseudomonas aeruginosa hemolytic phospholipase C suppresses neutrophil respiratory burst activity. Infect Immun. 1999;67(5):2371–6. doi: 10.1128/iai.67.5.2371-2376.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luberto C, Stonehouse MJ, Collins EA, et al. Purification, characterization, and identification of a sphingomyelin synthase from Pseudomonas aeruginosa. PlcH is a multifunctional enzyme. J Biol Chem 20032783532733–43. [DOI] [PubMed] [Google Scholar]
- 42.Parkhill J, Sebaihia M, Preston A, et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet. 2003;35(1):32–40. doi: 10.1038/ng1227. [DOI] [PubMed] [Google Scholar]
- 43.Glaser P, Sakamoto H, Bellalou J, Ullmann A, Danchin A. Secretion of cyclolysin, the calmodulin-sensitive adenylate cyclase-haemolysin bifunctional protein of Bordetella pertussis. EMBO J. 1988;7(12):3997–4004. doi: 10.1002/j.1460-2075.1988.tb03288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gallay J, Vincent M, Li de la Sierra IM, et al. Insight into the activation mechanism of Bordetella pertussis adenylate cyclase by calmodulin using fluorescence spectroscopy. Eur J Biochem. 2004;271(4):821–33. doi: 10.1111/j.1432-1033.2004.03987.x. [DOI] [PubMed] [Google Scholar]
- 45.Bumba L, Masin J, Fiser R, Sebo P. Bordetella adenylate cyclase toxin mobilizes its β2 integrin receptor into lipid rafts to accomplish translocation across target cell membrane in two steps. PLoS Pathog. 2010;6(5):e1000901. doi: 10.1371/journal.ppat.1000901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith AM, Guzmán CA, Walker MJ. The virulence factors of Bordetella pertussis: a matter of control. FEMS Microbiol Rev. 2001;25(3):309–33. doi: 10.1111/j.1574-6976.2001.tb00580.x. [DOI] [PubMed] [Google Scholar]
- 47.Locht C, Coutte L, Mielcarek N. The ins and outs of pertussis toxin. FEBS J. 2011;278(23):4668–82. doi: 10.1111/j.1742-4658.2011.08237.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. This table shows the tested major pathogenic virulence factors.