Abstract
High-throughput genomic assays are performed using tissue samples with the goal of classifying the samples as normal < pre-malignant < malignant or by stage of cancer using a small set of molecular features. In such cases, molecular features monotonically associated with the ordinal response may be important to disease development; that is, an increase in the phenotypic level (stage of cancer) may be mechanistically linked through a monotonic association with gene expression or methylation levels. Though traditional ordinal response modeling methods exist, they assume independence among the predictor variables and require the number of samples (n) to exceed the number of covariates (P) included in the model. In this paper, we describe our ordinalgmifs R package, available from the Comprehensive R Archive Network, which can fit a variety of ordinal response models when the number of predictors (P) exceeds the sample size (n). R code illustrating usage is also provided.
Keywords: ordinal response, high-dimensional features, penalized models, R
Introduction
Though there are traditional methods for modeling an ordinal response, these methods assume independence among the predictor variables and require that the number of samples (n) exceed the number of covariates (P) included in the model. For high-throughput genomic data, P >> n, so the traditional ordinal response models cannot be estimated. Penalized models have been demonstrated to have excellent performance when applied to high-throughput genomic datasets in fitting linear, logistic, and Cox proportional hazards models. However, penalized methods have not been fully extended to the ordinal response setting even though ordinal responses are pervasive in medicine. For example, tissue samples may be collected with the goal of classifying them as normal < pre-malignant < malignant or by stage of cancer. In these cases, molecular features monotonically associated with the ordinal response may be important to disease development; that is, an increase in the phenotypic level (stage of cancer) may be mechanistically linked through a monotonic association with gene expression. While one can apply nominal response classification methods to ordinal response data, in so doing some information is lost that may improve the predictive performance of the classifier. We previously developed software for the R programming environment that uses the coordinate descent algorithms of Refs. 1 and 2 for fitting penalized constrained continuation ratio models.3 These packages, glmpathcr and glmnetcr, are capable of modeling an ordinal response in settings where P >> n. Unfortunately, extension to the cumulative link, adjacent category, stereotype logit models, and other link functions using this framework is not straightforward. Other algorithms can be used for obtaining solutions for the least absolute shrinkage and selection operator (LASSO)4,5 and elastic net penalized models.6 In the linear regression setting, the incremental forward stagewise (IFS) is a penalized solution that enforces monotonicity.7 IFS can be generalized to problems involving those other than squared error loss, and the adaption is called the generalized monotone incremental forward stagewise (GMIFS) method.7 Herein, we extended the GMIFS method7 to ordinal response models and describe our ordinalgmifs R package. The ordinal.gmifs function can be used to fit traditional and penalized cumulative link, forward continuation ratio, and backward continuation ratio models using either a logit, probit, or complementary log–log link. It can also be used to fit adjacent category and stereotype logit models.
In the following sections, we provide an overview of the LASSO, IFS, and GMIFS methods for the linear and logistic regression settings. We then describe our implementation of the GMIFS method for modeling an ordinal response. Our implementation includes methods for initializing the ordinal thresholds, methods for updating estimates for a no penalty subset of predictors if specified by the user, derivatives for identifying which covariate to update, and convergence criteria. Because the GMIFS method requires the derivatives of each log-likelihood, where the log-likelihood function is derived from the traditional ordinal response models, including the cumulative logit, adjacent category, continuation ratio, and stereotype logit models, we also provide an overview of traditional ordinal response models. We then illustrate the functions in the ordinalgmifs R package using a dataset where we were interested in predicting normal < pre-neoplastic < neoplastic states of liver disease using high-throughput methylation data.8
Brief Overview of Penalized Methods
Traditional variable selection methodologies, including best subsets, and forward selection and backward elimination procedures, often fail to provide feasible and stable results because of their strong assumptions of covariate independence as well as their discrete procedures. Regularization methods provide a more continuous process and yield statistical models having coefficients with non-zero estimates for “important” covariates, while many coefficients are shrunk to be exactly zero. There are several algorithms that yield a penalized solution that are applicable for continuous responses.
Penalized methods for linear regression
The LASSO4 is a widely used method for deriving a parsimonious model for high-dimensional data. It produces penalized estimates of the unknown regression parameters by including the L1 norm of the coefficients as a constraint in the least-squares estimate
hence, the terms LASSO and L1 penalized regression are often used synonomously. Here λ is the tuning parameter that controls the amount of shrinkage: if λ = 0, the solution is the OLS estimates; as λ increases, the amount of shrinkage of the parameter estimates increases. Because the L1 model shrinks some coefficients to be exactly zero, it is better than ridge regression in terms of model parsimony and interpretability. Algorithms for fitting penalized linear regression models include the least angle regression (LAR) algorithm9 and the IFS regression method.7 The advantage of IFS is that it can be generalized to problems involving other than squared error loss. Therefore, we present the IFS method for the linear regression setting as a prelude to the GMIFS method.
IFS method for linear regression
The forward stagewise method is a greedy procedure similar to forward stepwise regression but is more cautious in that the coefficient updates are made using very small increments. It was historically considered to be inefficient because of its slow-fitting nature but later showed to be competitive in terms of variable selection stability and prediction accuracy.7 The IFS method7 proceeds according to the following steps to admit the solution:
Standardize the predictors, and then initialize the residuals to and initialize the p = 1, …, P parameter estimates to
Find the predictor xm most correlated with
Update , where and ε is some small positive constant such as 0.001.
Update r = r − δmxm, and repeat steps 2–3 until no predictor has any correlation with r.
At the end of this iterative algorithm, the final are taken as the penalized solution. It was previously shown that the penalized solution can be obtained using the expanded predictor space .7 When using the expanded predictor space, the absolute value of the correlation estimates is unnecessary in step 2, while the coefficient and residual updates in steps 3 and 4 let δm = ε. Finally, the P × 1 vector of coefficient estimates corresponding to the original representation of the covariate matrix X can be obtained using
Extending IFS for logistic regression
For observations i = 1, …, n, let yi represent the dichotomous dependent variable and
represent the conditional probability of experiencing the event given the independent predictor variables xi. Then the likelihood function is simply the product of the n independent binomial terms
Mathematically, it is easier to maximize the log-likelihood, which is given by
When the response is discrete, minimizing the residual sum of squares is not reasonable; so a modified penalized model that maximizes the penalized log-likelihood is needed. For an L1 penalized logistic regression model, this is expressed as
Obviously, the residual, r, in the IFS procedure is no longer appropriate for a categorical response, so it cannot be used in the logistic regression setting. However, the GMIFS regression method, where the coefficient to be updated is selected based on the gradient of the log-likelihood, can be used to provide the coefficient estimates for a penalized logistic regression model.7 Starting with the expanded covariate space , the GMIFS algorithm is
Standardize the predictors and initialize the p = 1, …, 2P parameter estimates to at step s = 0.
Find the predictor xm, where at the current estimate .
Update the corresponding coefficient to yield a new vector of parameter estimates .
Repeat steps 2 and 3 many times.
At the end of the iterative procedure, the P × 1 vector of coefficient estimates is obtained by .
Proposed GMIFS algorithm for ordinal response models
Although the GMIFS algorithm has been described, there was no specific stopping criteria, but rather step 4 in Ref. 7 recommends to repeat steps 2 and 3 many times. Therefore, we implemented the criteria to stop the iterative process when the difference between two successive log-likelihoods is smaller than a pre-specified tolerance τ. Further, recall that for the linear regression scenario, the constant term β0 term is commonly omitted because the response is centered. However, for ordinal response models having K different ordinal levels, we require K – 1 separate threshold estimates be included in the model. These K – 1 α intercept terms are essential for an ordinal response model that assumes proportional odds, because under this assumption, the β estimates do not have category-specific effects.10 Therefore, the log-odds ratio, which measures the degree of association between two ordinal levels, can be explained by the difference between the corresponding thresholds, logit(γj) – logit(γj) = αj – αj, such that the α estimates are highly important for distinguishing between ordinal classes. Moreover, there are some research problems for which a subset of predictor variables is to be included in the model (ie, not penalized). That is, in certain modeling contexts, it is desired to include relevant variables based on subject-specific knowledge. Therefore, we have included in our algorithm the ability to include a set of covariates as a no penalty subset of predictor variables. For K ordinal classes and P predictors, our GMIFS algorithm is:
-
0a.
Let be our standardized predictors that are to be penalized and, if applicable, W be our predictor(s) that is(are) not to be penalized (no penalty subset). Let β represent the coefficients associated with X and θ represent the coefficients associated with W.
-
0b.Initialize α and θ based on the specific ordinal response model (see the following ordinal response models section).
- Initialize the components of at step s = 0 as .
- Find at the current estimate
- Update .
- Update α’s and θ by maximum likelihood considering the vector of coefficients from step 3 as fixed.
- Repeat steps 2–4 until log L(s+1) − log L(s) < τ, where τ is pre-specified tolerance.
Therefore, for each ordinal response model, we need to initialize the α1, …, αK–1 and θ estimates, provide expressions for δ log L/δβp for updating the β’s, and provide maximum likelihood equations for updating the α1, …, αK–1 and θ estimates after the β’s have been updated. These equations are presented in the subsequent sections for the cumulative link, adjacent category, forward and backward continuation ratio model, and stereotype logit model. While our ordinalgmifs R package additionally allows fitting cumulative link, and forward and backward continuation ratio models using a probit or complementary log–log link, we have omitted the details regarding their derivatives and initial α estimates here.
Ordinal Response Models
Let Yi represent the ordinal response for observation i that can take on one of K ordinal levels. Denote the n × P covariate matrix as x so that xi represents a P × 1 vector for observation i and xp represents the n × 1 vector for covariate p. For observations i = 1, …, n, the response Yi can be reformatted as a response matrix consisting of n rows and K columns where
Therefore, yj is an n × 1 vector representing class j membership. Letting πj(xi) represent the probability that observation i with covariates xi belongs to class j, the likelihood for an ordinal response model with K ordinal levels can be expressed as
| (1) |
Similar to logistic regression, it is more convenient to work with the log-likelihood, which be expressed as
| (2) |
Cumulative logit model
The cumulative logit model models K − 1 logits of the form.
where αj denotes the class-specific intercept and β is a p × 1 vector of coefficients associated with explanatory variables xi.11 Note that the class-specific probabilities can be calculated by subtracting successive cumulative logits,
Therefore, for any class j, we can express the class-specific probabilities by
Because we require α0 = – ∞, for j = 1, this expression simplifies to
and because we require αK = ∞, for j = K, this expression simplifies to
Therefore, the derivative of the log-likelihood with respect to βp is
| (3) |
In the GMIFS algorithm, the alpha terms are initialized to If a no penalty subset is included, we can consider x to be the expanded matrix of w: x, and the parameter vector β to be the expanded vector θ: β for calculating the class-specific probabilities. However, the derivative is only estimated with respect to the variables in the penalized portion of the model. Also, when a no penalty subset is included, α and θ are estimated by maximum likelihood at the initial step and after each β update. Because the class-specific probabilities are obtained by subtracting successive cumulative logits, we require α1 ≤ α2 ≤ … ≤ αK–1. Therefore, the maximum likelihood estimation of α and θ includes that constraint on the threshold estimates.
Adjacent category model
The adjacent category model models the logits of ordinal categories that are adjacent to one another. That is, for any ordinal level j = 1, …, K − 1, its logit is
| (4) |
where
| (5) |
Plugging equation 5 into the logit in equation 4, we have the simplification,
so that
The class-specific probabilities for the adjacent category model can be calculated using the baseline category framework. Suppose we let the first class be our baseline category (note that the baseline category is arbitrary). Then for any other class j ≤ K, we can express its baseline category logit as
Adding log(π1) to both sides of this equation and then exponentiating yields
and
The derivative of the log-likelihood with respect to βp is the sum of the derivatives of the K class components
| (6) |
In the GMIFS algorithm, the α terms are initialized as the logits of the adjacent class probabilities: α1 = log(π2/π1), α2 = log (π3/π2), …, αK–1 = log(πK/πK–1). If a no penalty subset is included, α and θ are estimated by maximum likelihood at the initial step and after each β update.
Continuation ratio models
Although the likelihood for continuation ratio models can be expressed using equation 1, it is more convenient to express the likelihood using the continuation ratios. There are different ways in which one can set up a continuation ratio model, so here we present the backward and forward continuation ratio models separately.
Backward continuation ratio model
The backward continuation ratio model models the logit of the j = 2, …, K conditional probabilities or
Here we have used the backward formulation, which is commonly used when progression through disease states from none, mild, moderate, and severe is represented by increasing integer values, and interest lies in estimating the odds of more severe disease compared to less severe disease.12 Letting δij represent the conditional probabilities,
such that for K ordinal classes, there are K – 1 logits. The likelihood can be expressed using these j = 2, …, K conditionally independent probabilities:
which can be factored as the product of K – 1 binomial likelihoods. Using this expression, the derivative of the log-likelihood with respect to βp for the backward continuation ratio is given by
| (7) |
In the GMIFS algorithm, the α terms are initialized as αj = log (Πj/(1 – Πj)), where If a no penalty subset is included, α and θ are estimated by maximum likelihood at the initial step and after each β update.
Forward continuation ratio model
The forward continuation ratio model models the logit of the conditional j = 1, …, K – 1 probabilities or
As with the backward continuation ratio model, the likelihood for the forward continuation ratio model can be expressed using the K – 1 conditionally independent probabilities,
The derivative of the log-likelihood is the same, which for the forward continuation ratio model is given by,
| (8) |
In the GMIFS algorithm the α terms are initialized as αj = log (Πj/(1 – Πj)), where . If a no penalty subset is included, α and θ are estimated by maximum likelihood at the initial step and after each β update.
Stereotype logit model
The proportional odds versions of the cumulative logit, adjacent category, and forward and backward continuation ratio models all assume that the odds at x = x1 compared to x = x2 are exp(β(x1 – x2)) regardless of the response category. The stereotype logit model is more flexible than the proportional odds versions of the cumulative logit, adjacent category, forward continuation ratio, and backward continuation ratio models, yet is more parsimonious in comparison to fully unconstrained versions of these models.13,14 From the definition of the stereotype logit model,13 the probability that observation i is from class j is defined as
| (9) |
and to impose an ordinality restriction, we let αK = 0 and ϕK = 0 such that for category K,
| (10) |
Note that these probabilities are equivalent to expressing the stereotype logit model using the Kth class as the baseline category; the j = 1, …, K – 1 logits are of the form
| (11) |
such that the class-specific probabilities for classes j = 1, …, K – 1 are expressed as
| (12) |
where
| (13) |
Substituting πj(xi) and πK(xi) into equation 2 and simplifying yields
| (14) |
Because log(1) = 0, our final log-likelihood is
| (15) |
or given the second form for πK(xi) in equation 10,
| (16) |
To ensure ordinality of the model,13 it was further required that ϕ1 = 1, ϕK = 0, and ϕ1 ≥ ϕ2 ≥ ϕ3 ≥ … ϕK–1 ≥ ϕK.
The first derivative of the log-likelihood with respect to βp is given by
| (17) |
The α terms are initialized as
| (18) |
while ϕ are initialized as ϕ1 = 1, ϕ2, …, ϕK–1 = 0.1. If a no penalty subset is included, α, ϕ, and θ are estimated by maximum likelihood at the initial step and after each β update.
Implementation
The ordinalgmifs package was written in the R programing environment.15 The ordinal.gmifs function allows the user to specify a model formula, identify the matrix of covariates to be penalized in the model fitting algorithm using the x parameter, and additionally specify the model type (probability. model) and link function (link). The default is to fit a cumulative logit model, though allowable probability models include Cumulative, ForwardCR, BackwardCR, AdjCategory, and Stereotype while allowable links include logit, probit, and cloglog for the first three and loge and logit for the last two, respectively. The defaults for updating the penalized coefficients are epsilon = 0.001 and tol = 1e – 5. Our likelihood functions were written in R and tested by comparing our R output to output produced by the vglm R VGAM package for cumulative link, adjacent category, and forward and backward continuation ratio models and to STATA’s slogit function and the rrvglm function in the R VGAM for the stereotype logit model using benchmark datasets for data where P < n.
Examples
The ordinalgmifs package includes example datasets having an ordinal response and a detailed tutorial as a vignette. The example data are a subset of subjects and CpG sites reported in the original paper where liver samples were assayed using the Illumina GoldenGate Methylation BeadArray Cancer Panel I.8 Technical replicate samples and matched cirrhotic samples from subjects with hepatocellular carcinoma (HCC) were removed to ensure all samples were independent. These data are in two formats: as a data.frame (hccframe) and as a BioConductor ExpressionSet (hccmethyl). However, for the demonstration provided here, we illustrate downloading the full dataset, GSE18081, from Gene Expression Omnibus, filtering on relevant criterion, and fitting a cumulative logit model to the independent subjects whose liver was either normal (n = 20) < cirrhotic but not having HCC (n = 16, cirrhosis non-HCC) < HCC (n = 20, HCC) using the CpG site methylation values as predictor variables. To download the GSE18081 dataset, we load the GEOquery R package before using the getGEO function.
> library("GEOquery")
> data<–getGEO("GSE18081") [[1]]
We then removed paired cirrhotic samples from patients that also contributed to tumor samples and replicate hybridizations so that only independent subjects are included in our dataset.
> data<–data [,ifelse (pData (data) $characteristics_ch1=="disease state: Cirrhosis", FALSE, TRUE)]
> data<–data [,–grep ("Replicate",pData (data) title)]
> hccCancerPanel<–data.frame(Tissue = factor(ifelse (pData (data) $characteristics_ch1=="disease state:Cirrhosis non-HCC", "Cirrhosis non-HCC", ifelse (pData(data) $characteristics_ch1=="disease state: Normal", "Normal", "HCC")), ordered=TRUE, levels=c("Normal", "Cirrhosis non—HCC", "HCC")), t(exprs(data)))
> rownames(hccCancerPanel)<–rownames (pData(data))
The class to be predicted (normal < cirrhosis non-HCC < HCC) is stored in our hccCancerPanel data.frame as Tissue along with the 1,505 CpG site methylation values. However, prior to model fitting, NA values should be imputed or removed from the data.frame. Our first filtering step applied removed 10 CpG sites that had at least one missing value. Additionally, we removed any CpG site that had a variance of 0 (n = 26), leaving 1,469 CpG sites for our predictive model.
> filter<–c(0,apply(hccCancerPanel[,–1],2,function(x) sum (is.na(x))))
> hccCancerPanel<–hccCancerPanel [,filter==0] > filter2<–c(1,apply(hccCancerPanel[,–1],2,function(x) sd(x)))
> hccCancerPanel<–hccCancerPanel [, (1: dim (hccCancerPanel) [2]) [filter2!=0]]
To fit a model where all predictors are penalized, the model formula is specified to fit an intercept only model and the predictors to be penalized are specified using the x parameter. When fitting a penalized model, it is expected that more than one variable is included in the x parameter. The x parameter can either be a vector naming columns in the data.frame specified by the data parameter or be a data.frame name with the columns to include (or exclude) indicated by their (negative) index. By default, a cumulative logit model is fit when neither probability.model nor link is specified by the user. Because Tissue is the first variable in hccCancerPanel, we fit a model penalizing all CpG sites by specifying x = hccCancerPanel[,–1], which simply removes our ordinal outcome.
> library("ordinalgmifs")
> cumulative.logit<–ordinal.gmifs(Tissue ~ 1, x = hccCancerPanel[,–1], data = hccCancerPanel)
Because the GMIFS procedure is incremental, the user may want to specify verbose = TRUE to print the step number in order to monitor the status of the model fitting procedure.
Methods including coef, plot, predict, fitted, print, and summary can be applied to ordinalgmifs model objects. Because the returned list differs depending on whether a no penalty subset is included or a stereotype logit model is fit, the print function returns the object names of the fitted object.
> print (cumulative.logit)
| [1] | “beta” | “alpha” | “zeta” |
| [4] | “x” | “y” | “w” |
| [7] | “scale” | “logLik” | “AIC” |
| [10] | “BIC” | “model.select” | “probability.model” |
| [13] | “link” |
By default coef, predict, and summary extracts the relevant information from the step in the solution path that attained the minimum AIC. Because most of the features will have a coefficient estimate of zero, it is more convenient to extract the coefficient estimates and then examine those features with non-zero estimates. Here we see that in the AIC selected model there are 15 CpG sites with non-zero coefficient estimates.
> head (summary(cumulative.logit))
Cumulative model using a logit link
| at step | = 5085 |
| logLik | = –12.164 |
| AIC | = 111578 |
| BIC | = 122035 |
| (Intercept):1 | (Intercept):2 | AATK_E63_R | AATK_P519_R | AATK_P709_R |
| –1.9115 | 1.9158 | 0.0000 | 0.0000 | 0.0000 |
| ABCA1_E120_R 0.0000 | ||||
> coefficients<–coef (cumulative.logit)
> coefficients [coefficients!=0]
| (Intercept):1 | (Intercept):2 | CDKN2B_seq_50_S294_F |
| –1.9115 | 1.9158 | –0.5730 |
| DDIT3_P1313_R | ERN1_P809_R | GML_E144_F |
| –0.6240 | 0.3620 | 0.6130 |
| HDAC9_P137_R | R HLA.DPA1_P205 | R HOXB2_P488_R |
| 0.0830 | 0.3540 | –0.0760 |
| IL16_P226_F | IL16_P93_R | IL8_P83_F |
| 0.3970 | 0.1460 | 0.1710 |
| MPO_E302_R | MPO_P883_R | PADI4_P1158_R |
| 0.3270 | 0.1390 | –0.0860 |
| SOX17_P287_R | TJP2_P518_F | |
| –0.8210 | –0.3130 |
Although the AIC is the default model selected, any step along the solution path can be extracted by specifying the step using the model.select parameter for these three functions. For example, the model attaining the minimum BIC can be extracted using summary(cumulative.logit, model.select = which.min (cumulative.logit$BIC)).
Alternatively, the 150th step can be extracted using summary (cumulative.logit, model.select = 150). Note that the αj thresholds are labeled as (Intercept): 1, …, (Intercept):K – 1.
When examining the non-zero coefficient estimates, SOX17 and DDIT3 had the largest absolute values. Boxplots of the β values by tissue type revealed a monotonic relationship for both (Fig. 1).
Figure 1.
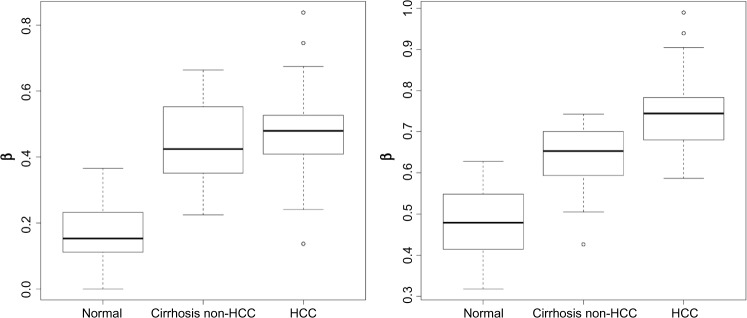
Boxplots of β values for SOX17 (left panel) and DDIT3 (right panel) by tissue type.
The plot function plots the solution path of the model fit. The vertical axis can be changed using the type parameter with allowable selections being trace (default), AIC, BIC, or logLik. Although there are default x-axis, y-axis, and titles provided for each plot, the user can modify these by supplying their own arguments to xlab, ylab, and main, respectively.
> plot(cumulative.logit)
The predict function (or equivalently, fitted) returns a list containing predicted, a matrix of the class probabilities from the fitted model, and class, the class having the maximum predicted probability from the fitted model. As with coef and summary, the predict function by default extracts the model that attained the minimum AIC, but predictions for any step along the solution path can be obtained by specifying the step using the model.select parameter.
> phat <– predict(cumulative.logit)
> table(phat$class, hccCancerPanel$Tissue)
| Normal | Cirrhosis | non-HCC | HCC | |
| Normal | 20 | 0 | 0 | |
| Cirrhosis non-HCC | 0 | 16 | 0 | |
| HCC | 0 | 0 | 20 |
> head(phat$predicted)
| [,1] | [,2] | [,3] | |
| [1,] | 0.00125343 | 0.0532523 | 0.94549 |
| [2,] | 0.01002675 | 0.3074904 | 0.68248 |
| [3,] | 0.00019167 | 0.0085375 | 0.99127 |
| [4,] | 0.00527017 | 0.1904609 | 0.80427 |
| [5,] | 0.01017666 | 0.3105981 | 0.67923 |
| [6,] | 0.01333243 | 0.3696477 | 0.61702 |
> boxplot(hccCancerPanel$SOX17_P287_R~hccCancerPanel$Tissue, xlab="", ylab=expression(beta))
> boxplot(hccCancerPanel$DDIT3_P1313_R~hccCancerPanel$Tissue, xlab="", ylab=expression(beta))
For the AIC selected model, there were no misclassification errors. However, a fair way of estimating generalization error should be applied. When there are small sample sizes in one or more groups, K-fold cross-validation (CV) methods may not perform well as a means to estimate generalization error because of the random inclusion of samples into each of the folds. That is, multiple folds may include few if any subjects from the small classes. Therefore, here we have demonstrated N-fold CV for this dataset. Note that we include the drop = FALSE argument to preserve the dimension format of the object when only one subject comprises the test set. Note that we used the foreach and doSNOW packages for parallel processing to speed up our computations.
> library("doSNOW")
> library("itertools")
> machines <– rep("localhost", each=4)
> cl <– makeCluster(machines, type="SOCK", outfile="test.txt")
> registerDoSNOW(cl)
> iter <– isplitIndices(nrow(hccCancerPanel), chunks =nrow(hccCancerPanel))
> nfold.class <– foreach(i = iter,.combine=c,.packages="ordinalgmifs")%dopar% {fit<–ordinal.gmifs(Tissue ~ 1, x=hccCancerPanel[–i,–1], data=hccCancerPanel [–i,]) return(predict(fit, newx=hccCancerPanel [i,–1,drop=FALSE])$class) }
> stopCluster(cl)
> table(hccCancerPanel$Tissue, nfold.class)
| nfold.class 1 | |||
| 1 | 2 | 3 | |
| Cirrhosis non-HCC | 14 | 2 | 0 |
| HCC | 3 | 14 | 3 |
| Normal | 0 | 0 | 20 |
There were 8 of 56 misclassifications from the N-fold CV procedure, which yields a generalized misclassification rate of 14.3%.
Aside from a logit link, a probit or complementary log–log link can be used in conjunction with the cumulative link probability model. These three links are also available for probability.model = "ForwardCR" and probability. model = "BackwardCR". A stereotype logit model only uses a logit link, while an adjacent category model only uses a loge link. Misspecifying the link for either a stereotype logit or adjacent category yields a warning that is printed to the R console, but only the correct link is used in the model fit.
Summary
Herein we presented an extension of the GMIFS method for predicting an ordinal response in high-dimensional covariates spaces. If interest lies in predicting an ordinal response and the number of covariates is small relative to the available sample size, we recommend using the VGAM package. However, our ordinalgmifs R package is capable of fitting models in high-dimensional covariate spaces and the penalization process better handles multicollinearity problems. We presented the flexibility of the GMIFS method by adapting it to fit cumulative link model, forward and backward continuation ratio models using either a logit, probit, or complementary log– log link, as well as the adjacent category and stereotype logit models. Functions for extracting coefficients, obtaining fitted probabilities, predicting class, plotting, and summarizing the fitted model object are also provided. Our ordinalgmifs package should be helpful when predicting an ordinal response for datasets where the number of covariates exceeds the number of available samples.
Footnotes
Author Contributions
Conceived and designed the methods: KJA, JH. Wrote the Software code: KJA, JH, QZ, KF, JGL, AEG. Wrote the first draft of the manuscript: KJA, JH. Contributed to the writing of the manuscript: QZ, KF, AEG. Agree with manuscript results and conclusions: KJA, JH, QZ, KF, AEG. Jointly developed the structure and arguments for the paper: KJA, JH. Made critical revisions and approved final version: KJA, JH, KF. All authors reviewed and approved of the final manuscript.
ACADEMIC EDITOR: J T Efird, Editor in Chief
FUNDING: Research reported in this publication was supported by the National Library of Medicine of the National Institutes of Health under Award Number R01LM011169. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review by minimum of two reviewers. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
REFERENCES
- 1.Park MY, Hastie T. L1-regularization path algorithm for generalized linear models. J R Stat Soc B. 2007;64:659–77. [Google Scholar]
- 2.Friedman JH, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 3.Archer KJ, Williams AAA. L1 penalized continuation ratio models for ordinal response prediction using high-dimensional datasets. Stat Med. 2012;31:1464–74. doi: 10.1002/sim.4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tibshirani R. Regression shrinkage and selection via the Lasso. J R Stat Soc B. 1996;58:267–88. [Google Scholar]
- 5.Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med. 1997;16:385–95. doi: 10.1002/(sici)1097-0258(19970228)16:4<385::aid-sim380>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 6.Zou H, Hastie T. Regularization and variable selection via the elastic net. J R Stat Soc B. 2005;67:301–20. [Google Scholar]
- 7.Hastie T, Taylor J, Tibshirani R, Walther G. Forward stagewise regression and the monotone lasso. Electron J Stat. 2007;1:1–29. [Google Scholar]
- 8.Archer Kellie J, Mas Valeria R, Maluf Daniel G, Fisher Robert A. High-throughput assessment of CpG site methylation for distinguishing between HCV-cirrhosis and HCV-associated hepatocellular carcinoma. Mol Genet Genomics. 2010;283:341–9. doi: 10.1007/s00438-010-0522-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Efron B, Hastie T, Johnstone I, Tibshirani R. Least angle regression. Ann Stat. 2004;32:407–99. [Google Scholar]
- 10.McCullagh P. Regression models for ordinal data. J R Stat Soc B. 1980;42:109–42. [Google Scholar]
- 11.Agresti A. Analysis of Ordinal Categorical Data. Hoboken, NJ: John Wiley & Sons; 2010. [Google Scholar]
- 12.Bender R, Benner A. Calculating ordinal regression models in SAS and S-Plus. Biom J. 2000;42:677–99. [Google Scholar]
- 13.Anderson JA. Regression and ordered categorical variables. J R Stat Soc Ser B. 1984;46:1–30. [Google Scholar]
- 14.Greenland S. Alternative models for ordinal logistic regression. Stat Med. 1994;13:1665–77. doi: 10.1002/sim.4780131607. [DOI] [PubMed] [Google Scholar]
- 15.R Core Team . R: A Language and Environment for Statistical Computing. Austria: R Foundation for Statistical Computing Vienna; 2013. [Google Scholar]


