Abstract
Malignant cancer cells utilize their intrinsic migratory ability to invade adjacent tissues and the vasculature, and ultimately to metastasize. Cell migration is the sum of multi-step processes initiated by the formation of membrane protrusions in response to migratory and chemotactic stimuli. The driving force for membrane protrusion is localized polymerization of submembrane actin filaments. Recently, several studies revealed that molecules that link migratory signals to the actin cytoskeleton are upregulated in invasive and metastatic cancer cells. In this review, we summarize recent progress on molecular mechanisms of formation of invasive protrusions used by tumor cells, such as lamellipodia and invadopodia, with regard to the functions of key regulatory proteins of the actin cytoskeleton; WASP family proteins, Arp2/3 complex, LIM-kinase, cofilin, and cortactin.
Keywords: Cell motility, Lamellipodia, Invadopodia, WASP family, Cofilin, Cortactin
1. Introduction
Cell migration is required for many biological processes, such as embryonic morphogenesis, immune surveillance, and tissue repair and regeneration. Aberrant regulation of cell migration drives progression of many diseases, including cancer invasion and metastasis [1-3]. Therefore, understanding the fundamental mechanisms of cell migration is critical for our understanding of both basic biology and the pathology of disease. Cell migration is a highly integrated multistep process that is initiated by the protrusion of the cell membrane [4]. Protrusive structures formed by migrating and invading cells were termed filopodia, lamellipodia, and invadopodia/podosomes, dependently on their morphological, structural, and functional characters (Fig. 1). Formation of these structures is driven by spatially- and temporally-regulated actin polymerization at the leading edge [5].
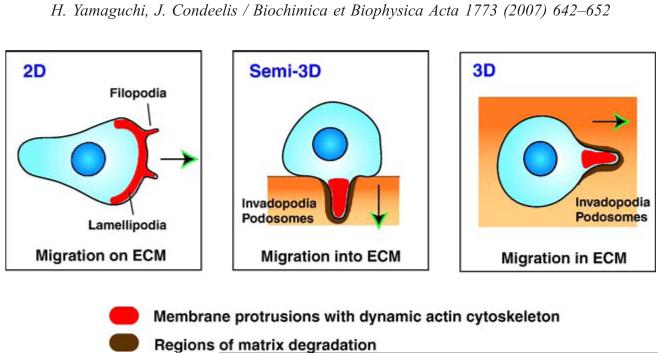
Fig. 1.
Cell migration and membrane protrusions in different environments. Cells migrating on 2D substrates form membrane protrusions called filopodia and lamellipodia at the leading edge. Cells entering into and migrating in a dense rigid ECM in 3D, such as tumor cells on top of a thick ECM and those found around blood vessels, need to form membrane protrusions at the invading front, such as invadopodia and podosomes that have an ECM remodeling activity. Formation of these structures is driven by localized actin polymerization. Proteins involved in formation of these protrusions are often upregulated in malignant cancer cells and associated with increased cell motility and invasion. Arrows indicate the direction of cell migration.
Cell migration and invasion are triggered by a number of chemoattractants. Upon binding to cell surface receptors, these chemoattractants stimulate intracellular signaling pathways that regulate reorganization of the actin cytoskeleton. To date, several important proteins that mediate the signaling pathways have been identified as overexpressed in several types of cancers [2] and in the subpopulation of invasive tumor cells in breast tumors [6]. Among them, Wiskott–Aldrich syndrome protein (WASP) family proteins/Arp2/3 complex, LIM-kinase/cofilin, and cortactin pathways have been studied extensively due to their apparent importance in cell migration and invasion (Fig. 2). In this review, we summarize recent findings on molecular mechanisms underlying formation of membrane protrusions and functions of these proteins particularly in cancer cell migration, invasion, and metastasis.
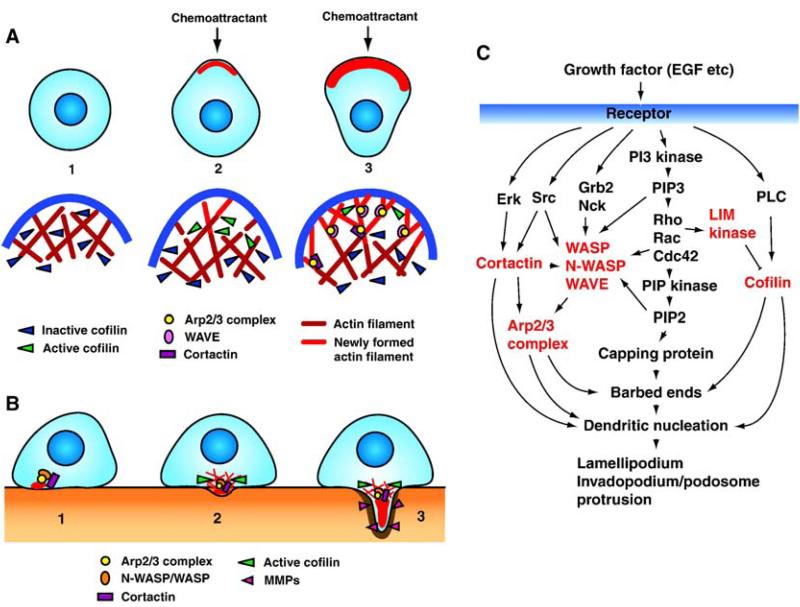
Fig. 2.
Model for lamellipodium and invadopodium/podosome formation. (A) 1: Unstimulated cells have non-polarized cell morphology in which molecular machinery for barbed end formation including cofilin is inactive. 2: Chemoattractant stimulation induces local activation of cofilin at the leading edge, which leads to severing of pre-existing actin filaments and formation of free barbed ends from which new actin filaments are assembled. This initiates membrane protrusions and sets the direction of cell migration. 3: Arp2/3 complex and WAVEs associate with newly formed actin filaments and induce formation of further barbed ends and the branched actin network. Subsequently, the branched actin filaments are stabilized by cortactin. This strengthens the protrusive force of lamellipodia and leads to cell movement. (B) 1: Invadopodium/podosome formation is triggered by N-WASP/WASP, Arp2/3 complex and cortactin, probably by coupled activation of growth factor receptor and integrin signaling. 2: This precursor is stabilized by further recruitment of invadopodium/podosome components and formation of actin network by cofilin. 3: Anchored precursor then gathers matrix-degrading proteinases to degrade ECM and protrude into matrix. N-WASP/Arp2/3 complex, cortactin, and cofilin continue to induce actin polymerization to maintain the structural core. In contrast to lamellipodia, the structure and organization of actin filaments are not yet determined in invadopodia/podosomes. (C) The signaling pathways leading to protrusion of lamellipodia and invadopodia/podosomes in response to growth factor stimulation. Molecules discussed in this review are highlighted in red.
2. Lamellipodia generate the driving force for cell migration
Lamellipodia are flat, sheet-like membrane protrusions formed at the leading edge of migrating cells. It is generally believed that lamellipodia have a major role in driving cell migration by attaching to the substrate and generating force to pull the cell body forward. In agreement with this, carcinoma cells crawling on extracellular matrix (ECM) fibers toward blood vessels in primary tumors extend pseudopodia (functionally equivalent to lamellipodia) that attach to the fibers at the migration front [7]. Lamellipodia contain dendritic arrays of actin filaments and the molecular machinery that controls polymerization/depolymerization and organization of actin filaments [8]. Filopodia are thin, finger-like projections consisting of bundled, crosslinked actin filaments and they are also observed at the migrating front of cells [9]. Although filopodia are proposed to sense external cues to set the direction of cell migration, the exact role of filopodia is still not understood. Therefore, we will not discuss filopodia in this review.
The protrusion of lamellipodia is initiated by localized polymerization of actin, and this requires generation of free barbed ends of actin filaments at the leading edge. There are three major mechanisms for generation of free barbed ends: (1) de novo nucleation by Arp2/3 complex and formins, (2) severing of pre-existing actin filaments by cofilin, and (3) uncapping of barbed ends on pre-existing actin filaments [10,11]. Many chemotactic factors have been shown to stimulate intracellular signaling pathways, which lead to barbed end formation through these mechanisms. Among them, EGF is a critical chemotactic, lamellipodia-inducing factor for breast cancer cells and activation of EGF signaling pathway is directly correlated with increased invasion, intravasation, and metastasis [12]. Gene expression analyses revealed that components involved in lamellipodium formation downstream of EGF signaling pathway, including the WAVE-Arp2/3 complex and the LIM kinase-cofilin pathways, are upregulated in invasive breast cancer cells [6]. Other studies also identified these proteins as overexpressed in several types of cancers (Table 1). These proteins coordinately regulate the formation of lamellipodia, thereby regulating migration and invasion of cancer cells.
Table 1.
Functions of WASP family proteins, Arp2/3 complex, cofilin, LIM kinase, and cortactin in cancer cell migration, invasion, and metastasis
| Protein | Overexpression in metastatic/invasive cancer (evidence) |
Protrusions involved |
Cancer cell motility and invasion in vitro |
Tumor metastasis in vivo |
|---|---|---|---|---|
| N-WASP | Colorectal (MA) [30] | IP formation FP formation |
KD blocks invadopodium formation and invasion by carcinoma cells [25] |
DN blocks lung metastasis of breast cancer*1 |
| WAVE1 | Melanoma (IB) [32] | LP formation | KD has little effect on melanoma cell invasion [32] |
KD has little effect on lung metastasis of melanoma cells [32] |
| WAVE2 | Melanoma (IB) [32] Lung (IH) [33] |
LP formation | KD blocks melanoma cell invasion [32] | KD blocks lung metastasis of melanoma cells [32] |
| WAVE3a | Breast (MA) [6] | LP formation | KD blocks carcinoma cell migration and invasion [56,57] |
ND |
| Arp2/3 | Breast (MA) [6] | LP formation | KD or DN blocks carcinoma cell | ND |
| complexa | Colorectal (IH) [31] Lung (IH) [33] |
IP formation | invasiveness [25] | |
| LIM | Breast (MA) [6] | LP formation | OE suppresses and DN increases | OE decreases and DN increases |
| kinasea | Breast (IB) [116] Prostate (IB) [116, 117] |
motility of neuroblastoma [118] and breast cancer cells [74,119] OE increases and DN decreases motility and invasion of breast cancer cells [116] Reduced expression by antisense abolishes invasion of prostate cancer cells [117] |
intravasation and metastasis of breast cancer [119] OE enhances and DN blocks bone metastasis of breast cancer [116] |
|
| Cofilina | Breast (MA) [6] Glioblastoma (SG) [77] |
LP formation IP dynamics |
Local activation induces lamellipodia formation and sets direction of cell motility [72] KD reduces invasion of carcinoma cells [25] Moderate OE enhances migration of glioblastoma cells [76] Extreme OE inhibits invasion of lung cancer cells [79] |
ND |
| Cortactin | Colorectal (IB, IH) [120] Breast (DS) [121] Head and neck (DP) [92] Liver (MA, IH) [93] Melanoma (IB) [122] |
LP dynamics IP formation |
OE promotes invasion of breast and liver cancer cells [93,94] OE enhances and KD inhibits migration and invasion of fibrosarcoma cells [98] KD*2, [102] or antibody injection [101] blocks invasion of breast cancer cells |
OE enhances bone metastasis of breast cancer [94] and intrahepatic metastasis of liver cancer [93] |
Abbreviations: MA, cDNA or oligonucleotide microarray; IB, immunoblotting; IH, immunohistochemistry; DP, differential PCR; DS, DNA slot-blot; SG, serial analysis of gene expression; LP, lamellipodia; IP, invadopodia, FP, filopodia; OE, overexpression; KD, knockdown by RNAi; DN, dominant-negative overexpression; ND, not determined.
H. Yamaguchi and J. Wyckoff
H. Yamaguchi, unpublished observations.
Components of the invasion signature of mammary carcinoma cells [6].
3. Invadopodia/podosomes promote cell migration through ECM
To migrate through a dense barrier of ECM, cells need to degrade and remodel ECM structures [13]. Invadopodia are ventral membrane protrusions with an ECM degradation activity formed by highly invasive cancer cells on thick physiological substrates [14]. Invadopodia are composed of a variety of proteins, such as actin and actin regulatory proteins, adhesion molecules, membrane remodeling and signaling proteins, and matrix degradation enzymes. Among them, N-WASP and cortactin are essential components that compose core structure of invadopodia and these proteins are upregulated in malignant cancer cells (Table 1). Carcinoma cells seem to utilize invadopodia type protrusions to migrate and invade through tumor stroma and into blood vessels in the process of metastasis [3,7]. Podosomes are similar to invadopodia in their appearance and molecular composition. Classic podosomes are formed by cell types of monocytic origin, such as macrophages, dendritic cells, and osteoclasts. Podosome-like structures have also been reported in other cell types, including smooth muscle and endothelial cells [15,16]. Importantly, the formation of podosomes (sometimes interchangeably called invadopodia) is induced by oncogenic transformation of fibroblasts by v-Src, suggesting the importance of these structures in oncogene-driven cell motility and invasion. Podosomes had been considered as dynamic adhesion sites required for chemotactic cell migration. Recent studies, however, demonstrated that podosomes are capable of degrading ECM of physiological substrates [17-20]. Therefore, invadopodia and podosomes are proposed to have the same physiological function, i.e., remodeling of ECM structures, even though their morphological appearance may vary among cell types.
Accumulating evidence suggests that the interaction between tumor cells and the stromal compartment, including stromal cells, ECM, chemokines, and blood vessels, have a major role in cancer progression. For example, macrophages within primary tumors facilitate tumor progression and metastasis [21,22]. Invasive carcinoma cells and tumor-associated macrophages have been shown to interact through a CSF-1/EGF paracrine loop, which enhance migration, invasion, and intravasation of carcinoma cells [23,24]. Since EGF and CSF-1 stimulate the formation of invadopodia in carcinoma cells and podosomes in macrophages, respectively [20,25], the CSF-1/EGF paracrine loop is likely to promote matrix remodeling required for tumor invasion. Angiogenic chemokines that exist in tumor stroma, such as VEGF, TNFα, and TGFβ, were also shown to induce the formation of podosomes in endothelial cells [19,26]. Given that angiogenesis requires extensive remodeling of ECM surrounding capillaries, podosomes formed by endothelial cells may be involved in this process.
4. WASP family proteins and Arp2/3 complex
The WASP family consists of five members in mammalian cells, including WASP, N-WASP (neural WASP), WAVE1 (WASP family verprolin homologous protein 1), WAVE2, and WAVE3, which can be divided into two subgroups, WASP/N-WASP and WAVEs [27,28]. WASP family proteins integrate multiple upstream signals to induce actin polymerization through the Arp2/3 complex, an activator of actin filament nucleation and branching [29]. Several lines of evidence indicate that these proteins are necessary for cell protrusive activity associated with cell migration and invasion. Moreover, the expression of WASP family proteins and Arp2/3 complex has been associated with malignant phenotypes of cancer cells, indicating the importance of these proteins in cancer cell migration and invasion [6,30-33].
4.1. WASP and N-WASP in invadopodium/podosome formation
WASP and N-WASP share functional domains that interact with upstream regulators, such as Cdc42, SH3 domain-containing proteins (e.g. Nck and Grb2), and phosphoinositides. WASP and N-WASP are also regulated through phosphorylation by Src family kinases. WASP is expressed exclusively in hematopoietic cells, while N-WASP is ubiquitously expressed and abundant in brain. N-WASP was originally implicated in filopodium formation downstream of Cdc42, a potent inducer of filopodia [34], while N-WASP may not be involved in the formation of lamellipodia in mammalian cells [35]. N-WASP was shown to be necessary for the invasion of epithelial cells into 3D collagen gels and to localize at the invasion front [36], suggesting that N-WASP is involved in formation of invasive membrane protrusions. Indeed, several studies recently revealed that WASP and N-WASP have a pivotal role in formation of matrix-degrading structures, podosomes/invadopodia.
N-WASP and Arp2/3 complex localize at invadopodia in highly metastatic MTLn3 rat mammary adenocarcinoma cells [25] and the activation of N-WASP was observed at invadopodia actively degrading ECM [37]. Knockdown of either N-WASP, or its upstream and downstream effectors, including Nck1, Cdc42, WIP (WASP-interacting protein), and Arp2/3 complex, suppressed invadopodium formation and matrix degradation activity in MTLn3 cells [25]. These results demonstrate that the N-WASP signaling pathway is necessary for invadopodium formation in carcinoma cells. Consequently, over expression or increased activity of N-WASP is likely to promote cancer cell invasion and metastasis. N-WASP is also involved in podosome formation in non-hematopoietic and non-cancer cells, including epithelial cells, endothelial cells, and v-Src transformed fibroblasts [17,19,38]. Similar to N-WASP, WASP is an essential component of podosomes in hematopoietic cells, such as macrophages and dendritic cells. These cell types derived from Wiskott–Aldrich syndrome patients, which is caused by mutation in the gene encoding WASP, are unable to form podosomes [39,40]. Therefore, WASP and N-WASP are involved in formation of podosomes and invadopodia in diverse cell types.
The precise function of WASP/N-WASP in invadopodium/podosome formation remains to be determined. Because these structures are shown to require dynamic rearrangement and continuous assembly of actin filaments [41], WASP/N-WASP may contribute to these processes by inducing actin filament nucleation through activation of the Arp2/3 complex. In addition, since WASP and N-WASP have been involved in endocytotic and phagocytotic processes [42,43], these proteins may promote internalization of degraded matrix components and/or recycling of components of invadopodia/podosomes, such as adhesion molecules and membrane type matrix metalloproteinases.
4.2. WAVEs in lamellipodium formation and more
The function of WAVEs in lamellipodium and membrane ruffle formation is well established. Although WAVEs bind to several signaling proteins and phosphoinositides, they are primarily regulated by Rac, a Rho family small GTPase, indirectly through intermediate molecules [44-48]. WAVE1 and WAVE3 are rich in brain and also expressed in other tissues at lower levels, while WAVE2 is ubiquitously expressed and rich in hematopoietic cells. Studies with fibroblasts derived form WAVE1 and WAVE2 knockout mice revealed distinct roles of these proteins in formation of membrane ruffling and lamellipodia [49-51]. WAVE2 regulates formation of peripheral lamellipodia, which are necessary for general cell migration, while WAVE1 seems to promote formation of dorsal membrane ruffling and stabilization of peripheral lamellipodia. Roles of WAVE1 and WAVE2 in cancer cells were studied in metastatic mouse melanoma cell lines, in which these proteins are overexpressed as they become metastatic [32]. WAVE2 knockdown by RNAi suppressed membrane ruffling, cell motility, invasion, and pulmonary metastasis of the melanoma cells, whereas WAVE1 knockdown had little effect on these processes. These results indicate that WAVE2 is a primary regulator of melanoma cell invasion and metastasis. Involvement of WAVE2 in tumor metastasis is also suggested by an immunohistochemical study showing that coexpression of WAVE2 and Arp2, a subunit of Arp2/3 complex, is observed in malignant human lung cancers and correlated with poor patient outcome [33]. Moreover, Huang et al. reported that overexpression of motility-related protein-1 (MRP-1/CD9), a potential suppressor of tumor metastasis that inhibits cancer cell migration, downregulates WAVE2 expression, which is associated with inhibition of lamellipodium formation in HT1080 human fibrosarcoma cells [52]. All these data indicate that WAVE2 is a key regulator of lamellipodium formation, and therefore, involved in cancer cell migration. Because WAVE2 is neither localized at invadopodia nor required for invadopodium formation in breast cancer cells [25], WAVE2 is likely to specifically regulate lamellipodium formation-driven general cell motility.
Although WAVE3 seems to be regulated by similar molecular mechanisms as WAVE1 and 2 [53], its physiological role has not been well studied. However, several recent reports described possible unique functions of WAVE3 in progression of cancer. Sossey-Alaoui et al. found that the gene encoding WAVE3 is truncated as a result of a chromosome translocation in a patient with ganglioneuroblastoma [54]. While it is unclear whether this truncation causes complete loss of WAVE3 function or results in production of unregulated product, this finding implicates WAVE3 in tumor progression. In search of binding partners of WAVE3, it was found that LDOC1 (a leucine zipper protein, down-regulated in cancer cells) preferentially associates with WAVE3 among WASP family proteins [55]. Ectopically expressed LDOC1 induces apoptosis of cells through accumulation of a tumor suppressor p53. Interestingly, WAVE3 co-expression inhibits the LDOC1-induced apoptosis by inducing translocation of LDOC1 from the nucleus to the cytoplasm. Therefore, WAVE3 may negatively regulate the LDOC1 function, which potentially promotes progression of tumors. In agreement with this idea, our group reported that WAVE3 is upregulated in invasive population of MTLn3 carcinoma cells [6]. Notably, WAVE3 expression is hardly detected in cultured MTLn3 cells [25], suggesting that WAVE3 expression is regulated by the in vivo tumor microenvironment. Additionally, knockdown studies with adenocarcinoma cells revealed that WAVE3 regulates cell migration and invasion [56,57], and interestingly, expression levels of several matrix metalloproteinases [56]. Thus, WAVE3 may not only have expected functions in lamellipodial protrusions, but also have potential roles in suppression of apoptosis and matrix metalloproteinase production, both hallmarks of malignant cancer cells.
5. Cofilin and LIM kinase
Cofilin, which belongs to a family of related proteins [ADF; Actophorin—A. castellanii; Coactosin—Dictyostelium discoidium; Twinstar—Drosophila melanogaster; unc-60A and unc-60B—Caenorhabditis elegans; XAC1 and XAC2—Xenopus; actophorin—Acanthamoeba], as reviewed in [58], is a small ubiquitous protein (~19 kDa) that is able to bind both monomeric and filamentous actin, and is an essential regulator of actin dynamics at the plasma membrane during cell migration through its ability to sever actin filaments. Cofilin can be regulated through different upstream effectors, as reviewed in [59]; LIM 1 and 2, and TES 1 and 2 kinases phosphorylate cofilin on the serine 3 residue, thus rendering it inactive [60-63]; while type 1, 2A, 2B, slingshot, and chronophin phosphatases dephosphorylate cofilin [64-67]. The protein phosphatase slingshot and the novel HAD-family phosphatase, chronophin, have been proposed to be the primary activators of cofilin by dephosphorylation at serine 3 in a variety of cell types [64,67]. However, the initiation of cofilin activation in response to EGF in invasive tumor cells is not coupled to dephosphorylation [68]. In addition, cofilin is inhibited when bound to phosphatydalinositol-4,5-bisphosphate (PIP2) [69,70]. In vivo studies suggest that PLC-mediated hydrolysis of PIP2 can release cofilin from this complex thereby activating it [71].
5.1. Cofilin is an essential regulator of cancer cell motility and invasion
There is little doubt that cofilin activity is required for cell motility and invasion. Local activation of cofilin by uncaging induces lamellipodia formation and sets the direction of cell motility [72]. Inhibition of cofilin activity in carcinoma cells with either siRNA [73] or expression of constitutively active LIM kinase domain [74] inhibits cell motility. The suppression of cofilin expression with siRNA reduces the invasion of carcinoma cells; in particular cofilin is involved in the assembly and stability of invadopodia [25]. The over expression of cofilin by 2–4 fold at the protein level increases the velocity of cell migration in Dictyostelium [75] and in human glioblastoma cells [76]. The spontaneous over expression of cofilin by 2–4 fold at the mRNA level has been detected in the invasive subpopulation of tumor cells in mammary tumors [6]. Cofilin is over expressed in the highly invasive C6 rat glioblastoma cell line [77], and the amount of phosphorylated, inactive cofilin is decreased in cell lines derived from T-lymphoma (Jurkat) and carcinomas from the cervix (HeLa), colon (KM12), liver (HepG2), and kidney (COS1) [78].
The activation of cofilin occurs as a discrete transient after the stimulation of carcinoma cell motility with EGF, indicating that the precise balancing of cofilin and LIM kinase activities is required for stimulated cell migration in tumor cells [71]. The transient presumably occurs as the result of the EGF-receptor’s activation of PLC which releases cofilin from its inactive complex with PIP2 [71]. Simultaneously, EGF-receptor activates LIM kinase which limits the spatial and temporal extent of cofilin activity leading to a localized burst of actin polymerization seen as a spatially and temporally discrete transient [68]. The loss of this balance by the hyper activation of either cofilin or LIM kinase inhibits cell motility and invasion as shown by the inhibitory effects on cell motility of huge un-physiological over expression of cofilin (15 fold) [79], microinjection of constitutively active cofilin (Mouneimne personal communication) and the expression of constitutively active LIM kinase domain [74].
The importance of the transient activation of cofilin in stimulated cell motility has been illustrated by work that has shown that both PLC and an early actin polymerization transient at 1 min are required by carcinoma cells for chemotaxis to a gradient of EGF. In particular, cofilin was shown to be responsible for this early actin polymerization transient and to require PLC activation for its activity [71]. In addition, uncaging of cofilin activity was shown to be sufficient for initiating both actin polymerization and protrusion in vivo, and in defining the direction of cell movement [72]. These results indicate that spatially and temporally localized cofilin activity is involved in chemotactic sensing during invasion [71].
A possible molecular basis for the transient activation of cofilin is the simultaneous activation of both LIM kinase and cofilin activities in response to stimulation. The activation of LIM kinase and phosphorylation of cofilin in response to the stimulation of cells with growth factors has been observed [71,80]. However, in some cases the phosphorylation was assumed, but not shown, to inactivate all cofilin within the cells and led to the interpretation that cofilin must be inhibited for cell protrusion and locomotion to occur [80]. This interpretation is inconsistent with the increases in cofilin activity observed simultaneously with an increase in cofilin phosphorylation [68,71,81], and the requirement of cofilin for cell motility [73]. The interpretation that cofilin must be inhibited for protrusion and locomotion to occur is also at odds with the observation that uncaging of cofilin activity in vivo is sufficient to cause actin polymerization, protrusion and locomotion [72]. We propose that a more accurate explanation for these results is that there are two different populations of cofilin that exist simultaneously in cells during stimulation of migration: one that is locally activated allowing localized protrusion, and one that is being phosphorylated to either recycle cofilin or to confine the cofilin activity to a discrete location in the cell.
These considerations illustrate the need to measure cofilin activity in cells during stimulation and migration in order to interpret the consequences of either cofilin phosphorylation or dephosphorylation on cofilin function in vivo. Furthermore, additional work is needed to determine the mechanism of cofilin activation in cells during stimulations that do not induce the de-phosphorylation of cofilin.
5.2. The function of cofilin in vivo can be explained by a simple severing mechanism
It is well established that cofilin functions to depolymerize filaments in cells so that actin can be recycled for another round of polymerization [73,82]. It is also clear that cofilin can induce the polymerization of actin in cells by virtue of its severing activity [71,72]. However, it has been confusing as to whether the ability of cofilin to depolymerize F-actin is linked to its severing activity. Kinetic experiments with recombinant cofilin have been interpreted to mean that cofilin can increase the off rate of monomers from the pointed end of filaments without severing [83]. The absence of severing was assumed in these studies because it was not possible to detect severing activity in preparations of recombinant cofilin. However, we now know that it is difficult to detect severing activity in recombinant cofilin due to the heterogeneity of the recombinant cofilin molecules compared to native cofilin and that both native and certain chromatographically purified fractions of recombinant cofilin show a high specific activity of severing [84]. Cofilin severing was detected by light microscopy [84-86] and by intrinsic fluorescence [87]. It is particularly interesting in this regard that cross linked actin filaments are severed by cofilin at 500 fold lower concentrations of cofilin (K50=9 nM) than when filaments are in solution (K50=5 μM) [86,88], demonstrating that cofilin is a much stronger severing protein than originally suspected. Since the concentration of cofilin in tumor cells is ~10 μM [81], these results suggest that only a small proportion of the cofilin in cells is sufficient to sever the cross linked actin filaments commonly found in vivo.
These considerations warrant a re-evaluation of certain assumptions regarding cofilin activity and its mechanism of depolymerization of F-actin that were made when it was thought that cofilin does not sever filaments. One assumption is that cofilin induces the depolymerization of actin filaments preferentially from their pointed ends. However, when the number of new pointed and barbed ends produced by cofilin severing are accounted for in the analysis of cofilin-induced depolymerization of F-actin, there is no bias of depolymerization to the pointed end, i.e. both barbed and pointed ends contribute to depolymerization, and there is no significant increase in the off rate of actin monomers from filament ends [84,87]. That is, the rate constants for actin assembly/disassembly are constant in the presence of cofilin. All of these results indicate that both the depolymerization and polymerization activities of cofilin can be explained by a single event, cofilin severing of filaments to increase the number of free pointed and barbed ends.
6. Cortactin
Cortactin is a ubiquitously expressed, actin-binding and scaffolding protein that plays crucial roles in the regulation of the actin cytoskeleton. Cortactin has been implicated in processes that require a dynamic actin cytoskeleton, including cell motility, endocytosis, and intracellular motility of several pathogens [89,90]. Since human cortactin (EMS1) is upregulated in several types of human cancers as a result of amplification of chromosome locus 11q13, cortactin is thought to have a role in tumor malignancy [91,92]. Although, the 11q31 region contains several genes, amplification of both the EMS1 and CCDN1 gene, which encodes cyclin D1, is often correlated with tumor progression [91]. This is supported by studies showing that ectopic overexpression of cortactin accelerates bone metastasis of breast cancer cells and intrahepatic metastasis of liver cancer cells [93,94].
Cortactin was originally shown to bind to and crosslink actin filaments. In addition to these activities, recent biochemical studies suggest that cortactin also contributes to the generation of barbed ends and branched arrays of actin filaments. Cortactin directly activates the actin nucleation activity of Arp2/3 complex through its N-terminal region, although the activity is relatively weak when compared with that of WASP family proteins [95]. Cortactin is also able to indirectly promote Arp2/3 complex-mediated actin polymerization by binding to N-WASP and activating it [96]. Another study demonstrated that cortactin also stabilizes branched actin filaments produced by Arp2/3 complex [97]. Moreover, cortactin preferentially associates with newly formed, ATP-bound actin filaments, rather than older ADP-actin filaments [98]. Taken together, cortactin seems to promote Arp2/3 complex-mediated actin nucleation and stabilizes newly formed branched actin filaments in the dynamic actin cytoskeleton.
Cortactin knockdown by RNAi in fibrosarcoma cells results in impaired cell migration and invasion [98]. These phenotypes seem to be due to defects in the persistence of lamellipodial protrusions [98,99]. Importantly, although cortactin localizes at leading edges of lamellipodia, cortactin knockdown does not inhibit protrusion of lamellipodia per se. Cortactin knockdown cells show a decrease in barbed end formation at the leading edge, which is consistent with the idea that cortactin promotes activation of Arp2/3 complex. Interestingly, these cells also show a defect in the assembly of new adhesions in lamellipodia. As Arp2/3 complex has been shown to associate with vinculin, cortactin may be involved in actin assembly at adhesion sites [100]. These results indicate that cortactin is not necessary for the protrusive force of lamellipodia, but important for proper barbed end formation and assembly of new adhesions, which are required for persistence of lamellipodia and cell migration.
In contrast to lamellipodium formation, invadopodium/podosome formation is clearly dependent on cortactin activity. In breast cancer cells, cortactin localizes at invadopodia and injection of an antibody against cortactin inhibits invasive activity of breast cancer cells [101]. Moreover, cortactin knockdown by RNAi blocks assembly of invadopodia and associated matrix degradation activity in breast cancer cells ([102] and H. Yamaguchi, unpublished observations), indicating that cortactin-mediated invadopodium formation is necessary for invasive activity of these cells. Time-lapse imaging of invadopodium assembly revealed that cortactin induces formation of actin-rich invadopodial core structures and this precedes recruitment of matrix proteinases and subsequent matrix degradation [102]. Podosome formation is also blocked by down regulation of cortactin in smooth muscle cells and osteoclasts [103,104]. Since cortactin can bind to several components of invadopodia/podosomes, such as N-WASP, WIP, Arp2/3 complex, and dynamin-2, cortactin may cooperate with these proteins to assemble actin structures at invadopodia [17,95,105,106]. It is also possible that cortactin is involved in protein transport from the Golgi apparatus to maintain the stability and structure of invadopodia, as cortactin–dynamin-2 interaction is necessary for Golgi function and invadopodia were shown to associate with the Golgi apparatus [107,108].
Cortactin also participates in receptor-mediated endocytosis, and a recent study showed that this function might contribute to tumor malignancy. Timpson et al. demonstrated that over-expression of cortactin in head and neck carcinoma cells is associated with attenuated ligand-induced down regulation of EGF receptor, resulting in a sustained activation of the EGF receptor signaling [109]. As described above, because EGF receptor signaling has been implicated in invadopodium formation and is also correlated with increased invasion and metastasis of carcinoma cells, this observation may add another role for cortactin in tumor malignancy.
It is not well understood how cortactin activity is regulated in lamellipodium and invadopodium formation. Cortactin was originally identified as a major phosphorylated protein in v-Src transformed cells [110]. Cortactin is also phosphorylated downstream of many biological stimuli, such as growth factor stimulation and cell adhesion. Biochemical studies determined three tyrosine phosphorylation sites by v-Src at the C-terminal proline-rich region [111]. Because v-Src transformed fibroblasts form prominent podosomes/invadopodia with matrix degradation activity and Src activity is necessary for podosome/invadopodium formation, cortactin phosphorylation by Src seems to regulate cortactin activity in the formation of these structures. However, at least in smooth muscle cells, over-expression of cortactin mutants, which have mutations in the Src phosphorylation sites, had no effect on formation of podosomes and these mutant proteins can localize at podosomes [112]. This result raises the possibility that cortactin phosphorylation has other functions, such as regulation of stability and/or turnover of these structures. Cortactin activity may be regulated by other or additional mechanisms, such as serine phosphorylation by ERK and cleavage by calpain [110,113]. Further work will be necessary to clarify the regulatory mechanisms of cortactin activity in invasive protrusions.
7. Conclusion
Key proteins involved in the actin cytoskeleton described above are linked to the invasive and metastatic phenotypes of malignant cancer cells. Recent progress has begun to clarify molecular functions of each protein in cancer cell migration and invasion in vitro. However, in vivo, invasive tumor cells often coordinately overexpress groups of cytoskeletal proteins in multiple signaling pathways, such as the N-WASP/Arp2/3 complex pathway in conjunction with the LIM kinase/cofilin pathway, as part of an invasion signature [114]. These pathways cooperate to amplify the nucleation activity of the Arp2/3 complex: e.g. Arp2/3 complex and cofilin synergize to amplify dendritic nucleation [115]. Also, cortactin obviously participates in this process by controlling the stability of branched actin filaments. Therefore, the next challenge is to understand how the coordinately regulated pathways of the invasion signature determine the metastatic phenotype. That is, rather than analyzing a single protein it will be necessary to analyze the integrated output of the entire pathway to predict motility phenotype and metastatic potential. Furthermore, it will also be important to explore possible master regulators that coordinate the expression and activity of key actin regulatory proteins.
Acknowledgments
We apologize to those authors whose work we were unable to cite because of space and date of publication limitations. We are grateful to all lab members and collaborators for their support and helpful discussions. This work was supported by grants from the NIH (GM38511 and CA100324).
References
- [1].Condeelis J, Singer R, Segall JE. The great escape: when cancer cells hijack the genes for chemotaxis and motility. Annu. Rev. Cell Dev. Biol. 2005;21:695–718. doi: 10.1146/annurev.cellbio.21.122303.120306. [DOI] [PubMed] [Google Scholar]
- [2].Sahai E. Mechanisms of cancer cell invasion. Curr. Opin. Genet. Dev. 2005;15:87–96. doi: 10.1016/j.gde.2004.12.002. [DOI] [PubMed] [Google Scholar]
- [3].Yamaguchi H, Wyckoff J, Condeelis J. Cell migration in tumors. Curr. Opin. Cell Biol. 2005;17:559–564. doi: 10.1016/j.ceb.2005.08.002. [DOI] [PubMed] [Google Scholar]
- [4].Bailly M, Condeelis J. Cell motility: insights from the backstage. Nat. Cell Biol. 2002;4:E292–E294. doi: 10.1038/ncb1202-e292. [DOI] [PubMed] [Google Scholar]
- [5].Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- [6].Wang W, Goswami S, Lapidus K, Wells AL, Wyckoff JB, Sahai E, Singer RH, Segall JE, Condeelis JS. Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res. 2004;64:8585–8594. doi: 10.1158/0008-5472.CAN-04-1136. [DOI] [PubMed] [Google Scholar]
- [7].Condeelis J, Segall JE. Intravital imaging of cell movement in tumours. Nat. Rev., Cancer. 2003;3:921–930. doi: 10.1038/nrc1231. [DOI] [PubMed] [Google Scholar]
- [8].Nicholson-Dykstra S, Higgs HN, Harris ES. Actin dynamics: growth from dendritic branches. Curr. Biol. 2005;15:R346–R357. doi: 10.1016/j.cub.2005.04.029. [DOI] [PubMed] [Google Scholar]
- [9].Faix J, Rottner K. The making of filopodia. Curr. Opin. Cell Biol. 2006;18:18–25. doi: 10.1016/j.ceb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- [10].Zigmond SH. Formin-induced nucleation of actin filaments. Curr. Opin. Cell Biol. 2004;16:99–105. doi: 10.1016/j.ceb.2003.10.019. [DOI] [PubMed] [Google Scholar]
- [11].Condeelis J. How is actin polymerization nucleated in vivo? Trends Cell Biol. 2001;11:288–293. doi: 10.1016/s0962-8924(01)02008-6. [DOI] [PubMed] [Google Scholar]
- [12].Xue C, Wyckoff J, Liang F, Sidani M, Violini S, Tsai KL, Zhang ZY, Sahai E, Condeelis J, Segall JE. Epidermal growth factor receptor overexpression results in increased tumor cell motility in vivo coordinately with enhanced intravasation and metastasis. Cancer Res. 2006;66:192–197. doi: 10.1158/0008-5472.CAN-05-1242. [DOI] [PubMed] [Google Scholar]
- [13].Even-Ram S, Yamada KM. Cell migration in 3D matrix. Curr. Opin. Cell Biol. 2005;17:524–532. doi: 10.1016/j.ceb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- [14].Buccione R, Orth JD, McNiven MA. Foot and mouth: podosomes, invadopodia and circular dorsal ruffles. Nat. Rev., Mol. Cell Biol. 2004;5:647–657. doi: 10.1038/nrm1436. [DOI] [PubMed] [Google Scholar]
- [15].Linder S, Kopp P. Podosomes at a glance. J. Cell Sci. 2005;118:2079–2082. doi: 10.1242/jcs.02390. [DOI] [PubMed] [Google Scholar]
- [16].Linder S, Aepfelbacher M. Podosomes: adhesion hot-spots of invasive cells. Trends Cell Biol. 2003;13:376–385. doi: 10.1016/s0962-8924(03)00128-4. [DOI] [PubMed] [Google Scholar]
- [17].Mizutani K, Miki H, He H, Maruta H, Takenawa T. Essential role of neural Wiskott–Aldrich syndrome protein in podosome formation and degradation of extracellular matrix in src-transformed fibroblasts. Cancer Res. 2002;62:669–674. [PubMed] [Google Scholar]
- [18].Burgstaller G, Gimona M. Podosome-mediated matrix resorption and cell motility in vascular smooth muscle cells. Am. J. Physiol.: Heart Circ. Physiol. 2005;288:H3001–H3005. doi: 10.1152/ajpheart.01002.2004. [DOI] [PubMed] [Google Scholar]
- [19].Osiak AE, Zenner G, Linder S. Subconfluent endothelial cells form podosomes downstream of cytokine and RhoGTPase signaling. Exp. Cell Res. 2005;307:342–353. doi: 10.1016/j.yexcr.2005.03.035. [DOI] [PubMed] [Google Scholar]
- [20].Yamaguchi H, Pixley F, Condeelis J. Invadopodia and podosomes in tumor invasion. Eur. J. Cell Biol. 2006;85:213–218. doi: 10.1016/j.ejcb.2005.10.004. [DOI] [PubMed] [Google Scholar]
- [21].Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat. Rev., Cancer. 2004;4:71–78. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- [22].Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- [23].Wyckoff J, Wang W, Lin EY, Wang Y, Pixley F, Stanley ER, Graf T, Pollard JW, Segall J, Condeelis J. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 2004;64:7022–7029. doi: 10.1158/0008-5472.CAN-04-1449. [DOI] [PubMed] [Google Scholar]
- [24].Goswami S, Sahai E, Wyckoff JB, Cammer M, Cox D, Pixley FJ, Stanley ER, Segall JE, Condeelis JS. Macrophages promote the invasion of breast carcinoma cells via a colony-stimulating factor-1/epidermal growth factor paracrine loop. Cancer Res. 2005;65:5278–5283. doi: 10.1158/0008-5472.CAN-04-1853. [DOI] [PubMed] [Google Scholar]
- [25].Yamaguchi H, Lorenz M, Kempiak S, Sarmiento C, Coniglio S, Symons M, Segall J, Eddy R, Miki H, Takenawa T, Condeelis J. Molecular mechanisms of invadopodium formation: the role of the N-WASP-Arp2/3 complex pathway and cofilin. J. Cell Biol. 2005;168:441–452. doi: 10.1083/jcb.200407076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Varon C, Tatin F, Moreau V, Van Obberghen-Schilling E, Fernandez-Sauze S, Reuzeau E, Kramer I, Genot E. Transforming growth factor beta induces rosettes of podosomes in primary aortic endothelial cells. Mol. Cell. Biol. 2006;26:3582–3594. doi: 10.1128/MCB.26.9.3582-3594.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Takenawa T, Miki H. WASP and WAVE family proteins: key molecules for rapid rearrangement of cortical actin filaments and cell movement. J. Cell Sci. 2001;114:1801–1809. doi: 10.1242/jcs.114.10.1801. [DOI] [PubMed] [Google Scholar]
- [28].Stradal TE, Rottner K, Disanza A, Confalonieri S, Innocenti M, Scita G. Regulation of actin dynamics by WASP and WAVE family proteins. Trends Cell Biol. 2004;14:303–311. doi: 10.1016/j.tcb.2004.04.007. [DOI] [PubMed] [Google Scholar]
- [29].Millard TH, Sharp SJ, Machesky LM. Signalling to actin assembly via the WASP (Wiskott–Aldrich syndrome protein)-family proteins and the Arp2/3 complex. Biochem. J. 2004;380:1–17. doi: 10.1042/BJ20040176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Yanagawa R, Furukawa Y, Tsunoda T, Kitahara O, Kameyama M, Murata K, Ishikawa O, Nakamura Y. Genome-wide screening of genes showing altered expression in liver metastases of human colorectal cancers by cDNA microarray. Neoplasia. 2001;3:395–401. doi: 10.1038/sj.neo.7900185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Otsubo T, Iwaya K, Mukai Y, Mizokami Y, Serizawa H, Matsuoka T, Mukai K. Involvement of Arp2/3 complex in the process of colorectal carcinogenesis. Mod. Pathol. 2004;17:461–467. doi: 10.1038/modpathol.3800062. [DOI] [PubMed] [Google Scholar]
- [32].Kurisu S, Suetsugu S, Yamazaki D, Yamaguchi H, Takenawa T. Rac-WAVE2 signaling is involved in the invasive and metastatic phenotypes of murine melanoma cells. Oncogene. 2005;24:1309–1319. doi: 10.1038/sj.onc.1208177. [DOI] [PubMed] [Google Scholar]
- [33].Semba S, Iwaya K, Matsubayashi J, Serizawa H, Kataba H, Hirano T, Kato H, Matsuoka T, Mukai K. Coexpression of actin-related protein 2 and Wiskott–Aldrich syndrome family verproline-homologous protein 2 in adenocarcinoma of the lung. Clin. Cancer Res. 2006;12:2449–2454. doi: 10.1158/1078-0432.CCR-05-2566. [DOI] [PubMed] [Google Scholar]
- [34].Miki H, Sasaki T, Takai Y, Takenawa T. Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP. Nature. 1998;391:93–96. doi: 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- [35].Snapper SB, Takeshima F, Anton I, Liu CH, Thomas SM, Nguyen D, Dudley D, Fraser H, Purich D, Lopez-Ilasaca M, Klein C, Davidson L, Bronson R, Mulligan RC, Southwick F, Geha R, Goldberg MB, Rosen FS, Hartwig JH, Alt FW. N-WASP deficiencyreveals distinct pathways for cell surface projections and microbial actin-based motility. Nat. Cell Biol. 2001;3:897–904. doi: 10.1038/ncb1001-897. [DOI] [PubMed] [Google Scholar]
- [36].Yamaguchi H, Miki H, Takenawa T. Neural Wiskott–Aldrich syndrome protein is involved in hepatocyte growth factor-induced migration, invasion, and tubulogenesis of epithelial cells. Cancer Res. 2002;62:2503–2509. [PubMed] [Google Scholar]
- [37].Lorenz M, Yamaguchi H, Wang Y, Singer RH, Condeelis J. Imaging sites of N-WASP activity in lamellipodia and invadopodia of carcinoma cells. Curr. Biol. 2004;14:697–703. doi: 10.1016/j.cub.2004.04.008. [DOI] [PubMed] [Google Scholar]
- [38].Spinardi L, Rietdorf J, Nitsch L, Bono M, Tacchetti C, Way M, Marchisio PC. A dynamic podosome-like structure of epithelial cells. Exp. Cell Res. 2004;295:360–374. doi: 10.1016/j.yexcr.2004.01.007. [DOI] [PubMed] [Google Scholar]
- [39].Calle Y, Chou HC, Thrasher AJ, Jones GE. Wiskott–Aldrich syndrome protein and the cytoskeletal dynamics of dendritic cells. J. Pathol. 2004;204:460–469. doi: 10.1002/path.1651. [DOI] [PubMed] [Google Scholar]
- [40].Linder S, Nelson D, Weiss M, Aepfelbacher M. Wiskott–Aldrich syndrome protein regulates podosomes in primary human macrophages. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9648–9653. doi: 10.1073/pnas.96.17.9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Destaing O, Saltel F, Geminard JC, Jurdic P, Bard F. Podosomes display actin turnover and dynamic self-organization in osteoclasts expressing actin-green fluorescent protein. Mol. Biol. Cell. 2003;14:407–416. doi: 10.1091/mbc.E02-07-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lorenzi R, Brickell PM, Katz DR, Kinnon C, Thrasher AJ. Wiskott–Aldrich syndrome protein is necessary for efficient IgG-mediated phagocytosis. Blood. 2000;95:2943–2946. [PubMed] [Google Scholar]
- [43].Innocenti M, Gerboth S, Rottner K, Lai FP, Hertzog M, Stradal TE, Frittoli E, Didry D, Polo S, Disanza A, Benesch S, Di Fiore PP, Carlier MF, Scita G. Abi1 regulates the activity of N-WASP and WAVE in distinct actin-based processes. Nat. Cell Biol. 2005;7:969–976. doi: 10.1038/ncb1304. [DOI] [PubMed] [Google Scholar]
- [44].Suetsugu S, Kurisu S, Oikawa T, Yamazaki D, Oda A, Takenawa T. Optimization of WAVE2 complex-induced actin polymerization by membrane-bound IRSp53, PIP(3), and Rac. J. Cell Biol. 2006;173:571–585. doi: 10.1083/jcb.200509067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Oikawa T, Yamaguchi H, Itoh T, Kato M, Ijuin T, Yamazaki D, Suetsugu S, Takenawa T. PtdIns(3,4,5)P3 binding is necessary for WAVE2-induced formation of lamellipodia. Nat. Cell Biol. 2004;6:420–426. doi: 10.1038/ncb1125. [DOI] [PubMed] [Google Scholar]
- [46].Steffen A, Rottner K, Ehinger J, Innocenti M, Scita G, Wehl J, Stradal TE. Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. EMBO J. 2004;23:749–759. doi: 10.1038/sj.emboj.7600084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature. 2002;418:790–793. doi: 10.1038/nature00859. [DOI] [PubMed] [Google Scholar]
- [48].Miki H, Yamaguchi H, Suetsugu S, Takenawa T. IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature. 2000;408:732–735. doi: 10.1038/35047107. [DOI] [PubMed] [Google Scholar]
- [49].Suetsugu S, Yamazaki D, Kurisu S, Takenawa T. Differential roles of WAVE1 and WAVE2 in dorsal and peripheral ruffle formation for fibroblast cell migration. Dev. Cell. 2003;5:595–609. doi: 10.1016/s1534-5807(03)00297-1. [DOI] [PubMed] [Google Scholar]
- [50].Yamazaki D, Suetsugu S, Miki H, Kataoka Y, Nishikawa S, Fujiwara T, Yoshida N, Takenawa T. WAVE2 is required for directed cell migration and cardiovascular development. Nature. 2003;424:452–456. doi: 10.1038/nature01770. [DOI] [PubMed] [Google Scholar]
- [51].Yamazaki D, Fujiwara T, Suetsugu S, Takenawa T. A novel function of WAVE in lamellipodia: WAVE1 is required for stabilization of lamellipodial protrusions during cell spreading. Genes Cells. 2005;10:381–392. doi: 10.1111/j.1365-2443.2005.00845.x. [DOI] [PubMed] [Google Scholar]
- [52].Huang CL, Ueno M, Liu D, Masuya D, Nakano J, Yokomise H, Nakagawa T, Miyake M. MRP-1/CD9 gene transduction regulates the actin cytoskeleton through the downregulation of WAVE2. Oncogene. 2006 doi: 10.1038/sj.onc.1209654. doi:10.1038/sj.onc.1209654. [DOI] [PubMed] [Google Scholar]
- [53].Stovold CF, Millard TH, Machesky LM. Inclusion of Scar/WAVE3 in a similar complex to Scar/WAVE1 and 2. BMC Cell Biol. 2005;6:11. doi: 10.1186/1471-2121-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Sossey-Alaoui K, Su G, Malaj E, Roe B, Cowell JK. WAVE3, an actin-polymerization gene, is truncated and inactivated as a result of a constitutional t(1;13)(q21;q12) chromosome translocation in a patient with ganglioneuroblastoma. Oncogene. 2002;21:5967–5974. doi: 10.1038/sj.onc.1205734. [DOI] [PubMed] [Google Scholar]
- [55].Mizutani K, Koike D, Suetsugu S, Takenawa T. WAVE3 Functions as a Negative Regulator of LDOC1. J. Biochem. (Tokyo) 2005;138:639–646. doi: 10.1093/jb/mvi160. [DOI] [PubMed] [Google Scholar]
- [56].Sossey-Alaoui K, Ranalli TA, Li X, Bakin AV, Cowell JK. WAVE3 promotes cell motility and invasion through the regulation of MMP-1, MMP-3, and MMP-9 expression. Exp. Cell Res. 2005;308:135–145. doi: 10.1016/j.yexcr.2005.04.011. [DOI] [PubMed] [Google Scholar]
- [57].Sossey-Alaoui K, Li X, Ranalli TA, Cowell JK. WAVE3-mediated cell migration and lamellipodia formation are regulated downstream of phosphatidylinositol 3-kinase. J. Biol. Chem. 2005;280:21748–21755. doi: 10.1074/jbc.M500503200. [DOI] [PubMed] [Google Scholar]
- [58].Bamburg JR. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu. Rev. Cell Dev. Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- [59].DesMarais V, Ghosh M, Eddy R, Condeelis J. Cofilin takes the lead. J. Cell Sci. 2005;118:19–26. doi: 10.1242/jcs.01631. [DOI] [PubMed] [Google Scholar]
- [60].Okano I, Hiraoka J, Otera H, Nunoue K, Ohashi K, Iwashita S, Hirai M, Mizuno K. Identification and characterization of a novel family of serine/threonine kinases containing two N-terminal LIM motifs. J. Biol. Chem. 1995;270:31321–31330. doi: 10.1074/jbc.270.52.31321. [DOI] [PubMed] [Google Scholar]
- [61].Yang N, Higuchi O, Ohashi K, Nagata K, Wada A, Kangawa K, Nishida E, Mizuno K. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature. 1998;393:809–812. doi: 10.1038/31735. [DOI] [PubMed] [Google Scholar]
- [62].Toshima J, Toshima JY, Amano T, Yang N, Narumiya S, Mizuno K. Cofilin phosphorylation by protein kinase testicular protein kinase 1 and its role in integrin-mediated actin reorganization and focal adhesion formation. Mol. Biol. Cell. 2001;12:1131–1145. doi: 10.1091/mbc.12.4.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Mizuno K, Okano I, Ohashi K, Nunoue K, Kuma K, Miyata T, Nakamura T. Identification of a human cDNA encoding a novel protein kinase with two repeats of the LIM/double zinc finger motif. Oncogene. 1994;9:1605–1612. [PubMed] [Google Scholar]
- [64].Niwa R, Nagata-Ohashi K, Takeichi M, Mizuno K, Uemura T. Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell. 2002;108:233–246. doi: 10.1016/s0092-8674(01)00638-9. [DOI] [PubMed] [Google Scholar]
- [65].Meberg PJ, Ono S, Minamide LS, Takahashi M, Bamburg JR. Actin depolymerizing factor and cofilin phosphorylation dynamics: response to signals that regulate neurite extension. Cell Motil. Cytoskeleton. 1998;39:172–190. doi: 10.1002/(SICI)1097-0169(1998)39:2<172::AID-CM8>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- [66].Ambach A, Saunus J, Konstandin M, Wesselborg S, Meuer SC, Samstag Y. The serine phosphatases PP1 and PP2A associate with and activate the actin-binding protein cofilin in human T lymphocytes. Eur. J. Immunol. 2000;30:3422–3431. doi: 10.1002/1521-4141(2000012)30:12<3422::AID-IMMU3422>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- [67].Gohla A, Birkenfeld J, Bokoch GM. Chronophin, a novel HAD-type serine protein phosphatase, regulates cofilin-dependent actin dynamics. Nat. Cell Biol. 2005;7:21–29. doi: 10.1038/ncb1201. [DOI] [PubMed] [Google Scholar]
- [68].Song X, Chen X, Yamaguchi H, Mouneimne G, Condeelis J, Eddy R. Initiation of cofilin activity in response to EGF is uncoupled from cofilin phosphorylation and dephosphorylation in carcinoma cells. J. Cell Sci. 2006;119:2871–2881. doi: 10.1242/jcs.03017. [DOI] [PubMed] [Google Scholar]
- [69].Yonezawa N, Homma Y, Yahara I, Sakai H, Nishida E. A short sequence responsible for both phosphoinositide binding and actin binding activities of cofilin. J. Biol. Chem. 1991;266:17218–17221. [PubMed] [Google Scholar]
- [70].Yonezawa N, Nishida E, Iida K, Yahara I, Sakai H. Inhibition of the interactions of cofilin, destrin, and deoxyribonuclease I with actin by phosphoinositides. J. Biol. Chem. 1990;265:8382–8386. [PubMed] [Google Scholar]
- [71].Mouneimne G, Soon L, DesMarais V, Sidani M, Song X, Yip SC, Ghosh M, Eddy R, Backer JM, Condeelis J. Phospholipase C and cofilin are required for carcinoma cell directionality in response to EGF stimulation. J. Cell Biol. 2004;166:697–708. doi: 10.1083/jcb.200405156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Ghosh M, Song X, Mouneimne G, Sidani M, Lawrence DS, Condeelis JS. Cofilin promotes actin polymerization and defines the direction of cell motility. Science. 2004;304:743–746. doi: 10.1126/science.1094561. [DOI] [PubMed] [Google Scholar]
- [73].Hotulainen P, Paunola E, Vartiainen MK, Lappalainen P. Actin-depolymerizing factor and cofilin-1 play overlapping roles in promoting rapid F-actin depolymerization in mammalian nonmuscle cells. Mol. Biol. Cell. 2005;16:649–664. doi: 10.1091/mbc.E04-07-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Zebda N, Bernard O, Bailly M, Welti S, Lawrence DS, Condeelis JS. Phosphorylation of ADF/cofilin abolishes EGF-induced actin nucleation at the leading edge and subsequent lamellipod extension. J. Cell Biol. 2000;151:1119–1128. doi: 10.1083/jcb.151.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Aizawa H, Sutoh K, Yahara I. Overexpression of cofilin stimulates bundling of actin filaments, membrane ruffling, and cell movement in Dictyostelium. J. Cell Biol. 1996;132:335–344. doi: 10.1083/jcb.132.3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Yap CT, Simpson TI, Pratt T, Price DJ, Maciver SK. The motility of glioblastoma tumour cells is modulated by intracellular cofilin expression in a concentration-dependent manner. Cell Motil. Cytoskeleton. 2005;60:153–165. doi: 10.1002/cm.20053. [DOI] [PubMed] [Google Scholar]
- [77].Gunnersen JM, Spirkoska V, Smith PE, Danks RA, Tan SS. Growth and migration markers of rat C6 glioma cells identified by serial analysis of gene expression. Glia. 2000;32:146–154. [PubMed] [Google Scholar]
- [78].Nebl G, Meuer SC, Samstag Y. Dephosphorylation of serine 3 regulates nuclear translocation of cofilin. J. Biol. Chem. 1996;271:26276–26280. doi: 10.1074/jbc.271.42.26276. [DOI] [PubMed] [Google Scholar]
- [79].Lee YJ, Mazzatti DJ, Yun Z, Keng PC. Inhibition of invasiveness of human lung cancer cell line H1299 by over-expression of cofilin. Cell Biol. Int. 2005;29:877–883. doi: 10.1016/j.cellbi.2005.07.005. [DOI] [PubMed] [Google Scholar]
- [80].Nishita M, Tomizawa C, Yamamoto M, Horita Y, Ohashi K, Mizuno K. Spatial and temporal regulation of cofilin activity by LIM kinase and Slingshot is critical for directional cell migration. J. Cell Biol. 2005;171:349–359. doi: 10.1083/jcb.200504029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Chan AY, Bailly M, Zebda N, Segall JE, Condeelis JS. Role of cofilin in epidermal growth factor-stimulated actin polymerization and lamellipod protrusion. J. Cell Biol. 2000;148:531–542. doi: 10.1083/jcb.148.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Arber S, Barbayannis FA, Hanser H, Schneider C, Stanyon CA, Bernard O, Caroni P. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature. 1998;393:805–809. doi: 10.1038/31729. [DOI] [PubMed] [Google Scholar]
- [83].Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 1997;136:1307–1322. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Ichetovkin I, Han J, Pang KM, Knecht DA, Condeelis JS. Actin filaments are severed by both native and recombinant dictyostelium cofilin but to different extents. Cell Motil. Cytoskeleton. 2000;45:293–306. doi: 10.1002/(SICI)1097-0169(200004)45:4<293::AID-CM5>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- [85].Maciver SK, Zot HG, Pollard TD. Characterization of actin filament severing by actophorin from Acanthamoeba castellanii. J. Cell Biol. 1991;115:1611–1620. doi: 10.1083/jcb.115.6.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Ichetovkin I, Grant W, Condeelis J. Cofilin produces newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex. Curr. Biol. 2002;12:79–84. doi: 10.1016/s0960-9822(01)00629-7. [DOI] [PubMed] [Google Scholar]
- [87].Du J, Frieden C. Kinetic studies on the effect of yeast cofilin on yeast actin polymerization. Biochemistry. 1998;37:13276–13284. doi: 10.1021/bi981117r. [DOI] [PubMed] [Google Scholar]
- [88].Condeelis J, Song X, Backer JM, Wyckoff J, Segall J. In: Cell Motility: from Molecules to Organisms. Ridley A, Peckham M, Clark P, editors. Wiley; London: 2003. pp. 175–186. [Google Scholar]
- [89].Selbach M, Backert S. Cortactin: an Achilles’ heel of the actin cytoskeleton targeted by pathogens. Trends Microbiol. 2005;13:181–189. doi: 10.1016/j.tim.2005.02.007. [DOI] [PubMed] [Google Scholar]
- [90].J R. Daly, Cortactin signalling and dynamic actin networks. Biochem. J. 2004;382:13–25. doi: 10.1042/BJ20040737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Ormandy CJ, Musgrove EA, Hui R, Daly RJ, Sutherland RL. Cyclin D1, EMS1 and 11q13 amplification in breast cancer. Breast Cancer Res. Treat. 2003;78:323–335. doi: 10.1023/a:1023033708204. [DOI] [PubMed] [Google Scholar]
- [92].Rodrigo JP, Garcia LA, Ramos S, Lazo PS, Suarez C. EMS1 gene amplification correlates with poor prognosis in squamous cell carcinomas of the head and neck. Clin. Cancer Res. 2000;6:3177–3182. [PubMed] [Google Scholar]
- [93].Chuma M, Sakamoto M, Yasuda J, Fujii G, Nakanishi K, Tsuchiya A, Ohta T, Asaka M, Hirohashi S. Overexpression of cortactin is involved in motility and metastasis of hepatocellular carcinoma. J. Hepatol. 2004;41:629–636. doi: 10.1016/j.jhep.2004.06.018. [DOI] [PubMed] [Google Scholar]
- [94].Li Y, Tondravi M, Liu J, Smith E, Haudenschild CC, Kaczmarek M, Zhan X. Cortactin potentiates bone metastasis of breast cancer cells. Cancer Res. 2001;61:6906–6911. [PubMed] [Google Scholar]
- [95].Uruno T, Liu J, Zhang P, Fan Y, Egile C, Li R, Mueller SC, Zhan X. Activation of Arp2/3 complex-mediated actin polymerization by cortactin. Nat. Cell Biol. 2001;3:259–266. doi: 10.1038/35060051. [DOI] [PubMed] [Google Scholar]
- [96].Martinez-Quiles N, Ho HY, Kirschner MW, Ramesh N, Geha RS. Erk/Src phosphorylation of cortactin acts as a switch on-switch off mechanism that controls its ability to activate N-WASP. Mol. Cell. Biol. 2004;24:5269–5280. doi: 10.1128/MCB.24.12.5269-5280.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Weaver AM, Karginov AV, Kinley AW, Weed SA, Li Y, Parsons JT, Cooper JA. Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr. Biol. 2001;11:370–374. doi: 10.1016/s0960-9822(01)00098-7. [DOI] [PubMed] [Google Scholar]
- [98].Bryce NS, Clark ES, Leysath JL, Currie JD, Webb DJ, Weaver AM. Cortactin promotes cell motility by enhancing lamellipodial persistence. Curr. Biol. 2005;15:1276–1285. doi: 10.1016/j.cub.2005.06.043. [DOI] [PubMed] [Google Scholar]
- [99].Kempiak SJ, Yamaguchi H, Sarmiento C, Sidani M, Ghosh M, Eddy RJ, Desmarais V, Way M, Condeelis JS, Segall JE. An N-WASP-mediated pathway for localized activation of actin polymerization which is regulated by cortactin. J. Biol. Chem. 2004 doi: 10.1074/jbc.M410713200. [DOI] [PubMed] [Google Scholar]
- [100].DeMali KA, Barlow CA, Burridge K. Recruitment of the Arp2/3 complex to vinculin: coupling membrane protrusion to matrix adhesion. J. Cell Biol. 2002;159:881–891. doi: 10.1083/jcb.200206043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Bowden ET, Barth M, Thomas D, Glazer RI, Mueller SC. An invasion-related complex of cortactin, paxillin and PKCmu associates with invadopodia at sites of extracellular matrix degradation. Oncogene. 1999;18:4440–4449. doi: 10.1038/sj.onc.1202827. [DOI] [PubMed] [Google Scholar]
- [102].Artym VV, Zhang Y, Seillier-Moiseiwitsch F, Yamada KM, Mueller SC. Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer Res. 2006;66:3034–3043. doi: 10.1158/0008-5472.CAN-05-2177. [DOI] [PubMed] [Google Scholar]
- [103].Zhou S, Webb BA, Eves R, Mak AS. Effects of tyrosine phosphorylation of cortactin on podosome formation in A7r5 vascular smooth muscle cells. Am. J. Physiol.: Cell Physiol. 2006;290:C463–C471. doi: 10.1152/ajpcell.00350.2005. [DOI] [PubMed] [Google Scholar]
- [104].Tehrani S, Faccio R, Chandrasekar I, Ross FP, Cooper JA. Cortactin has an essential and specific role in osteoclast actin assembly. Mol. Biol. Cell. 2006;17:2882–2895. doi: 10.1091/mbc.E06-03-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].McNiven MA, Kim L, Krueger EW, Orth JD, Cao H, Wong TW. Regulated interactions between dynamin and the actin-binding protein cortactin modulate cell shape. J. Cell Biol. 2000;151:187–198. doi: 10.1083/jcb.151.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Kinley AW, Weed SA, Weaver AM, Karginov AV, Bissonette E, Cooper JA, Parsons JT. Cortactin interacts with WIP in regulating Arp2/3 activation and membrane protrusion. Curr. Biol. 2003;13:384–393. doi: 10.1016/s0960-9822(03)00107-6. [DOI] [PubMed] [Google Scholar]
- [107].Baldassarre M, Pompeo A, Beznoussenko G, Castaldi C, Cortellino S, McNiven MA, Luini A, Buccione R. Dynamin participates in focal extracellular matrix degradation by invasive cells. Mol. Biol. Cell. 2003;14:1074–1084. doi: 10.1091/mbc.E02-05-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Cao H, Weller S, Orth JD, Chen J, Huang B, Chen JL, Stamnes M, McNiven MA. Actin and Arf1-dependent recruitment of a cortactin-dynamin complex to the Golgi regulates post-Golgi transport. Nat. Cell Biol. 2005;7:483–492. doi: 10.1038/ncb1246. [DOI] [PubMed] [Google Scholar]
- [109].Timpson P, Lynch DK, Schramek D, Walker F, Daly RJ. Cortactin overexpression inhibits ligand-induced down-regulation of the epidermal growth factor receptor. Cancer Res. 2005;65:3273–3280. doi: 10.1158/0008-5472.CAN-04-2118. [DOI] [PubMed] [Google Scholar]
- [110].Lua BL, Low BC. Cortactin phosphorylation as a switch for actin cytoskeletal network and cell dynamics control. FEBS Lett. 2005;579:577–585. doi: 10.1016/j.febslet.2004.12.055. [DOI] [PubMed] [Google Scholar]
- [111].Huang C, Liu J, Haudenschild CC, Zhan X. The role of tyrosine phosphorylation of cortactin in the locomotion of endothelial cells. J. Biol. Chem. 1998;273:25770–25776. doi: 10.1074/jbc.273.40.25770. [DOI] [PubMed] [Google Scholar]
- [112].Zhou S, Webb BA, Eves R, Mak AS. Effects of Tyr-phosphorylation of cortactin on podosome-formation in A7r5 vascular smooth muscle cells. Am. J. Physiol.: Cell Physiol. 2006;290:C463–C471. doi: 10.1152/ajpcell.00350.2005. [DOI] [PubMed] [Google Scholar]
- [113].Perrin BJ, Amann KJ, Huttenlocher A. Proteolysis of cortactin by calpain regulates membrane protrusion during cell migration. Mol. Biol. Cell. 2006;17:239–250. doi: 10.1091/mbc.E05-06-0488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Wang W, Goswami S, Sahai E, Wyckoff JB, Segall JE, Condeelis JS. Tumor cells caught in the act of invading: their strategy for enhanced cell motility. Trends Cell Biol. 2005;15:138–145. doi: 10.1016/j.tcb.2005.01.003. [DOI] [PubMed] [Google Scholar]
- [115].DesMarais V, Macaluso F, Condeelis J, Bailly M. Synergistic interaction between the Arp2/3 complex and cofilin drives stimulated lamellipod extension. J. Cell Sci. 2004;117:3499–3510. doi: 10.1242/jcs.01211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Yoshioka K, Foletta V, Bernard O, Itoh K. A role for LIM kinase in cancer invasion. Proc. Natl. Acad. Sci. U. S. A. 2003;100:7247–7252. doi: 10.1073/pnas.1232344100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Davila M, Frost AR, Grizzle WE, Chakrabarti R. LIM kinase 1 is essential for the invasive growth of prostate epithelial cells: implications in prostate cancer. J. Biol. Chem. 2003;278:36868–36875. doi: 10.1074/jbc.M306196200. [DOI] [PubMed] [Google Scholar]
- [118].Meyer G, Kim B, van Golen C, Feldman EL. Cofilin activity during insulin-like growth factor I-stimulated neuroblastoma cell motility. Cell. Mol. Life Sci. 2005;62:461–470. doi: 10.1007/s00018-004-4456-6. [DOI] [PubMed] [Google Scholar]
- [119].Wang W, Mouneimne G, Sidani M, Wyckoff J, Chen X, Makris A, Goswami S, Bresnick AR, Condeelis JS. The activity status of cofilin is directly related to invasion, intravasation, and metastasis of mammary tumors. J. Cell Biol. 2006;173:395–404. doi: 10.1083/jcb.200510115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Zhang LH, Tian B, Diao LR, Xiong YY, Tian SF, Zhang BH, Li WM, Ren H, Li Y, Ji JF. Dominant expression of 85-kDa form of cortactin in colorectal cancer. J. Cancer Res. Clin. Oncol. 2005:1–8. doi: 10.1007/s00432-005-0046-8. [DOI] [PubMed] [Google Scholar]
- [121].Hui R, Campbell DH, Lee CS, McCaul K, Horsfall DJ, Musgrove EA, Daly RJ, Seshadri R, Sutherland RL. EMS1 amplification can occur independently of CCND1 or INT-2 amplification at 11q13 and may identify different phenotypes in primary breast cancer. Oncogene. 1997;15:1617–1623. doi: 10.1038/sj.onc.1201311. [DOI] [PubMed] [Google Scholar]
- [122].Huang J, Asawa T, Takato T, Sakai R. Cooperative roles of Fyn and cortactin in cell migration of metastatic murine melanoma. J. Biol. Chem. 2003;278:48367–48376. doi: 10.1074/jbc.M308213200. [DOI] [PubMed] [Google Scholar]