Abstract
Background
The PPARα gene encodes the peroxisome proliferator-activator receptor alpha, a central regulator of expression of other genes involved in fatty acid metabolism. The purpose of this study was to determine the prevalence of G allele of the PPARα intron 7 G/C polymorphism (rs4253778) in professional Italian soccer players.
Methods
Sixty professional soccer players and 30 sedentary volunteers were enrolled in the study. Samples of venous blood were obtained at rest, in the morning, by conventional clinical procedures; blood serum was collected and total cholesterol, high-density lipoprotein cholesterol, low-density lipoprotein cholesterol, and triglycerides were measured. An aliquot of anticoagulant-treated blood was used to prepare genomic DNA from whole blood. The G/C polymorphic site in PPARα intron 7 was scanned by using the PCR-RFLP (polymerase chain reaction restriction fragment length polymorphism) protocol with TaqI enzyme.
Results
We found variations in genotype distribution of PPARα polymorphism between professional soccer players and sedentary volunteers. Particularly, G alleles and the GG genotype were significantly more frequent in soccer players compared with healthy controls (64% versus 48%). No significant correlations were found between lipid profile and genotype background.
Conclusion
Previous results demonstrated an association of intron 7 G allele as well as the GG genotype in endurance athletes. Our result suggests that this is the case also in professional soccer players.
Keywords: PCR-RFLP, gene variants, endurance athlete, G allele
Introduction
High-level athletic performance is at least partly influenced by genetic components.1,2 Even though it is difficult to determine the exact genetic fundaments of performance there are many critical gene polymorphisms that have been reported to have a physiological impact on human performance. These gene variants have been called performance-enhancing polymorphisms and it was suggested that their simultaneous presence might affect the phenotype of elite athletes.3
Although the allelic/genotypic frequency of some polymorphisms do not significantly differ among populations, in recent years, an increasing body of evidence indicates that there are a number of genetic variants that do influence sports performance.4 A paradigmatic example of the complexity of gene/performance interaction is the Z-disk protein α-actinin 3 gene (ACTN3). The null allele (XX) of this gene is related to a lower sprint/power performance, whilst RR and RX are associated to a higher sprint/power performance.5 The frequency of the XX-null genotype appears to be higher in endurance athletes than in controls5 but with contradictory results.6
It is important to underline that differences in factors like sex, ethnic origin, or competition level of the study subjects as well as the size of population samples and statistical approaches make comparison between studies difficult.4 As we stated in a previous paper:
from the data so far we can conclude that genes do influence physiological processes that contribute to power vs endurance potential but that in themselves their predictive power is not sufficient to determine what sports a particular individual may excel in.7
Nevertheless, the genetic information could be used as an aid to design an appropriate training schedule and there is a good possibility that eventually genetic variation will be able to discriminate a predisposition towards specific types of sports.
Some genes that show allelic differences that appear positively associated with endurance athlete status are those encoding the peroxisome proliferator-activated receptors (PPARs). PPARs are members of the nuclear hormone receptor superfamily involved in lipid metabolism and glucose homeostasis. In particular, PPARα regulates expression of genes involved in multiple steps of the lipid metabolism such as fatty acid uptake, transport, and oxidation.8 Consequently, PPARα shows increased expression in tissues involved in fatty acids utilization such as liver, skeletal muscle, and cardiac muscle.9,10 Moreover, the regulation of expression of this gene may enhance skeletal muscle oxidative capacity in relation to endurance training.11 So, it is not surprising that PPARα appears to be related to some cardiovascular risk factors, and the rs4253778 C allele in the intronic polymorphism of the PPARα gene is thought to be associated with higher plasma lipid levels,12 cardiac growth,13 and increased risk of coronary artery disease.14 Hence, the aim of our study is to analyze the frequency distribution of PPARα G/C (rs4253778) polymorphisms in professional soccer players compared with healthy sedentary individuals together with blood lipid profile.
Materials and methods
Subjects
Sixty male professional (series B and C of the Italian Soccer Championship) soccer players (age 22.5±2.2 years) were recruited for this study. Control samples were prepared from 30 unrelated sedentary volunteers (male students of University of Palermo, aged 21.2±2.3 years).
The athletes and controls were all Caucasian to avoid potential problems of population stratification. The local ethics committee approved the procedures followed in the study. All participants gave informed consent to genotyping on the understanding that it was anonymous and that the results would have confidential status.
Measurements
All venous blood samples were taken at rest between 8 am and 10 am by conventional clinical methods, using heparin as anticoagulant. Fasting total cholesterol, high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), and triglycerides (TG) were measured by an enzymatic colorimetric method using a Modular D2400 (Hoffman-La Roche Ltd, Basel, Switzerland). LDL-C fraction was calculated from Friedewald’s formula:
| (1) |
The interassay coefficients of variations for total cholesterol, HDL-C, and triacylglycerol concentrations were 2.9%, 1.8%, and 2.4%, respectively. An aliquot of blood was used for the extraction of genomic DNA (K182018-01; Thermo Fisher Scientific, Waltham, MA, USA).
The polymorphic site in PPARα intron 7 was scanned by using PCR-RFLP protocol with TaqI enzyme. PCR primers were: forward ACAATCACTCCTTAAATATGGTGG and reverse AAGTAGGGACAGACAGGACCAGTA, generating a fragment of 266 base pair (bp) digested by TaqI to 216 bp and 50 bp. PCR reactions were performed in 10 μL containing: 1× reaction buffer, 0.75 nmol each deoxyribonucleotide, 15 mmol MgCl2, 3 pmol each primer, and 0.5 U Taq polymerase. TaqI digestion was performed by adding 6 U of TaqI (Thermo Fisher Scientific), adding 1× restriction enzyme buffer in a volume of 20 μL, and incubation for 2 hours at 65°C.
Maximal VO2 consumption was evaluated through a maximal incremental ramp test consisting of 5 minutes of priming exercise at 50 W, followed by a continuous increase in the workload by 20 W/min until exhaustion. The accepted criteria for maximal effort was: respiratory exchange ratio >1.1, and heart rate >90% of the predicted maximum based on age.15
Body composition was assessed using bioelectrical impedance analysis (BIA; Akern Bioresearch, Pontassieve, FI, Italy) which is a noninvasive and portable method for the estimation of fluid compartments, fat, and fat-free mass.16 Weight was measured to the nearest 0.1 kg using an electronic scale (BWB-800 Medical Scales, Tanita Corporation, Arlington Heights, IL, USA), and height to the nearest 1 cm using a Harpenden portable stadiometer (Holtain Ltd, Crosswell, UK). General characteristics of the subjects are shown in Table 1.
Table 1.
Demographics and physiological characteristics of subjects
| Soccer players | Controls | Significance | |
|---|---|---|---|
| Age (years) | 22.5±2.2 | 21.3±3.1 | NS |
| Weight (kg) | 78.3±4.5 | 75.3±5.9 | NS |
| Height (cm) | 178.2±4.7 | 174.6±3.6 | NS |
| % Body fat | 14.6±1.7 | 13.9±2.2 | NS |
| Waist–hip ratio | 0.96±0.1 | 0.84±0.03 | P<0.05 |
| Resting heart rate (bpm) | 62.6±8.4 | 56.3±3.5 | NS |
| Maximal heart rate (bpm) | 194.5±9.2 | 185.5±3.4 | NS |
| VO2max (mL/kg/min) | 57.5±4.3 | 54.1±4.5 | NS |
Abbreviations: NS, not significant; VO2max, maximal oxygen consumption.
Statistical analysis
The STATISTICA statistical package, version 8.0, was used to perform all statistical evaluations. The values from the professional soccer players were compared with those from the control subjects using unpaired Student’s t-test. Allele frequencies were determined by gene counting. A Pearson χ2 test or Yates corrected χ2 test was used to compare the PPARα intron 7 G/C alleles and genotype frequencies between athletes and control subjects. The level of statistical significance was set at P<0.05.
Results
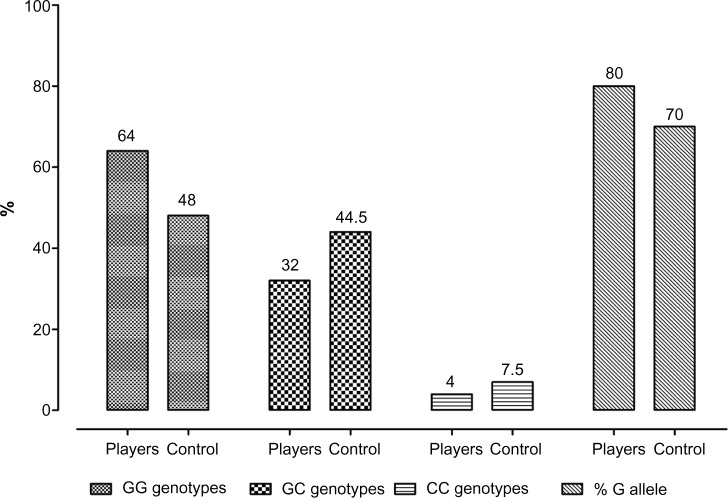
Our data showed distribution differences of PPARα gene G/C polymorphism in Italian soccer players compared with control subjects; these genotype differences are summarized in Table 2. The genotype frequency of GG, GC, and CC among two groups was: controls 48%, 44,5% and 7,5%; and soccer players 64%, 32%, and 4%; respectively (Figure 1 and Table 2). No significant difference was found in C allele frequencies within two groups. The observed distribution of the 7 G/C polymorphism is consistent with reports from Russian athletes,17 Polish rowers,18 and nonathletic young men.19 A significant excess of G allele was noted in the Italian soccer players (80% compared with 70% in control subjects; P=0.04). The PPARα genotype distribution was in agreement with Hardy–Weinberg equilibrium.
Table 2.
Polymorphism of PPARα in soccer players and controls
| Group | Number |
PPARα genotype
|
Controls versus players by Fisher’s exact test
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| GG % | GC % | CC % | G allele % | Allele | df | Pearson’s chi-squared | P | ||
| Controls | 27 | 48 | 44,5 | 7.5 | 70 | GG | 1 | 3.86 | 0.04 |
| Soccer players | 56 | 64 | 32 | 4 | 80 | GC | 1 | 2.85 | 0.09 |
| CC | 1 | 0.29 | 0.59 | ||||||
Figure 1.
PPARα genotype frequencies in 60 Italian soccer players and 30 healthy volunteers.
As shown in Table 3, serum triglyceride, total cholesterol, and LDL-C concentration were increased in the soccer player group but the change was not significant.
Table 3.
Serum values of triglycerides, total cholesterol, LDL, and HDL from samples of venous blood
| Lipid profile (mg/dL) | Soccer players | Controls | Significance |
|---|---|---|---|
| Total cholesterol | 175.1±5.32 | 164.0±6.95 | NS |
| Triglycerides | 97.37±6.52 | 88.37±13.50 | NS |
| HDL cholesterol | 62.27±2.66 | 66.85±2.42 | NS |
| LDL cholesterol | 93.63±4.79 | 80.52±6.69 | NS |
Abbreviations: HDL, high-density lipoprotein; LDL, low-density lipoprotein; NS, not significant.
Although HDL-C levels were reduced in the soccer player group compared with the healthy volunteers group, the changes were also not statistically significant (Table 3).
Discussion
PPARα is a transcription factor and a major regulator of lipid and glucose metabolism in the liver, skeletal muscle, and heart. It is also involved in energy homeostasis. Activation of PPARα promotes uptake, utilization, and catabolism of fatty acids by upregulation of genes involved in fatty acid transport as well as in peroxisomal and mitochondrial fatty acid metabolism. In this study we found some variation in genotype distribution of the rs4253778 PPARα polymorphism between professional soccer players and sedentary volunteers. Specifically, the G allele was significantly more frequent in soccer players compared with healthy controls, as was the GG genotype; on the contrary, no significant associations were found between lipid profile and genotype background.
Several studies have tested the plausible effect of PPARα on athletic performance. Ahmetov et al reported that the G allele was significantly higher in Russian endurance-orientated athletes (80.3%) compared to power-oriented athletes (50.6%) whereas the C allele could be an advantage in power-type activities.17 They also reported that GG homozygotes had significantly higher percentages of slow-twitch fibers (55.5%±2.0% versus 38.5%±2.3%) than CC homozygotes. They concluded that PPARα intron 7 G/C polymorphism was associated with physical performance in Russian athletes, and this may be explained, in part, by the association between PPARα genotype and muscle-fiber-type composition. The same authors also found that the frequencies of the PPARα G alleles in 205 elite rowers were significantly greater than in control subjects (90.1% versus 83.6%), plus the G alleles were associated with higher values of aerobic performance.20 Ginevičienė reported that, in 193 elite Lithuanian athletes, the PPARα C allele was more common in the mixed athlete group (endurance, mixed sports, speed/power, and team sports) than in the general population, plus the PPARα CC genotype was associated with muscle mass and single muscular contraction power, whereas the GG genotype was more related to endurance sports.21 Recently, Maciejewska et al described PPARα genotype distribution among a group of elite Polish athletes and they found a significantly higher frequency of the GG genotype (87% versus 63%) and G allele in the rowers compared to sedentary controls.18 Also, Eynon found that a higher frequency of the PPARα GG genotype is associated with top-level endurance athletes in the study population of 155 Israeli athletes (endurance athletes and sprinters) and 240 healthy controls.11 In addition, they suggested that the above PPARα single-nucleotide polymorphism may belong to a group of several genetic variations that influence top-level endurance performance. In line with this, Ahmetov et al reported that a number of variants, including the PPARα G allele, were overrepresented in a large cohort of 1,423 Russian athletes compared to 1,132 controls.22
The exact function of this single-nucleotide polymorphism is speculative. Regulatory regions have been found in introns; however, it is unlikely that the intron 7 variant is itself functional as it is not close to the splice branch point sequence.23 Of interest, there are also previous studies indicating that the intron 7 C allele is in allelic association with the V162 allele, which encodes a more transcriptionally active PPARα and attenuates the effect of the intron 7 C allele, eg, on exercise-induced left ventricular hypertrophy.13 It is suggested that the C allele of the PPARα intron 7 G/C polymorphism is in linkage disequilibrium with a functional variant in the promoter or enhancer element of the PPARα gene that results in reduced PPARα gene expression.
While the association with endurance has been repeated in several studies with consistent results, other analysis on this polymorphism indicated that the PPARα intron 7 G/C polymorphism has little or no effect on strength-related phenotypes in nonathletic young men. Isometric and concentric peak torque was found to be similar between GG homozygotes and C allele (GC + CC). Also, no differences were found in muscular endurance ratio between the two groups. The impact of this individual polymorphism seems to be of insufficient magnitude to influence muscle characteristics in a nonathletic population.19
The C allele has been associated with cardiac disorders including higher left-ventricular growth after training and the effect was more pronounced in hypertensives.13 Horowitz et al found that the protein content of PPARα in human muscle, as well as that of some of its target enzymes, is increased after 12 weeks of endurance training.24 Activation of PPARα also directly upregulates transcription of long-chain fatty acid acetyl-CoA synthase and other enzymes of the β-oxidation pathway.25 Finally, in an extensive review, Cresci discussed the evidence for the association of PPARα allelic polymorphism with cardiovascular disorders.26
Conclusion
Genotype distribution and allele frequency examined in our study showed a significantly higher frequency of the GG genotype and G allele in soccer players compared to controls in accordance with previous results.27 The novelty of our study is that even in a team sport (like professional soccer) it is possible to find this kind of association. Soccer requires variable talents but endurance is an important asset as, during a 90-minute game, elite-level players run about 10 km at an average intensity close to the anaerobic threshold (80%–90% of maximal heart rate). Within this endurance context, numerous explosive bursts of activity are required, including jumping, kicking, tackling, turning, sprinting, changing pace, and sustaining forceful contractions to maintain balance and control of the ball against defensive pressure.27,28 But GG genotype, related to endurance performance, appears to be more represented in elite soccer players despite the mixed characteristics of effort in soccer.
Thus, we want to stress that athletic performance is influenced by multifactorial events: environmental, gene–gene, and gene–environmental interaction factors are also important contributors to the “complex trait” of being an athletic champion. Such traits are likely not reducible to defined genetic polymorphisms.7
Acknowledgments
This study was supported by Scuola dello Sport of National Olympic Committee (CONI; Comitato Provinciale Palermo). We would also like to thank all the participants in this study. Patrizia Saladino was supported by a fellowship of the University of Palermo.
Footnotes
Author contributions
Patrizia Proia, Antonino Bianco, Giovanni Caramazza, Marcello Traina, Antonio Palma and Antonio Paoli made substantial contributions to conception and design. Patrizia Proia, Gabriella Schiera, Valentina Contrò, Patrizia Saladino and Giovanni Caramazza, contributed to acquisition of data. Antonino Bianco and Keith A Grimaldi performed statistical analysis and interpretation of data. Patrizia Proia, Gabriella Schiera and Patrizia Saladino drafted the article with the help of Antonio Paoli, Antonio Palma, Marcello Traina and Keith A Grimaldi. Antonino Bianco, Valentina Contrò, Giovanni Caramazza, Marcello Traina, Keith A Grimaldi, Antonio Palma and Antonio Paoli revised the article critically for important intellectual content. All authors approved the final version to be published and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy and integrity of any part of the work are appropriately investigated and resolved.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Macarthur DG, North KN. Genes and human elite athletic performance. Hum Genet. 2005;116(5):331–339. doi: 10.1007/s00439-005-1261-8. [DOI] [PubMed] [Google Scholar]
- 2.Rankinen T, Bray MS, Hagberg JM, et al. The human gene map for performance and health-related fitness phenotypes: the 2005 update. Med Sci Sports Exerc. 2006;38(11):1863–1888. doi: 10.1249/01.mss.0000233789.01164.4f. [DOI] [PubMed] [Google Scholar]
- 3.Ostrander EA, Huson HJ, Ostrander GK. Genetics of athletic performance. Annu Rev Genomics Hum Genet. 2009;10:407–429. doi: 10.1146/annurev-genom-082908-150058. [DOI] [PubMed] [Google Scholar]
- 4.Buxens A, Ruiz JR, Arteta D, et al. Can we predict top-level sports performance in power vs endurance events? A genetic approach. Scand J Med Sci Sports. 2011;21(4):570–579. doi: 10.1111/j.1600-0838.2009.01079.x. [DOI] [PubMed] [Google Scholar]
- 5.Berman Y, North KN. A gene for speed: the emerging role of alpha-actinin-3 in muscle metabolism. Physiology (Bethesda) 2010;25(4):250–259. doi: 10.1152/physiol.00008.2010. [DOI] [PubMed] [Google Scholar]
- 6.Lucia A, Gómez-Gallego F, Santiago C, et al. ACTN3 genotype in professional endurance cyclists. Int J Sports Med. 2006;27(11):880–884. doi: 10.1055/s-2006-923862. [DOI] [PubMed] [Google Scholar]
- 7.Grimaldi KA, Paoli A, Smith GJ. Personal genetics: sports utility vehicle? Recent Pat DNA Gene Seq. 2012;6(3):209–215. doi: 10.2174/187221512802717312. [DOI] [PubMed] [Google Scholar]
- 8.Ordovas JM. Genetic influences on blood lipids and cardiovascular disease risk: tools for primary prevention. Am J Clin Nutr. 2009;89(5):1509S–1517S. doi: 10.3945/ajcn.2009.27113E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lemberger T, Braissant O, Juge-Aubry C, et al. PPAR tissue distribution and interactions with other hormone-signaling pathways. Ann N Y Acad Sci. 1996;804:231–251. doi: 10.1111/j.1749-6632.1996.tb18619.x. [DOI] [PubMed] [Google Scholar]
- 10.Braissant O, Foufelle F, Scotto C, Dauça M, Wahli W. Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-alpha, -beta, and -gamma in the adult rat. Endocrinology. 1996;137(1):354–366. doi: 10.1210/endo.137.1.8536636. [DOI] [PubMed] [Google Scholar]
- 11.Eynon N, Meckel Y, Sagiv M, et al. Do PPARGC1A and PPARalpha polymorphisms influence sprint or endurance phenotypes? Scand J Med Sci Sports. 2010;20(1):e145–e150. doi: 10.1111/j.1600-0838.2009.00930.x. [DOI] [PubMed] [Google Scholar]
- 12.Flavell DM, Pineda Torra I, Jamshidi Y, et al. Variation in the PPARalpha gene is associated with altered function in vitro and plasma lipid concentrations in Type II diabetic subjects. Diabetologia. 2000;43(5):673–680. doi: 10.1007/s001250051357. [DOI] [PubMed] [Google Scholar]
- 13.Jamshidi Y, Montgomery HE, Hense HW, et al. Peroxisome proliferator-activated receptor alpha gene regulates left ventricular growth in response to exercise and hypertension. Circulation. 2002;105(8):950–955. doi: 10.1161/hc0802.104535. [DOI] [PubMed] [Google Scholar]
- 14.Flavell DM, Jamshidi Y, Hawe E, et al. Peroxisome proliferator-activated receptor alpha gene variants influence progression of coronary atherosclerosis and risk of coronary artery disease. Circulation. 2002;105(12):1440–1445. doi: 10.1161/01.cir.0000012145.80593.25. [DOI] [PubMed] [Google Scholar]
- 15.Doria C, Toniolo L, Verratti V, et al. Improved VO2 uptake kinetics and shift in muscle fiber type in high-altitude trekkers. J Appl Physiol (1985) 2011;111(6):1597–1605. doi: 10.1152/japplphysiol.01439.2010. [DOI] [PubMed] [Google Scholar]
- 16.Paoli A, Bianco A, Grimaldi KA, Lodi A, Bosco G. Long term successful weight loss with a combination biphasic ketogenic Mediterranean diet and Mediterranean diet maintenance protocol. Nutrients. 2013;5(12):5205–5217. doi: 10.3390/nu5125205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahmetov II, Mozhayskaya IA, Flavell DM, et al. PPARalpha gene variation and physical performance in Russian athletes. Eur J Appl Physiol. 2006;97(1):103–108. doi: 10.1007/s00421-006-0154-4. [DOI] [PubMed] [Google Scholar]
- 18.Maciejewska A, Sawczuk M, Cieszczyk P. Variation in the PPARα gene in Polish rowers. J Sci Med Sport. 2011;14(1):58–64. doi: 10.1016/j.jsams.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 19.Broos S, Windelinckx A, De Mars G, et al. Is PPARα intron 7 G/C polymorphism associated with muscle strength characteristics in nonathletic young men? Scand J Med Sci Sports. 2013;23(4):494–500. doi: 10.1111/j.1600-0838.2011.01406.x. [DOI] [PubMed] [Google Scholar]
- 20.Akhmetov II, Popov DV, Mozhaĭskaia IA, et al. Association of regulatory genes polymorphisms with aerobic and anaerobic performance of athletes. Ross Fiziol Zh Im I M Sechenova. 2007;93(8):837–843. [PubMed] [Google Scholar]
- 21.Ginevičienė V, Pranckevičienė E, Milašius K, Kučinskas V. Relating fitness phenotypes to genotypes in Lithuanian elite athletes. Acta Med Litu. 2010;17(1–2):1–10. [Google Scholar]
- 22.Ahmetov II, Williams AG, Popov DV, et al. The combined impact of metabolic gene polymorphisms on elite endurance athlete status and related phenotypes. Hum Genet. 2009;126(6):751–761. doi: 10.1007/s00439-009-0728-4. [DOI] [PubMed] [Google Scholar]
- 23.Foucher C, Rattier S, Flavell DM, et al. Response to micronized fenofibrate treatment is associated with the peroxisome-proliferator-activated receptors alpha G/C intron7 polymorphism in subjects with type 2 diabetes. Pharmacogenetics. 2004;14(12):823–829. doi: 10.1097/00008571-200412000-00005. [DOI] [PubMed] [Google Scholar]
- 24.Horowitz JF, Leone TC, Feng W, Kelly DP, Klein S. Effect of endurance training on lipid metabolism in women: a potential role for PPARalpha in the metabolic response to training. Am J Physiol Endocrinol Metab. 2000;279(2):E348–E355. doi: 10.1152/ajpendo.2000.279.2.E348. [DOI] [PubMed] [Google Scholar]
- 25.Berger J, Moller DE. The mechanisms of action of PPARs. Annu Rev Med. 2002;53:409–435. doi: 10.1146/annurev.med.53.082901.104018. [DOI] [PubMed] [Google Scholar]
- 26.Cresci S. The PPAR genes, cardiovascular disease and the emergence of PPAR pharmacogenetics. Expert Opin Pharmacother. 2005;6(15):2577–2591. doi: 10.1517/14656566.6.15.2577. [DOI] [PubMed] [Google Scholar]
- 27.Proia P, Bianco A, Schiera G, et al. The effects of a 3-week training on basal biomarkers in professional soccer players during the preseason preparation period. J Sports Med Phys Fitness. 2012;52(1):102–106. [PubMed] [Google Scholar]
- 28.Stolen T, Chamari K, Castagna C, Wisløff U. Physiology of soccer: an update. Sports Med. 2005;35(6):501–536. doi: 10.2165/00007256-200535060-00004. [DOI] [PubMed] [Google Scholar]