Abstract
Background/Objective:
The non-invasive prenatal detection of fetal microdeletions becomes increasingly challenging as the size of the mutation decreases, with current practical lower limits in the range of a few Mb. Our goals were to explore the lower limits of microdeletion size detection via NIPT using MINK and introduce/evaluate a novel statistical approach we recently developed called the GC Content Random Effect Model (GCREM).
Methods:
Maternal plasma was obtained from a pregnancy affected by a 4.2Mb fetal microdeletion and three normal controls. Plasma DNA was subjected to capture of an 8Mb sequence spanning the breakpoint region and sequenced. Data were analyzed with our published method, Minimally Invasive Karyotyping (MINK) and a new method called GCREM.
Results:
The 8Mb capture segment was divided into either 38 or 76 non-overlapping regions of 200Kb and 100Kb respectively. At 200Kb resolution, using GCREM (but not MINK) we obtained significant adjusted p values for all 20 regions overlapping the deleted sequence, and non-significant p values for all 18 reference regions. At 100Kb resolution, GCREM identified significant adjusted p values for all but one 100Kb region located inside the deleted region.
Conclusion:
Targeted sequencing and GCREM analysis may enable cost effective detection of fetal microdeletions and microduplications at high resolution.
Keywords: DNA Sequencing, Deletion, Prenatal, Genetics, Fetus, Plasma, Region Capture, Hybridization
Introduction
Definitive prenatal diagnosis of genetic disease is performed via amniocentesis (AF) or chorionic villus sampling (CVS). These are invasive procedures that carry an inherent risk of miscarriage, fetal morbidity and parental anxiety. 1-7 There has been considerable progress in the development of non-invasive prenatal tests (NIPT) and rapid translation of these methods in the clinical setting 8-11. The most commonly used approach involves the analysis of cell-free fetal DNA isolated from a maternal blood sample taken in early gestation. Current NIPT methods are focused towards the detection of common aneuploidies. However, recent progress in NIPT of fetal microdeletions has been reported 12-15 which is significant given that the incidence of aneuploidy in human pregnancy is around 1-2%, whereas the collective incidence of microdeletions and microduplications is 3.6%. 16, 17
The majority of microdeletions and microduplications are <5Mb, which is below the resolution (5-10Mb) of traditional metaphase chromosome analysis. Because of the clinical significance of these disorders there has been considerable effort directed towards the development and validation of methods for their diagnosis. Array comparative genomic hybridization (aCGH) is a powerful tool for the high-resolution evaluation of many microdeletions/microduplications in parallel. aCGH is now considered to be the diagnostic standard of care for pregnancies with an abnormal ultrasound and it is rapidly becoming the standard of care for prenatal diagnosis. However, aCGH must be performed using cellular material obtained via either AF or CVS. It is therefore essential that NIPT technologies be further developed to enable the detection of a broad spectrum of microdeletions and microduplications.
In 2011 we published a proof of concept study in which we were able to detect a 4Mb fetal microduplication via whole genome next-generation DNA sequencing of maternal plasma DNA. 13 Three more recent studies have further demonstrated the significant potential of whole genome DNA sequencing of maternal plasma DNA for the NIPT of fetal microdeletions and microduplications. 12, 14, 15 However, these whole genome sequencing methods become more challenging and expensive as the mutation size gets smaller, with the need to acquire more sequence reads, the potential for troublesome false positive results and the challenge of reporting unanticipated clinical findings (Krier and Green).
In light of this, the goals of this study were to explore the potential of genome capture and targeted sequencing of plasma DNA for high resolution microdeletion detection and the evaluation, in this context, of two statistical approaches. One of these, MINK, has been previously published. The second, called GC Content Random Effect Model (GCREM), is introduced herein.
Methods
Human DNA Samples
The University of Pittsburgh IRB approved the patient consenting process and collection of all samples used in this study. Written informed consent was obtained in every case. A family presented to the Center for Medical Genetics at the Magee-Womens Hospital of UPMC with a history of cognitive delay and dysmorphia. The father was diagnosed with Asperger syndrome, short stature, poor dentition, broad thumbs, brachydactyly, short metacarpals, and facial dysmorphia. His wife had no known medical problems. Their first pregnancy resulted in a daughter with a single-copy loss at the proximal region of the short arm of chromosome 12 involving 12p12.1-p11.22 region, encompassing approximately 4.2Mb in size (chr12: 24,346,835-28,542,656; hg18 coordinates; www.genome.ucsc.edu). FISH analysis using BAC clone RP11-105M17 (12p11.23) confirmed the interstitial deletion in the proband and identified the same deletion in her father. The microdeletion encompasses 37 genes and causes haploin sufficiency in the PTHLH, the gene coding for parathyroid hormone related protein, and implicated in premature differentiation of chondrocytes, brachydactyly and short stature 18. Maternal FISH analysis using the same BAC probe showed a normal hybridization pattern.
The couple conceived again, and after genetic counseling regarding the risk of paternal transmission of the microdeletion to the fetus, amniocentesis was performed at 21 weeks of gestation. Prenatal GTG-banded chromosome analysis on cultured amniocytes showed a normal male chromosome complement: 46,XY in 15 cells analyzed. Microarray analysis performed on DNA extracted from cultured amniocytes identified single-copy 12p12.1-p11.22 loss, the same deletion as identified in sibling and father. FISH analysis using BAC clone RP11-105M17 (12p11.23) confirmed the interstitial deletion in the fetus. The fetal karyotype was designated as 46,XY,del(12)(p11.22p12.1)pat. Multiple prenatal ultrasound evaluations noted appropriate fetal growth, normal gross anatomy and increased amniotic fluid volume in the third trimester. A maternal blood sample was drawn at 35 gestational weeks and a DNA extracted from the plasma. Using Taqman based quantification of SRY gene sequence we determined that the fetal DNA frequency was 5.7% (not shown). This is relatively low, particularly considering the gestational age at which the sample was obtained 19, 20, but is within a range that suggests this approach will have utility earlier in gestation. 21 This plasma DNA sample was previously analyzed as the focus of a proof of concept report of the use of whole genome sequencing for NIPD of the same fetal microdeletion 13.
Preparation of Targeted Sequencing Libraries
Plasma was separated from whole blood by centrifugation at 1,600 x g for 10 minutes, followed by a second centrifugation to remove contaminating nucleated cells at 16,000 x g for 10 minutes. DNA was extracted from 5.4mL plasma using the QIAamp DNA Blood Mini kit (Qiagen, Valencia, CA). Plasma DNA libraries were prepared using standard Illumina TruSeq protocols (Illumina, San Diego, CA). An initial 15 cycle PCR reaction was carried and 500ng of the resulting product was incubated with SureSelect biotinylated probes for 24 hours as described in the Agilent SureSelect protocol (Agilent, Santa Clara, CA). Baits spanning a region between chr12:22,455,568-30,651,389 (hg19) were designed for this purpose by Agilent. Targets were captured using Dynal MyOne Streptavidin T1 beads (Invitrogen, Carlsbad, CA) and a final library amplification of 12 cycles was carried out as described in the Illumina TruSeq protocol. Libraries were quantified via real time PCR and sequenced on a HiSeq2000 (Illumina) using 100bp paired-end reads.
Analysis of Sequencing Data
We developed a new statistical procedure, GC content random effect model (GCREM) to detect the presence of insertion/deletion in the captured region. The most important feature of the GCREM algorithm is that it can automatically correct the GC bias in the sequencing data. It is well known that DNA sequencing data produced by the current high throughput sequencing technologies, including the Illumina technology used in this study, are subject to the bias caused by different GC content level over different genomic regions. 13, 19, 22 In particular, the bias caused by the uneven GC content is not constant over all libraries, but specific to each individual library 22. In Chu et al 2009 22, a statistical method MINK was proposed to address this library specific bias, where the ratio of a target library to a reference library is used to remove the library specific GC bias. While the MINK method has been successfully applied to tests of aneuploidy 22 and a 4Mb microdeletion deletion 13, it is designed to work in a pair wise fashion. The library to be tested is always compared to a single reference library. Using MINK, when multiple reference libraries are available, multiple test results will be generated, and a follow up step would be required to summarize all the results. The GCREM method described in this study is based on the same observation of the library specific GC bias, but is designed to test a target library against a group of reference libraries.
Briefly, we propose a linear mixed effect model for the tag counts of different genomic regions in a DNA library, where the GC content is an independent variable with a library specific random coefficient. This linear mixed effect model is fitted, using a set of libraries with known normal genomes, and estimates the region specific corrections for a list of genomic regions of interest. Then we apply the region specific corrections to the library from the individual sample to be tested and fit a linear model derived from the original linear mixed effect model using only the tag count data from regions that are believed to be normal in the new patient. We then predict the tag count for the regions that may carry insertion/deletion. By comparing the observed tag counts with the predicted tag counts, we can test if the suspected regions indeed carry insertion/deletion.
GCREM Algorithm
Consider reference sequencing libraries L1, …, LN, all of which are from normal maternal plasma, and targeting genomic regions R1, …, RM. The basic assumption of the GCREM model is that the log tag count in the ith library, jth region can be represented by the following linear mixed effect model:
| (1) |
where m is the overall mean, Rj is the region specific effect, Gj the GC content of the jth region, Ti0 and Ti1 the library specific random effects of the ith library, and eij is the random error associated with Yij. Note that here the tag count could be measured in different ways: It could be measured as a library’s median or mean of the coverage over each region, or the sum of tags from a library in each region.
We can fit the model in Equation (1) using the reference libraries, where we believe no mutation occurs in those M regions, and estimate the values of fixed region effect Rj for each of the M regions. Let ^Rj be the estimation for the jth region Rj using the reference libraries. The formula for ^Rj can be found in Rao et al 2010, p. 174 (Equation 4.163).
Now consider a new library, the target library Lt. Suppose that we believe that the regions R1, …, RK, called the control regions, are normal in the target library, but would like to test if mutation occurs in the other M-K regions. Subtract the estimation of Rj from the model for the target target library, we have:
| (2) |
Let , , and , Equation (2) becomes:
| (3) |
Written in vector notation, we then have:
| (4) |
where D is the design matrix [1 G], where G = [G1, …, GM]T, and B the parameter vector [1 Tt1]T.
Let and DK = matrix [1 GK], where GK = [G1, …, GK]T. Fit the data for the regions R1, …, RK from the target library to the model represented by Equation (4), we can get the estimation of the parameter vector , and the fitted value . Under the null hypothesis that the region Rk, where k = K+1, …, M, of the target library is normal, the predicted value of Y’tk then is , and the estimated variance of is:
^s’tk2 = ^s2(1 + 1/(K−2) + (Gk−^G)2 / j=1,…,K(Gj−^G)2), where ^s2 = (YKt − ^YKt)T(YKt − ^YKt)/(K-2) is the estimated variance of e’tj, and ^G = (j=1,...,K Gj)/K. The statistic Zk = (Y’kt − ^Y’kt)/^s’tk then has a Student’s t distribution with K-2 degrees of freedom under the null hypothesis that the region Rk is normal in the target library.
Compared to the MINK algorithm, we believe the GCREM algorithm should have higher sensitivity in detecting microdeletion/microduplication. We notice that the GCREM and MINK algorithms use similar test statistics, where the difference between the predicted value for log tag count (for GCREM) or log tag count ratio (for MINK) and the observed value is divided by the square root of the variance of the prediction. In the GCREM algorithm, the variance of the prediction for region j is proportional to the variance of the error term e’tj in equation (3), which is equal to the sum of the variance of the error term etj and the variance of ^Rj. (The formula for the variance of ^Rj can be found in Rao et al 2010, p. 179, Equation 4.193). On the other hand, in the MINK algorithm, when performing a pairwise test between the target library t and a reference library r, the variance of the prediction for region j is proportional to the variance of the term (etj-erj), which is equal to the sum of variance of the error terms etj and erj. Clearly, when the variance of ^Rj, which is inversely proportional to the number of reference libraries used in the GCREM algorithm, becomes smaller than the variance of erj, the GCREM algorithm will have a higher power (i.e., is more sensitive) than the MINK algorithm.
Results
Maternal plasma-derived DNA from the affected sample (PL565) and three further maternal plasma samples, all of which were from confirmed karyotypically normal pregnancies, were subjected to targeted region capture of an 8Mb sequence on chromosome 12p12.1-p11.22 followed by next generation DNA sequencing on the HiSeq2000 (Illumina). We obtained 45 million paired reads (approximately 15% of a single flow cell on the HiSeq2000) for the affected sample (PL565), and between 75 million to 97 million paired reads for the three normal samples. Around 70~75% of the paired reads in each case were aligned to the targeted 8Mb region. The 8Mb target region was divided into 36 non-overlapping segments of 200Kb each. Among the 36 regions, 18 are located entirely inside the 4.2Mb deleted region, 2 cover the junctions at the deleted region and the flanking normal regions, and 16 are located entirely in the flanking normal regions (Figures 1-3). All the following statistical analyses were performed using statistical computing language R.
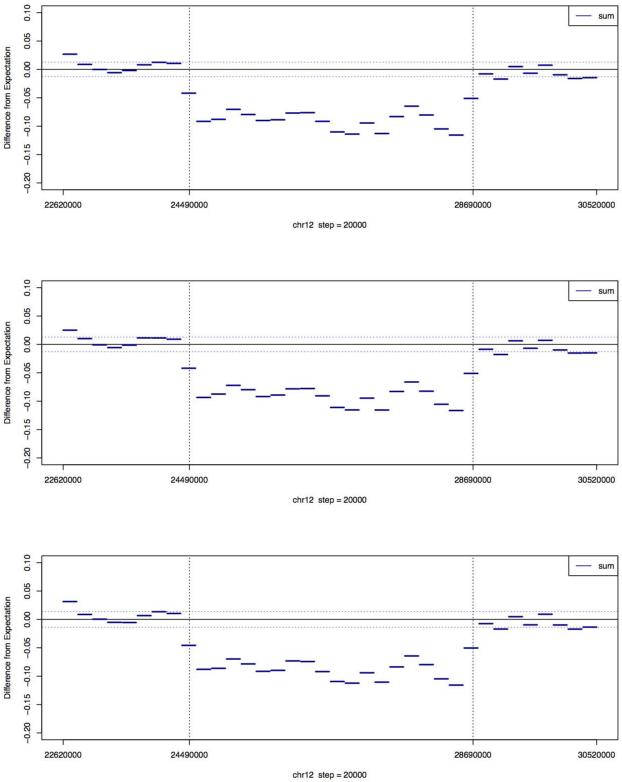
Figure 1. Difference between the fitted and observed normalized log tag count in PL565 for reads aligned to the 36 regions of chr12 using the GCREM algorithm with 3 reference libraries.
Each region is 200kb long. The regions between the two vertical dashed lines indicate the position where the microdeletion occurs. The two horizontal dashed lines represent the standard deviation of the difference. For each region with complete or partial deletion (chr12:24380000~28760000), the GCREM test is using the 16 regions covering chr12:22620000~24360000 and chr12:28780000~30520000 as the control regions. For each of the 16 regions with no deletion, the test is using the other 15 regions with no deletion as control regions. The three plots, from top to bottom, are based on the full, one-half, and one-quarter of the PL565 sequence reads respectively.
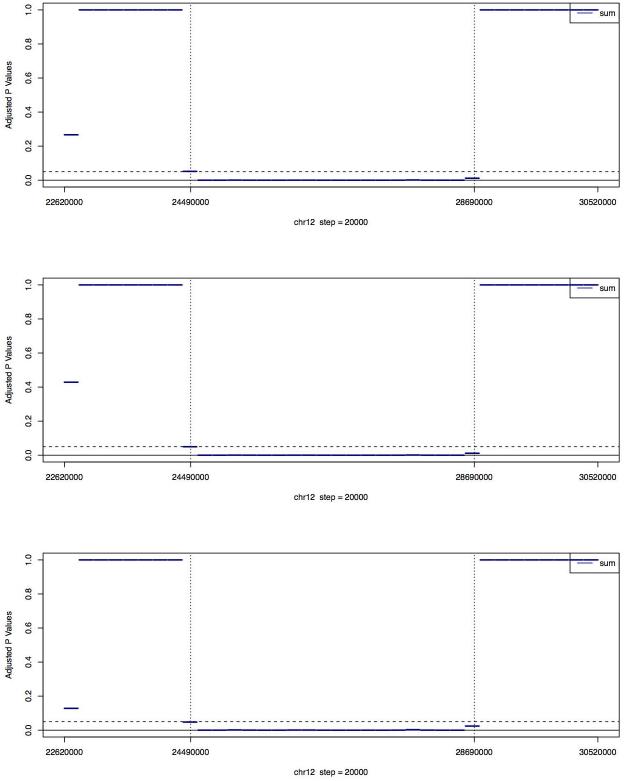
Figure 3A. Medians of adjusted p values (using Holm’s method) of the MINK tests for PL565 on the 36 regions of chr12.
The regions between the two vertical dashed lines are where the microdeletion occurs. Each region is 200kb long. For each region with complete or partial deletion (chr12:24380000~28760000), three MINK tests, each against a different reference library, were performed, using the 16 regions covering chr12:22620000~2436000 and chr12:28780000~30520000 as the control regions. For each of the 16 regions with no deletion, three MINK tests are using the other 15 regions with no deletion as control regions. For each region, the median of the adjusted p values reported by the three MINK tests is plotted. The three plots, from top to bottom, are based on the full, one-half, and one-quarter of the PL565 sequence reads respectively.
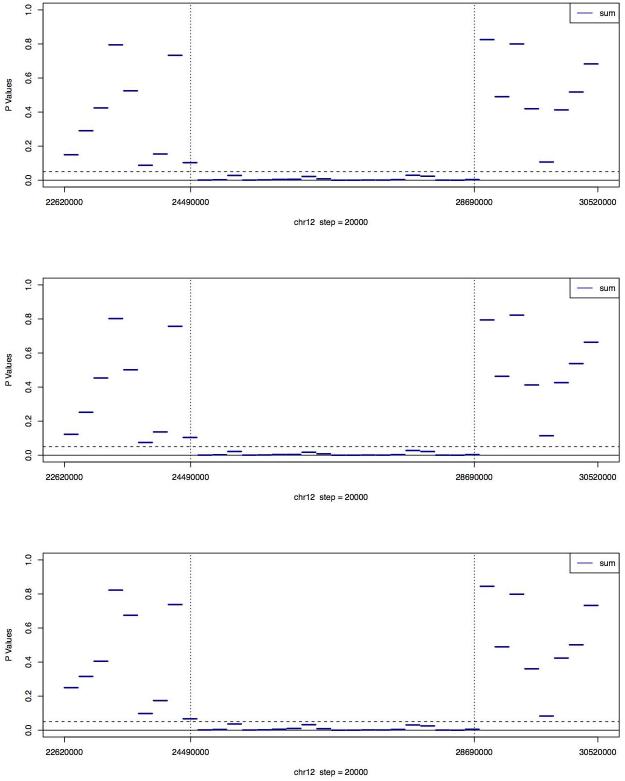
Using the 16 regions that aligned outside the deleted region as reference regions, and another 3 normal libraries as reference libraries, the GCREM algorithm was applied to test, for each of the 18 regions located inside the deletion area and the 2 regions covering the junctions, whether that region in the PL565 library carried deletion/insertion. For PL565, the observed log tag counts within the boundaries of the deleted region were clearly reduced relative to the predicted log tag counts (Figure 1). The resulting p values were adjusted using the Holm’s method to control the family-wise error rate 23. Significant adjusted p values (<= 0.05) were found for all 18 regions inside the deleted sequence, and non-significant p values for all 16 reference regions (Figure 2), demonstrating that the targeted capture method, combined with the GCREM algorithm, is able to report significant adjusted p values for all the deleted regions and non-significant adjusted p values for all normal regions at a resolution of 200Kb. Use of the MINK algorithm, without adjustment of p values correctly identifies all the deletion regions and all the normal regions (Fig 3B). However, the adjusted MINK p values for the deletion regions are not always significant (Fig 3A), indicating a lower sensitivity of MINK compared to the GCREM algorithm.
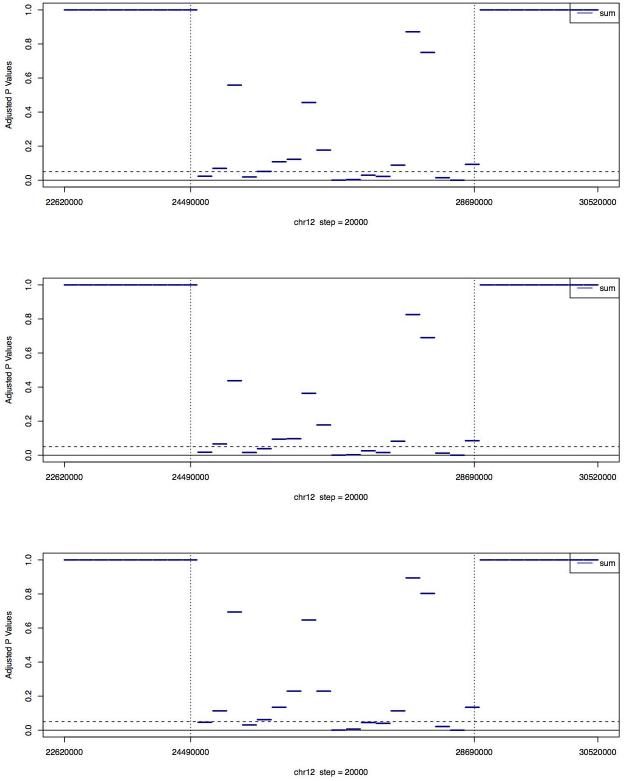
Figure 2. Adjusted p values (using Holm’s method) of the GCREM tests for PL565 on the 36 regions of chr12.
Each region is 200kb long. The regions between the two vertical dashed lines are where the microdeletion occurs. For each region with complete or partial deletion (chr12:24380000~28760000), the GCREM test is using the 16 regions covering chr12:22620000~2436000 and chr12:28780000~30520000 as the control regions. For each of the 16 regions with no deletion, the test is using the other 15 regions with no deletion as control regions. The three plots, from top to bottom, are based on the full, one-half, and one-quarter of the PL565 sequence reads respectively.
To explore whether we could detect the deleted regions using fewer sequencing reads, we randomly sampled one-half and one-quarter of the reads in library PL565 and found that GCREM was able to report significant adjusted p values for all the deleted regions and non-significant adjusted p values for all normal regions, even when using only 50% or 25% of the total number of PL565 sequence reads (Figure 2). In contrast, MINK was unable to return significant adjusted p values for all deleted regions using either 50% or 25% of the PL565 data (Figure 3A). In our experience, 25% of the PL565 reads corresponds to approximately 5% of the capacity of a single HiSeq2000 paired-end v3 flow cell.
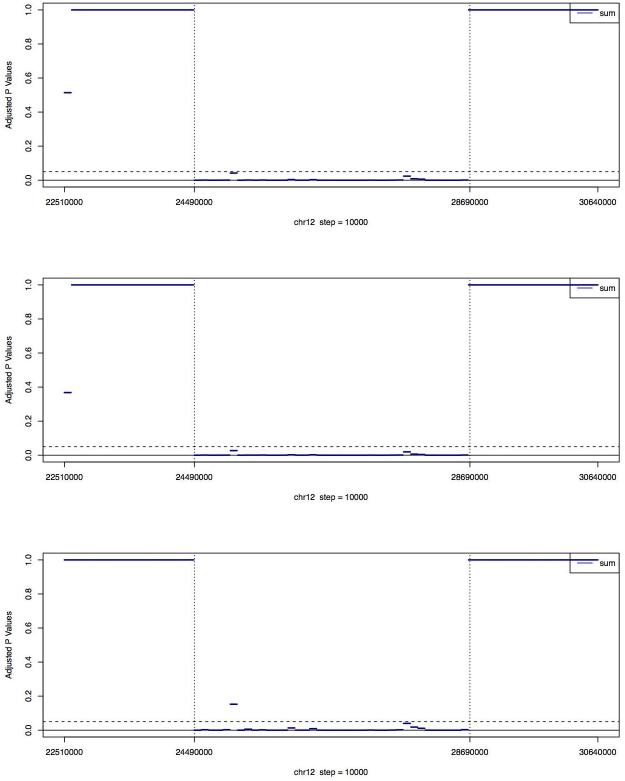
We further investigated the sensitivity of the GCREM method by dividing the 8Mb target region into 76 non-overlapping regions of approximately 100Kb. When using all reads or half of the reads in PL565, we were able to report significant adjusted p values for all deleted regions, and non-significant adjusted p values for all normal regions. When using only one-quarter of PL565, we got almost the same results, except for one deleted region, for which we reported a non-significant adjusted p value (Figure 4). This suggests that our algorithm can reach a practical level of resolution of 100kb at a reasonable read depth using approximately 11 million sequence reads. Moreover, even using only one-quarter of PL565, the adjusted GCREM p values for the deletion regions and normal regions were well separated. More precisely, the GCREM algorithm’s Area Under the receiver operating characteristic Curve (AUC) was 1. In contrast, the adjusted MINK p values for the deletion regions were no longer well separated from the adjusted p values for the normal regions at the 100kb resolution, indicating a lower sensitivity of MINK compared to the GCREM algorithm. Using DeLong’s test for AUC 24 the AUC of the GCREM algorithm was significant higher than the AUC of the MINK algorithm at the 100k resolution (p value <= 0.05), regardless whether the full, or one-half, or one-quarter of the PL565 library was used.
Figure 3B. Medians of unadjusted p values of the MINK tests for PL565 on the 36 regions of chr12.
The regions between the two vertical dashed lines are where the microdeletion occurs. The three plots, from top to bottom, are based on the full, one-half, and one-quarter of the PL565 sequence reads respectively.
Discussion
We present a novel statistical algorithm (GCREM) for fetal copy number mutation testing that can automatically correct for the GC bias in whole genome sequencing data from maternal plasma DNA. We also provide proof of concept for the non-invasive prenatal diagnosis (NIPD) of a fetal microdeletion at high resolution via the targeted sequence capture of maternal plasma DNA. Specifically, we are able to reliably detect the presence of 100Kb of a heterozygous deletion using approximately 5% of the available reads from a single HiSeq2000 paired-end v3 flow cell. Notably, our previously published statistical method, which works adequately for the NIPT of aneuploidy 22 was not as effective in this context. While using the MINK p values without adjustment we can correctly identify all the deletion regions and all the normal regions (Fig 3B), whereas the adjusted MINK p values for the deletion regions are not always significant (Fig 3A) indicating a lower sensitivity compared to the GCREM algorithm.
The development of NIPD assays for genomic imbalances such as microdeletions is an important goal because current DNA sequencing based methods for NIPD are specifically focused on aneuploidy 10, 11, 19, 22, 25, despite the fact that a wide range of other genomic imbalances are a major cause of perinatal morbidity and mortality 26, 27. Currently, such anomalies can only be diagnosed in utero via the use of invasive approaches, followed by karyotyping or comparative genomic hybridization (CGH).
One disadvantage of targeted approaches is the requirement that regions of interest must be identified in advance. However, there are certain advantages to focusing only on specific genomic regions of interest, including the fact that this reduces the complication of reporting incidental findings or those of unknown clinical significance (Krier and Green). As we have shown, a second advantage is that the resolution of the targeted approach is far higher than can currently be achieved via a whole genome shotgun approach. Another negative element of the approach we describe is the need to subject plasma DNA to solution phase targeted capture. This adds a number of steps to the sample preparation and requires financial investment in the capture reagents. However, these are offset by the relatively small amount of sequence data required. Specifically, our data show that around 5% or less of the sequence reads required by other recently published methods are sufficient for detection of a single microdeletion at this resolution. Importantly, the read requirement would increase only slightly when multiple loci are interrogated in parallel because we do not need to increase the size of the control region.
The approach described herein, coupled with our previously described whole genome approach (Peters et al., 2011), has the potential to dramatically extend the diagnostic utility of emerging tests for NIPT and represents a key step in the development of risk-free non-invasive alternatives to conventional karyotyping and CGH. Significantly, the combination of targeted capture and the GCREM algorithm makes it possible to detect relatively small fetal mutations with low sequencing cost and this is potentially scalable. We anticipate therefore that, in future, a multiplexed targeted NIPT approach, coupled with sophisticated statistical analysis such as that provided by GCREM, will enable the development of routine non-invasive diagnostic tests for a range of structural chromosomal anomalies and related genetic disorders.
What is already known about this topic?
Previously demonstrated that whole genome sequencing of maternal plasma DNA can be used for the non-invasive detection of fetal microdeletions and microduplications with resolution >1Mb.
What does this study add?
We show targeted sequencing can accurately increase the resolution of microdeletion / microduplication detection to 100Kb.
Significantly fewer sequencing reads are required compared to whole genome approaches.
The method is scalable so that multiple genomic regions of interest can be included.
Figure 4. Adjusted p values (using Holm’s method) of the GCREM tests for PL565 on the 74 regions of chr12.
Each region is 100kb long. The regions between the two vertical dashed lines are where the microdeletion occurs. For each region with complete or partial deletion, the GCREM test is using the 35 regions with no deletion, covering chr12:22510000~24370000 and chr12:28670000~30640000, as the control regions. For each of the 35 regions with no deletion, the test is using the other 34 regions with no deletion as control regions. The three plots, from top to bottom, are based on the full, one-half, and one-quarter of the PL565 sequence reads respectively.
Table 1.
NIPD of a fetal microdeletion on chromosome 12 via targeted region capture. Boundaries of the 36 regions of interest at 200Kb resolution are shown, along with region-specific tag counts and mutation status. Clear and shaded rows represent reference and deleted regions respectively.
| Region | Start | End | PL565 Tag Count |
Fetal Status |
|---|---|---|---|---|
| chr12.2 | 22620000 | 22820000 | 196828 | Normal |
| chr12.3 | 22840000 | 23040000 | 220892 | Normal |
| chr12.4 | 23060000 | 23260000 | 216660 | Normal |
| chr12.5 | 23280000 | 23480000 | 199991 | Normal |
| chr12.6 | 23500000 | 23700000 | 166721 | Normal |
| chr12.7 | 23720000 | 23920000 | 230286 | Normal |
| chr12.8 | 23940000 | 24140000 | 218677 | Normal |
| chr12.9 | 24160000 | 24360000 | 252043 | Normal |
| chr12.10 | 24380000 | 24580000 | 222943 | Partial Deletion |
| chr12.11 | 24600000 | 24800000 | 209772 | Deletion |
| chr12.12 | 24820000 | 25020000 | 209038 | Deletion |
| chr12.13 | 25040000 | 25240000 | 165276 | Deletion |
| chr12.14 | 25260000 | 25460000 | 175128 | Deletion |
| chr12.15 | 25480000 | 25680000 | 168384 | Deletion |
| chr12.16 | 25700000 | 25900000 | 164063 | Deletion |
| chr12.17 | 25920000 | 26120000 | 194770 | Deletion |
| chr12.18 | 26140000 | 26340000 | 201582 | Deletion |
| chr12.19 | 26360000 | 26560000 | 186357 | Deletion |
| chr12.20 | 26580000 | 26780000 | 204741 | Deletion |
| chr12.21 | 26800000 | 27000000 | 184468 | Deletion |
| chr12.22 | 27020000 | 27220000 | 173967 | Deletion |
| chr12.23 | 27240000 | 27440000 | 171138 | Deletion |
| chr12.24 | 27460000 | 27660000 | 183489 | Deletion |
| chr12.25 | 27680000 | 27880000 | 204760 | Deletion |
| chr12.26 | 27900000 | 28100000 | 175254 | Deletion |
| chr12.27 | 28120000 | 28320000 | 190028 | Deletion |
| chr12.28 | 28340000 | 28540000 | 192406 | Deletion |
| chr12.29 | 28560000 | 28760000 | 201233 | Partial Deletion |
| chr12.30 | 28780000 | 28980000 | 188014 | Normal |
| chr12.31 | 29000000 | 29200000 | 230399 | Normal |
| chr12.32 | 29220000 | 29420000 | 177159 | Normal |
| chr12.33 | 29440000 | 29640000 | 203176 | Normal |
| chr12.34 | 29660000 | 29860000 | 213385 | Normal |
| chr12.35 | 29880000 | 30080000 | 178934 | Normal |
| chr12.36 | 30100000 | 30300000 | 195746 | Normal |
| chr12.37 | 30320000 | 30520000 | 176586 | Normal |
Acknowledgements
This work was supported by grants from the NIH (R01 HD068578) and the Magee-Womens Research Institute and Foundation (to DGP). The funders had no role or influence regarding study design, data collection, data analysis, manuscript preparation and/or publication decision.
Footnotes
Conflict of Interest: The authors have no conflict of interest.
References
- 1.Hertling-Schaal E, Perrotin F, de Poncheville L, et al. Maternal anxiety induced by prenatal diagnostic techniques: detection and management. Gynecol Obstet Fertil. 2001 Jun;29(6):440–6. doi: 10.1016/s1297-9589(01)00158-8. [DOI] [PubMed] [Google Scholar]
- 2.Hewison J, Nixon J, Fountain J, et al. Amniocentesis results: Investigation of anxiety. The ARIA trial. Health Technol Assess. 2006 Dec;10(50):iii. doi: 10.3310/hta10500. ix-x, 1-78. [DOI] [PubMed] [Google Scholar]
- 3.Hewison J, Nixon J, Fountain J, et al. A randomised trial of two methods of issuing prenatal test results: the ARIA (Amniocentesis Results: Investigation of Anxiety) trial. BJOG. 2007 Apr;114(4):462–8. doi: 10.1111/j.1471-0528.2007.01276.x. [DOI] [PubMed] [Google Scholar]
- 4.Mujezinovic F, Alfirevic Z. Procedure-related complications of amniocentesis and chorionic villous sampling: a systematic review. Obstet Gynecol. 2007 Sep;110(3):687–94. doi: 10.1097/01.AOG.0000278820.54029.e3. [DOI] [PubMed] [Google Scholar]
- 5.Odibo AO, Gray DL, Dicke JM, et al. Revisiting the fetal loss rate after second-trimester genetic amniocentesis: a single center's 16-year experience. Obstet Gynecol. 2008 Mar;111(3):589–95. doi: 10.1097/AOG.0b013e318162eb53. [DOI] [PubMed] [Google Scholar]
- 6.Tabor A, Alfirevic Z. Update on procedure-related risks for prenatal diagnosis techniques. Fetal Diagn Ther. 2010;27(1):1–7. doi: 10.1159/000271995. [DOI] [PubMed] [Google Scholar]
- 7.Tabor A, Philip J, Madsen M, et al. Randomised controlled trial of genetic amniocentesis in 4606 low-risk women. Lancet. 1986 Jun 7;1:8493, 1287–93. doi: 10.1016/s0140-6736(86)91218-3. [DOI] [PubMed] [Google Scholar]
- 8.Bianchi DW, Platt LD, Goldberg JD, et al. Genome-wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet Gynecol. 2012 May;119(5):890–901. doi: 10.1097/AOG.0b013e31824fb482. [DOI] [PubMed] [Google Scholar]
- 9.Ehrich M, Deciu C, Zwiefelhofer T, et al. Noninvasive detection of fetal trisomy 21 by sequencing of DNA in maternal blood: a study in a clinical setting. Am J Obstet Gynecol. 2011 Mar;204(3):205. doi: 10.1016/j.ajog.2010.12.060. e1-11. [DOI] [PubMed] [Google Scholar]
- 10.Palomaki GE, Deciu C, Kloza EM, et al. DNA sequencing of maternal plasma reliably identifies trisomy 18 and trisomy 13 as well as Down syndrome: An international collaborative study. Genet Med. 2012 Mar;14(3):296–305. doi: 10.1038/gim.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palomaki GE, Kloza EM, Lambert-Messerlian GM, et al. DNA sequencing of maternal plasma to detect Down syndrome: An international clinical validation study. Genet Med. 2011 Nov;13(11):913–20. doi: 10.1097/GIM.0b013e3182368a0e. [DOI] [PubMed] [Google Scholar]
- 12.Jensen TJ, Dzakula Z, Deciu C, et al. Detection of microdeletion 22q11.2 in a fetus by next-generation sequencing of maternal plasma. Clin Chem. 2012 Jul;58(7):1148–51. doi: 10.1373/clinchem.2011.180794. [DOI] [PubMed] [Google Scholar]
- 13.Peters D, Chu T, Yatsenko SA, et al. Noninvasive prenatal diagnosis of a fetal microdeletion syndrome. N Engl J Med. 2011 Nov 10;365(19):1847–8. doi: 10.1056/NEJMc1106975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Srinivasan A, Bianchi DW, Huang H, et al. Noninvasive detection of fetal subchromosome abnormalities via deep sequencing of maternal plasma. Am J Hum Genet. 2013 Feb 7;92(2):167–76. doi: 10.1016/j.ajhg.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu SC, Jiang P, Choy KW, et al. Noninvasive prenatal molecular karyotyping from maternal plasma. PLoS One. 2013;8(4):e60968. doi: 10.1371/journal.pone.0060968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hillman SC, Pretlove S, Coomarasamy A, et al. Additional information from array comparative genomic hybridization technology over conventional karyotyping in prenatal diagnosis: a systematic review and meta-analysis. Ultrasound Obstet Gynecol. 2011 Jan;37(1):6–14. doi: 10.1002/uog.7754. [DOI] [PubMed] [Google Scholar]
- 17.Shaffer LG, Dabell MP, Fisher AJ, et al. Experience with microarray-based comparative genomic hybridization for prenatal diagnosis in over 5000 pregnancies. Prenat Diagn. 2012 Oct;32(10):976–85. doi: 10.1002/pd.3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klopocki E, Hennig BP, Dathe K, et al. Deletion and point mutations of PTHLH cause brachydactyly type E. Am J Hum Genet. 2010 Mar 12;86(3):434–9. doi: 10.1016/j.ajhg.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan HC, Blumenfeld YJ, Chitkara U, et al. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci U S A. 2008 Oct 21;105(42):16266–71. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo YM, Tein MS, Lau TK, et al. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet. 1998 Apr;62(4):768–75. doi: 10.1086/301800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiu RW, Cantor CR, Lo YM. Non-invasive prenatal diagnosis by single molecule counting technologies. Trends Genet. 2009 Jul;25(7):324–31. doi: 10.1016/j.tig.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 22.Chu T, Bunce K, Hogge WA, et al. Statistical model for whole genome sequencing and its application to minimally invasive diagnosis of fetal genetic disease. Bioinformatics. 2009 May 15;25(10):1244–50. doi: 10.1093/bioinformatics/btp156. [DOI] [PubMed] [Google Scholar]
- 23.Holm S. A simple sequentially rejective multiple test procedure. Scand J Statistic. 1979;6:65–70. [Google Scholar]
- 24.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988 Sep;44(3):837–45. [PubMed] [Google Scholar]
- 25.Chiu RW, Chan KC, Gao Y, et al. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc Natl Acad Sci U S A. 2008 Dec 23;105(51):20458–63. doi: 10.1073/pnas.0810641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lupski JR, Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005 Dec;1(6):e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaffer LG, Lupski JR. Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu Rev Genet. 2000;34:297–329. doi: 10.1146/annurev.genet.34.1.297. [DOI] [PubMed] [Google Scholar]