SUMMARY
Precise control of gene expression plays fundamental roles in brain development, but the roles of chromatin regulators in neuronal connectivity have remained poorly understood. We report that depletion of the NuRD complex by in vivo RNAi and conditional knockout of the core NuRD subunit Chd4 profoundly impairs the establishment of granule neuron parallel fiber/Purkinje cell synapses in the rodent cerebellar cortex in vivo. By interfacing genome-wide sequencing of transcripts and ChIP-Seq analyses, we uncover a network of repressed genes and distinct histone modifications at target gene promoters that are developmentally regulated by the NuRD complex in the cerebellum in vivo. Finally, in a targeted in vivo RNAi screen of NuRD target genes, we identify a program of NuRD-repressed genes that operate as critical regulators of presynaptic differentiation in the cerebellar cortex. Our findings define NuRD-dependent promoter decommissioning as a developmentally-regulated programming mechanism that drives synaptic connectivity in the mammalian brain.
INTRODUCTION
Control of gene expression plays fundamental roles in brain development and disease. Besides DNA sequence-specific transcription factors, global regulators of chromatin robustly influence transcription at the genome level and are prime candidates for triggering long-lasting cell-intrinsic changes in neuronal connectivity during critical stages of brain development. However, the functions of chromatin regulators in neuronal connectivity in the developing brain have remained poorly understood.
Synapse differentiation is an essential step in the assembly of neuronal circuits during brain development. Accordingly, defects in synapse differentiation are thought to play a critical role in developmental disorders of cognition (Abrahams and Geschwind, 2008; Ebert and Greenberg, 2013; Kelleher and Bear, 2008; Sudhof, 2008; Zoghbi, 2003). Recent advances in human genetics reveal that among global regulators of chromatin, ATP-dependent chromatin remodeling enzymes including the Chd and SWI/SNF families of ATPases are critical targets of mutations in autisms and intellectual disability (Neale et al., 2012; O’Roak et al., 2012a; Ronan et al., 2013; Talkowski et al., 2012), raising the fundamental question of whether these global chromatin regulators orchestrate synaptogenesis during brain development.
Among ATP-dependent chromatin remodeling enzymes, the nucleosome remodeling and deacetylation (NuRD) complex controls the programming of cell states during development including the differentiation of embryonic stem (ES) cells and progenitor cells (Hong et al., 2005; Kaji et al., 2006; Reynolds et al., 2012; Yoshida et al., 2008; Zhang et al., 2012). Notably, inhibition of the NuRD complex removes a rate-limiting barrier in somatic cells for their re-programming into induced pluripotent (iPS) cells (Rais et al., 2013). However, the role of the NuRD complex in programming cellular states in post-mitotic tissues including the brain has remained poorly understood.
In this study, we report that the NuRD chromatin remodeling complex programs the differentiation of presynaptic sites in the mammalian brain. Depletion of the NuRD complex by in vivo RNAi and conditional knockout strategies profoundly impairs the differentiation of presynaptic sites in the rodent cerebellar cortex in vivo. By interfacing genome-wide sequencing of transcripts (RNA-Seq) and ChIP-Seq analyses, we uncover a network of repressed genes and decommissioned promoters that are developmentally orchestrated by the NuRD complex in the cerebellum in vivo. Finally, in a targeted in vivo RNAi screen of NuRD target genes, we identify a program of NuRD-repressed genes that operate as critical regulators of presynaptic differentiation in the cerebellar cortex. Our findings define NuRD-dependent promoter decommissioning and transcriptional repression as a fundamental developmentally regulated programming mechanism that drives synaptic connectivity in the mammalian brain.
RESULTS
The NuRD complex is expressed in the developing cerebellar cortex
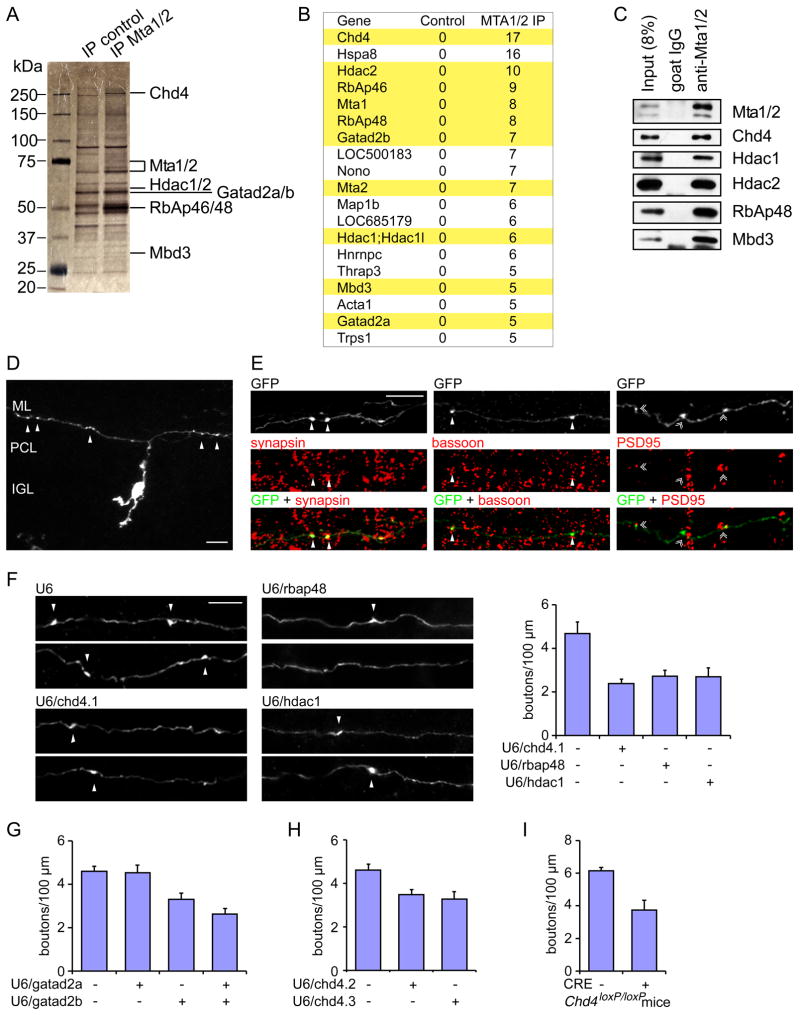
To characterize the role of the NuRD chromatin remodeling complex in brain, we first purified the endogenous NuRD complex from rat cerebella. Immunoprecipitation of the core NuRD complex protein, the metastasis associated proteins Mta1/2, in nuclear lysates of cerebella followed by mass spectrometry analyses led to the identification of the ATPase Chd4, Mta1/2, the zinc finger domain containing protein Gatad2a/b, the histone deacetylase Hdac1/2, the histone-binding protein RbAp46/48 and the methyl CpG-binding domain protein Mbd3 (Figures 1A and 1B) (Denslow and Wade, 2007; Xue et al., 1998; Zhang et al., 1998). In coimmunoprecipitation analyses, we confirmed that endogenous Mta1/2 forms a complex with Chd4, Hdac1/2, RbAp48 and Mbd3 in the rat cerebellum (Figure 1C). These results show that an intact NuRD complex is expressed in the rodent cerebellum.
Figure 1. The NuRD complex assembles in the cerebellar cortex and promotes granule neuron presynaptic differentiation in vivo.
(A, B) The NuRD complex was affinity purified from nuclear lysates of rat cerebella using the Mta1/2 antibody, followed by silver staining (A) and mass spectrometry analyses (B). The spectral count for each protein in mass spectrometry analyses is shown. The subunits of the NuRD complex are highlighted in yellow. (C) Lysates of rat cerebella were subjected to immunoprecipitation with the Mta1/2 antibody followed by immunoblotting with the Mta1/2, Chd4, Hdac1/2, RbAp48, or Mbd3 antibody. Core subunits of the NuRD complex assemble in the cerebellum. (D) P4 rat pups were electroporated with the GFP expression plasmid and sacrificed 8 days later. Cerebella were removed, sectioned, and subjected to immunohistochemistry using the GFP antibody. A representative image of a granule neuron is shown. The soma resides in the internal granule layer (IGL) and the parallel fiber axon spans the molecular layer (ML). Arrowheads denote presynaptic boutons along the parallel fiber. (E) Cerebellar sections prepared as in (D) were immunolabeled with the GFP antibody together with the synapsin, bassoon, or PSD95 antibody. Synapsin and bassoon co-localize in presynaptic boutons in transfected granule neuron axons (arrowheads), and PSD95 puncta were adjacent to the presynaptic boutons (double arrowheads), indicating that presynaptic boutons represent sites of presynaptic axonal differentiation. (F) P4 rat pups were electroporated with the U6/chd4.1, U6/rbap48, U6/hdac1, or control U6 RNAi plasmid together with pCAG-GFP plasmid and analyzed as in (D). Left panels: Representative images of Chd4 knockdown, RbAp48 knockdown, Hdac1 knockdown, and control neurons are shown. Right panel: Knockdown of Chd4, RbAp48, Hdac1 reduced the density of presynaptic boutons in transfected neurons (p<0.005, ANOVA followed by Fisher’s PLSD post hoc test, n=3). Scale bars: 10μm. (G, H) P4 rat pups were electroporated with the U6/gatad2a, U6/gatad2b, U6/chd4.2, U6/chd4.3, or control U6 RNAi plasmid together with pCAG-GFP plasmid and analyzed as in (D). Knockdown of Gatad2a/b (G) or Chd4 using two additional shRNAs targeting distinct regions (H) reduced the density of presynaptic boutons (p<0.005, ANOVA followed by Fisher’s PLSD post hoc test, n=3). (I) P6 Chd4loxP/loxP mice were electroporated with the Cre expression plasmid or the control vector together with the GFP expression plasmid and analyzed as in (D). Cre-induced knockout of Chd4 in granule neurons reduced presynaptic bouton density (p<0.005, t-test, n=4).
We next characterized the expression profile of the NuRD complex in the mouse cerebellar cortex. Immunohistochemical analyses revealed abundant Chd4 and Hdac1 expression in the internal granule layer (IGL), where granule neurons reside (Figures S1A and S1B). The subunits of the NuRD complex including Chd4, RbAp48, Mbd3, Hdac1, and Mta1/2 were all highly expressed in the cerebellum throughout the second and third postnatal weeks in mice (Figure S1C), when granule neurons form synapses and integrate into cerebellar circuits (Altman and Bayer, 1997). These results suggest that the NuRD complex may have roles in neuronal connectivity in the developing cerebellar cortex.
The NuRD complex drives granule neuron presynaptic differentiation in vivo
To determine the functions of the NuRD complex in synapse differentiation in the cerebellar cortex, we first employed an in vivo electroporation approach to characterize the morphogenesis of granule neurons in the postnatal rodent brain (Figures 1D and 1E) (Konishi et al., 2004; Shalizi et al., 2006; Yang et al., 2009). We electroporated postnatal day 4 (P4) rat pups with the GFP expression plasmid, and sacrificed the animals 8 days later at P12. In immunohistochemical analyses of the cerebellar cortex using the GFP antibody, we visualized granule neuron somas and their dendrites and parallel fiber axons in the internal granule layer and molecular layer, respectively (Figure 1D). Along granule neuron parallel fiber axons, boutons were observed that harbored the presynaptic proteins synapsin and bassoon (Figure 1E). The presynaptic boutons were closely apposed to the postsynaptic protein PSD95 (Figure 1E). These data suggest that parallel fiber axon boutons represent presynaptic sites in vivo.
We next assessed the function of the NuRD complex in the cerebellum by depleting components of the NuRD complex in vivo using two distinct genetic approaches. Knockdown of the NuRD complex subunit Chd4, RbAp48, Hdac1, or Gatad2a/b by RNA interference (RNAi) in postnatal rat pups profoundly reduced the density of presynaptic parallel fiber boutons in the cerebellar cortex in vivo (Figures 1F–H, S1D-I). We also induced conditional knockout of the core NuRD subunit Chd4 in granule neurons in the mouse cerebellar cortex in vivo. Knockout of Chd4, achieved by delivery of the recombinase Cre in Chd4loxP/loxP mice by electroporation, substantially reduced the density of granule neuron presynaptic boutons in the cerebellar cortex in vivo, phenocopying the effects of knockdown of Chd4, RbAp48, Hdac1, or Gatad2a/b in the rat cerebellar cortex in vivo (Figure 1I). Together, these results suggest that the NuRD complex plays an essential cell-autonomous role in presynaptic differentiation in the mammalian cerebellar cortex in vivo.
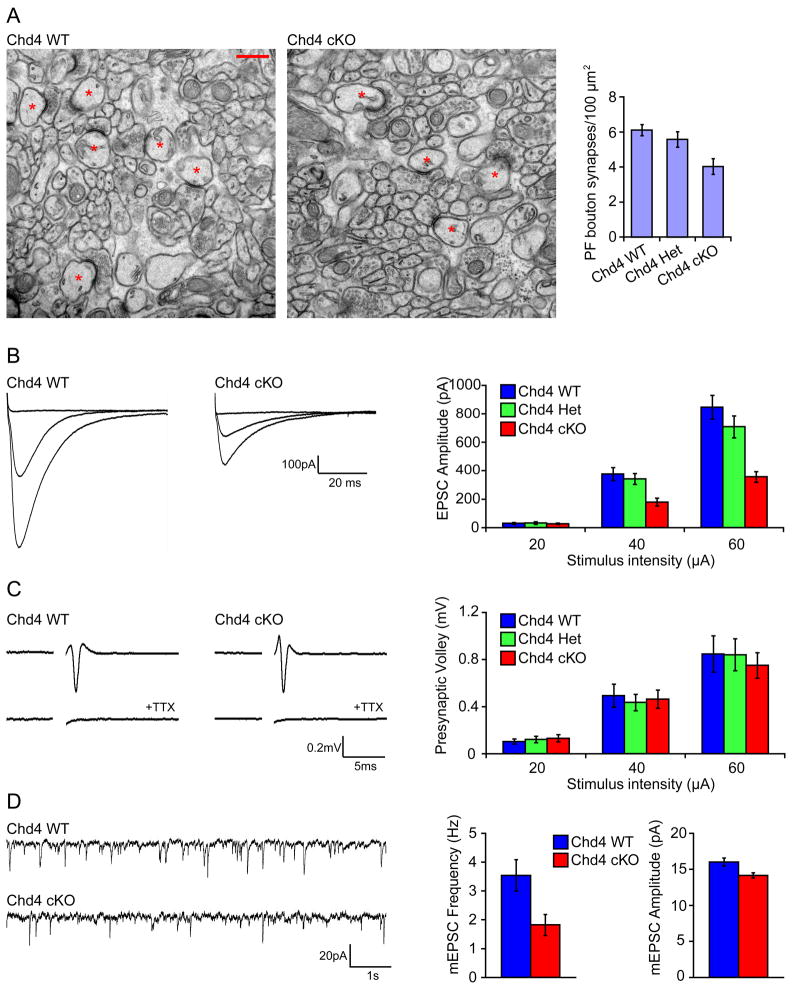
Granule neuron parallel fiber boutons synapse onto dendritic spines of Purkinje neurons in the cerebellar cortex (Altman and Bayer, 1997; Palay and Chan-Palay, 1974). We therefore assessed the effect of conditional knockout of Chd4 in granule neurons on the development of synapses between parallel fibers and Purkinje cell dendrites. To induce conditional knockout of Chd4 selectively in granule neurons in the cerebellar cortex, we crossed Chd4loxP/loxP mice with a transgenic mouse line in which the expression of Cre is driven by the GABA(A)α6 receptor promoter (G6R-Cre) (Funfschilling and Reichardt, 2002). Expression of Chd4 was reduced specifically in the IGL within the cerebellar cortex in Chd4 conditional knockout mice during the third postnatal week (Figures S1B and S1C). The anatomical architecture of the cerebellar cortex was not altered in Chd4 conditional knockout mice (Figures S1B and S2A). In electron microscopy analyses, the density of synapses between parallel fibers and Purkinje cell dendrites in vivo was substantially lower in Chd4 conditional knockout mice compared to control Chd4loxP/loxP mice (Figures 2A and S2B). Mice heterozygous for the disrupted Chd4 allele displayed little difference in synapse density as compared to control Chd4loxP/loxP mice (Figure 2A). Notably, depletion of Chd4 had little or no effect on the size of boutons or the number of presynaptic vesicles (Figures S2C and S2D). Taken together, these results suggest that the NuRD complex promotes synaptogenesis in the cerebellar cortex in vivo.
Figure 2. Knockout of the core NuRD complex subunit Chd4 impairs granule neuron/Purkinje cell synaptogenesis and neurotransmission in the cerebellar cortex.
(A) Cerebella from P22 Chd4 conditional knockout mice, Chd4 heterozygous mice, and control Chd4loxP/loxP mice were subjected to electron microscopy analyses. Left panels: representative electron micrographs of the molecular layer of the cerebellar cortex from Chd4 conditional knockout mice (Chd4 cKO) and control Chd4loxP/loxP mice (Chd4 WT) are shown. Synapses comprising of parallel fiber presynaptic boutons apposed to Purkinje cell postsynaptic spines are denoted with asterisks. Scale bar: 500nm. Right panel: quantification of the density of granule parallel fiber/Purkinje cell synapses in Chd4 conditional knockout (Chd4 cKO), Chd4 heterozygous (Chd4 Het), and control Chd4loxP/loxP (Chd4 WT) mice. The density of synapses is reduced in Chd4 conditional knockout mice compared to control Chd4loxP/loxP mice (p<0.005, ANOVA followed by Fisher’s PLSD post hoc test, n=10–12 regions, 2 brains). (B) Acute sagittal cerebellar slices were prepared from P20–P24 Chd4 conditional knockout, Chd4 heterozygous, and Chd4loxP/loxP mice and parallel fiber-evoked Purkinje cell currents (PF-EPSCs) were recorded in response to increasing stimulus intensities (20, 40, and 60μA). Representative current traces (left panels) and quantification of the PF-EPSC amplitude (right panel) are shown. Chd4 conditional knockout mice (Chd4 cKO) have reduced evoked EPSC amplitude compared to control Chd4loxP/loxP mice (Chd4 WT) (p<0.001, ANOVA followed by Fisher’s PLSD post hoc test, n=21–26 neurons, 5 brains). (C) Acute coronal cerebellar slices were prepared as in (B), and parallel fiber axons were stimulated at sites 400 μm away from an extracellular recording electrode. A representative trace of the stimulus-evoked presynaptic waveform before and after application of TTX is shown in the left panels. The stimulus artifact was removed for clarity. Quantification of presynaptic volley amplitude is shown in the right panel. Conditional knockout of Chd4 has little or no effect on the presynaptic volley amplitude. (D) Acute sagittal slices cerebellar were prepared as in (B) and Purkinje cell mEPSCs were recorded in the presence of TTX. Representative traces of mEPSCs from Chd4 conditional knockout and control Chd4loxP/loxP mice are shown in the left panel. Quantification of the mEPSC frequency and amplitude are shown in the right panels. Chd4 conditional knockout mice had reduced mEPSC frequency and amplitude compared to control Chd4loxP/loxP mice (p<0.05, t-test, n=24–27 neurons, 7 brains).
The NuRD complex promotes granule neuron to Purkinje cell neurotransmission in the cerebellar cortex
The requirement for the NuRD complex in synaptogenesis in the cerebellar cortex led to the prediction that synaptic transmission at parallel fiber/Purkinje cell synapses might be impaired in Chd4 conditional knockout mice. Consistent with this prediction, electrophysiological analyses in acute cerebellar slices revealed that evoked excitatory postsynaptic currents (EPSCs) at parallel fiber/Purkinje cell synapses were substantially reduced in Chd4 conditional knockout mice compared to littermate Chd4 heterozygous mice or Chd4loxP/loxP mice (Figure 2B). There was little or no difference in the amplitude of the presynaptic volley in mice with the different Chd4 genotypes (Figure 2C), suggesting that the impairment of evoked EPSCs in conditional Chd4 knockout mice is not secondary to changes in axon excitability and instead reflects synaptic dysfunction.
Because the NuRD complex promotes synaptogenesis, we assessed if the neurotransmission defects in Chd4 conditional knockout mice might be secondary to changes in synapse number. The frequency of miniature EPSCs (mEPSCs) in Purkinje cells was reduced in acute cerebellar slices from Chd4 conditional knockout mice compared to control Chd4loxP/loxP mice (Figure 2D), consistent with the conclusion that synapse number is reduced in Chd4 conditional knockout animals. The amplitude of mEPSCs was modestly reduced in Chd4 conditional knockout mice. Collectively, our data suggest that the NuRD complex programs the structural and functional maturation of synapses in the cerebellar cortex.
The NuRD complex decommissions promoters and thereby represses a network of genes in the developing cerebellum in vivo
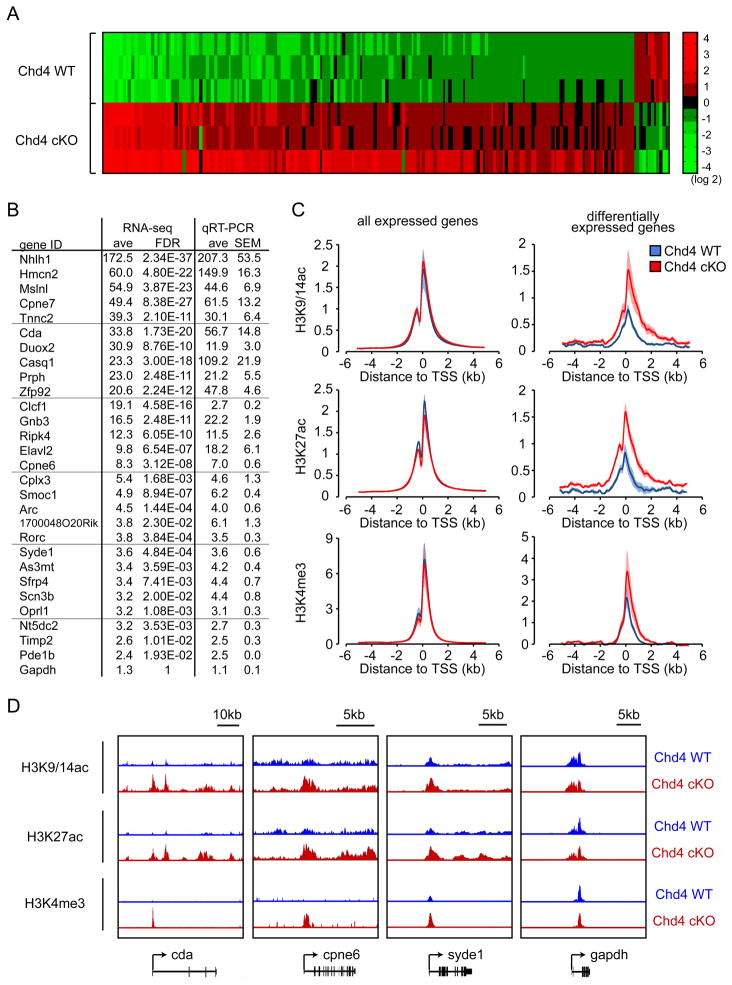
We next determined the mechanisms by which the NuRD complex coordinates synapse differentiation. Because the NuRD complex is a chromatin regulatory enzyme (Xue et al., 1998; Zhang et al., 1998), we reasoned that depletion of Chd4 might alter the expression of a large set of genes in neurons. We therefore assessed the effect of Chd4 knockout on the transcriptome in the cerebellum in vivo. We analyzed RNA from cerebella of Chd4 conditional knockout and control Chd4loxP/loxP mice at P22, when synapse development and function are impaired in Chd4 knockout animals. The RNA-Seq analyses led to the identification of significant alterations in the expression of 199 genes in the cerebellum of Chd4 conditional knockout mice as compared to control Chd4loxP/loxP mice (Figure 3A and Table S1). Remarkably, 93% of the significantly altered genes in Chd4 conditional knockout cerebella were upregulated, suggesting that Chd4 operates primarily as a transcriptional repressor in granule neurons of the developing cerebellum (Figure 3A and Table S1). Independent microarray analyses of RNA revealed that 94% of the differentially expressed genes in the cerebellum of Chd4 conditional knockout mice as compared to control Chd4loxP/loxP mice were upregulated (Figure S3A).
Figure 3. The NuRD complex decommissions the promoters of a specific set of genes and thereby represses their expression in the cerebellum in vivo.
(A) RNA was extracted from cerebella of P22 control Chd4loxP/loxP mice and Chd4 conditional knockout mice and subjected to RNA-Seq analyses. A heatmap of the expression levels of significantly differentially expressed genes between control Chd4loxP/loxP and Chd4 conditional knockout cerebella is shown (FDR<0.05, 3 independent brains per condition, base2 log-transformed mean centered). The vast majority (93%) of differentially expressed genes identified by RNA-Seq were derepressed in Chd4 conditional knockout cerebella. (B) RNA from P22 control Chd4loxP/loxP mice and Chd4 conditional knockout mice were subjected to qRT-PCR using primers specific to Chd4-regulated genes identified by RNA-Seq. Fold change of gene expression by RNA-Seq and qRT-PCR are shown. Changes in gene expression between Chd4loxP/loxP mice and Chd4 conditional knockout mice as measured by RNA-Seq are in good agreement with qRT-PCR analyses. The gapdh gene was included as a negative control. (C) Cerebella of P22 control Chd4loxP/loxP mice and Chd4 conditional knockout mice were subjected to ChIP-Seq analyses. The profiles of the transcriptionally-active histone marks H3K9/14ac, H3K27ac, and H3K4me3 surrounding the transcription start site (TSS) of all expressed genes (left panels) and NuRD-repressed target genes (right panels) are shown. The abundance of H3K9/14ac, H3K27ac, and H3K4me3 marks was increased at the promoters of NuRD-repressed target genes in Chd4 conditional knockout mice compared to control Chd4loxP/loxP mice. There were little or no differences in the genome-wide level of H3K9/14ac, H3K27ac, and H3K4me3 marks between Chd4 conditional knockout mice (Chd4 cKO) and control Chd4loxP/loxP mice (Chd4 WT). The shading denotes standard error. (D) Representative genomic regions of NuRD-regulated target genes are shown. The abundance of H3K9/14ac, H3K27ac, and H3K4me3 histone marks was increased at the promoters of the NuRD-targets cda, cpne6, and syde1, but not at the promoter of the control gene gapdh, in Chd4 conditional knockout mice.
We validated putative Chd4 target genes by quantitative RT-PCR (qRT-PCR) analyses, and found good agreement between RNA-Seq and qRT-PCR analyses (Figure 3B). In other qRT-PCR analyses, knockdown of Chd4, Hdac1, and Gatad2a/b in primary granule neurons led to upregulation of the distinct target genes Nhlh1, Elavl2, Scn3b, and Necab1 (Figure S3B). These results suggest that the NuRD complex functions primarily as a transcriptional repressor in neurons and thereby specifically inhibits the expression of a set of genes in granule neurons in the cerebellar cortex.
The acetylation of lysine residues on histone tails including histone H3 lysine 9 (H3K9), H3K14, and H3K27 at the promoters of genes tightly correlates with active transcription (Kouzarides, 2007; Li et al., 2007; Shahbazian and Grunstein, 2007; Wang et al., 2009b; Wang et al., 2008). Because the NuRD complex acts primarily as a transcriptional repressor in granule neurons, we asked whether the NuRD complex might regulate these histone modifications at the promoters of actively transcribed genes in post-mitotic neurons. We performed chromatin immunoprecipitation followed by massive parallel sequencing (ChIP-Seq) analyses using the histone H3 acetylated-K9/14 or acetylated-K27 antibody in the cerebellum of Chd4 conditional knockout mice and control Chd4loxP/loxP mice (Figures 3C, 3D, S3C, and S3D). Interestingly, depletion of Chd4 in granule neurons of the cerebellum in Chd4 conditional knockout mice had little or no effect on enrichment of H3K9/14 or H3K27 acetylation at the promoters of expressed genes on a global scale (Figure 3C). Strikingly, however, intersection of RNA-Seq and ChIP-Seq analyses revealed that the acetylation of H3K9/14 and H3K27 was substantially enhanced in the cerebellum of Chd4 conditional knockout mice selectively on the promoters of the set of genes repressed by the NuRD complex (Figures 3C, 3D, S3C, and S3D). In control analyses, there was little or no change in the occupancy of total histone H3 and H4 at the promoters of NuRD-repressed target genes in Chd4 conditional knockout mice compared to control Chd4loxP/loxP mice (Figure S3C). These data suggest that the NuRD complex triggers the deacetylation of histone H3 lysine residues at the promoters of a specific set of genes in post-mitotic neurons in the mammalian brain.
In addition to the acetylation of histone H3 K9/14 and K27, the trimethylation of histone H3K4 is also associated with transcriptional activation (Heintzman et al., 2007; Kouzarides, 2007; Li et al., 2007; Wang et al., 2008). We found that trimethylation of H3K4 was stimulated on the promoters of NuRD target genes in the cerebellum in Chd4 conditional knockout mice compared to control Chd4loxP/loxP mice (Figures 3C, 3D, S3C, and S3D). In contrast, H3K4 trimethylation failed to increase at expressed gene promoters globally in the cerebellum of Chd4 conditional knockout mice (Figure 3C). Thus, in addition to inducing the deacetylation of histone H3K9/14 and K27, the NuRD complex triggers the demethylation of histone H3K4 at distinct set of gene promoters in neurons. Taken together, our data suggest that the NuRD complex decommissions the promoters of a specific set of genes by turning off histone modifications associated with transcriptional activation at these genes and thereby triggers their repression in post-mitotic neurons in the mammalian brain in vivo.
The NuRD complex represses developmentally downregulated genes in the developing cerebellar cortex in vivo
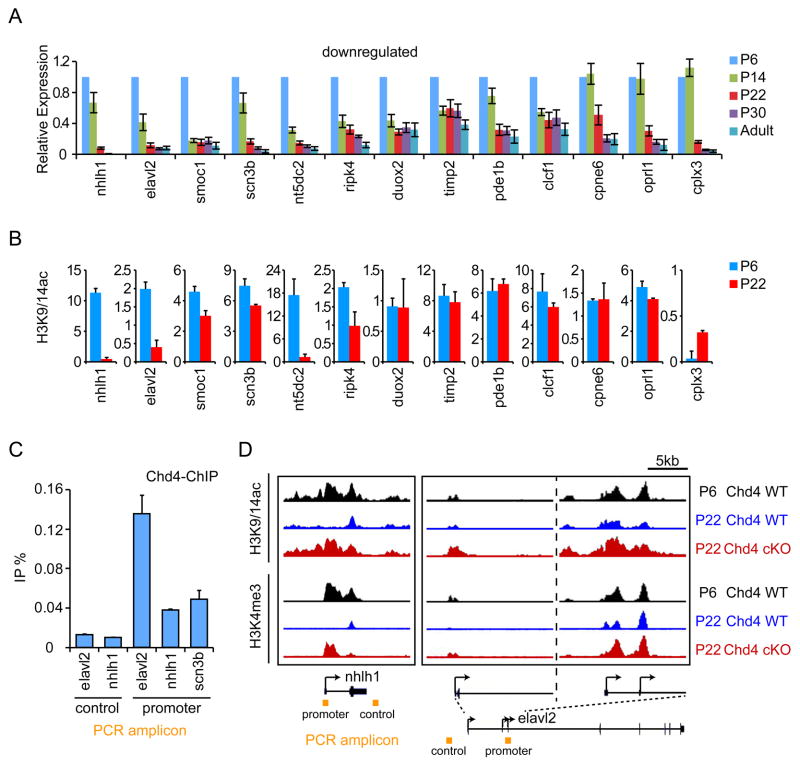
Temporal regulation of gene expression is necessary for the proper differentiation of neurons (de la Torre-Ubieta and Bonni, 2011; Ronan et al., 2013). Presynaptic differentiation at parallel fiber/Purkinje cell synapses in the rodent cerebellar cortex occurs during the first postnatal month (Altman and Bayer, 1997), and synapse number increased progressively during this developmental period in vivo (Figure S4A). Because the NuRD complex programs synapse development, we reasoned that repression of NuRD target genes upon neuronal maturation may facilitate synaptogenesis. To test this model, we first characterized the developmental expression profile of a panel of 24 NuRD-regulated genes, comprised of the robustly derepressed genes in Chd4 knockout mice as well as genes implicated in synapse differentiation and function (Figure 3B). In qRT-PCR analyses, among this panel of NuRD target genes, more than half were progressively downregulated during the second or third week of postnatal development (Figure 4A and S4B).
Figure 4. The NuRD complex orchestrates developmental repression of a program of genes that inhibit presynaptic differentiation in vivo.
(A) Total RNA of cerebella from rat pups at P6, P14, P22, P30, and during adulthood were subjected to qRT-PCR analyses using primers to a panel of NuRD-regulated genes. Gene expression was normalized to Gapdh expression. The majority of NuRD-regulated genes were progressively downregulated during cerebellar development. (B) Cerebella of P6 and P22 mice were subjected to ChIP-Seq analyses as in Figure 3C using the H3K9/14ac antibody. The sum of normalized H3K9/14ac reads in a 1 kb window centered at the +500 position relative to the TSS of NuRD-repressed target genes is shown. The abundance of histone H3K9/14ac is reduced at the promoters of the majority of developmentally downregulated NuRD target genes at P22 compared to P6. Error bars denote standard error of two biological ChIP-seq replicates. (C) P14 mice cerebella were subjected to ChIP-qPCR analyses using the Chd4 antibody and primers specific to the promoters of elavl2, nhlh1, and scn3b and control regions. Chd4 is enriched at target gene promoters (p<0.05, ANOVA followed by Fisher’s PLSD post hoc test, n=4). (D) Representative genomic regions of the nhlh1 and elavl2 genes in cerebella of control P6 and P22 Chd4loxP/loxP mice (Chd4 WT) and P22 Chd4 conditional knockout mice (Chd4 cKO). The abundance of H3K9/14ac (top panels) and H3K4me3 (bottom panels) marks at the nhlh1 and elavl2 gene promoters was decreased at P22 (blue) relative to P6 (black) in the cerebellum in control Chd4loxP/loxP mice. By contrast, the abundance of H3K9/14ac (top panels) and H3K4me3 (bottom panels) marks in Chd4 conditional knockout mice at P22 (red) was similar to that in control Chd4loxP/loxP mice at P6 (black).
We next determined the role of chromatin regulatory mechanisms in orchestrating the temporal expression profile of NuRD-regulated genes in the developing cerebellum. Remarkably, the majority of the NuRD-repressed and developmentally downregulated genes also had reduced H3K9/14 acetylation at their promoters at P22 compared to P6 (Figures 4A and 4B). By contrast, NuRD-regulated genes whose expression was not robustly downregulated during development had similar or higher levels of H3K9/14 acetylation at their promoters at P22 as compared to P6 (Figures S4B and S4C). In other experiments, H3K4 trimethylation was also reduced at the promoters of NuRD-repressed and developmentally downregulated genes at P22 compared to P6 (Figures S4D and S4E). These data suggest that promoter decommissioning and transcriptional repression of a substantial subset of NuRD target genes in post-mitotic neurons occurs in a developmentally-regulated fashion in the cerebellum in vivo.
We next asked if the NuRD complex directly controls the developmental expression profile of NuRD-regulated genes. In ChIP followed by quantitative PCR (ChIP-qPCR) analyses, Chd4 occupied the promoters of distinct NuRD-regulated genes encoding the transcription factor Nhlh1, the RNA-binding protein Elavl2, and the voltage-sensitive sodium channel Scn3b in the mouse cerebellum at P14 (Figure 4C), a time when these genes are in transition or poised to move from an active to repressed transcription state. In other ChIP-qPCR analyses, we found that Chd4 occupies the promoters of another set of NuRD-regulated genes encoding Scn3b, the calcium-binding protein Cpne9, the cyclic nucleotide phosphodiesterase Pde1b, and the metallopeptidase inhibitor Timp2 in the cerebellum in control Chd4loxP/loxP mice but not Chd4 knockout mice (Figure S4F). Together, these data suggest that the NuRD complex directly binds NuRD-regulated and developmentally repressed target genes.
We next determined whether the NuRD complex is required for promoter decommissioning of developmentally downregulated genes. In control Chd4loxP/loxP mice, the acetylation of H3K9/14 and trimethylation of H3K4 were robustly reduced in the mouse cerebellum in vivo during development at the promoters of NuRD target genes, including the nhlh1 and elavl2 genes (Figures 4B, 4D, S4D, and S4G). Strikingly, conditional knockout of Chd4 in granule neurons of the cerebellum blocked the downregulation of histone acetylation and methylation at the promoters of these NuRD target genes in vivo (Figures 4D and S4G). Collectively, these data suggest that the NuRD complex plays a critical and direct role in promoter decommissioning of a subset of developmentally regulated genes in the brain.
In vivo RNAi screen of NuRD target genes implicates Nhlh1, Elavl2, and Cplx3 in the regulation of presynaptic differentiation in the cerebellar cortex
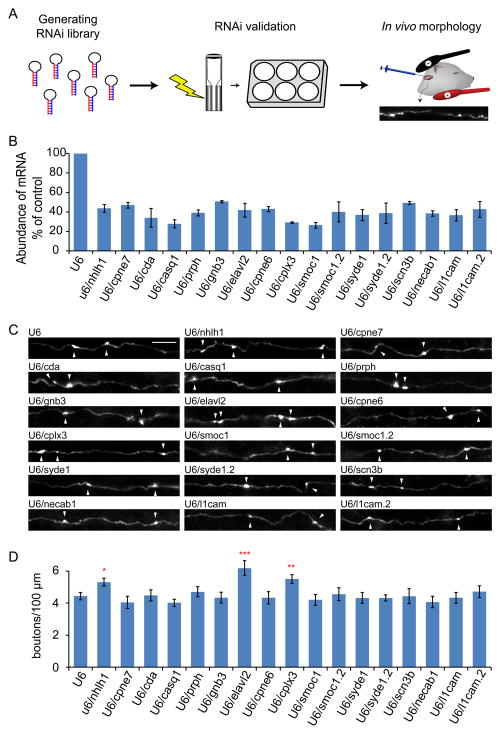
The identification of a program of NuRD-repressed genes that are downregulated with neuronal maturation in the mammalian brain led us next to determine the role of these genes in presynaptic differentiation in vivo. To address this question, we performed an in vivo RNAi screen of NuRD target genes in the cerebellar cortex (Figure 5A). The criteria for selection of NuRD targets in the in vivo screen included genes that were robustly derepressed upon conditional knockout of Chd4 in granule neurons in the cerebellar cortex (Figures 3A, 3B, and S3A), developmentally downregulated in the cerebellum (Figure 4A), or demonstrated to function in synapse development. Using these criteria, we identified 24 candidate NuRD target genes for further study and generated 64 shRNAs targeting these genes. In qRT-PCR analyses in primary granule neurons, the endogenous expression of 14 target genes was reduced over 50% by 17 shRNAs (Figure 5B). Since the NuRD complex drives the formation of presynaptic boutons and represses target gene expression, knockdown of physiologically relevant NuRD-repressed genes would be predicted to stimulate presynaptic differentiation. We found that knockdown of the transcription factor Nhlh1, the RNA binding protein Elavl2, and the presynaptic regulator Cplx3 increased the density of presynaptic boutons in granule neurons in vivo (Figures 5C and 5D). These results suggest that that the NuRD complex represses a program of genes with distinct cellular and biochemical functions to drive presynaptic differentiation in vivo. Collectively, our findings define a novel chromatin regulatory pathway that drives promoter decommissioning and hence repression of a program of genes in post-mitotic neurons culminating in synapse differentiation in the mammalian brain.
Figure 5. In vivo RNAi screen of NuRD complex target genes implicates Nhlh1, Elavl2, and Cplx3 in the suppression of presynaptic differentiation in the cerebellar cortex in vivo.
(A) The experimental design for the in vivo screen of regulators of presynaptic differentiation is shown. We first generated a plasmid library containing RNAi short hairpin RNAs (shRNAs) targeting NuRD-repressed genes (left panel). We next validated the efficacy of the RNAi vectors using primary granule neurons (middle panel). Finally, we electroporated validated RNAi plasmids together with GFP in rat pups and analyzed their effects on granule neuron presynaptic differentiation in vivo (right panel). (B) Granule neurons were transfected with the indicated RNAi or control U6 RNAi plasmid and subjected to qRT-PCR analyses using the primers specific to the gene indicated, along with Gapdh, the latter serving as a control. The knockdown efficiency of the RNAi plasmids are shown relative to the control U6 condition. The expression of 14 NuRD-repressed genes was downregulated over 50% by 17 shRNAs (p<0.001, ANOVA followed by Fisher’s PLSD post hoc test, n=3). (C, D) P4 rat pups were electroporated with the validated RNAi plasmids indicated in (B) together with pCAG-GFP plasmid and analyzed as in Figure 1D. Knockdown of Nhlh1, Elavl2, and Cplx3 increased the density of presynaptic boutons along granule neuron parallel fibers in the cerebellum in vivo (p<0.05, ANOVA followed by Fisher’s PLSD post hoc test, n=3–8 brains). Scale bar: 10μm.
DISCUSSION
In this study, we have discovered a novel chromatin regulatory pathway that programs synaptic connectivity in the mammalian brain. Depletion of the NuRD chromatin remodeling complex by in vivo RNAi in rats and conditional knockout in mice dramatically impairs the development of granule neuron parallel fiber/Purkinje cell synapses in the cerebellar cortex. We have also elucidated the mechanism underlying the novel function of the NuRD complex in post-mitotic neurons. The NuRD complex decommissions the promoters of nearly 200 genes by turning off histone modifications associated with transcriptional activation at these genes, thereby triggering their repression in the cerebellum in vivo. A targeted RNAi screen revealed that the NuRD complex represses a program of genes that operate as negative regulators of presynaptic differentiation in vivo. Collectively, our findings define NuRD-dependent promoter decommissioning as a developmentally regulated programming mechanism in post-mitotic neurons that drives synaptic connectivity in the mammalian brain.
The identification of a function for the NuRD complex in synapse differentiation in the cerebellar cortex unveils a novel epigenetic role for regulators of chromatin in the establishment of neuronal connectivity in the brain. Our findings suggest that the NuRD complex triggers promoter decommissioning and transcriptional repression of a specific set of genes in post-mitotic neurons. The expression of a subset of these genes is downregulated with neuronal maturation in the cerebellum in a NuRD-dependent manner, suggesting that promoter decommissioning and transcriptional repression triggers the transition from an immature newly post-mitotic state to mature neuronal state, leading to the integration of neurons into circuits.
How does the NuRD complex orchestrate the differentiation of presynaptic sites in the mammalian cerebellar cortex? Our results in the targeted in vivo RNAi screen provide significant insights into this question. Among the three newly identified NuRD-repressed target genes that regulate presynaptic differentiation, Nhlh1 encodes a transcription factor (Uittenbogaard et al., 1999), and Elavl2 encodes an RNA-binding protein that may control mRNA splicing and stability (Darnell, 2013). Because Nhlh1 and Elavl2 regulate the expression of other genes, the NuRD complex may operate at the apex of a gene expression program that governs presynaptic development in the cerebellar cortex. The in vivo knockdown and conditional knockout studies suggest that the NuRD complex operates throughout the second and third weeks of postnatal rodent cerebellar development. These findings further strengthen the conclusion that the NuRD complex plays a pivotal role in presynaptic connectivity, perhaps regulating multiple stages of presynaptic development.
The NuRD complex plays essential roles in programming cellular states during development including the differentiation of ES and progenitor cells (Fujita et al., 2004; Hong et al., 2005; Kaji et al., 2006; Whyte et al., 2012; Yoshida et al., 2008), highlighting fundamental and conserved functions for transcriptional repression in fate specification of ES and progenitor cells and the maturation of post-mitotic neurons in the brain. Notably, the NuRD complex imposes a rate-limiting barrier in somatic cells for their re-programming into iPS cells (Rais et al., 2013). Because iPS cells are widely used to model neurodevelopmental and neurodegenerative diseases in which synaptic impairment is a prominent pathological feature (Marchetto et al., 2010; Shcheglovitov et al., 2013), our findings suggest that engineering the restoration of NuRD function in NuRD-deficient reprogrammed iPS cells would be necessary for the study of synaptic dysfunction in these cells.
Although the NuRD complex has been studied in ES cells and progenitors, prior to our study, the composition, roles, and mechanism of NuRD function in the brain remained unexplored. The core subunit Chd3 and Chd4 are thought to form distinct NuRD complexes (Ivanov et al., 2007; Schultz et al., 2001). Notably, in our IP-mass spectrometry analyses, although Mta1/2 formed a complex with Chd4, an Mta1/2-Chd3 interaction was not detected in the cerebellum, suggesting that the NuRD complex contains Chd4 but not Chd3 in the cerebellar cortex.
Our knockout and in vivo RNAi studies suggest that both the chromatin remodeling enzyme encoded by Chd4 and the deacetylase activity encoded by Hdac1 within the NuRD complex are required for the differentiation of presynaptic sites in the cerebellar cortex. In addition to the increased histone H3 acetylation, H3K4 trimethylation at the promoters of NuRD target genes increases in the cerebellum in Chd4 conditional knockout mice. It will be important to determine whether the NuRD complex associates with histone demethylases in post-mitotic neurons to trigger the coordinate demethylation of histone H3K4 at NuRD targets in the brain. Consistent with this possibility, the NuRD complex may interact with the histone demethylases LSD1 and JARID1b to induce the demethylation of histone H3K4 in ES and breast cancer cells (Li et al., 2011; Wang et al., 2009a; Whyte et al., 2012). Another mutually non-exclusive mechanism that might underlie NuRD-regulation of histone H3K4 trimethylation is that the NuRD complex may inhibit the recruitment of a histone methyltransferase to the promoters of NuRD targets in neurons.
Although we have focused our study on the role and mechanisms of the NuRD complex in the establishment of synaptic connectivity in the brain, our findings have implications in the study of developmental disorders of neuronal connectivity. Notably, mutations of Chd4-related ATP-dependent chromatin remodeling enzymes have been implicated in syndromic and non-syndromic autism spectrum disorders (Jiang et al., 2013; O’Roak et al., 2012a; O’Roak et al., 2012b; Vissers et al., 2004). However, the roles of these enzymes in neuronal connectivity in the brain have remained to be characterized. The elucidation of the function and mechanisms of the NuRD complex in synapse differentiation in this study provides a roadmap for the study of other Chd family chromatin remodeling complexes in neuronal connectivity in vivo, thus facilitating our understanding of neurodevelopmental disorders of cognition.
EXPERIMENTAL PROCEDURES
Animals
Rodents were purchased or maintained under pathogen-free conditions. All animal experiments were done according to protocols approved by the Animal Studies Committee of Washington University School of Medicine and the Harvard Medical Area Standing Committee on Animals and in accordance with the National Institutes of Health guidelines. Chd4loxP/loxP and GABA(A)α6-CRE mice have been previously described (Funfschilling and Reichardt, 2002; Yoshida et al., 2008). Chd4 knockout was confirmed by PCR analysis of genomic DNA and by qRT–PCR.
Antibodies
Antibodies to synapsin (Millipore), bassoon (Assay Designs), PSD95 (Neuromab), Flag (Sigma M2), Chd4 (Abcam ab72418), Hdac1 (Abcam ab7028), Hdac2 (Abcam ab51832), Mta1/2 (Santa Cruz Biotechnology, sc9447), RbAp48 (Abcam ab488), Mbd3 (Cell Signaling #3896) ERK1/2 (Cell Signaling), histone H3K9/14ac (Millipore 06–599), histone H3K27ac (Abcam ab4729), histone H3K4me3 (Abcam ab8580), total H3 (active motif #39163), total H4 (Millipore, 05–858), goat serum (Sigma), and rabbit IgG (Millipore) were purchased.
Purification of the NuRD complex
Purification of the NuRD complex was performed using P8 rat cerebellar nuclear lysates. Cerebella were homogenized with hypotonic buffer (10 mM Hepes (pH 7.9), 10 mM KCl, 1.5 mM MgCl2) to remove the cytoplasmic fraction, and nuclear lysates were prepared with an extraction buffer (20 mM Hepes (pH 7.9), 20% glycerol, 250 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA). The Mta1/2 antibody coupled to protein G beads (GE healthcare) using disuccinimidyl suberate (DSS) (Pierce) according to the manufacture’s protocol was incubated overnight at 4°C with cerebellar nuclear lysate and washed extensively with wash buffer (20 mM Hepes (pH 7.9), 20% glycerol, 700 mM KCl, 0.2 mM EDTA, 0.5% NP-40) (Zhang et al., 1997). The bound proteins were eluted with 0.2 M glycine (pH 2.6), neutralized with 1 M Tris–HCl (pH 8.0), concentrated by TCA precipitation, and analyzed by mass spectrometry. A portion of the eluate was analyzed by SDS-PAGE followed by silver staining (Invitrogen).
Immunoprecipitation
Rat cerebella were homogenized with lysis buffer (50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1% Triton X-100, proteinase inhibitor cocktail) 25 times and incubated on ice for 20 minutes. Lysates were incubated with the Mta1/2 antibody overnight at 4°C and mixed with protein G beads (GE healthcare). The beads were washed with lysis buffer four times, and the immunoprecipitates were subjected to immunoblotting analyses.
Cerebellar granule neuron cultures
Granule neurons were prepared from cerebella of P6 Long Evans rat pups as described (Bilimoria and Bonni, 2008). High-efficiency transfection of granule neurons (maximum efficiency 80%) for biochemical analyses was achieved using a nucleofection method with the Amaxa electroporation device as described (Yamada et al., 2013).
In vivo electroporation and immunohistochemistry
In vivo electroporation of postnatal rat pups was performed as described (Konishi et al., 2004; Shalizi et al., 2006; Yang et al., 2009). The indicated plasmids were injected into the anterior cerebellum of P4 Sprague-Dawley rat pups or P6 mouse pups, and were then subjected to five electric pulses of 175 mV (rat) or 135 mV (mouse) with 950 ms intervals. Electroporated pups were returned to moms and examined 8 days later following immunohistochemistry analyses. Rat or mouse pups were fixed with 4% PFA and 4% sucrose and labeled with the relevant antibodies. For synapsin, bassoon, or PSD95 antibody, the sections were pre-treated with pepsin (Dako) for antigen retrieval. Images of transfected neurons were taken in a blinded manner on an Olympus Fluoview FV1000 confocal microscope and analyzed using the FV10-ASW and SPOT imaging softwares.
Electron microscopy
P22 mice were perfused with 2% formaldehyde and 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4. Cerebella were collected, postfixed overnight, and washed and stored in 0.1 M cacodylate buffer. The cerebella were then embedded in 4% agar and 0.75 mm sections were cut sagittally on a tissue chopper (McIlwain). The small sections were fixed with 1% osmium tetroxide/1.5% potassium ferrocyanide for 1 hr, washed in water 3x, and incubated in 1% aqueous uranyl acetate for 1 hr followed by 2 washes in water and subsequent dehydration in grades of alcohol (10 min each; 50%, 70%, 90%, 2× 10 min 100%). The samples were then incubated with propyleneoxide for 1 hr and infiltrated overnight in a 1:1 mixture of propyleneoxide and TAAB Epon (Marivac Canada Inc. St. Laurent, Canada). The following day the samples were embedded in TAAB Epon and polymerized at 60 C for 48 hrs. Ultrathin sections (about 75–80nm) were cut on a Reichert Ultracut-S microtome, picked up on to copper grids stained with lead citrate and examined in a TecnaiG2 Spirit BioTWIN transmission electron microscope. Images were recorded with an AMT 2k CCD camera.
Electrophysiology
Acute 250 μm sagittal and coronal slices were prepared from the cerebella of P20–P24 control Chd4loxP/loxP, Chd4 heterozygous, and Chd4 conditional knockout mice. Slices were cut in dissecting solution containing: 83 mM NaCl, 65 mM sucrose, 26 mM NaHCO3, 25 mM glucose, 6.8 mM MgCl2, 2.5 mM KCl, 1.25 NaH2PO4 and 0.5 CaCl2. Slices were incubated at 35°C for 1 hour in artificial cerebrospinal fluid (ACSF) containing: 125 mM NaCl, 26 mM NaHCO3, 1.25 mM NaH2PO4, 2.5 mM KCl, 1 mM MgCl2, 2 mM CaCl2, and 25 mM glucose, and switched to room temperature prior to recording. Slice solutions were constantly bubbled with 95% O2 and 5% CO2. 20 μM picrotoxin was added to the bath ACSF recording solution to block inhibitory currents. Electrophysiological signals were acquired with a Multiclamp 700B amplifier, digitized at 10 kHz with a Digidata 1440A D-A converter, and Bessel filtered at 2 kHz.
Whole-cell patch-clamp recordings in Purkinje cells were obtained with recording electrodes (1.5–2 MΩ) filled with intracellular solution containing: 130 mM Cs-methanesulfonate, 5 mM CsCl, 10 mM HEPES, 0.5 mM EGTA, 2 mM MgCl2, 2 mM Na-ATP, 0.5 mM Na-GTP. Purkinje neurons were voltage-clamped at −70 mV. For mEPSC recordings, 1 μM tetrodotoxin was added to the bath solution. Data analysis was performed using MiniAnalysis software (Synaptosoft) with an amplitude threshold of 10 pA. mEPSC traces were additionally high-pass filtered at 0.5 Hz and low-pass filtered at 1 kHz. For evoked EPSC and presynaptic volley recordings, the molecular layer was stimulated with a bipolar concentric electrode (FHC) using brief (0.1 ms) current pulses. Evoked EPSCs were measured using the whole-cell recording electrodes as described above. Extracellular presynaptic volley recordings using 1 MΩ electrodes filled with 3 MNaCl were made 400 μm away from the site of stimulation (Sabatini and Regehr, 1997). The fiber volley amplitude was derived from the negative-going phase of the extracellular field potential.
RNA-Seq and qRT-PCR
For RNA-Seq, total RNA was extracted from cerebella at P22 mice using Trizol (Invitrogen) according to the manufacturer’s instructions. RNA was reverse-transcribed with oligo-dT priming and the cDNA was sequenced on an Illumina HiSeq 2000. The raw sequence reads were converted to basecalls, demultiplexed, and aligned to a reference sequence with Tophat v2.0.9 and Bowtie2 v2.1.0. Gene and exon-level abundances were derived by HTSeq. Gene and exon level differential expression was estimated through pair-wise negative binomial tests with EdgeR and DEXSeq, respectively. The Benjamini-Hochberg false discovery rate (FDR) was calculated for all genes. For qRT-PCR, reverse transcription reactions were performed with Superscript III (Invitrogen) according to the manufacturer’s protocol. Real time PCR reactions were performed using Lightcycler 480 SYBR Green I Master (Roche). In the qRT-PCR and RNA-Seq analyses in Figure 3B, we used one common set for qRT-PCR and RNA-Seq analyses and two independent sets of samples in each type of analysis.
ChIP-qPCR and ChIP-Seq
ChIP-qPCR and ChIP-Seq assays were performed with mouse cerebella fixed with 1.1% formaldehyde solution, homogenized, and incubated for 15 mins at room temperature. Reactions were stopped by adding glycine solution (final 125 mM glycine). The lysates were further homogenized with lysis buffer (10 mM Tris-HCl, 0.25% Triton X-100, 10 mM EDTA and 0.5 mM EGTA). After spin down, the supernatant was removed and the nuclear pellet was resuspended with sonication buffer (10 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA and 0.5 mM EGTA) and sonicated to shear crosslinked DNA. After sonication, SDS was added to the lysate (final 1%) and incubated at room temperature for 1hr. The lysate was sonicated again to shear DNA until the size of DNA was between 100 to 500 bps. Immunoprecipitation was performed in RIPA buffer (10 mM Tris-HCl (pH 8.0), 140 mM NaCl, 0.1% SDS, 1% Triton-X, 0.1% DOC, 1 mM EDTA, 0.5 mM EGTA) using the indicated antibodies with protein G sepharose beads (GE healthcare) as described (Yamada et al., 2013). ChIP-Seq was performed using libraries prepared with the Illumina ChIP-Seq DNA Sample Prep Kit as per the manufacturer’s instructions and sequenced on the Illumina HiSeq 2000 platform (Beijing Genomics Institute). Two biological ChIP-Seq replicates and three independent ChIP-qPCR replicates were performed in all experiments.
ChIP-Seq analyses
For each experiment, the ChIP-Seq reads were normalized to 10M reads and subsequently, the number of input reads at each locus was subtracted. To calculate the average profile at promoters, we aligned the 5′-most TSS for each RefSeq expressed gene and calculated the density of reads as a function of the distance to the TSS. We required a minimum RPKM geometric mean density of 0.01 for the three WT replicates for a gene to be considered expressed, and this left us with a total of 13096 genes. A similar strategy was used for the subset of genes that were categorized as misregulated in the RNA-Seq analyses. After a log-transform, the mean and standard deviation of the differentially expressed genes in the WT cerebellum were 1.7 and 0.76, respectively. The 13096 genes that were used as control had a mean of 2.25 and a standard deviation of 0.99. This suggests that the differentially expressed genes are comparable to the set of all expressed genes. For the subset of misregulated genes, the enrichment of histone modifications was quantified by summing the total number of normalized and input subtracted reads in the [0, 1000] bps region relative to the TSS.
Statistics
Statistical analyses were done using GraphPad Prism 6.0 software. Bar graphs are presented as the mean ± SEM. For experiments in which only two groups were analyzed, the t-test was used. Pairwise comparisons within multiple groups were done by analysis of variance (ANOVA) followed by the Fischer’s PLSD post-hoc test.
Supplementary Material
Acknowledgments
We thank members of the Bonni laboratory for helpful discussions and critical reading of the manuscript. Supported by NIH grant NS041021 (A.B.), the Mathers Foundation (A.B.), the Japan Society for the Promotion of Science (T.Yamada), NIH training grant AG000222 (Y.Y.), NIH grant NS032405 (W.G.R.), and NIH training grant NS007484 (D.F.). We thank the Genome Technology Access Center at Washington University, which is supported by NCI P30 CA91842 to the Siteman Cancer Center and by ICTS/CTSA UL1TR000448.
Footnotes
Accession Numbers
The GEO accession ID for the ChIP-seq data is GSE57758 (www.ncbi.nlm.nih.gov/geo/).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Development of the cerebellar system : in relation to its evolution, structure, and functions. Boca Raton: CRC Press; 1997. [Google Scholar]
- Bilimoria PM, Bonni A. Cultures of cerebellar granule neurons. CSH Protoc. 2008;2008 doi: 10.1101/pdb.prot5107. pdb prot5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell RB. RNA protein interaction in neurons. Annual review of neuroscience. 2013;36:243–270. doi: 10.1146/annurev-neuro-062912-114322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre-Ubieta L, Bonni A. Transcriptional regulation of neuronal polarity and morphogenesis in the mammalian brain. Neuron. 2011;72:22–40. doi: 10.1016/j.neuron.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–5438. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- Ebert DH, Greenberg ME. Activity-dependent neuronal signalling and autism spectrum disorder. Nature. 2013;493:327–337. doi: 10.1038/nature11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Jaye DL, Geigerman C, Akyildiz A, Mooney MR, Boss JM, Wade PA. MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell. 2004;119:75–86. doi: 10.1016/j.cell.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Funfschilling U, Reichardt LF. Cre-mediated recombination in rhombic lip derivatives. Genesis. 2002;33:160–169. doi: 10.1002/gene.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature genetics. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Hong W, Nakazawa M, Chen YY, Kori R, Vakoc CR, Rakowski C, Blobel GA. FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. EMBO J. 2005;24:2367–2378. doi: 10.1038/sj.emboj.7600703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov AV, Peng H, Yurchenko V, Yap KL, Negorev DG, Schultz DC, Psulkowski E, Fredericks WJ, White DE, Maul GG, et al. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell. 2007;28:823–837. doi: 10.1016/j.molcel.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YH, Yuen RK, Jin X, Wang M, Chen N, Wu X, Ju J, Mei J, Shi Y, He M, et al. Detection of Clinically Relevant Genetic Variants in Autism Spectrum Disorder by Whole-Genome Sequencing. Am J Hum Genet. 2013 doi: 10.1016/j.ajhg.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol. 2006;8:285–292. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- Kelleher RJ, 3rd, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135:401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Konishi Y, Stegmuller J, Matsuda T, Bonni S, Bonni A. Cdh1-APC controls axonal growth and patterning in the mammalian brain. Science. 2004;303:1026–1030. doi: 10.1126/science.1093712. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Li Q, Shi L, Gui B, Yu W, Wang J, Zhang D, Han X, Yao Z, Shang Y. Binding of the JmjC demethylase JARID1B to LSD1/NuRD suppresses angiogenesis and metastasis in breast cancer cells by repressing chemokine CCL14. Cancer Res. 2011;71:6899–6908. doi: 10.1158/0008-5472.CAN-11-1523. [DOI] [PubMed] [Google Scholar]
- Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, Chen G, Gage FH, Muotri AR. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, Sabo A, Lin CF, Stevens C, Wang LS, Makarov V, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, Carvill G, Kumar A, Lee C, Ankenman K, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012a;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012b;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palay SL, Chan-Palay V. Cerebellar cortex: cytology and organization. New York: Springer; 1974. [Google Scholar]
- Rais Y, Zviran A, Geula S, Gafni O, Chomsky E, Viukov S, Mansour AA, Caspi I, Krupalnik V, Zerbib M, et al. Deterministic direct reprogramming of somatic cells to pluripotency. Nature. 2013;502:65–70. doi: 10.1038/nature12587. [DOI] [PubMed] [Google Scholar]
- Reynolds N, Latos P, Hynes-Allen A, Loos R, Leaford D, O’Shaughnessy A, Mosaku O, Signolet J, Brennecke P, Kalkan T, et al. NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment. Cell Stem Cell. 2012;10:583–594. doi: 10.1016/j.stem.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet. 2013;14:347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini BL, Regehr WG. Control of neurotransmitter release by presynaptic waveform at the granule cell to Purkinje cell synapse. J Neurosci. 1997;17:3425–3435. doi: 10.1523/JNEUROSCI.17-10-03425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Friedman JR, Rauscher FJ., 3rd Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev. 2001;15:428–443. doi: 10.1101/gad.869501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- Shalizi A, Gaudilliere B, Yuan Z, Stegmuller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW, Bonni A. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- Shcheglovitov A, Shcheglovitova O, Yazawa M, Portmann T, Shu R, Sebastiano V, Krawisz A, Froehlich W, Bernstein JA, Hallmayer JF, et al. SHANK3 and IGF1 restore synaptic deficits in neurons from 22q13 deletion syndrome patients. Nature. 2013 doi: 10.1038/nature12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkowski ME, Rosenfeld JA, Blumenthal I, Pillalamarri V, Chiang C, Heilbut A, Ernst C, Hanscom C, Rossin E, Lindgren AM, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149:525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uittenbogaard M, Peavy DR, Chiaramello A. Expression of the bHLH gene NSCL-1 suggests a role in regulating cerebellar granule cell growth and differentiation. J Neurosci Res. 1999;57:770–781. [PubMed] [Google Scholar]
- Vissers LE, van Ravenswaaij CM, Admiraal R, Hurst JA, de Vries BB, Janssen IM, van der Vliet WA, Huys EH, de Jong PJ, Hamel BC, et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nature genetics. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, Liang J, Sun L, Yang X, Shi L, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009a;138:660–672. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009b;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nature genetics. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AE, Greenberg ME. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb Perspect Biol. 2011:3. doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- Yamada T, Yang Y, Huang J, Coppola G, Geschwind DH, Bonni A. Sumoylated MEF2A coordinately eliminates orphan presynaptic sites and promotes maturation of presynaptic boutons. J Neurosci. 2013;33:4726–4740. doi: 10.1523/JNEUROSCI.4191-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Kim AH, Yamada T, Wu B, Bilimoria PM, Ikeuchi Y, de la Iglesia N, Shen J, Bonni A. A Cdc20-APC ubiquitin signaling pathway regulates presynaptic differentiation. Science. 2009;326:575–578. doi: 10.1126/science.1177087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Hazan I, Zhang J, Ng SY, Naito T, Snippert HJ, Heller EJ, Qi X, Lawton LN, Williams CJ, et al. The role of the chromatin remodeler Mi-2beta in hematopoietic stem cell self-renewal and multilineage differentiation. Genes Dev. 2008;22:1174–1189. doi: 10.1101/gad.1642808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Jackson AF, Naito T, Dose M, Seavitt J, Liu F, Heller EJ, Kashiwagi M, Yoshida T, Gounari F, et al. Harnessing of the nucleosome-remodeling-deacetylase complex controls lymphocyte development and prevents leukemogenesis. Nat Immunol. 2012;13:86–94. doi: 10.1038/ni.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- Zoghbi HY. Postnatal neurodevelopmental disorders: meeting at the synapse? Science. 2003;302:826–830. doi: 10.1126/science.1089071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.