Abstract
Actinobacteria comprise a substantial fraction of the bacterioplankton in freshwater lakes and streams. Numerous cultivation-independent investigations have retrieved actinobacterial 16S rRNA gene sequences from such habitats. The taxa detected in freshwater habitats are usually absent from terrestrial and marine systems. So far, none of the indigenous freshwater lineages is represented by a taxon with a validly published name. The seven organisms for which Candidatus status is described here were isolated from freshwater lakes and ponds located in tropical, subtropical and temperate climatic zones. Phylogenetic analyses demonstrated that they are affiliated with one of the actinobacterial lineages indigenous to freshwater bacterioplankton. The seven novel taxa could only be cultivated to date as mixed cultures that also contain non-actinobacterial strains. Due to the lack of pure cultures, I propose to establish the candidate species ‘Candidatus Planktoluna difficilis’, ‘Candidatus Aquiluna rubra’, ‘Candidatus Flaviluna lacus’, ‘Candidatus Rhodoluna limnophila’, ‘Candidatus Rhodoluna planktonica’, ‘Candidatus Rhodoluna lacicola’ and ‘Candidatus Limnoluna rubra’ for these taxa.
Until the mid-1990s, it was believed that freshwater lakes and ponds do not harbour indigenous actinobacteria (e.g. Rheinheimer, 1980). This notion was based exclusively on results of cultivation experiments, which demonstrated the presence of small numbers of Gram-positive strains in freshwater habitats. The taxa isolated from freshwater habitats were well known from terrestrial environments, indicating a non-indigenous (i.e. allochthonous) origin of the cultured strains. It was assumed that the cell-wall architecture of Gram-positive bacteria represents an adaptation against desiccation, which is not an advantageous trait in a permanently aqueous environment. Cultivation-independent investigations on the diversity of freshwater bacterioplankton changed radically the notion of the non-existence of indigenous actinobacteria (Glöckner et al., 2000; Zwart et al., 2002). It became clear that indigenous actinobacteria comprise an integral part of freshwater bacterioplankton, typically comprising 10–60 % of the total numbers of bacteria (e.g. Glöckner et al., 2000; Sekar et al., 2003; Allgaier & Grossart, 2006). A couple of actinobacterial lineages indigenous to freshwater systems were identified (Zwart et al., 2002; Warnecke et al., 2004; Allgaier & Grossart, 2006; Wu et al., 2007). In 2003, the first cultivated actinobacterial strains affiliated with such an indigenous freshwater lineage were presented (Hahn et al., 2003). The cultivated strains were characterized by remarkably small cells and thin cell walls. Both traits were interpreted as adaptations to a planktonic lifestyle in the water column of freshwater habitats. Currently, the causes of the enormous ecological success of indigenous actino-bacteria in freshwater habitats are not understood. An important ecological trait of at least some freshwater actinobacteria is a strong resistance against predation by bacterivorous protists (Hahn et al., 2003; Jezbera et al., 2005). Recently, Sharma et al. (2008) discovered by metagenomic data mining that freshwater actinobacteria encode actinorhodopsins, which are variants of the light-energy-harvesting proteorhodopsins. These transmembrane proteins were previously known from marine members of the Alphaproteobacteria, Gammaproteobacteria and Bacteroidetes, but not from freshwater actinobacteria. The genes that encode actinorhodopsins could be assigned to actinobacteria by phylogenetic analysis of neighbouring genes; however, the identity of the taxa that harbour these genes could not be yet established (Sharma et al., 2008).
Here, seven candidate species representing actinobacteria indigenous to freshwater habitats are described. The 16S rRNA genes of the novel taxa show sequence similarities of 93–94 % with genes of the closest described actinobacterial species. The distant relationship to species with validly published names justifies the proposal of novel taxa. Due to the lack of pure cultures, the establishment of candidate species is proposed (Murray & Stackebrandt, 1995).
Cultivation
A large number of actinobacterial strains could be isolated from freshwater habitats by using the filtration-acclimatization method (Hahn et al., 2003, 2004; Hahn & Pöckl, 2005). In brief, water samples from ponds and lakes were filtered through sterile 0.2 μm filters, inoculated in inorganic IBM medium (Hahn et al., 2004) and acclimatized stepwise to relatively high concentrations [3 g l−1 (w/v)] of organic NSY medium (Hahn et al., 2004) by adding increasing doses of this medium. After completion of the acclimatization procedure, the cultures were transferred onto NSY agar plates. Cultures were purified by inoculation of single colonies in liquid NSY medium and plating of dilution series of grown cultures on agar plates. This process was repeated three times. Plating of several cultures did not result in independently growing colonies (see below). These isolation experiments resulted in cultures of yellow- and red-pigmented actinobacteria. Previous phylogenetic analyses demonstrated that the cultivated actinobacteria formed two clusters (Hahn et al., 2003). The cluster harbouring red-pigmented strains was previously designated the ‘Luna-1 cluster’ and the cluster exclusively containing yellow-pigmented strains was designated the ‘Luna-2 cluster’. All previously established yellow cultures represented pure cultures, while many red cultures were obviously mixed cultures of red-pigmented actinobacteria and other non-pigmented bacteria. These mixed cultures grew well in liquid medium and displayed intense red colours, but transfer of several cultures to solidified NSY medium resulted mainly in growth of the non-actinobacterial strains. The red-pigmented actinobacteria also grew on the agar plates, but only in direct contact with or in the immediate surroundings of non-actinobacterial colonies (see Supplementary Fig. S1, available in IJSEM Online). Experiments with other complex media and supplementation of media with vitamins did not result in independent growth of the actinobacteria contained in the mixed cultures. Only a few cultures of red-pigmented strains, namely MWH-Dar1, MWH-Dar4 and MWH-VicMua1, seemed to represent pure cultures (Hahn et al., 2004). These cultures formed exclusively actinobacterial colonies on agar plates; however, recent detailed investigation with fluorescent in situ hybridization (FISH) probes revealed the presence of small numbers of betaproteobacteria in these cultures. Thus, all cultures described here have to be considered to be mixed cultures. The non-actinobacterial strains present in the cultures were affiliated with Spirochaeta, Hylemonella, Polynucleobacter or other taxa known to be able to penetrate 0.2 μm filters (Hahn et al., 2004).
The mixed cultures of the candidate species could be preserved in glycerol-supplemented NSY medium at −70 °C. Reactivation of most cultures worked well, even after a few years of storage; however, some strains seem to be more sensitive to this kind of preservation. For example, reactivation of the culture containing ‘Candidatus Planktoluna difficilis’ strain MWH-7Well8 was problematic and sometimes failed completely.
Phenotypic traits
All seven actinobacterial strains described here have selenoid cell morphologies. They have small cells, with diameters <0.5 μm and lengths <1.2 μm, with red or yellow pigmentation (Table 1). All these strains resemble the previously characterized, distantly related actinobacterium MWH-Ta1 in morphology and cell size (Hahn et al., 2003). It seems that all the novel actinobacteria form circular, slightly concave colonies with smooth and shiny surfaces when growing on agar plates; however, growth of colonies was frequently disturbed by neighbouring nonactinobacterial strains (Supplementary Fig. S1).
Table 1.
Characteristics of cultures representing the proposed candidate species
| Candidate species | Strain | 16S rRNA gene sequence accession number | Pigmentation | Non-actinobacteria in liquid culture |
|
|---|---|---|---|---|---|
| Proportion (%) | Affiliation | ||||
| ‘Candidatus Planktoluna difficilis’ | MWH-7Well8 | AM939566 | Red | >10 | Spirochaeta |
| ‘Candidatus Aquiluna rubra’ | MWH-Dar4 | AJ565416 | Red | <1–15 | Betaproteobacteria |
| ‘Candidatus Limnoluna rubra’ | MWH-EgelM2-3 | AM943659 | Red | >20 | Polynucleobacter |
| ‘Candidatus Flaviluna lacus’ | MWH-Creno3D3 | AM939567 | Yellow | <1 | Betaproteobacteria |
| ‘Candidatus Rhodoluna lacicola’ | MWH-Ta8 | AM182889 | Red | <1 | Betaproteobacteria |
| ‘Candidatus Rhodoluna limnophila’ | MWH-VicMua1 | AJ565417 | Red | <1 | Betaproteobacteria |
| ‘Candidatus Rhodoluna planktonica’ | MWH-Dar1 | AJ565415 | Red | <1 | Betaproteobacteria |
Phylogeny
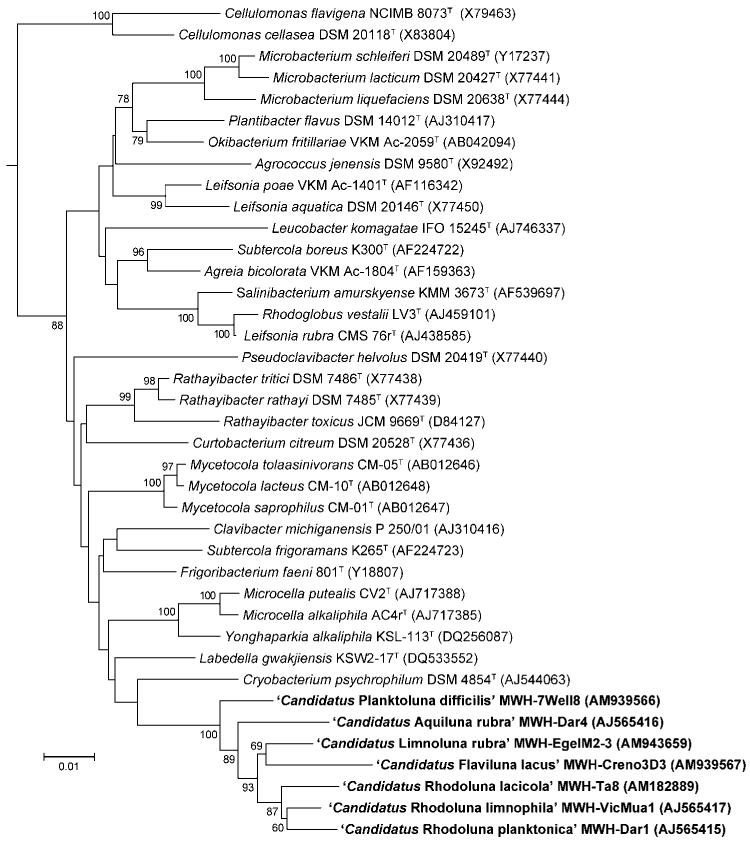
Almost full-length 16S rRNA gene sequences could be obtained by amplification either with universal primers (27F and 1492R) or, in the cases of cultures with high nonactinobacterial content, with primers LEAD1 and 1492R (Hahn & Pöckl, 2005). The primer LEAD1 has been developed for specific amplification of 16S rRNA genes of strains affiliated with the Luna clusters and related bacteria. Phylogenetic analyses revealed that the sequences of the candidate species form a monophyletic cluster within the actinobacterial family Microbacteriaceae (Fig. 1). However, bootstrap support is insufficient for identification of the closest relatives of the novel candidate species. Interestingly, the sequence of the yellow-pigmented ‘Candidatus Flaviluna lacus’ strain MWH-Creno3D3 did not cluster with the previously described, yellow-pigmented strains that form the ‘Luna-2 cluster’ (Hahn et al., 2003). Instead, the sequence of this strain clustered together with the red-pigmented strains within the ‘Luna-1 cluster’. blast searches revealed that six of the seven novel taxa are closely related to uncultured bacteria, but all are only distantly related to described species. Strains representing six of the candidate species described here share 16S rRNA gene sequence similarities in the range 98.0–99.6 % with uncultured actinobacteria. In contrast, the representative of the seventh candidate species, ‘Candidatus Flaviluna lacus’ strain MWH-Creno3D3, showed only 96.8 % similarity to the sequence of the closest uncultured bacterium. Strains of all seven taxa showed 16S rRNA gene sequence similarities of only 93–94 % with species with validly published names. Comparison of the 16S rRNA genes of the seven novel taxa revealed similarity values in the range 95.5–98.3 % (Supplementary Table S1). Thus, none of the strains showed similarity >98.7 % to any other strain, which makes it unlikely that any pair of strains would show DNA–DNA reassociation values of >70 % (Stackebrandt & Ebers, 2006). Consequently, each strain has to be considered to represent a distinct species (Rosselló-Mora & Amann, 2001). In order to evaluate whether the novel taxa should be placed in one or more candidate genera, 16S rRNA gene sequence similarity between members of closely related genera with validly published names within the family Microbacteriaceae was analysed. This revealed a separation of genera with validly published names by divergence in the range 0.4–4.3 % (similarity of 95.7–99.6 %) (10 pairings were analysed). The mean similarity between these genera was 96.8 % (SD 1.2 %). Based on the observed similarity values that typically separate closely related genera within the family Microbacteriaceae and the reconstructed phylogeny shown in Fig. 1, I propose to establish five candidate genera and seven candidate species for the novel actinobacterial strains presented. Three of the strains that possess 16S rRNA gene sequence similarity of >97.8 % were placed in the same genus. All other strains possess lower similarity to the closest other strain.
Fig. 1.
Neighbour-joining tree based on almost full-length 16S rRNA gene sequences (E. coli positions 50–1458) of strains affiliated with the phylum Actinobacteria. Bootstrap values represent percentage support of nodes based on 1000 resamplings (only values ≥60 % are shown). Bar, 1 % estimated sequence divergence. The sequence of Brevibacterium linens DSM 20425T (GenBank accession no. X77451) served as an outgroup (not shown).
A tree presenting the closest uncultured relatives of the candidate species, as well as other cultivated strains closely related to the strains representing the candidate species, is available as Supplementary Fig. S2.
Ecology
All cultures were isolated from surface-water samples of freshwater lakes and ponds (Supplementary Table S2). Furthermore, almost all of the closest related sequences found by blast search also originate from the water column of freshwater habitats. This indicates clearly that these actinobacteria represent indigenous freshwater strains with a planktonic lifestyle. The seven cultures reported in this study were isolated from habitats located in tropical, subtropical and temperate climatic zones (Supplementary Table S1). One culture was isolated from tropical Lake Victoria, the world’s second largest freshwater lake by area. Two strains were isolated from a small tropical pond located at the university campus in Dar es Salaam, Tanzania. Two cultures were obtained from two separate samples from eutrophic to hypertrophic Lake Taihu (Wu & Hahn, 2006). This large (2250 km2) but shallow (maximum depth 2–3 m) lake is located in the subtropical part of China. One strain was isolated from Lake Creno, a mountain lake located at an altitude of 1300 m on the Mediterranean island of Corsica (France). The seventh culture was isolated from a eutrophic lake located in Austria. These habitats differ widely in climatic conditions, surface area, depth, water transparency, water chemistry and trophic status.
A specific relationship, for example a tight symbiosis, between the actinobacteria and the non-actinobacterial strains in the established mixed cultures is unlikely. Various non-actinobacterial strains occurred in the cultures, but no taxon-specific pattern of co-occurrence was observed (data not shown). The non-actinobacterial strains probably provide substances that are lacking in the medium but essential for the actinobacteria. A broad range of bacteria, from spirochaetes to betaproteobacteria, seems to be able to supplement the medium in a suitable way.
Description of ‘Candidatus Flaviluna lacus’
‘Candidatus Flaviluna lacus’ [Fla.vi.lu’na. L. adj. flavus yellow; L. fem. n. luna the moon; N.L. fem. n. Flaviluna yellow moon, referring to the yellow pigmentation and selenoid morphology of the first strain. la’cus. L. gen. n. lacus of a lake]. Represented by the aerobic, yellow-pigmented strain MWH-Creno3D3, which was isolated from Lake Creno (Corsica, France). The strain is contained in a mixed culture with only a small fraction of a non-actinobacterial strain. This mixed culture grows in liquid NSY medium and on NSY agar. Plating on agar plates did not provide hints of the presence of a non-actinobacterial strain; however, FISH revealed the presence of small numbers of betaproteobacteria. Basis of assignment is the nucleic acid sequence of the 16S rRNA gene of strain MWH-Creno3D3 (GenBank accession no. AM939567) and the unique oligonucleotide sequence 5′-CCCTAGGGGCATCCCTGGGGATGGA-3′ within the 16S rRNA gene (Escherichia coli positions 183–195). ‘Candidatus Flaviluna lacus’ differs in pigmentation from the other six taxa described here.
Description of ‘Candidatus Aquiluna rubra’
‘Candidatus Aquiluna rubra’ [A’qui.lu’na. L. fem. n. aqua water; L. fem. n. luna the moon; N.L. fem. n. Aquiluna water moon, referring to the aquatic habitat of the first strain and its selenoid morphology. rub’ra. L. fem. adj. rubra red]. Represented by the aerobic, red-pigmented strain MWH-Dar4, which was isolated from a eutrophic pond in Tanzania. The strain is contained in a mixed culture with a variable fraction of a non-actinobacterial strain. Basis of assignment is the nucleic acid sequence of the 16S rRNA gene of strain MWH-Dar4 (GenBank accession no. AJ565416). This strain and closely related strains (Supplementary Fig. S2) can be distinguished from other bacteria by the combined presence of the two specific sequences 5′-AAACTGTAGCTAATACCGGATACTTTCC-3′ (E. coli positions 160–183) and 5′-AATATTGCACAATGGGCGCAAGCCT-3′ (E. coli positions 363–387) within the 16S rRNA gene. These two oligonucleotide sequences are also present in almost full-length 16S rRNA sequences of three cultivated, red-pigmented strains and one uncultured bacterium, which all share sequence similarities >98.6 % with strain MWH-Dar4 (Supplementary Fig. S2). Furthermore, a blast search revealed many short sequences of 300–500 nt that also contain these two oligonucleotide sequences. Preliminary phylogenetic analyses indicated a close relationship between these short sequences and the above-mentioned almost full-length sequences (data not shown). The three cultivated strains were isolated from freshwater habitats in Tanzania, France and China, while the sequence of the uncultured bacterium PH-F7 was retrieved from a seawater sample taken in Poole Harbour, UK.
Description of ‘Candidatus Rhodoluna limnophila’
‘Candidatus Rhodoluna limnophila’ [Rho.do.lu’na. Gr. adj. rhodeos rosy; L. fem. n. luna the moon; N.L. fem. n. Rhodoluna rosy (red) moon, referring to the red pigmentation and the selenoid morphology of the first strain. lim.no’phi.la. Gr. n. limnos lake; N.L. fem. adj. phila from Gr. fem. adj. philê loving; N.L. fem. adj. limnophila lake loving]. Represented by the aerobic, red-pigmented strain MWH-VicMua1, which was isolated from Lake Victoria (East Africa). The strain is contained in a mixed culture with only a small fraction of non-actinobacterial cells. Basis of assignment is the nucleic acid sequence of the 16S rRNA gene of strain MWH-VicMua1 (GenBank accession no. AJ565417) and the combined presence of the two oligonucleotide sequences 5′-GTTCCATTCCACGGAATCC-3′ (E. coli positions 836–852) and 5′-AATACCGCAAGGTGGAGCGAATCCCATAA-3′ (E. coli positions 1260–1288) within the 16S rRNA gene. This trait is also present in four environmental sequences (Supplementary Fig. S2), which were obtained from a freshwater reservoir in the Czech Republic (two clones), Lake Niegocin in Poland (Warnecke et al., 2004) and an upland stream in Germany (Beier et al., 2008). These four environmental sequences and the strain MWH-VicMua1 form together a cluster with sequence similarities >98.8 % (Supplementary Fig. S2).
Description of ‘Candidatus Rhodoluna planktonica’
‘Candidatus Rhodoluna planktonica’ [plank.to’ni.ca. N.L. fem. adj. planktonica living in the plankton, planktonic (from Gr. adj. planktos wandering)]. Represented by the aerobic, red-pigmented strain MWH-Dar1, which was isolated from a eutrophic pond in Tanzania. The strain is contained in a mixed culture with only a small fraction of a non-actinobacterial strain. Basis of assignment is the nucleic acid sequence of the 16S rRNA gene of strain MWH-Dar1 (GenBank accession no. AJ565415) and the presence of the oligonucleotide sequence 5′-CGAGACCTGAAGGCATCTTTGGGTCT-3′ (E. coli positions 183–193) within the 16S rRNA gene. This diagnostic oligonucleotide sequence is also present in two partial (430 nt) 16S rRNA gene sequences of uncultured bacteria (GenBank accession nos EF149338 and EF149356), which were retrieved from surface microlayers of freshwater habitats populated by mosquito larvae (Anopheles spp.) in Kenya. These two short sequences share similarities of 98.3 % with the sequence of strain MWH-Dar1.
Description of ‘Candidatus Rhodoluna lacicola’
‘Candidatus Rhodoluna lacicola’ [la.ci’co.la. L. masc. n. lacus lake; L. suff. n. -cola from incola the inhabitant; N.L. masc. n. lacicola inhabitant of lakes]. Represented by the aerobic, red-pigmented strain MWH-Ta8. This strain was isolated from the hypertrophic Mailing Bay, Lake Taihu, China. The strain is cultured as a mixed culture containing only a small fraction of betaproteobacteria, which were observed by FISH but did not appear as colonies on agar plates. Basis of assignment is the nucleic acid sequence of the 16S rRNA gene of strain MWH-Ta8 (GenBank accession no. AM182889) and the combined presence of the two oligonucleotide sequences 5′-CTTGCTCCGGTGGATTAGTGG-3′ (E. coli positions 83–105) and 5′-ACGACACCTTGGGGCATCCCAGGGTGTGGAA-3′ (E. coli positions 181–196) within the 16S rRNA gene. This diagnostic combination of oligonucleotide sequences is also present in two other cultivated strains, isolated from ponds in Australia (strain MWH-Aus1W7Actino) and subtropical China (strain MWH-SuzW20). All three cultivated strains share red pigmentation, small cells and a selenoid cell morphology. The diagnostic sequences are also present in ribosomal sequences of two uncultured actinobacteria, which were retrieved from running water in the north-eastern USA (Zwart et al., 2002; Crump & Hobbie, 2005). These five sequences form a cluster characterized by sequence similarities >99.4 % (Supplementary Fig. S2). Furthermore, blast searches revealed the presence of the combined diagnostic oligonucleotide sequences in shorter partial 16S rRNA gene sequences of uncultured bacteria retrieved from Lake Taihu (Wu et al., 2007), an urban freshwater creek in the USA and East African freshwater habitats populated by Anopheles spp. larvae. These partial sequences share sequence similarities >99 % with the five almost fulllength sequences presented in Supplementary Fig. S2.
Description of ‘Candidatus Limnoluna rubra’
‘Candidatus Limnoluna rubra’ [Lim’no.lu’na. Gr. n. limnos lake; L. fem. n. luna the moon; N.L. fem. n. Limnoluna lake moon, referring to the habitat occupied by the first strain and its selenoid morphology. rub’ra. L. fem. adj. rubra red, referring to the red pigmentation of the strain]. Represented by the aerobic, red-pigmented strain MWH-EgelM2-3, which was isolated from a small lake near Salzburg, Austria. The strain is cultured as a mixed culture containing a relatively large fraction of a non-actinobacterial strain (Supplementary Fig. S1). Basis of assignment is the nucleic acid sequence of the 16S rRNA gene of strain MWH-EgelM2-3 (GenBank accession no. AM943659) and the combined presence of the oligonucleotide sequences 5′-CTTGCTCYAGTGGATTAGTGG-3′ (E. coli positions 83–105) and 5′-ACGACACCTTGGGGCATCCCRRGGTGTGGAA-3′ (E. coli positions 181–196) within the 16S rRNA gene. This combined oligonucleotide sequence is also present in two environmental sequences of uncultured actinobacteria (Supplementary Fig. S2) originating from a lake in Germany (Warnecke et al., 2004). The sequences of strain MWH-EgelM2-3 and the two uncultured actinobacteria form a cluster characterized by sequence similarity values >99.2 % (Supplementary Fig. S2).
Description of ‘Candidatus Planktoluna difficilis’
‘Candidatus Planktoluna difficilis’ [Plank’to.lu’na. Gr. adj. planktos wandering, drifting; L. fem. n. luna the moon; N.L. fem. n. Planktoluna planktonic moon, referring to the planktonic lifestyle and the solenoid morphology of the first strain. dif.fi.ci’lis. L. fem. adj. difficilis difficult, referring to difficulties in cultivation of the first strain]. Represented by the aerobic, red-pigmented strain MWH-7Well8, which was isolated from Mailing Bay, Lake Taihu, China. The strain is cultured as a mixed culture containing a relatively large fraction of a spirochaete. Basis of assignment is the nucleic acid sequence of the 16S rRNA gene of strain MWH-7Well8 (GenBank accession no. AM939566) and the presence of the specific oligonucleotide sequence 5′-CTTGGAGGCATCTCCAGG-3′ (E. coli positions 183–193) within the 16S rRNA gene. This diagnostic oligonucleotide sequence is also present in five environmental sequences of uncultured actinobacteria (Supplementary Fig. S2) originating from four different European lakes (Warnecke et al., 2004). The sequences of strain MWH-7Well8 and the five uncultured actinobacteria form a cluster characterized by sequence similarity values >98.9 % (Supplementary Fig. S2). Furthermore, the diagnostic sequence is also present in three partial sequences (GenBank accession nos EF150062, EF150044 and AF141476) that show sequence similarities in the range 98.3–99.4 % with the sequence of strain MWH-7Well8. These sequences were retrieved from the Columbia River, USA, and from freshwater habitats in Kenya populated by Anopheles spp. larvae.
Supplementary Material
Acknowledgements
I am grateful to Hans G. Trüper and one anonymous reviewer for etymological advice and to several colleagues for providing water samples used for isolation of the candidate species. Matthias Pöckl and Ulrike Brandt are acknowledged for excellent technical assistance. This study was supported by the Austrian Science Fund (project P15655) and the Austrian Academy of Sciences.
Abbreviation
- FISH
fluorescent in situ hybridization
Footnotes
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene sequences of strains representing ‘Candidatus Rhodoluna planktonica’, ‘Candidatus Aquiluna rubra’, ‘Candidatus Rhodoluna limnophila’, ‘Candidatus Rhodoluna lacicola’, ‘Candidatus Planktoluna difficilis’, ‘Candidatus Flaviluna lacus’ and ‘Candidatus Limnoluna rubra’ are AJ565415–AJ565417, AM182889, AM939566, AM939567 and AM943659, respectively.
A photo of growth of one of the novel strains, a 16S rRNA gene sequence-based neighbour-joining tree, further details of strain isolation sites and a 16S rRNA gene sequence similarity matrix are available as supplementary material with the online version of this paper.
References
- Allgaier M, Grossart H-P. Diversity and seasonal dynamics of Actinobacteria populations in four lakes in northeastern Germany. Appl Environ Microbiol. 2006;72:3489–3497. doi: 10.1128/AEM.72.5.3489-3497.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier S, Witzel KP, Marxsen J. Bacterial community composition in Central European running waters examined by temperature gradient gel electrophoresis and sequence analysis of 16S rRNA genes. Appl Environ Microbiol. 2008;74:188–199. doi: 10.1128/AEM.00327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump BC, Hobbie JE. Synchrony and seasonality in bacterioplankton communities of two temperate rivers. Limnol Oceanogr. 2005;50:1718–1729. [Google Scholar]
- Glöckner FO, Zaichikov E, Belkova N, Denissova L, Pernthaler J, Pernthaler A, Amann R. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria. Appl Environ Microbiol. 2000;66:5053–5065. doi: 10.1128/aem.66.11.5053-5065.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Pöckl M. Ecotypes of planktonic actinobacteria with identical 16S rRNA genes adapted to thermal niches in temperate, subtropical, and tropical freshwater habitats. Appl Environ Microbiol. 2005;71:766–773. doi: 10.1128/AEM.71.2.766-773.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Lünsdorf H, Wu Q, Schauer M, Höfle MG, Boenigk J, Stadler P. Isolation of novel ultramicrobacteria classified as actinobacteria from five freshwater habitats in Europe and Asia. Appl Environ Microbiol. 2003;69:1442–1451. doi: 10.1128/AEM.69.3.1442-1451.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Stadler P, Wu QL, Pöckl M. The filtration-acclimatization-method for isolation of an important fraction of the not readily cultivable bacteria. J Microbiol Methods. 2004;57:379–390. doi: 10.1016/j.mimet.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Jezbera J, Horňák K, Šimek K. Food selection by bacterivorous protists: insight from the analysis of the food vacuole content by means of fluorescence in situ hybridization. FEMS Microbiol Ecol. 2005;52:351–363. doi: 10.1016/j.femsec.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Murray RGE, Stackebrandt E. Taxonomic note: implementation of the provisional status Candidatus for incompletely described procaryotes. Int J Syst Bacteriol. 1995;45:186–187. doi: 10.1099/00207713-45-1-186. [DOI] [PubMed] [Google Scholar]
- Rheinheimer G. Aquatic Microbiology. 2nd edn Wiley; New York: 1980. [Google Scholar]
- Rosselló-Mora R, Amann R. The species concept for prokaryotes. FEMS Microbiol Rev. 2001;25:39–67. doi: 10.1111/j.1574-6976.2001.tb00571.x. [DOI] [PubMed] [Google Scholar]
- Sekar R, Pernthaler A, Pernthaler J, Warnecke F, Posch T, Amann R. An improved protocol for quantification of freshwater Actinobacteria by fluorescence in situ hybridization. Appl Environ Microbiol. 2003;69:2928–2935. doi: 10.1128/AEM.69.5.2928-2935.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma AK, Zhaxybayeva O, Papke RT, Doolittle WF. Actinorhodopsins: proteorhodopsin-like gene sequences found predominantly in non-marine environments. Environ Microbiol. 2008;10:1039–1056. doi: 10.1111/j.1462-2920.2007.01525.x. [DOI] [PubMed] [Google Scholar]
- Stackebrandt E, Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152–155. [Google Scholar]
- Warnecke F, Amann R, Pernthaler J. Actinobacterial 16S rRNA genes from freshwater habitats cluster in four distinct lineages. Environ Microbiol. 2004;6:242–253. doi: 10.1111/j.1462-2920.2004.00561.x. [DOI] [PubMed] [Google Scholar]
- Wu QL, Hahn MW. Differences in structure and dynamics of Polynucleobacter communities in a temperate and a subtropical lake revealed at three phylogenetic levels. FEMS Microbiol Ecol. 2006;57:67–79. doi: 10.1111/j.1574-6941.2006.00105.x. [DOI] [PubMed] [Google Scholar]
- Wu QL, Zwart G, Wu J, Kamst-van Agterveld MP, Liu S, Hahn MW. Submersed macrophytes play a key role in structuring bacterioplankton community composition in the large, shallow, subtropical Taihu Lake, China. Environ Microbiol. 2007;9:2765–2774. doi: 10.1111/j.1462-2920.2007.01388.x. [DOI] [PubMed] [Google Scholar]
- Zwart G, Crump BC, Kamst-van Agterveld MP, Hagen F, Han S-K. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat Microb Ecol. 2002;28:141–155. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.