Abstract
Background
A meta-analysis was performed to estimate the association between HIF-1α polymorphism (C1772T) and breast cancer risk.
Material/Methods
The relevant published literature was retrieved from PubMed, Web of Knowledge, and Embase. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated to estimate the strength of the associations.
Results
Six case-control studies, including 2043 cases and 2146 controls were identified. Meta-analysis showed that there was no marked association between C1772T polymorphism and breast cancer risk in the overall population in the dominant model. The subgroup analysis showed an increased breast cancer risk in Asians based on homozygote comparison and the recessive model. There were no associations between C1772T polymorphism with clinicopathological parameters and habits.
Conclusions
The present meta-analysis suggests that HIF-1α C1772T polymorphism is a risk factor for susceptibility to breast cancer in Asians.
MeSH Keywords: Breast Disease; Hypoxia-inducible Factor 1, alpha subunit; Polymorphism, Genetic; Meta-Analysis
Background
Breast cancer is the leading cause of death by cancer for females worldwide [1]. Although there are some well-established risk factors identified for breast cancer [2,3], unfortunately, the mechanisms have not been clearly identified. Hypoxia has been confirmed as an important factor in most solid tumors, including breast cancer. A hypoxic microenvironment triggers multiple cellular responses, such as angiogenesis, and involving cell-cycle control proteins, metastasis, and poor prognosis. These responses are highly dependent on the activation of hypoxia-inducible factor-1 (HIF-1). HIF-1 is a key transcription factor that regulates the cellular adaptation to hypoxia [4]. The HIF-1 gene is located on chromosome 14q21–24 and consists of 15 exons and codes the cDNA of 3919 bps [5]. HIF-1 consists of HIF-1α and HIF-1β subunits (also known as aryl hydrocarbon nuclear translocation, ARNT). The HIF-1β subunit is constitutively expressed, while HIF-1α protein expression and transcriptional activity are tightly regulated by the oxygen level [6]. Under normoxic conditions, HIF-1α is rapidly degraded, binding to the von Hippel-Lindau tumor suppressor protein (pVHL), a recognition component for ubiquitination and proteasomal degradation of HIF-1α. In hypoxic conditions, HIF-1α degradation is limited and rapidly accumulated in the nucleus to drive transcription of many survival genes [7]. A number of polymorphisms and mutations within the HIF-1α gene have been identified. The most widely studied single-nucleotide polymorphism (SNP) in HIF-1α, C1772T (P582S, rs11549465), has been identified in exon 12 of the gene within the oxygen-dependent degradation (ODD) domain 11 [8]. The C>T change at 1772 leads rise to Pro/Ser variation at codon 582 [8]. The presence of the polymorphic variant was shown to cause a markedly higher transcriptional activity than the wild type under both normoxic and hypoxic conditions in in vitro studies [9,10]. HIF-1α 1772 C/T genetic polymorphism has been considered to influence risk for many types of cancer, but the results of epidemiological studies are conflicting. C1772T polymorphism was found to confer increased risk of developing glioma [11], cervical cancer [12], endometrial cancer [12], and pancreatic cancer [13] but not liver cancer [14], oral cancer [15], or esophageal cancer [16]. Even in studies on breast cancer, the results were inconsistent. Several studies showed no marked differences between patients and controls in terms of the distribution of C1772T polymorphism [17–19], and others showed opposite conclusions [20–22].
The inconsistent conclusions may due to the relatively small sample sizes and differences in patient ethnic backgrounds. To clarify the association of HIF-1α C1772T polymorphism with breast cancer risk, we estimated the overall cancer risk of this polymorphism by performing a meta-analysis on all eligible case-control studies In addition, we stratified the subgroup analysis according to ethnic backgrounds, clinicopathological parameters, and habits.
Material and Methods
Publication search
Computer searches were performed independently by 2 authors in PubMed (published before June 2013 in English) to collect articles about the association of HIF-1α C1772T polymorphism and breast cancer risk. The following keywords were used: breast cancer, hypoxia-induced factor 1/HIF-1/HIF-1α, C1772T/P582S/rs11549465 and polymorphism/genotype/SNP. We also reviewed reference lists of main reports and review articles by a manual search to identify additional relevant articles.
Inclusion criteria and data extraction
The following criteria were used to choose studies for the meta-analysis: (1) use of a case-control design; (2) evaluated the associations between C1772T polymorphism and breast cancer risk; (3) contained at least 2 comparison groups (cancer group and control group); (4) included detailed data. Accordingly, articles lacking data or containing data inappropriate for meta-analysis, review articles without original data, and case reports were excluded. Two reviewers independently extracted data and all authors discussed disputable data. The following characteristics were recorded from each study: the first author’s surname, year of publication, country of origin, patient ethnicity, source of control, number of cases and controls, genotyping methods, and number of various genotypes. Subjects were extracted separately according to ethnicity, clinicopathological parameters, and habits for subgroup analyses. The clinicopathological parameters included menopausal status, clinical stage, lymph nodes, histological grade, and estrogen receptor. The habits included alcohol consumption and cigarette smoking.
Statistical analysis
The associations between HIF-1α C1772T polymorphism and breast cancer risk were measured using odds ratio (OR) with 95% confidence interval (CI). We used the Z test to assess the significance of the Ors, and I2 and Q statistics to determine the statistical heterogeneity among studies. We used a random-effects model when P value of heterogeneity tests was no more than 0.1 (P≤0.1); if not, we used a fixed-effects model. Sensitivity analyses were performed to assess the stability of the results by removing 1 study at a time to reflect the influence of the individual data set on the pooled OR. Publication bias was evaluated by funnel plot and Egger’s test. We investigated the strength of the association in the allele model (T vs. C), the dominant model (TT+CT vs. CC), and the recessive model (TT vs. CT+CC), respectively. P value <0.05 were considered significant. All statistical analyses were performed using Review Manager version 5.1 (Revman; The Cochrane Collaboration, Oxford, UK).
Results
Study characteristics
After searching 13 articles meeting the search criteria, we identified 6 relevant publications, including 2143 cases and 2046 controls, to assess the relationship of HIF-1α C1772T polymorphism and breast cancer (Figure 1) [17–22]. The characteristics of the included studies are listed in Table 1. Three studies [17–19] were performed in Caucasians and 3 [20–22] were based in Asians. All studies were case-control, including 3 population-based studies and 3 hospital-based studies.
Figure 1.
Flow chart of study selection.
Table 1.
Characteristics of the studies included in the meta-analysis.
| First author | Year | Country | Ethnicity | Study design | Genotyping medthod | Source of control | Total sample size (case/control) |
|---|---|---|---|---|---|---|---|
| Zagouri [18] | 2012 | Greece | Caucasian | CC | PCR-RELP | Hospital | 113/124 |
| Ribeiro [17] | 2013 | Sweden | Caucasian | CC | PCR–RELP | Hospital | 96/74 |
| Naidu [20] | 2009 | Malaysia | Asian | CC | PCR-RELP | population | 410/275 |
| Lee [22] | 2008 | Korea | Asian | CC | PCR | Hospital | 1332/1369 |
| Kim [21] | 2008 | Korea | Asian | CC | Sequencing | population | 90/102 |
| Apaydin [19] | 2008 | Turkey | Caucasian | CC | PCR-RELP | population | 102/102 |
CC – case-control; PCR-RFLP – polymerase chain reaction-restriction fragment length polymorphism.
Quantitative data synthesis
For HIF-1α C1772T polymorphism, a variation in the T allele frequency was identified across the 6 studies, ranging from 0.04 to 0.19 (Table 2). The mean frequency of the T allele in Caucasian populations was 0.11, which was higher than that in Asian populations (0.08). There was no significant difference between Asians and Caucasians (P>0.05).
Table 2.
The genotype and allele frequencies of HIF-1α C1772T polymorphism in cases and controls.
| First author | Genotype (N) | Allele frequency (N) | MAF | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | ||||||||||
| Total | CC | CT | TT | Total | CC | CT | TT | C | T | C | T | ||
| Ribeiro | 96 | 74 | 21 | 1 | 74 | 61 | 9 | 4 | 169 (0.88) | 23 (0.12) | 131 (0.89) | 17 (0.11) | 12.0/11.5 |
| Zagouri | 113 | 98 | 15 | 0 | 124 | 107 | 17 | 0 | 211 (0.93) | 15 (0.07) | 231 (0.93) | 17 (0.07) | 6.6/6.9 |
| Apaydin | 102 | 79 | 21 | 2 | 102 | 68 | 29 | 5 | 179 (0.88) | 25 (0.12) | 165 (0.81) | 39 (0.19) | 12.3/19.1 |
| Naidu | 410 | 294 | 100 | 16 | 275 | 222 | 50 | 3 | 688 (0.84) | 132 (0.16) | 494 (0.90) | 56 (0.10) | 16.1/10.2 |
| Kim | 90 | 81 | 8 | 1 | 102 | 93 | 9 | 0 | 170 (0.94) | 10 (0.06) | 195 (0.96) | 9 (0.04) | 5.6/4.4 |
| Lee | 1332 | 1207 | 119 | 6 | 1369 | 1245 | 123 | 1 | 2533 (0.95) | 131 (0.05) | 2613 (0.95) | 125 (0.05) | 4.9/4.6 |
MAF – minor allele frequencies.
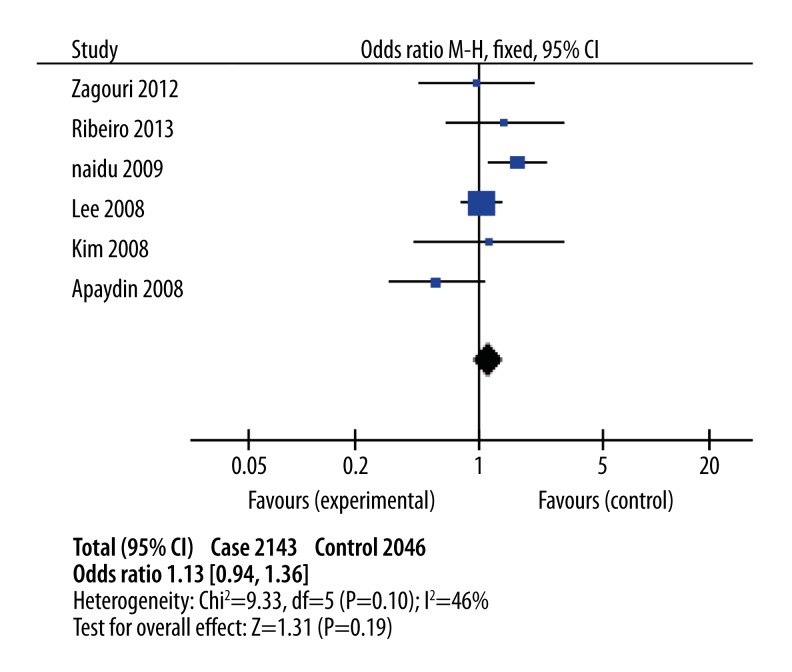
As shown in Table 3, there was no significant association between C1772T polymorphism and breast cancer risk in the overall population in the dominant model (Figure 2, OR 1.13; 95% CI, 0.94–1.36; Pheterogeneity=0.10, P=0.06) and the recessive model (OR1.62; 95% CI 0.83–3.15; Pheterogeneity=0.04; P=0.16).
Table 3.
Meta-analysis results.
| Comparisons | OR | 95%CI | P value | Heterogeneity | Effects model | |
|---|---|---|---|---|---|---|
| I2 | P value | |||||
| T vs. C | ||||||
| Overall | 1.10 | 0.93–1.30 | 0.28 | 60% | 0.03 | R |
| Caucasian | 0.79 | 0.55–1.14 | 0.21 | 4% | 0.35 | F |
| Asian | 1.21 | 0.99–1.46 | 0.06 | 70% | 0.06 | R |
| TT vs. CC | ||||||
| Overall | 1.64 | 0.85–3.19 | 0.14 | 63% | 0.03 | R |
| Caucasian | 0.27 | 0.07–1.01 | 0.05 | 0% | 0.66 | F |
| Asian | 4.42 | 1.60–12.21 | 0.004 | 0% | 0.93 | F |
| CT vs. CC | ||||||
| Overall | 1.05 | 0.87–1.27 | 0.58 | 44% | 0.11 | F |
| Caucasian | 0.96 | 0.63–1.45 | 0.85 | 53% | 0.12 | F |
| Asian | 1.08 | 0.87–1.33 | 0.48 | 44% | 0.11 | F |
| CT+TT vs. CC | ||||||
| Overall | 1.13 | 0.94–1.36 | 0.19 | 46% | 0.10 | F |
| Caucasian | 0.86 | 0.58–1.29 | 0.47 | 36% | 0.21 | F |
| Asian | 1.21 | 0.99–1.49 | 0.06 | 51% | 0.13 | F |
| TT vs. CC+CT | ||||||
| Overall | 1.62 | 0.83–3.15 | 0.16 | 60% | 0.04 | R |
| Caucasian | 0.29 | 0.08–1.09 | 0.07 | 0% | 0.60 | F |
| Asian | 4.16 | 1.51–11.48 | 0.006 | 0% | 0.91 | F |
F – fixed effects model; R – random effects model.
Figure 2.
Combined meta-analyses of association between HIF-1α C1772T polymorphism and risk of breast cancer in the dominant model.
Subgroup analyses were extracted separately according to ethnic background (Table 3) and clinicopathological parameters and habits (Table 4). The subgroup analysis showed C1772T polymorphism was significantly associated with increased breast cancer risk in Asians based on homozygote comparison (OR 4.42; 95% CI 1.60–12.21; Pheterogeneity=0.93 for TT vs. CC; P=0.004) and the recessive model(OR 4.16; 95% CI 1.51–11.48; Pheterogeneity=0.91; P=0.006). Interestingly in Caucasians, we found the opposite result (OR 0.27; 95% CI 0.07–1.01; Pheterogeneity=0.66 for TT vs. CC; P=0.05). Furthermore, when stratified by clinicopathological parameters of breast cancer, there were no associations between C1772T polymorphism and the parameters in the dominant model, such as menopausal status (OR 0.72; 95% CI 0.38–1.34; P=0.50), clinical stage (OR 1.01; 95% CI 0.69–1.48; P=0.34), lymph nodes (OR 1.15; 95% CI 0.86–1.53; P=0.10), histological grade (OR 0.69; 95% CI 0.46–1.05; P=0.34), and estrogen receptor (OR 1.10; 95% CI 0.81–1.50; P=0.62). When data from breast cancer patients were stratified by habits, no significant associations were found between the genotypic distribution and cigarette smoking (P=0.95) and alcohol consumption (P=0.45).
Table 4.
Association between C1772T polymorphism and clinico-pathological parameters and habits of the breast cancer patients based on the dominant models.
| Clinic index | Genotype (N) | Association | Heterogeneity test | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| CC | CT+TT | OR (95% CI) | P | I2 | df | P | |
| Menopausal status | |||||||
| Premenopausal | 93 | 21 | 0.72(0.38–1.34) | 0.30 | 0 | 2 | 0.50 |
| Postmenopausal | 121 | 38 | |||||
|
| |||||||
| Clinical stage | |||||||
| I | 502 | 61 | 1.01(0.69–1.48) | 0.97 | 0 | 1 | 0.34 |
| II+III | 472 | 59 | |||||
|
| |||||||
| Lymph nodes | |||||||
| Negative | 767 | 131 | 1.15(0.86–1.53) | 0.35 | 51 | 3 | 0.10 |
| Positive | 567 | 107 | |||||
|
| |||||||
| Histological grade | |||||||
| I+II | 237 | 68 | 0.69(0.46–1.05) | 0.08 | 45 | 2 | 0.16 |
| III | 164 | 60 | |||||
|
| |||||||
| ER status | |||||||
| Negative | 491 | 95 | 1.10(0.81–1.50) | 0.52 | 0 | 2 | 0.62 |
| Positive | 735 | 116 | |||||
|
| |||||||
| Cigarette smoking | |||||||
| Yes | 15 | 9 | 2.24(0.90–5.55) | 0.08 | 0 | 1 | 0.95 |
| No | 131 | 35 | |||||
|
| |||||||
| Alcohol intake | |||||||
| Yes | 17 | 8 | 1.81(0.69–4.76) | 0.23 | 0 | 1 | 0.45 |
| No | 129 | 35 | |||||
We used Begg’s funnel plot and Egger’s test to assess the publication bias. The funnel plots appeared to be symmetrical in all models and Egger’s test did not reveal any evidence of publication bias (P>0.05).
Discussion
HIF-1α has an important role in carcinogenesis; polymorphisms affecting its activity might conceivably influence tumor progression and aggressiveness. Consequently, multiple studies have been performed assessing the role of HIF-1α polymorphisms as risk factors in cancer. It was reported that HIF-1α1772 C/T polymorphism was a risk factor for many types of cancers, including head and neck, esophageal, lung, colonic, breast, gastric, pancreatic, kidney, prostatic, and endometrial cancer [7,23,24]. In breast cancer, HIF-1α expression has been shown to be associated with poor overall and disease-free survival [25,26] and progression of pathological stage [27,28].
HIF-1α 1772 C/T polymorphism, which induces proline-to-serine amino acid substitutions, is considered to increase tumor microvessel density, which result in cancer progression [29]. It has previously been shown to be associated with the risk for breast cancer. The present study included 2143 cases and 2046 controls for C1772T polymorphism, and investigated the association of HIF-1α C1772T polymorphism with breast cancer risk. Subgroup analyses by ethnicity, clinicopathological characteristics, and habits were also extracted, showing that there was no marked association between C1772T polymorphism and breast cancer risk in the overall population. In the subgroup analysis of ethnicity, we found an increased breast cancer risk in Asians based on homozygote comparison and the recessive model. However, in the TT vs. CC model of Caucasian populations, we found the opposite result (TT vs. CC OR=0.27, P=0.05). Ethnicity may be an essential biological factor that influences C1772T polymorphism through gene-gene interactions, which was particularly prominent in breast cancer. It is believed that breast cancers with different clinicopathological characteristics have widely divergent prognoses; therefore, we analyzed the association between HIF-1α C1772T polymorphism and clinicopathological characteristics of breast cancer patients such as menopausal status, lymph node metastasis, clinical stage, histologic grade, and ER Status. Ultimately, we found no association between them. We also stratified the association between HIF-1α C1772T polymorphism and habits, also obtaining negative results.
When interpreting the results of the current study, certain limitations of the meta-analysis must be considered. Firstly, this meta-analysis contained a relatively small sample size, deficient control populations, and especially, the limitation of clinicopathological data may have affected our final conclusion. Secondly, the meta-analysis was based on unadjusted estimates, and a more precise analysis is required when more detailed individual data become available; therefore, we could not estimate the risk of cancer according to gene-gene and gene-environment interactions.
Conclusions
The present meta-analysis provides clear evidence that HIF-1α C1772T polymorphism is a risk factor for susceptibility to breast cancer in Asians. There was no association between HIF-1α C1772T polymorphism with clinicopathological characteristics and habits of breast cancer patients. Large studies involving more detailed individual data are required to validate the results of the current study.
Footnotes
Source of support: National Natural Science Foundation of China (No. 81471670, 81472822); International Cooperative Project of Shaanxi province, People’s Republic of China (No. 2013KW-32-01)
References
- 1.Jemal A, Bray F, Center MM, et al. Global cancer statistics. Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Hulka BS, Moorman PG. Breast cancer: hormones and other risk factors. Maturitas. 2001;38:103–13. doi: 10.1016/s0378-5122(00)00196-1. [DOI] [PubMed] [Google Scholar]
- 3.Yager JD, Davidson NE. Estrogen carcinogenesis in breast cancer. N Engl J Med. 2006;354:270–82. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- 4.Carmeliet P, Dor Y, Herbert JM, et al. Role of HIF.1 alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature. 1998;394:485–90. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- 5.Semenza GL, Rue EA, Iyer NV, et al. Assignment of the hypoxia-inducible factor 1alpha gene to a region of conserved synteny on mouse chromosome 12 and human chromosome 14q. Genomics. 1996;34:437–39. doi: 10.1006/geno.1996.0311. [DOI] [PubMed] [Google Scholar]
- 6.Wang GL, Jiang BH, Rue EA, et al. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA. 1995;92:5510–14. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhong H, De Marzo AM, Laughner E, et al. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 1999;59:5830–35. [PubMed] [Google Scholar]
- 8.Clifford SC, Astuti D, Hooper L, et al. The pVHL-associated SCF ubiquitin ligase complex: molecular genetic analysis of elongin B and C, Rbx1 and HIF-1alpha in renal cell carcinoma. Oncogene. 2001;20:5067–74. doi: 10.1038/sj.onc.1204602. [DOI] [PubMed] [Google Scholar]
- 9.Smaldone MC, Maranchie JK. Clinical implications of hypoxia inducible factor in renal cell carcinoma. Urol Oncol. 2009;27(3):238–45. doi: 10.1016/j.urolonc.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 10.Tanimoto K, Yoshiga K, Eguchi H, et al. Hypoxia-inducible factor-1alpha polymorphisms associated with enhanced transactivation capacity, implying clinical significance. Carcinogenesis. 2003;24:1779–83. doi: 10.1093/carcin/bgg132. [DOI] [PubMed] [Google Scholar]
- 11.Xu G, Wang M, Xie W, et al. Hypoxia-inducible factor-1 alpha C1772T gene polymorphism and glioma risk: a hospital-based case-control study from China. Genet Test Mol Biomarkers. 2011;15:461–64. doi: 10.1089/gtmb.2010.0265. [DOI] [PubMed] [Google Scholar]
- 12.Konac E, Onen HI, Metindir J, et al. An investigation of relationships between hypoxia-inducible factor-1 alpha gene polymorphisms and ovarian, cervical and endometrial cancers. Cancer Detect Prev. 2007;31:102–9. doi: 10.1016/j.cdp.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Liu Y, Ren H, et al. Polymorphisms in the hypoxia-inducible factor-1alpha gene confer susceptibility to pancreatic cancer. Cancer Biol Ther. 2011;12:383–87. doi: 10.4161/cbt.12.5.15982. [DOI] [PubMed] [Google Scholar]
- 14.Hsiao PC, Chen MK, Su SC, et al. Hypoxia inducible factor-1alpha gene polymorphism G1790A and its interaction with tobacco and alcohol consumptions increase sus-ceptibility to hepatocellular carcinoma. J Surg Oncol. 2010;102:163–69. doi: 10.1002/jso.21539. [DOI] [PubMed] [Google Scholar]
- 15.Chen MK, Chiou HL, Su SC, et al. The association between hypoxia inducible factor-1alpha gene polymorphisms and increased susceptibility to oral cancer. Oral Oncol. 2009;45:e222–26. doi: 10.1016/j.oraloncology.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 16.Ling TS, Shi RH, Zhang GX, et al. Common single nucleotide polymorphism of hypoxia-inducible factor-1alpha and its impact on the clinicopathological features of esophageal squamous cell carcinoma. Chin J Dig Dis. 2005;6:155–58. doi: 10.1111/j.1443-9573.2005.00223.x. [DOI] [PubMed] [Google Scholar]
- 17.Ribeiro AL, Gaspar JF, Pereira T, et al. Lack of relevance of HIF-1α polymorphisms in breast cancer in a Portuguese population. Anticancer Res. 2013;33(6):2549–55. [PubMed] [Google Scholar]
- 18.Zagouri F, Sergentanis TN, Gazouli M, et al. HSP90, HSPA8, HIF-1 alpha and HSP70-2 polymorphisms in breast cancer: a case-control study. Mol Biol Rep. 2012;39(12):10873–79. doi: 10.1007/s11033-012-1984-2. [DOI] [PubMed] [Google Scholar]
- 19.Apaydin I, Konac E, Onen HI, et al. Single nucleotide polymorphisms in the hypoxia-inducible factor-1alpha (HIF-1alpha) gene in human sporadic breast cancer. Arch Med Res. 2008;39(3):338–45. doi: 10.1016/j.arcmed.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 20.Naidu R, Har YC, Taib NA. Associations between hypoxia-inducible factor-1alpha (HIF-1alpha) gene polymorphisms and risk of developing breast cancer. Neoplasma. 2009;56(5):441–47. doi: 10.4149/neo_2009_05_441. [DOI] [PubMed] [Google Scholar]
- 21.Kim HO, Jo YH, Lee J, et al. The C1772T genetic polymorphism in human HIF-1alpha gene associates with expression of HIF-1alpha protein in breast cancer. Oncol Rep. 2008;20(5):1181–87. [PubMed] [Google Scholar]
- 22.Lee JY, Choi JY, Lee KM, et al. Rare variant of hypoxia-inducible factor-1alpha (HIF-1A) and breast cancer risk in Korean women. Clin Chim Acta. 2008;389(1–2):167–70. doi: 10.1016/j.cca.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 23.Bos R, Zhong H, Hanrahan CF, et al. Levels of hypoxia-inducible factor-1 alpha during breast carcinogenesis. J Natl Cancer Inst. 2001;93:309–14. doi: 10.1093/jnci/93.4.309. [DOI] [PubMed] [Google Scholar]
- 24.Koukourakis MI, Giatromanolaki A, Skarlatos J, et al. Hypoxia inducible factor (HIF-1α and HIF-2α) expression in early esophageal cancer and response to photodynamic therapy and radiotherapy. Cancer Res. 2001;61:1830–32. [PubMed] [Google Scholar]
- 25.Dales JP, Beaufils N, Silvy M, et al. Hypoxia inducible factor 1α gene (HIF-1α) splice variants: Potential prognostic biomarkers in breast cancer. BMC Med. 2012;8:44. doi: 10.1186/1741-7015-8-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mabjeesh N, Amir S. Hypoxia-inducible factor (HIF) in human tumorigenesis. Histol Histopathol. 2007;22:559–72. doi: 10.14670/HH-22.559. [DOI] [PubMed] [Google Scholar]
- 27.Naidu R, Har YC, Taib NA. Associations between hypoxia-inducible factor-1α (HIF-1α) gene polymorphisms and risk of developing breast cancer. Neoplasma. 2009;56:441–47. doi: 10.4149/neo_2009_05_441. [DOI] [PubMed] [Google Scholar]
- 28.Bos R, van Diest PJ, van der Groep P, et al. Protein expression of B-cell lymphoma gene 6 (BCL-6) in invasive breast cancer is associated with cyclin D1 and hypoxia-inducible factor-1α (HIF-1α) Oncogene. 2003;22:8948–51. doi: 10.1038/sj.onc.1206995. [DOI] [PubMed] [Google Scholar]
- 29.Horrée N, Groot AJ, van Hattem WA, et al. HIF-1A gene mutations associated with higher microvessel density in endometrial carcinomas. Histopathology. 2008;52:637–39. doi: 10.1111/j.1365-2559.2008.02991.x. [DOI] [PubMed] [Google Scholar]