Abstract
Background
The aim of this study was to determine whether miR-210 can affect the apoptosis and proliferation of human U251 glioma cells from down-regulating SIN3A.
Material/Methods
The expression of miRNA-210 was detected by quantitative real-time PCR in normal brain tissue and glioma samples. The apoptosis and proliferation ability of U251 cells were analyzed by MTT and flow cytometry assay after anti-miR-210 transfection. For the regulation mechanism analysis of miR-210, TargetScan, PicTar, and microRNA were selected to predict some potential target genes of miR-210. The predicted gene was identified to be the direct and specific target gene of miR-210 by luciferase activities assay and Western blot. RNA interference technology was used to confirm that the apoptosis and proliferation effects of miR-210 were directly induced by SIN3A.
Results
The expression of miR-210 increased significantly in glioma in comparison with normal brain tissue. The silence of miR-210 expression could inhibit the proliferation of U251 cells and induce the apoptosis. Mechanism analysis revealed that SIN3A was a specific and direct target gene of miR-210. The siRNA-SIN3A could down-regulate the expression of SIN3A protein, which was up-regulated in U251 cells by anti-miR-210 transfection, and our experiments found that silence of SIN3A could inhibit the apoptosis and sharply increase the proliferation of U251 cells. The regulation effects of anti-miR-210 on apoptosis and proliferation can be reversed respectively by the expression silence of SIN3A.
Conclusions
Aberrantly expressed miR-210 regulates human U251 glioma cells apoptosis and proliferation partly through directly down-regulating SIN3A protein expression. This might offer a new potential therapeutic stratagem for glioma.
MeSH Keywords: Glioma, MicroRNAs, Neoplastic Stem Cells
Background
Glioma is the most general primary tumor in nervous system, and has the highest mortality and mortality among endocranial tumors, because of the marked characteristics of malignant proliferation and invasion [1,2]. Gliomas are rarely curable. Treatment for a glioblastoma is customized to the individual patient and may include surgery, radiation therapy, chemotherapy, or observation. So far, surgery is the most important initial approach. Chemotherapy and radiotherapy after initial surgical resection is regarded as an effective treatment plan to prevent recurrence and metastasis [3]. However, chemotherapy does not work for everyone with a glioma and it only helps about half the people treated. For this reason, the prognosis for patients, especially with high-grade glioma, is still gloomy, despite being given prompt and comprehensive treatment. The median survival time for adults with an anaplastic astrocytoma is about 2–3 years, and for those with more aggressive glioblastomas, median survival drops off to about 12~14.6 months, with a 2-year median survival rate of 30% [4].
MicroRNAs (miRNAs) are small endogenous RNAs that are noncoding. MiRNAs display abnormal expression and functions in various kinds of malignancies, which act as different kinds of tumor-related genes [5,6]. MiR-210 is an oncogene that is high-expressed in some kinds of malignant tumors, including glioblastomas. MiR-210 can specifically inhibit many target genes helpful to apoptosis and/or inhibitive to proliferation [7,8]. Evidence suggests that miR-210 is an anti-apoptotic factor and functions at least in part by targeting a host of pro-apoptotic genes [9,10]. MiR-210 could silence the expression of some genes helpful to apoptosis and inhibit apoptosis in cancer cells.
Our previous microarray assays found miR-210 had higher expression in glioma samples than in controls, suggesting that miR-210 might be an oncogene in tumorigenesis of glioma. In this study, we investigated how the molecular mechanism of miR-210 regulates glioma cells apoptosis and proliferation. Further related research might help the development of new therapeutic stratagems against glioblastoma.
Material and Methods
Clinical specimens
The study was approved by the Ethics Committees of Chian Medical University, and we obtained patient permission before surgery. A total of 46 glioma and corresponding paracancerous tissues were obtained through surgery. The patients had not experienced radiation or chemotherapy before surgery. The patient group included 31 men and 15 women (mean age: 53.9±3.7 years, age range: 47–68 years); and included 20 cases of astrogliomas (Grade I–II), 15 cases of anaplastic gliomas (Grade III), and 11 cases of glioblastomas (GBM, Grade IV). After surgery, the tissue samples were stored at −80°C, and the pathological information was obtained soon after.
Quantitative real time-PCR (qRT-PCR)
After total RNA was extracted from tissue and cell samples, cDNA was synthesized and used to detect the mRNA expression with an miRNA detection kit (Invitrogen, USA) [11]. The samples were normalized to 18s and the 18<CT<30 were calculated with 2−ΔΔCT using the Applied Biosystems 7500.
Transfection
Twenty-four hours before transfection, an appropriate concentration (about 80%) of resuspended U251 cells were seeded on 6-well plates. The miRNAs (Life Technologies, USA) were transfected into U251 cells with Lipofectamine™ 2000 reagent (Invitrogen, USA), and the final concentration of miRNAs was 60 nM. The transfected cells were incubated for 4 h, and normal media was added. After 48 h, the cells were harvested for further detection.
Cell proliferation assay
Cells were seeded in 96-well plate with 2×104 cells per well. Twenty μl of 0.5 mg/ml MTT solution (Sigma, USA) was added to each well, and the 96-well plate was incubated at 37°C. We cleaned up the media after 4 h, and added 0.2 ml DMSO to each well. The 96-well plate was incubated 30 min, and read on an enzyme-labeled instrument (Bio-Rad, USA) with 570-nm wavelength. The obtained numerical values were used to construct the cell viability. The cell viability=(OD value of test group − OD value of blank group)/(OD value of control group − OD value of blank group).
Flow cytometry detection
Cells (5×106) were harvested and the Annexin V-FITC apoptosis detection kit (Biosea, China) was used to examine the apoptosis rate in accordance with the manufacturer’s instruction. The cells were mixed with 10 μL Annexin and incubated at room temperature away from light for 15 min, and then 5 μL propidium iodide (PI, 10 mg/L) was added to each. Then the cells were read by flow cytometry (BD, USA) (Ex 488 nm, Em 635 nm) and the obtained numerical values were analyzed with CELLQuest 3.0 software (BD, USA). Annexin V-positive cells were regarded as apoptosis cells. The cells were counted by a dual-color flow cytometric method.
Target prediction
The prediction of the SIN3A 3′-UTR as a miRNA binding target was determined using TargetScan (www.targetscan.org), microRNA (www.microrna.org), and PicTar (pictar.mdc-berlin.de). MiRNAs that were simultaneously predicted by all 3 programs were selected for this study.
Vector construction and Luciferase reporter assay
The luciferase reporter vectors were constructed by Invitrogen. The SIN3A 3′-UTR, which included miR-210 seed binding sites, was cloned into the pGL3-promotor vector to construct the wide type (WT) vector, and the predicted seed zones with miR-210 were replaced by nonsense sequences in mutation type (MuT) vector.
In luciferase reporter assay, 100 ng SIN3A 3′UTR luciferase construct and 400 ng microRNA mimics were cotransfected into U251 cells, and Dual-Glo Luciferase assay was applied to quantitated the relative luciferase activity, which was computed by luciferase activity of firefly to Renilla. To normalize the transfection efficiency, the β-galactosidase expression vector was used in every transfection.
Western blot
Cells were harvested and extracted protein. SDS-PAGE electrophoresis and antibody hybridization were practiced as describe previously [12]. The ECL analysis system (Santa Cruz, USA) was used for detection in accordance with the manufacture’s protocol. Western blot quantification was performed using Image Processing and Analysis software. GAPDH was selected as the reference protein.
Small RNA interference
The specific small interfering RNAs (siRNA) to SIN3A gene and matching negative control oligonucleotides were synthesized by Invitrogen and transfected into U251 cells. At 48 h post-transfection, the SIN3A protein was detected to identify the silencing efficiency.
Statistical analysis
All experiments were repeated 3 times. All numerical data are presented as mean ± standard deviation (SD) and analyzed using with SPSS 13.0 software (SPSS, USA). One-way analysis of variance was used to analyze statistical significance. When P value was lower than 0.05, there was statistical significance, marked by (*). If P value was lower than 0.001, the statistical significance existed, indicated by (**).
Results
MiR-210 up-regulated in glioma specimens
To research the potential effect of miR-210 in glioma, the expression of miR-210 was first detected in 46 glioma and corresponding paracancerous tissues samples. In contrast to matching paracancerous tissues, the miR-210 expression in glioma increased remarkably (p<0.001) (Figure 1). The miR-210 expression in glioma U251 cells was similar to that in glioma tissue, and much higher than that in paracancerous tissues. Further, the expression of miR-210 was positively correlated with the pathologic grades (r=0.197, P=0.016, Pearson’s correlation coefficient; ie, negatively correlated with the differentiation) of glioma (P<0.05) (Table 1).
Figure 1.
qRT-PCR analysis for the expression of miR-210 and SIN3A in brain glioma tissue and U251 cells. Compared to paracancerous tissue, the expression of miR-210 was higher in brain glioma tissue and U251 cells (P<0.05), and SIN3A showed significant down-regulation (P<0.05). *, # P<0.05 vs. paracancerous tissue.
Table 1.
Correlation between miR-210 expression in brain glioma tissue and pathological differentiation.
| Differentiation (Grade) | Number of cases | miR-210 expression relative quantification (gliomas/peri-cancerous tissues) | P value |
|---|---|---|---|
| Medium-well differentiated (Grade I–II) | 20 | 1.425±0.152 | <0.05 |
| Anaplastic glioma (Grade: III) | 15 | 1.529±0.157 | |
| Glioblastoma (Grade: IV) | 11 | 1.739±0.161 |
A one-way ANOVA showed that miR-210 expression in brain glioma tissue was statistically different among the three groups with different pathological differentiation of glioma (P<0.05).
LSD test showed significant difference between groups of medium-well differentiated glioma (Grade I–II) and anaplastic glioma (Grade: III) (P=0.03), as well as groups of medium-well differentiated glioma (Grade I–II) and Glioblastoma (GBM, Grade: IV) (P=0.01).
MiR-210 functions as an oncogene in glioma cells
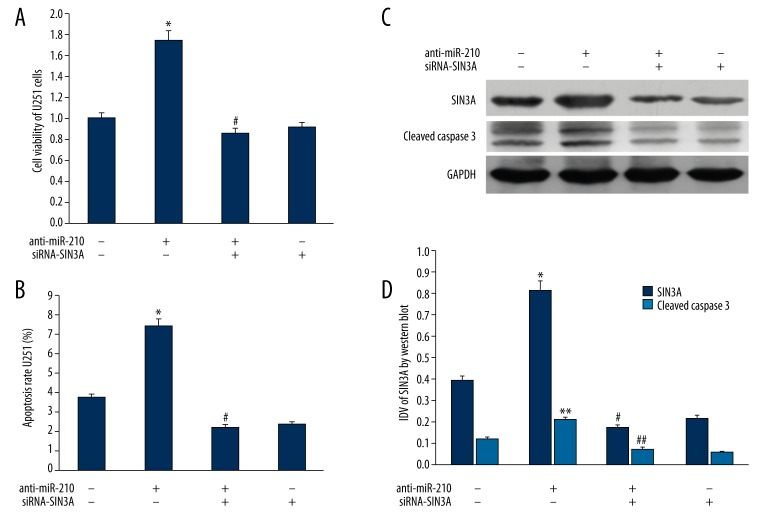
We investigated whether miR-210 influences the apoptosis and proliferation using U251 cells. The anti-miR-210 was transfected into U251 cells to inhibit the miR-210 expression. In comparison to control U251 cells, the proliferation ability of U251 cells transfected with anti-miR-210 was markedly reduced by MTT assay (Figure 2A). The flow cytometry showed that the apoptosis rate of U251 cells transfected with anti-miR-210 was 6.41% and that of control U251 cells was 3.27%; the statistical analysis revealed that the apoptosis rate increased significantly (Figure 2B). The expression of cleaved caspase3 showed a significant increasing after anti-miR-210 transfection (Figure 2C, 2D).
Figure 2.
(A) Impact of SIN3A gene expression changes on the proliferation ability of U251 cells. (B) Impact of SIN3A gene expression changes the apoptosis rate of U251 cells. (C) Western blot analysis for SIN3A and cleaved Caspase3 expression of U251 cells treated with anti-miR-210 and siRNA-SIN3A transfection. GAPDH was used as a reference control. (D) quantitative analysis of the relative protein levels of SIN3A and cleaved Caspase3 normalized to those of GAPDH was shown. *, ** P <0.05 vs. control U251 cells; #, ## P <0.05 vs. U251 cells with anti-miR-210. IDV is the abbreviation for “integrated density values”, which is calculated by computerized image analysis system (Fluor Chen 2.0) and normalized with that of GAPDH.
SIN3A was a direct and specific target of miR-210 in U251 cells
We then investigated the mechanisms by which miR-210 affects the apoptosis and proliferation phenotype in U251 cells. Online search to the target genes of miR-210 by PicTar, microRNA, and TargetScan indicated that SIN3A gene was a possible target of miR-210 (Figure 3A).
Figure 3.
(A) 3′UTR of SIN3A is a target of miR-210 predicted by TargetScan, microRNA and PicTar. (B) The luciferase reporter assay results with each bar representing values from three independent experiments. The transfection efficiency was normalized by co-transfected renilla luciferase and the light units were calculated by relative luciferase activity of firefly to renilla. * P<0.05. (C) Representative image of the protein level of SIN3A. GAPDH was used as a reference control. (D) quantitative analysis of the relative protein levels of SIN3A normalized to those of GAPDH was shown. Data were mean ± SD of three independent experiments. * P<0.05. # P<0.05.
qRT-PCR was used to detect that the expression of SIN3A gene down-regulated significantly in glioma tissues, in contrast with matching paracancerous tissues (Figure 1). Pearson’s correlation analysis showed a negative relationship between the expression of miR-210 and SIN3A.
The luciferase reporter assay was set-up to identify the direct miR210-SIN3A interaction. The pre-miR-210 and different reporter vectors were cotransfected into U251 cells. As shown in Figure 3B, the relative luciferase activity was much lower in U251 cells cotransfected with WT vector and pre-miR-210 than in other transfected U251 cells. This indicated that miR-210could specifically bind to seed zone of SIN3A 3′UTR to inhibit its expression, but MuT vector could not combine with miR-210 to decrease the relative luciferase activity. SIN3A is a specific and direct target gene of miR-210.
To make sure the regulative effects of miR-210 to endogenous SIN3A expression, the c results certified that pre-miR-210 could increase miR-210 expression level efficiently, and anti-miR-210 showed a contradictory regulative effect. The Western blot result showed that compared with control, the SIN3A protein level was significant suppressed with miR-210 enhancement and was up-regulated significantly in U251 cells transfected with anti-miR-210 (Figure 3C, 3D), whereas the qRT-PCR result showed that endogenous SIN3A expression was not significant alteration in U251 cells (data not shown). The conclusion can be made that miR-210 negatively regulate endogenous SIN3A expression at the post-translational level.
SIN3A expression mediates the effect of miR-210 on U251 cells apoptosis and proliferation
Because miR-210 could regulate the apoptosis and proliferation of U251 cells, and SIN3A was the target gene of miR-210, we studied the physiological role of miR-210-target SIN3A in U251 cells after RNA interference SIN3A. As shown in Figure 2C and 2D, combined with siRNA-control, after siRNA-SIN3A treatment, protein expression of SIN3A was significantly reduced in U251 cells transfected with anti-miR-210.
After the expression of SIN3A protein was inhibited, combined with the U251 cells transfected with anti-miR-210, the proliferation ability was markedly increased (Figure 2A), and the apoptosis radio of U251 cells decreased from 6.41% to 2.52%, which was statistically significant (Figure 2B).
Combined with our substantial evidence that miR-210 inversely regulated SIN3A expression, we believe that miR-210-controlled apoptosis and proliferation of U251 cells is mediated to a large extent through SIN3A down-regulation.
Discussion
Glioma cells have vigorous proliferation ability and low apoptosis rate, and have powerful resistance to radiation, chemotherapy, and biological treatment [13–15]. Those characteristics contribute to malignant growth of tumors and might be a vital reason for the inevitable recurrence of glioma [16].
Aberrant miRNA expression can regulate critical biological behaviors, such as apoptosis and proliferation, which may promote glioma cells development and lead to poor prognosis [17,18]. Our research discovered miR-210 was also over-expressed in brain glioma. Knockdown of miR-210 could increase the apoptosis rate of U251 cells and suppress the proliferation. Thus, we showed that miR-210 is very important in the genesis and development of glioma.
Because miRNA acts by targeting various genes, such as miR-22 and CBP, miR-21 and FASLG, miR-10b and E-cadherin [19–21], we speculate that miR-210 regulated some single genes to modify the regulation network and trigger cell apoptosis and proliferation in U251 cells in our study. Computational algorithms are effective tools to predict and validate the miRNA gene targets. Through analysis using TargetScan, PicTar, and microRNA, a number of important candidate targets for miR-210 were predicted. Among these potential targets, SIN3A is the most intriguing target. The SIN3A gene is a member of the SIN3 transcription regulator family and acts as a transcriptional repressor. SIN3A have been regarded as very important effectors of cell biological characteristics, such as cell cycle and apoptosis and invasion [22–24]. SIN3A gene is thought to modify gene expression through its role as histone deacetylases (HDACs). SIN3A could interact with MXI1 to repress MYC responsive genes and antagonize MYC oncogenic activities [25,26]. Few previous studies have demonstrated the abnormal expression and function of SIN3A in tumors, and there is not a conclusive report about SIN3A in glioma.
Our results obtained from gain-of-function approaches confirmed that SIN3A is miR-210’s direct target gene. First, SIN3A gene is down-regulated in glioma tissues, and its expression showed a negative relationship with the expression of miR-210. Second, over-expression of miR-210 remarkably reduces the relative luciferase activity of WT vector containing SIN3A 3′UTR. Third, mutation at the miR-210 target site in the 3′UTR of SIN3A could significant decrease the miR-210 regulation effect. Fourth, over-expression of miR-210 inhibits the expression of SIN3A protein at the post-translational level, and knockdown of miR-210 shows contrary effects. In summary, we conclude that SIN3A is a direct and pivotal target gene of miR-210.
Because knockdown of miR-210 could induce apoptosis and inhibit proliferation in U251 cells, and because SIN3A is a direct and pivotal target gene of miR-210, we suggest that miR-210 might regulate apoptosis and proliferation through directly down-regulating SIN3A. To verify this hypothesis, the siRNA-SIN3A was transfected to down-regulate the expression of SIN3A, which was up-regulated in U251 cells by anti-miR-210 transfection, and the following experiments found that silence of SIN3A could inhibit the apoptosis and increase the proliferation of U251 cells sharply. The regulation effects of anti-miR-210 on apoptosis and proliferation can be reversed by the expression silence of SIN3A. Accordingly, the verification of SIN3A as a target gene of miR-210 offers a probable explanation for how the over-expression of miR-210 can function as an oncogene in U251 cells.
Conclusions
In summary, aberrantly expressed miR-210 regulates human U251 glioma cells apoptosis and proliferation, partly through directly down-regulating SIN3A protein expression; this might offer a new potential therapeutic stratagem for glioma. Nevertheless, further research is essential to identify the detailed molecular mechanism of miR-210 in the genesis and development of glioma.
Footnotes
Competing interests
All authors announce that they have no competing interests.
Source of support: This work was supported by the National Nature Science Foundation of China (81172408, 81301862)
References
- 1.Kuhnt D, Becker A, Ganslandt O, et al. Correlation of the extent of tumor volume resection and patient survival in surgery of glioblastoma multiforme with high-field intraoperative MRI guidance. Neuro Oncol. 2011;13(12):1339–48. doi: 10.1093/neuonc/nor133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Houdek Z, Cendelín J, Kulda V, et al. Intracerebellar application of P19-derived neuroprogenitor and naive stem cells to Lurcher mutant and wild type B6CBA mice. Med Sci Monit. 2012;18(5):BR174–80. doi: 10.12659/MSM.882726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mrugala MM. Advances and challenges in the treatment of glioblastoma: a clinician’s perspective. Discov Med. 2013;15(83):221–30. [PubMed] [Google Scholar]
- 4.Nieder C, Astner ST, Mehta MP, et al. Improvement, clinical course, and quality of life after palliative radiotherapy for recurrent glioblastoma. Am J Clin Oncol. 2008;31:300–5. doi: 10.1097/COC.0b013e31815e3fdc. [DOI] [PubMed] [Google Scholar]
- 5.Shang C, Lu YM, Meng LR. MicroRNA-125b down-regulation mediates endometrial cancer invasion by targeting ERBB2. Med Sci Monit. 2012;18(4):BR149–55. doi: 10.12659/MSM.882617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H, Guo Y, Shang C, et al. miR-21 downregulated TCF21 to inhibit KISS1 in renal cancer. Urology. 2012;80(6):1298–302. doi: 10.1016/j.urology.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 7.Wang F, Xiong L, Huang X, et al. miR-210 suppresses BNIP3 to protect against the apoptosis of neural progenitor cells. Stem Cell Res. 2013;11(1):657–67. doi: 10.1016/j.scr.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 8.He J, Wu J, Xu N, et al. MiR-210 disturbs mitotic progression through regulating a group of mitosis-related genes. Nucleic Acids Res. 2013;41(1):498–508. doi: 10.1093/nar/gks995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z, Yin B, Wang B, et al. MicroRNA-210 promotes proliferation and invasion of peripheral nerve sheath tumor cells targeting EFNA3. Oncol Res. 2014;21(3):145–54. doi: 10.3727/096504013X13841340689573. [DOI] [PubMed] [Google Scholar]
- 10.Chio CC, Lin JW, Cheng HA, et al. MicroRNA-210 targets antiapoptotic Bcl-2 expression and mediates hypoxia-induced apoptosis of neuroblastoma cells. Arch Toxicol. 2013;87(3):459–68. doi: 10.1007/s00204-012-0965-5. [DOI] [PubMed] [Google Scholar]
- 11.Shang C, Zhang H, Guo Y, et al. MiR-320a down-regulation mediates bladder carcinoma invasion by targeting ITGB3. Mol Biol Rep. 2014;41(4):2521–27. doi: 10.1007/s11033-014-3110-0. [DOI] [PubMed] [Google Scholar]
- 12.Kumar M, Singh M, Singh SB. Optimization of conditions for expression of recombinant interferon-γ in E. coli. Mol Biol Rep. 2014;41(10):6537–43. doi: 10.1007/s11033-014-3537-3. [DOI] [PubMed] [Google Scholar]
- 13.Dahlrot RH. The prognostic value of clinical factors and cancer stem cell-related markers in gliomas. Dan Med J. 2014;61(10):B4944. [PubMed] [Google Scholar]
- 14.Lim YC, Roberts TL, Day BW, et al. Increased sensitivity to ionizing radiation by targeting the homologous recombination pathway inglioma initiating cells. Mol Oncol. 2014;8(8):1603–15. doi: 10.1016/j.molonc.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukai J, Koizumi F, Nakao N. Enhanced anti-tumor effect of zoledronic acid combined with temozolomide against human malignantglioma cell expressing O6-methylguanine DNA methyltransferase. PLoS One. 2014;9(8):e104538. doi: 10.1371/journal.pone.0104538. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Wang F, Zamora G, Sun CH, et al. Increased sensitivity of glioma cells to 5-fluorocytosine following photo-chemical internalization enhanced nonviral transfection of the cytosine deaminase suicide gene. J Neurooncol. 2014;118(1):29–37. doi: 10.1007/s11060-014-1410-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen T, Wang XY, Li C, Xu SJ. Downregulation of microRNA-124 predicts poor prognosis in glioma patients. Neurol Sci. 2014 doi: 10.1007/s10072-014-1895-1. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 18.Wang L, Shi ZM, Jiang CF, et al. MiR-143 acts as a tumor suppressor by targeting N-RAS and enhances temozolomide-inducedapoptosis in glioma. Oncotarget. 2014;5(14):5416–27. doi: 10.18632/oncotarget.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J, Chen L, Yang J, et al. MicroRNA-22 targeting CBP protects against myocardial ischemia-reperfusion injury through anti-apoptosis in rats. Mol Biol Rep. 2014;41(1):555–61. doi: 10.1007/s11033-013-2891-x. [DOI] [PubMed] [Google Scholar]
- 20.Shang C, Guo Y, Hong Y, et al. MiR-21 up-regulation mediates glioblastoma cancer stem cells apoptosis and proliferation by targeting FASLG. Mol Biol Rep. 2014 doi: 10.1007/s11033-014-3820-3. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Zhao J, Zhang PY, et al. MicroRNA-10b targets E-cadherin and modulates breast cancer metastasis. Med Sci Monit. 2012;18(8):BR299–308. doi: 10.12659/MSM.883262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji Q, Hu H, Yang F, et al. CRL4B interacts and coordinates with SIN3A/HDAC complex to repress CDKN1A in driving cell cycle progression. J Cell Sci. 2014;127(21):4679–91. doi: 10.1242/jcs.154245. [DOI] [PubMed] [Google Scholar]
- 23.Farhana L, Dawson MI, Dannenberg JH, et al. SHP and Sin3A expression are essential for adamantyl-substituted retinoid-related molecule-mediated nuclear factor-kappaB activation, c-Fos/c-Jun expression, and cellular apoptosis. Mol Cancer Ther. 2009;8(6):1625–35. doi: 10.1158/1535-7163.MCT-08-0964. [DOI] [PubMed] [Google Scholar]
- 24.Das TK, Sangodkar J, Negre N, et al. Sin3a acts through a multi-gene module to regulate invasion in Drosophila and human tumors. Oncogene. 2013;32(26):3184–97. doi: 10.1038/onc.2012.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao G, Alland L, Guida P, et al. Mouse Sin3A interacts with and can functionally substitute for the amino-terminal repression of theMyc antagonist Mxi1. Oncogene. 1996;12(5):1165–72. [PubMed] [Google Scholar]
- 26.Garcia-Sanz P, Quintanilla A, Lafita MC, et al. Sin3b interacts with Myc and decreases Myc levels. J Biol Chem. 2014;289(32):22221–36. doi: 10.1074/jbc.M113.538744. [DOI] [PMC free article] [PubMed] [Google Scholar]