Abstract
Cellular memory is crucial to many natural biological processes and for sophisticated synthetic-biology applications. Existing cellular memories rely on epigenetic switches or recombinases, which are limited in scalability and recording capacity. Here, we use the DNA of living cell populations as genomic ‘tape recorders’ for the analog and distributed recording of long-term event histories. We describe a platform for generating single-stranded DNA (ssDNA) in vivo in response to arbitrary transcriptional signals. When co-expressed with a recombinase, these intracellularly expressed ssDNAs target specific genomic DNA addresses, resulting in precise mutations that accumulate in cell populations as a function of the magnitude and duration of the inputs. This platform could enable long-term cellular recorders for environmental and biomedical applications, biological state machines, and enhanced genome engineering strategies.
Due to its high storage capacity, durability, ease of duplication, and high-fidelity maintenance of information, DNA has garnered much interest as an artificial storage medium (1, 2). However, existing technologies for in vivo autonomous recording of information in cellular memory are limited in their storage capacity and scalability (3). Epigenetic memory devices such as bi-stable toggle switches (4–7) and positive-feedback loops (8) require orthogonal transcription factors and can lose their digital state due to environmental fluctuations or cell death. Recombinase-based memory devices enable the writing and storage of digital information in the DNA of living cells (9–12), where binary bits of information are stored in the orientation of large stretches of DNA. However, these devices do not efficiently exploit the full capacity of DNA for information storage – recording a single bit of information with these devices often requires at least a few hundred base-pairs of DNA, overexpression of a recombinase protein to invert the target DNA, and engineering recombinase recognition sites into target loci in advance. The scalability of this type of memory is further limited by the number of orthogonal recombinases that can be used in a single cell. Finally, epigenetic and recombinase-based memory devices described to-date store digital information and their recording capacity is exhausted within a few hours of induction. Thus, these devices have not been adapted to record analog information, such as magnitude and time course of inputs over extended periods of time (i.e., multiple days or more).
Here, we introduce SCRIBE (Synthetic Cellular Recorders Integrating Biological Events), a compact, modular strategy for producing single-stranded DNA (ssDNA) inside of living cells in response to a range of regulatory signals, such as small chemical inducers and light. These ssDNAs uniquely address specific target loci based on sequence homology and introduce precise mutations into genomic DNA. The memory device can be easily reprogrammed to target different genomic locations by changing the ssDNA template. SCRIBE memory does not just record the absence or presence of arbitrary inputs (digital signals represented as binary ‘0s’ or ‘1s’). Instead, by encoding information into the collective genomic DNA of cell populations, SCRIBE can track the magnitude and long-term temporal behavior of inputs, which are analog signals since they can vary over a wide range of continuous values. This analog memory architecture leverages the large number of cells in bacterial cultures for distributed information storage and archives event histories in the fraction of cells in a population that carry specific mutations.
Single-Stranded DNA Expression in Living Cells
Previously, it was shown that synthetic oligonucleotides delivered by electroporation into cells that overexpress Beta recombinase (from bacteriophage λ) in Escherichia coli (E. coli) are specifically and efficiently recombined into homologous genomic sites (13–16). Thus, oligonucleotide-mediated recombination offers a powerful way to introduce targeted mutations in a bacterial genome (17, 18). However, this technique requires the exogenous delivery of ssDNAs and cannot be used to couple arbitrary signals into genetic memory. To overcome these limitations, we developed a genome-editing platform based on expressing ssDNAs inside of living cells by taking advantage of a widespread class of bacterial reverse transcriptases called retrons (19, 20).
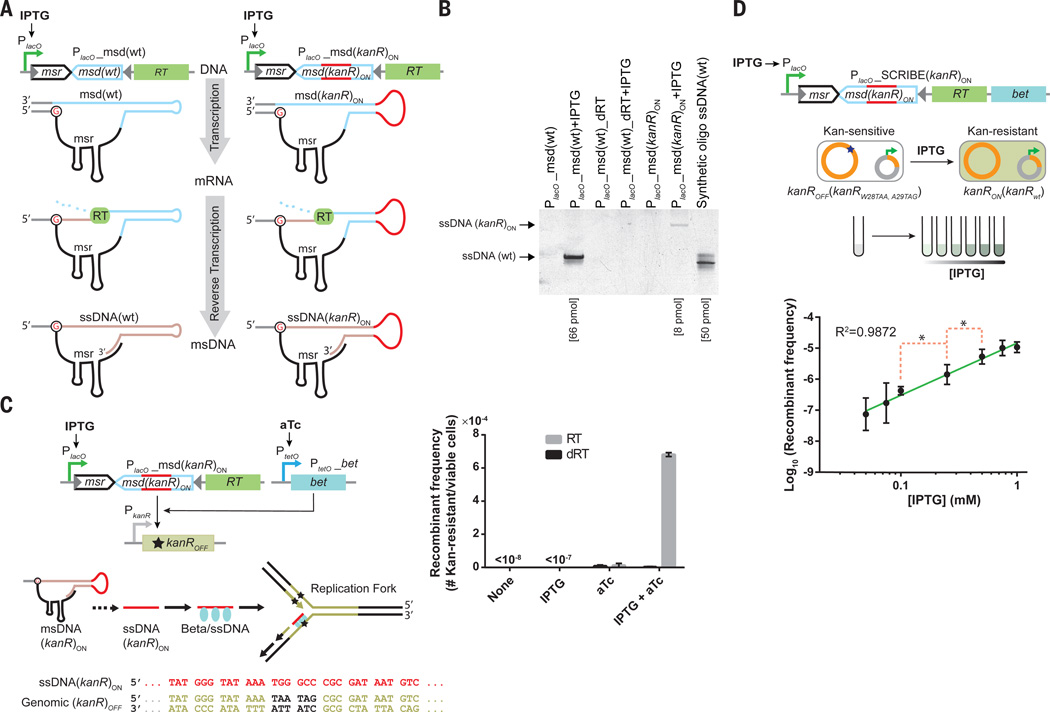
The wild-type retron cassette encodes three components in a single transcript – a reverse transcriptase protein (RT), and two RNA moieties, msr and msd, which act as the primer and the template for the reverse transcriptase, respectively (Fig. 1A, left). The msr-msd sequence in the retron cassette is flanked by two inverted repeats. Once transcribed, the msr-msd RNA folds into a secondary structure guided by the base-pairing of the inverted repeats and the msr-msd sequence. The RT recognizes this secondary structure and uses a conserved guanosine residue in the msr as a priming site to reverse transcribe the msd sequence and produce a hybrid RNA-ssDNA molecule called msDNA (20, 21). To couple the expression of ssDNA to an external input, the wild-type Ec86 retron cassette from E. coli BL21 (21) was placed under the control of an Isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible promoter (PlacO) in E. coli DH5αPRO cells (22), which expresses high levels of the LacI and TetR repressors (Fig. 1A). The wild-type retron ssDNA (ssDNA(wt)) was readily detected in IPTG-induced cells while no ssDNA was detected in non-induced cells (Fig. 1B). The identity of the detected ssDNA band was further confirmed by DNA sequencing (Fig. S1). To verify that ssDNA expression depended on RT activity, point mutations (D197A and D198A) were introduced to the active site of the RT to make a catalytically dead RT (dRT) (23). This modification completely abolished ssDNA production (Fig. 1B).
Fig. 1. SCRIBE system for recording inputs in the distributed genomic DNA of bacterial populations.
(A) Synthetic ssDNA (red line) generation inside of living cells by retrons. (B) Visualization of retron-mediated ssDNAs produced in living bacteria. The amount of ssDNA in each sample (shown in brackets) was calculated by densitometry. (C) A kanR reversion assay was used to measure the efficiency of DNA writing within living cells, where the msd(kanR)ON cassette and the bet gene were inducible by IPTG and aTc, respectively. (D) Demonstration of analog memory achieved via SCRIBE to record the magnitude of an input into genomic DNA. The green line is a linear regression fit. The red dashed brackets marked with asterisks connect the closest data points that are statistically significant with respect to each other (p-value < 0.05 based on one-tailed Welch’s t-test). Error bars indicate the standard error of the mean for three independent biological replicates.
To engineer the msd template to express synthetic ssDNAs of interest, we initially tried to replace the whole msd sequence with a desired template. However, no ssDNA was detected, suggesting that some features of msd are required for ssDNA expression, as was previously noted for another retron (24). A variant in which the flanking regions of the msd stem remained intact (Fig. 1A, right) produced detectable amounts of ssDNA when induced by IPTG (Fig. 1B, PlacO_msd(kanR)ON + IPTG). The correct identity of the detected ssDNA band was further confirmed by DNA sequencing (Fig. S1). Thus, the lower part of the msd stem is essential for reverse transcription while the upper part of the stem and the loop are dispensable and can be replaced with desired templates to produce ssDNAs of interest in vivo.
Regulated Genome Editing with In vivo ssDNAs
To demonstrate that intracellularly expressed ssDNAs can be recombined into target genomic loci by concomitant expression of Beta, we developed a selectable marker reversion assay (Fig. 1C). The kanR gene, which encodes neomycin phosphotransferase II and confers resistance to kanamycin (Kan), was integrated into the galK locus. Two stop codons were then introduced into the genomic kanR to make a Kan-sensitive kanROFF reporter strain (DH5αPRO galK::kanRW28TAA, A29TAG). These premature stop codons could be reverted back to the wild-type sequence via recombination with engineered ssDNA(kanR)ON, thus conferring kanamycin resistance (Fig. 1C). Specifically, ssDNA(kanR)ON contains 74 base-pairs (bps) of homology to the regions of the kanROFF locus flanking the premature stop codons, and replaces the stop codons with the wild-type kanR gene sequence (Fig. 1C).
We cloned the Beta gene (bet) into a plasmid under the control of the anhydrotetracycline (aTc)-inducible PtetO promoter and introduced it along with the IPTG-inducible msd(kanR)ON construct into the kanROFF strain (Fig. 1C). Induction of cultures harboring these two plasmids with either IPTG (1 mM) or aTc (100 ng/ml) resulted in a slight increase in the number of the Kan-resistant cells (Fig. 1C). However, co-expression of both ssDNA(kanR)ON and Beta with IPTG and aTc resulted in a >104-fold increase in the recombinant frequency relative to the non-induced cells. This corresponded to a >103-fold increase relative to cells induced with IPTG only and a 60-fold increase relative to cells induced with aTc only. This increase in the recombinant frequency was dependent on the RT activity, as it was largely abolished with dRT. The genotypes of randomly selected Kan-resistant colonies were further confirmed by DNA sequencing to contain precise reversions of the two codons to the wild-type sequence (Fig. S1). No Kan-resistant colonies were detected when a non-specific ssDNA (ssDNA(wt)) was co-expressed with Beta in the kanROFF reporter cells, confirming that Kan-resistant cells were not produced due to spontaneous mutations. In additional experiments, high-throughput sequencing (Illumina HiSeq) on the bacterial populations was used to analyze the genomically encoded memory ( Supplementary Materials, Fig. S2). Comparable recombinant frequencies were obtained from both the plating assay and sequencing, confirming that genomically encoded memory can be read without the need for functional assays and reporters.
Recording Input Magnitudes into Genomic Memory
We reasoned that the rate of recombination between engineered ssDNAs and genomic DNA could be effectively modulated by changing expression levels of the engineered retron cassette and Beta. This feature would enable the recording of analog information, such as the magnitude of an input signal, in the proportion of cells in a population with a specific mutation in genomic DNA. To demonstrate this, both the ssDNA(kanR)ON expression cassette and bet were placed into a single synthetic operon (hereafter referred to as the SCRIBE(kanR)ON cassette) under the control of PlacO (Fig. 1D). The kanROFF reporter cells harboring this synthetic operon were induced with different concentrations of IPTG. The fraction of Kan-resistant recombinants increased linearly with the input inducer concentration on a log-log plot over a range of ~10−7 to ~10−5 (Fig. 1D). Statistical tests showed that at least four different concentrations of the inducer (including 0 mM IPTG) could be resolved in this experiment. Thus, the efficiency of genome writing in a population can be quantitatively tuned with external inputs.
Writing and Rewriting Genomic Memory
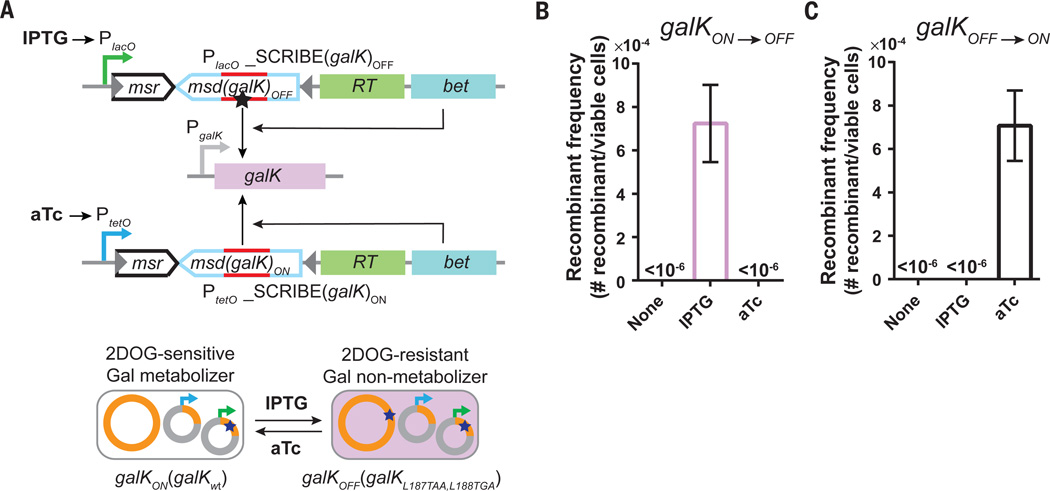
We next created a complementary set of SCRIBE cassettes to write and erase (rewrite) information in the genomic galK locus using two different chemical inducers. Cells expressing galK can metabolize and grow on galactose as the sole carbon source. However, these galK-positive (galKON) cells cannot metabolize 2-deoxy-galactose (2DOG) and cannot grow on plates containing glycerol (carbon source) + 2DOG. On the other hand, galK-negative (galKOFF) cells cannot grow on galactose as the sole carbon source but can grow on glycerol + 2DOG plates (25). We transformed DH5αPRO galKON cells with plasmids expressing IPTG-inducible SCRIBE(galK)OFF and aTc-inducible SCRIBE(galK)ON cassettes (Fig. 2A). Induction of SCRIBE(galK)OFF by IPTG resulted in the writing of two stop codons into galKON, leading to galKOFF cells that could grow on glycerol + 2DOG plates (Fig. 2B). Induction of SCRIBE(galK)ON in these galKOFF cells with aTc reversed the IPTG-induced modification, leading to galKON cells that could grow on galactose plates (Fig. 2C). These results show that writing on genomic DNA with SCRIBE is reversible and that distinct information can be written and rewritten into the same locus.
Fig. 2. SCRIBE can write multiple different DNA mutations into a common target loci (galK).
(A) Schematic of the procedure (see text for details). (B) galKON cells harboring the circuits shown in A) were induced with either IPTG (1 mM) or aTc (100 ng/ml) for 24 hours and the galKOFF frequencies in the population were determined by plating the cells on appropriate selective conditions. (C) galKOFF cells (obtained from the experiment described in B)) were induced with IPTG (1 mM) or aTc (100 ng/ml) for 24 hours and the galKON frequencies in the population were determined by plating the cells on appropriate selective conditions. Error bars indicate the standard error of the mean for three independent biological replicates.
Writing Multiple Mutations into Independent Loci
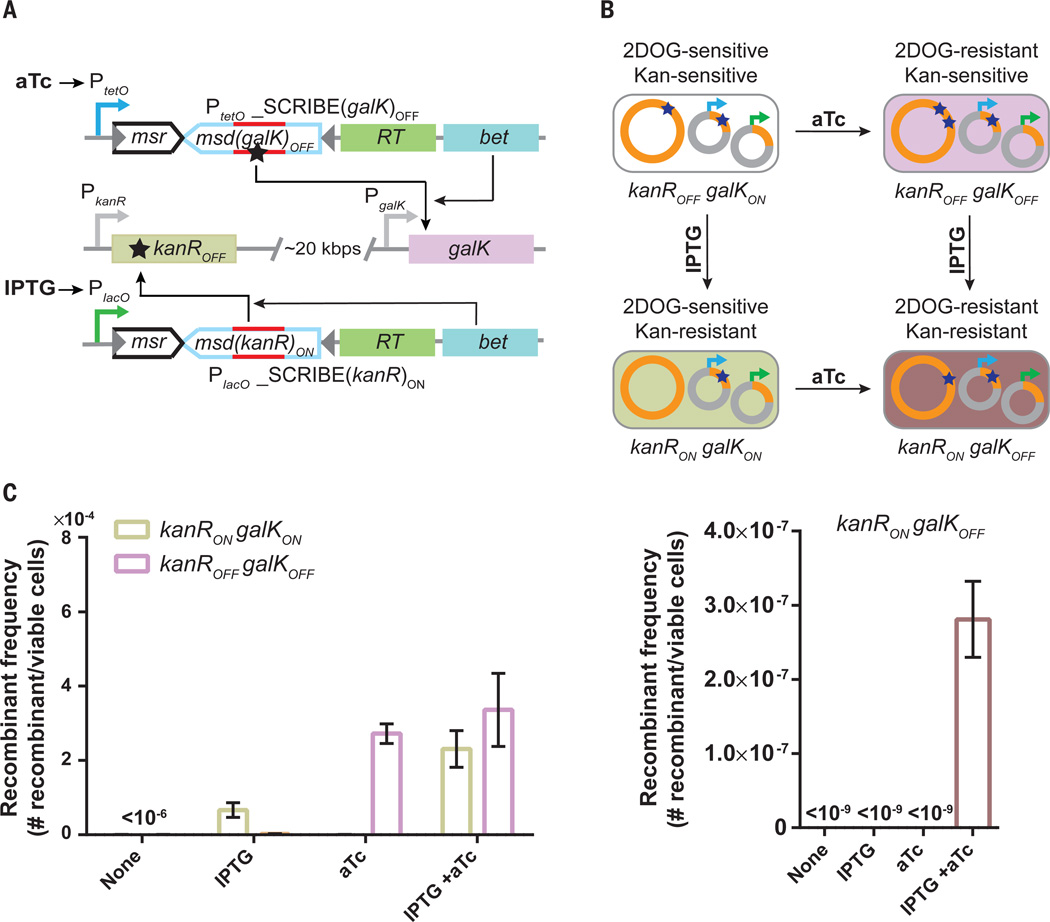
Scaling the capacity of previous memory devices is challenging since each additional bit of information requires new orthogonal proteins (e.g., recombinases or transcription factors). In contrast, orthogonal SCRIBE memory devices are potentially easier to scale because they can be built by simply changing the ssDNA template (msd). To demonstrate this, we used SCRIBE to record multiple independent inputs into different genomic loci of bacterial population. We integrated the kanROFF reporter gene into the bioA locus of DH5αPRO to create a kanROFFgalKON strain. These cells were then transformed with plasmids expressing IPTG-inducible SCRIBE(kanR)ON and aTc-inducible SCRIBE(galK)OFF cassettes (Fig. 3A). Induction of these cells with IPTG or aTc resulted in the production of cells with phenotypes corresponding to kanRON galKON or kanROFFgalKOFF genotypes, respectively (Fig. 3B and C). Comparable numbers of kanRON galKON and kanROFFgalKOFF cells (~2*10−4 and ~3*10−4 recombinant/viable cells, respectively) were produced when the cultures were induced with both aTc and IPTG (Fig. 3C). Furthermore, very few individual colonies (~3*10−7 recombinant/viable cells) containing both writing events (kanRON galKOFF) were obtained in the cultures that were induced with both aTc and IPTG (Figure 3C). These data suggest that while multiplexed writing at single-cell level is rare with SCRIBE’s current level of recombination efficiency, multiple independent inputs can be successfully recorded into the distributed genomic DNA of bacterial subpopulations.
Fig. 3. Writing multiple mutations into independent target loci within population.
(A) Constructs used to target genomic kanROFF and galKON loci with IPTG-inducible and aTc-inducible SCRIBE cassettes, respectively. (B) Induction of kanROFF galKON cells with IPTG or aTc generates cells with the kanRON galKON or kanROFF galKOFF genotypes, respectively. Induction of kanROFF galKON cells with IPTG and aTc generates cells with the kanRON galKOFF genotype. (C) kanROFF galKON reporter cells containing the circuits in A) were induced with different combinations of IPTG (1 mM) and aTc (100 ng/ml) for 24 h at 30°C and the fraction of cells with the various genotypes were determined by plating the cells on appropriate selective media. Error bars indicate the standard error of the mean for three independent biological replicates.
Optogenetic Genome Editing for Light-to-DNA Memory
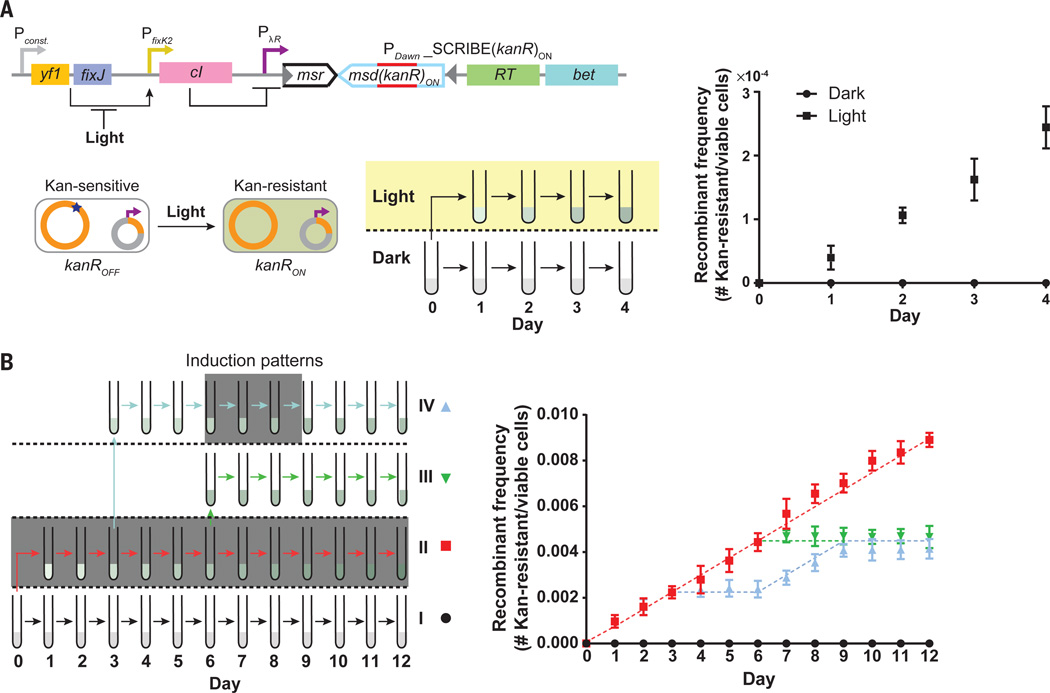
In SCRIBE, the expression of each individual ssDNA can be triggered by any endogenous or exogenous signal that can be coupled into transcriptional regulation, thus recording these inputs into long-lasting DNA storage. In addition to small-molecule chemicals we showed that light can be used to trigger specific genome editing for genomically encoded memory. We placed the SCRIBE(kanR)ON cassette under the control of a previously described light-inducible promoter (PDawn, (26)) within kanROFF cells (Fig. 4A). These cultures were then grown for 4 days in the presence of light or in the dark (Fig. 4A). As Beta-mediated recombination is reportedly replication-dependent (27–29), dilutions of these cultures were made into fresh media at the end of each day to maintain active replication in the cultures. At the end of each day, samples were taken to determine the number of Kan-resistant and viable cells (Fig. 4A). Cultures grown in the dark yielded undetectable levels of Kan-resistant cells (Fig. 4A). In contrast, the number of Kan-resistant colonies increased steadily over time in the cultures that were grown in the presence of light, indicating the successful recording of light input into long-lasting DNA memory. The analog memory faithfully stored the total time of light exposure, rather than just the digital presence or absence of light.
Fig. 4. Optogenetic genome editing and analog memory for long-term recording of input signal exposure times in the genomic DNA of living cell populations.
(A) We coupled expression of SCRIBE(kanR)ON to an optogenetic system (PDawn). The yf1/fixJ synthetic operon was expressed from a constitutive promoter – its products cooperatively activate the PfixK2 promoter, which drives lambda repressor (cI) expression, which subsequently represses the SCRIBE(kanR)ON cassette. Light inhibits the interaction between yf1 and fixJ, leading to the generation of ssDNA(kanR)ON and Beta expression, and thus the conversion of kanROFF to kanRON. Cells harboring this circuit were grown overnight at 37°C in the dark, diluted 1:1000, and then incubated for 24 h at 30°C in the dark (no shading) or in the presence of light (yellow shading). Subsequently, cells were diluted by 1:1000 and grown for another 24 h at 30°C in the dark or in the presence of light. The dilution/regrowth cycle was performed for four consecutive days. The kanR allele frequencies in the populations were determined by sampling the cultures after each 24-hour period. (B) SCRIBE analog memory records the total time exposure to a given input, regardless of the underlying induction pattern. Cells harboring the circuit shown in Fig. 1C were grown in four different patterns (I-IV) over a twelve-day period, where induction by IPTG (1 mM) and aTc (100 ng/mL) is represented by dark gray shading. At the end of each 24 h incubation period, cells were diluted by 1:1000 into fresh media. The number of Kan-resistant cells in the cultures was determined at the end of each day. Dashed lines represent the recombinant allele frequencies predicted by the model (see Supplementary Materials). Error bars indicate the standard error of the mean for three independent biological replicates.
Recording the Time Exposure of Inputs
The linear increase in the number of Kan-resistant colonies over time due to exposure to light indicates that the duration of inputs can be recorded into population-wide DNA memory using SCRIBE. To further explore population-wide genomically encoded memory whose state is a function of input exposure time, we used the kanROFF strain harboring the constructs shown in Fig. 1C, where expression of ssDNA(kanR)ON and Beta are controlled by IPTG and aTc, respectively. These cells were subjected to four different patterns of the inputs for 12 successive days (patterns I-IV, Fig. 4B). Kan-resistant cells did not accumulate in the negative control (pattern I), which was never exposed to the inducers. The fraction of Kan-resistant cells in the three other patterns (II, III, and IV) increased linearly over their respective induction periods and remained relatively constant when the inputs were removed. These data indicate that the genomically encoded memory was stable in the absence of the inputs over the course of the experiment. The recombinant frequencies in patterns III and IV, which were induced for the same total amount of time but with different temporal patterns, reached comparable levels at the end of the experiment. These data demonstrate that the genomic memory integrates over the total induction time and is independent of the input pattern, and therefore can be used to stably record event histories over many days.
The linear increase in the fraction of recombinants in the induced cell populations over time was consistent with a deterministic model (dashed lines in Fig. 4B, see Supplementary Materials). Specifically, when triggered by inputs, SCRIBE can significantly increase the rate of recombination events at a specific target site above the wild-type rate (which is reportedly <10−10 events/generation in recA− background (30)). When recombination rates are ~10−4 events/generation, which is consistent with the recombination rate estimated for SCRIBE from data in Fig. 4B, a simple deterministic model as well as a detailed stochastic simulation both predict a linear increase in the total number of recombinant alleles in a population over time, as long as the frequency of recombinants in the population is less than a few percent and cells in the population are equally fit over the time-scale of interest (Supplementary Materials, Fig. S3 and S4). These models enable one to determine the ideal range of recombination efficiencies for a given application, which depends on parameters such as the frequency of dilution, the sensitivity of the method used for reading the memory, the desired input duration to be recorded, and so forth. For example, recombination rates that are too low would be challenging to quantify and could result in loss of memory if the cultures were diluted. Moreover, higher recombination rates lead to more rapid saturation of memory capacity in which the system is unable to provide a straightforward linear relationship between the time exposure of an input and the state of the memory (Fig. S3). Thus, intermediate levels of recombination efficiency are desirable for population-level analog memory units that can record the time-span of exposure to inputs (see Supplementary Materials).
Decoupling Memory Operations
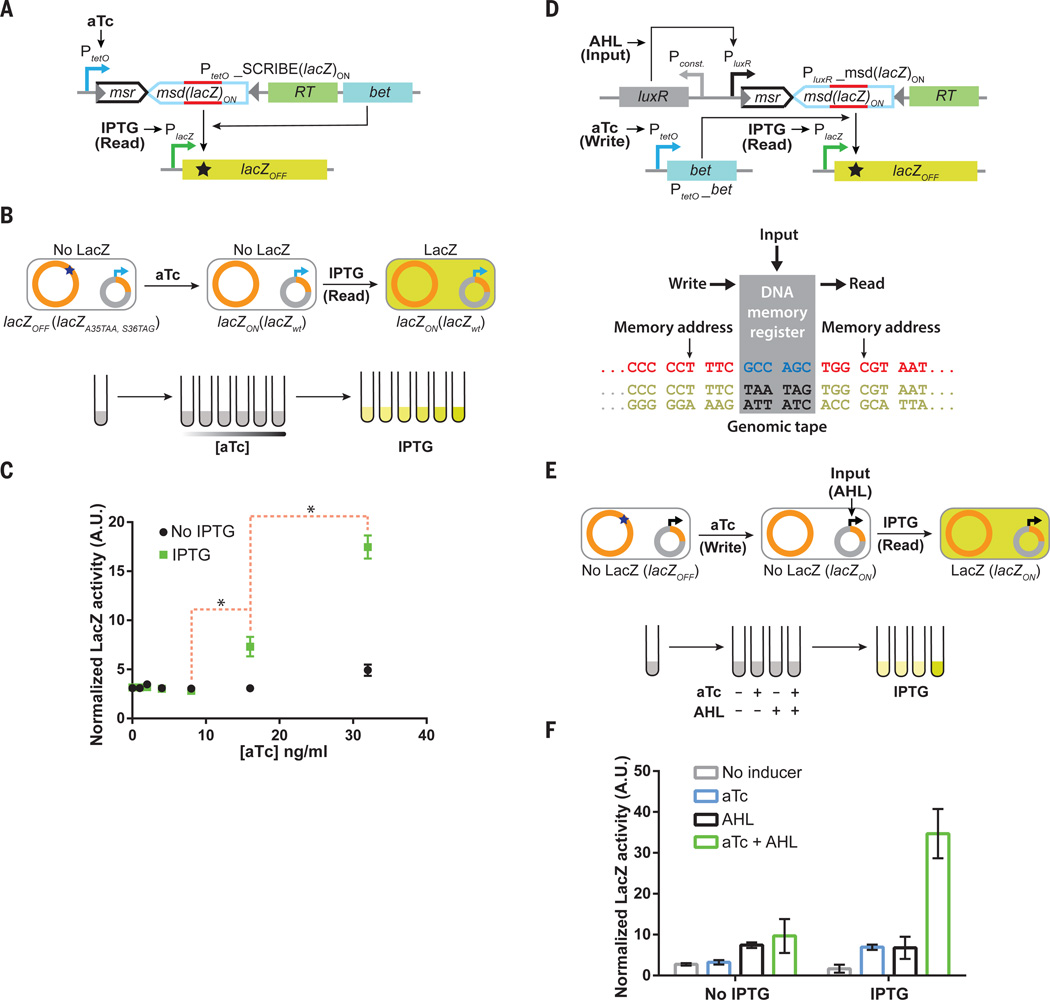
SCRIBE memory can be used to create more complex synthetic memory circuits. To demonstrate this, we first built a synthetic gene circuit that can record different input magnitudes into DNA memory. The memory state can then be read out later (after the initial input is removed) upon addition of a secondary signal. Specifically, we built an IPTG-inducible lacZOFF (lacZA35TAA, S36TAG) reporter construct in DH5αPRO cells (Fig. 5A). Expression of this reporter is normally repressed except when IPTG (“Read” signal, Fig. 5A) is added as an inducer, thus enabling a convenient and switchable population-level readout of the memory based on total LacZ activity (Fig. 5B). The lacZOFF reporter cells were transformed with a plasmid encoding an aTc-inducible SCRIBE(lacZ)ON cassette (Fig. 5A). Overnight cultures were diluted and induced with various amounts of aTc to write the genomic memory (Fig. 5B). These cells were grown up to saturation and then diluted into fresh media in the presence or absence of IPTG to read the genomic memory (Fig. 5B). In the absence of IPTG, the total LacZ activity remained low, regardless of the aTc concentration. In the presence of IPTG, cultures that had been exposed to higher aTc concentrations had greater total LacZ activity. These results show that population-level reading of genomically encoded memory can be decoupled from writing and controlled externally. Furthermore, this circuit enables the magnitude of the inducer (aTc) to be stably recorded in the distributed genomic memory of a cellular population. Independent control over the “Read” memory operation as shown in this experiment could help to minimize fitness costs associated with the expression of reporter genes until needed.
Fig. 5. SCRIBE memory operations can be decoupled into independent Input, Write, and Read operations, thus facilitating greater control over addressable memory registers in genomic tape recorders and the creation of sample-and-hold circuits.
(A) We built a circuit where information about the first inducer (aTc) is recorded in the population, which can then be read later upon addition of a second inducer (IPTG) that triggers a “Read” operation. We created an IPTG-inducible lacZOFF locus in the DH5αPRO background, which contains the full-length lacZ gene with two premature stop codons inside the open-reading frame. Expression of ssDNA(lacZ)ON from the aTc-inducible SCRIBE(lacZ)ON cassette results in the reversion of the stop codons inside lacZOFF to yield the lacZON genotype. (B) Cells harboring the circuit shown in A) were grown in the presence of different levels of aTc for 24 h at 30°C to enable recording into genomic DNA. Subsequently, cell populations were diluted into fresh media without or with IPTG (1 mM) and incubated at 37°C for 8 hours. (C) Total LacZ activity in these cultures was measured using a fluorogenic lacZ substrate (FDG) assay. The red dashed brackets marked with asterisks connect the closest data points of IPTG-induced samples that are statistically significant (p-value < 0.05 based on one-tailed Welch’s t-test). (D) We extended the circuit in A) to create a sample-and-hold circuit where “Input”, “Write”, and “Read” operations are independently controlled. This feature enables the creation of addressable Read/Write memory registers in the genomic DNA tape. Induction of cells with the “Input” signal (AHL) produces ssDNA(lacZ)ON, which targets the genomic lacZOFF locus for reversion to the wild-type sequence. In the presence of the “Write” signal (aTc), which expresses Beta, ssDNA(lacZ)ON is recombined into the lacZOFF locus and produces the lacZON genotype. Thus, the “Write” signal enables the “Input” signal to be sampled and held in memory. The total LacZ activity in the cell populations is retrieved by adding the “Read” signal (IPTG). (E) Cells harboring the circuit shown in D) were induced with different combinations of aTc (100 ng/ml) and AHL (50 ng/ml) for 24 h, after which the cultures were diluted in fresh media with or without IPTG (1 mM). These cultures were then incubated at 37°C for 8 hours and assayed for total LacZ activity with the FDG assay. (F) Cell populations that received both the “Input” and “Write” signals, followed by the “Read” signal exhibited enhanced levels of total LacZ activity. Error bars indicate the standard error of the mean for three independent biological replicates.
We have shown that both ssDNA expression and Beta are required for writing into genomic memory (Fig. 1C), multiple ssDNAs can be used to independently address different memory units (Fig. 3), and genomic memory is stably recorded into DNA and can be used to modify functional genes whose expression can be controlled by external inducers (Fig. 1–4). Thus, SCRIBE memory units can be conceptually decomposed into separate “Input”, “Write”, and “Read” operations to facilitate greater control and the integration of logic with memory. The separation of these signals could enable master control over the writing of multiple independent inputs into genomic memory. To achieve this, we placed the msd(lacZ)ON cassette under the control of an AHL-inducible promoter (PluxR) (31) and co-transformed this plasmid with an aTc-inducible Beta-expressing plasmid into the lacZOFF reporter strain (Fig. 5D). Using this design, information on the “Input” (ssDNA expression via addition of AHL) can be written into DNA memory only in the presence of the “Write” signal (Beta expression via addition of aTc). The information recorded in the memory register (i.e., the state of lacZ across the population) can be retrieved by adding the “Read” signal (IPTG).
To demonstrate this, overnight lacZOFF cultures harboring the circuit shown in Fig. 5D were diluted and then grown to saturation in the presence of all four possible combinations of AHL and aTc (Fig. 5E). The saturated cultures were then diluted into fresh media in the absence or presence of IPTG. As shown in Fig. 5F, only cultures that had been exposed to both the “Input” and “Write” signals simultaneously showed significant LacZ activity, and only when they were induced with the “Read” signal. These results indicate that short stretches of DNA of living organisms can be used as addressable read/write memory registers to record transcriptional inputs. Furthermore, SCRIBE memory can be combined with logic, such as the AND function between the “Input” and “Write” signals shown here. The logic in Fig. 5D enables this circuit to act as a “sample-and-hold” system in which information about an input can be recorded in the presence of another signal and read out at will. Additional “Inputs” in the form of orthogonal ssDNAs under the control of other inducible inputs (e.g., Fig. 3), could be written into genomic memory only when the “Write” signal (Beta expression) is present. Thus, SCRIBE memory units can be readily reprogrammed, integrated with logic circuits, and decomposed into independent “Input”, “Write”, and “Read” operations. We anticipate that more complex logic circuits could be combined with SCRIBE-based memory to create analog-memory-and-computation systems capable of storing the results of multi-input calculations (32, 33).
Discussion
We described a scalable platform that uses genomic DNA for analog, rewritable, and flexible memory distributed across living cell populations. One current limitation is the number of orthogonal inducible promoters that can be used as inputs, but this could be addressed by coupling ssDNA expression to endogenous promoters to record native cellular events and the development of additional inducible transcriptional regulatory devices (34). Although we primarily targeted mutations into functional genes to facilitate convenient functional and reporter assays in this paper, natural or synthetic non-coding DNA segments could also be used to record memory within genomic DNA. The recorded memory could then be read by high-throughput sequencing (Fig. S2). A potential benefit of using synthetic DNA segments as memory registers is the ability to introduce mutations for memory storage that are neutral in terms of fitness costs.
SCRIBE enables conditional increases in the recombination rate at specific loci beyond background levels. The maximum observed recombination rate of the current SCRIBE platform (~10−4 recombination events/generation) is suitable for long-term recording of analog memory distributed across the collective genomes of cellular populations (Fig. S3). However, it is not high enough to allow recording of digital information and efficient genome editing at the single-cell level. In principle, population-level analog memory could be achieved by other types of DNA memory switches, such as site-specific recombinases, if they were tuned to achieve intermediate recombination efficiencies. Further investigation is required to determine the exact mechanisms involved in processing retron-based ssDNAs for recombination into genomic DNA and the effects of different growth conditions on SCRIBE memory. Since Beta-mediated recombination is replication-dependent (27–29) and ssDNA is believed to be recombined into the genome during passage of the replication fork (27), we speculate that only actively dividing cells are likely to participate in the described population-level memory. Future optimization of SCRIBE (e.g., by modulating the mismatch repair system (14) and cellular exonucleases (35)) could lead to more efficient single-cell digital memories. This could enable other useful applications, including recording extracellular and intracellular events at the single cell level for biological studies, dynamic engineering of cellular phenotypes, experimental evolution and population dynamics studies, single-cell computation and memory, the construction of complex cellular state machines and biological Turing machines, and enhanced genome engineering techniques.
Additionally, since retrons have been found in a diverse range of microorganisms (20), in vivo ssDNA expression could be extended to hard-to-transform organisms where SCRIBE plasmids could be introduced by conjugation or transduction. Since retrons have also been shown to be functional in eukaryotes (24, 36, 37), they could be potentially used with other genome-editing tools for memory. Moreover, by using error-prone RNA polymerases (38) and reverse transcriptases (39, 40), we anticipate that mutagenized ssDNA libraries could be generated inside cells for in vivo continuous evolution (41) and cellular barcoding applications. Finally, in vivo ssDNA generation could be potentially used to create DNA nanosystems (42–48) and ssDNA-protein hybrid nanomachines in living cells (49), or could be optimized and scaled-up to create an economical source of ssDNAs for DNA nanotechnology (50). In summary, we envision that in vivo ssDNA production and SCRIBE platforms will open up a broad range of new capabilities for engineering biology.
Materials and Methods
Strains and Plasmids
Conventional cloning methods were used to construct the plasmids. Lists of strains and plasmids used in this study and the construction procedures are provided in Tables S1 and S2, respectively. The sequences for the synthetic parts and primers are provided in Tables S3 and S4.
Cells and Antibiotics
Chemically competent E. coli DH5α was used for cloning. Unless otherwise noted, antibiotics were used at the following concentrations: carbenicillin (50 µg/ml), kanamycin (20 µg/ml), chloramphenicol (30 µg/ml) and spectinomycin (100 µg/ml). In the experiment shown in Fig. 2, kanamycin (15 µg/ml), and chloramphenicol (15 µg/ml) were used.
Detection of Single-Stranded DNA
Overnight cultures harboring IPTG-inducible plasmids encoding msd(wt), msd(wt) with deactivated RT (msd(wt)_dRT), or msd(kanR)ON were grown overnight with or without IPTG (1 mM). Total RNA samples were prepared from non-induced or induced cultures using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. 10 µg total RNA from each sample was treated with RNase A (37°C, 2 hours) to remove RNA species and the msr moiety. The samples were then resolved on 10% TBE-Urea denaturing gel and visualized with SYBR-Gold. 50 pmol of a PAGE-purified synthetic oligo (FF_oligo347) with the same sequence as ssDNA(wt) was used as a molecular size marker. The band intensities were measured by Fiji software (51). The intensities were normalized to the intensity of the marker oligo and normalized intensities were used to calculate the amount of ssDNA in each sample.
Induction of Cells and Plating Assays
For each experiment, three transformants were separately inoculated in Luria Broth (LB) media + appropriate antibiotics and grown overnight (37°C, 700 RPM) to obtain seed cultures. Unless otherwise noted, inductions were performed by diluting the seed cultures (1:1000) in 2 ml of pre-warmed LB + appropriate antibiotics ± inducers followed by 24 hours incubation (30°C, 700 RPM). Aliquots of the samples were then serially diluted and appropriate dilutions were plated on selective media to determine the number of recombinants and viable cells in each culture. For each sample, the recombinant frequency was reported as the mean of the ratio of recombinants to viable cells for three independent replicates.
In all the experiments, the number of viable cells was determined by plating aliquots of cultures on LB + spectinomycin plates. LB + kanamycin plates were used to determine the number of recombinants in the kanR reversion assay. For the galK reversion assay (Fig. 2), the numbers of galKON recombinants were determined by plating the cells on MOPS EZ rich defined media (Teknova) + galactose (0.2%). The numbers of galKOFF recombinants were determined by plating the cells on MOPS EZ rich defined media + glycerol (0.2%) + 2-DOG (2%). For the experiment shown in Fig. 3, the numbers of kanRON galKON and kanROFF galKOFF cells were determined by using LB + kanamycin plates and MOPS EZ rich defined media + glycerol (0.2%) + 2-DOG (2%) + D-biotin (0.01%), respectively. The numbers of kanRON galKOFF cells were determined by plating the cells on MOPS EZ rich defined media + glycerol (0.2%) + 2-DOG (2%) + kanamycin + D-biotin (0.01%).
For the light-inducible SCRIBE experiment (Fig. 4A), induction was performed with white light (using the built-in fluorescent lamp in a VWR 1585 shaker incubator). The “dark” condition was achieved by wrapping aluminum foil around the tubes. Growth of these cultures and sampling from these cultures were performed as described earlier.
LacZ Assay
Overnight seed cultures were diluted (1:1000) in pre-warmed LB + appropriate antibiotics and inducers (with different concentrations of aTc or without aTc in Fig. 5A-C, and with all the four possible combinations of aTc (100 ng/ml) and AHL (50 ng/ml) in Fig. 5D-F) and incubated for 24 hours (30°C, 700 RPM). These cultures then were diluted (1:50) in pre-warmed LB + appropriate antibiotics with or without IPTG (1 mM) and incubated for 8 hours (37°C, 700 RPM). To measure LacZ activity, 60 µl of each culture was mixed with 60 µl of B-PER II reagent (Pierce Biotechnology) and Fluorescein Di-ß-D-Galactopyranoside (FDG, 0.05 mg/ml final concentration). The fluorescence signal (absorption/emission: 485/515) was monitored in a plate reader with continuous shaking for 2 hours. The LacZ activity was calculated by normalizing the rate of FDG hydrolysis (obtained from fluorescence signal) to the initial OD. For each sample, LacZ activity was reported as the mean of three independent biological replicates.
Supplementary Material
Acknowledgements
Supported by the NIH New Innovator Award (1DP2OD008435), NIH National Centers for Systems Biology (1P50GM098792), the Office of Naval Research (N000141310424), and the Defense Advanced Research Projects Agency.
Footnotes
Supplementary Materials:
Supplementary Text
Figs. S1 to S6
Tables S1 to S4
References (52–61)
References and Notes
- 1.Goldman N, et al. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. Nature. 2013 Feb 7;494:77. doi: 10.1038/nature11875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Church GM, Gao Y, Kosuri S. Next-generation digital information storage in DNA. Science. 2012 Sep 28;337:1628. doi: 10.1126/science.1226355. [DOI] [PubMed] [Google Scholar]
- 3.Inniss MC, Silver PA. Building synthetic memory. Curr Biol. 2013 Sep 9;23:R812. doi: 10.1016/j.cub.2013.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000 Jan 20;403:339. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 5.Kotula JW, et al. Programmable bacteria detect and record an environmental signal in the mammalian gut. Proc Natl Acad Sci U S A. 2014 Apr 1;111:4838. doi: 10.1073/pnas.1321321111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burrill DR, Inniss MC, Boyle PM, Silver PA. Synthetic memory circuits for tracking human cell fate. Genes Dev. 2012 Jul 1;26:1486. doi: 10.1101/gad.189035.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kramer BP, et al. An engineered epigenetic transgene switch in mammalian cells. Nat Biotechnol. 2004 Jul;22:867. doi: 10.1038/nbt980. [DOI] [PubMed] [Google Scholar]
- 8.Ajo-Franklin CM, et al. Rational design of memory in eukaryotic cells. Genes Dev. 2007 Sep 15;21:2271. doi: 10.1101/gad.1586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siuti P, Yazbek J, Lu TK. Synthetic circuits integrating logic and memory in living cells. Nat Biotechnol. 2013 Feb 10; doi: 10.1038/nbt.2510. [DOI] [PubMed] [Google Scholar]
- 10.Bonnet J, Subsoontorn P, Endy D. Rewritable digital data storage in live cells via engineered control of recombination directionality. Proc Natl Acad Sci U S A. 2012 Jun 5;109:8884. doi: 10.1073/pnas.1202344109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonnet J, Yin P, Ortiz ME, Subsoontorn P, Endy D. Amplifying Genetic Logic Gates. Science. 2013 Mar 28; doi: 10.1126/science.1232758. [DOI] [PubMed] [Google Scholar]
- 12.Friedland AE, et al. Synthetic gene networks that count. Science. 2009 May 29;324:1199. doi: 10.1126/science.1172005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellis HM, Yu D, DiTizio T, Court DL. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc Natl Acad Sci U S A. 2001 Jun 5;98:6742. doi: 10.1073/pnas.121164898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costantino N, Court DL. Enhanced levels of lambda Red-mediated recombinants in mismatch repair mutants. Proc Natl Acad Sci U S A. 2003 Dec 23;100:15748. doi: 10.1073/pnas.2434959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu D, et al. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A. 2000 May 23;97:5978. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sawitzke JA, et al. Probing cellular processes with oligo-mediated recombination and using the knowledge gained to optimize recombineering. J Mol Biol. 2011 Mar 18;407:45. doi: 10.1016/j.jmb.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swingle B, et al. Oligonucleotide recombination in Gram-negative bacteria. Mol Microbiol. 2010 Jan;75:138. doi: 10.1111/j.1365-2958.2009.06976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Datta S, Costantino N, Zhou X, Court DL. Identification and analysis of recombineering functions from Gram-negative and Gram-positive bacteria and their phages. Proc Natl Acad Sci U S A. 2008 Feb 5;105:1626. doi: 10.1073/pnas.0709089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yee T, Furuichi T, Inouye S, Inouye M. Multicopy single-stranded DNA isolated from a gram-negative bacterium, Myxococcus xanthus. Cell. 1984 Aug;38:203. doi: 10.1016/0092-8674(84)90541-5. [DOI] [PubMed] [Google Scholar]
- 20.Lampson BC, Inouye M, Inouye S. Retrons, msDNA, and the bacterial genome. Cytogenet Genome Res. 2005;110:491. doi: 10.1159/000084982. [DOI] [PubMed] [Google Scholar]
- 21.Lim D, Maas WK. Reverse transcriptase-dependent synthesis of a covalently linked, branched DNA-RNA compound in E. coli B. Cell. 1989 Mar 10;56:891. doi: 10.1016/0092-8674(89)90693-4. [DOI] [PubMed] [Google Scholar]
- 22.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997 Mar 15;25:1203. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma PL, Nurpeisov V, Schinazi RF. Retrovirus reverse transcriptases containing a modified YXDD motif. Antivir Chem Chemother. 2005;16:169. doi: 10.1177/095632020501600303. [DOI] [PubMed] [Google Scholar]
- 24.Mao JR, Shimada M, Inouye S, Inouye M. Gene regulation by antisense DNA produced in vivo. J Biol Chem. 1995 Aug 25;270:19684. doi: 10.1074/jbc.270.34.19684. [DOI] [PubMed] [Google Scholar]
- 25.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohlendorf R, Vidavski RR, Eldar A, Moffat K, Moglich A. From dusk till dawn: one-plasmid systems for light-regulated gene expression. J Mol Biol. 2012 Mar 2;416:534. doi: 10.1016/j.jmb.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 27.Huen MS, et al. The involvement of replication in single stranded oligonucleotide-mediated gene repair. Nucleic Acids Res. 2006;34:6183. doi: 10.1093/nar/gkl852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poteete AR. Involvement of DNA replication in phage lambda Red-mediated homologous recombination. Mol Microbiol. 2008 Apr;68:66. doi: 10.1111/j.1365-2958.2008.06133.x. [DOI] [PubMed] [Google Scholar]
- 29.Poteete AR. Involvement of DNA Replication Proteins in Phage Lambda Red-Mediated Homologous Recombination. PLoS One. 2013;8:e67440. doi: 10.1371/journal.pone.0067440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dutra BE, Sutera VA, Jr., Lovett ST. RecA-independent recombination is efficient but limited by exonucleases. Proc Natl Acad Sci U S A. 2007 Jan 2;104:216. doi: 10.1073/pnas.0608293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005 Apr 28;434:1130. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- 32.Auslander S, Auslander D, Muller M, Wieland M, Fussenegger M. Programmable single-cell mammalian biocomputers. Nature. 2012 Jul 5;487:123. doi: 10.1038/nature11149. [DOI] [PubMed] [Google Scholar]
- 33.Daniel R, Rubens JR, Sarpeshkar R, Lu TK. Synthetic analog computation in living cells. Nature. 2013 May 30;497:619. doi: 10.1038/nature12148. [DOI] [PubMed] [Google Scholar]
- 34.Olson EJ, Hartsough LA, Landry BP, Shroff R, Tabor JJ. Characterizing bacterial gene circuit dynamics with optically programmed gene expression signals. Nat Methods. 2014 Apr;11:449. doi: 10.1038/nmeth.2884. [DOI] [PubMed] [Google Scholar]
- 35.Mosberg JA, Gregg CJ, Lajoie MJ, Wang HH, Church GM. Improving lambda red genome engineering in Escherichia coli via rational removal of endogenous nucleases. PLoS One. 2012;7:e44638. doi: 10.1371/journal.pone.0044638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyata S, Ohshima A, Inouye S, Inouye M. In vivo production of a stable single-stranded cDNA in Saccharomyces cerevisiae by means of a bacterial retron. Proc Natl Acad Sci U S A. 1992 Jul 1;89:5735. doi: 10.1073/pnas.89.13.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mirochnitchenko O, Inouye S, Inouye M. Production of single-stranded DNA in mammalian cells by means of a bacterial retron. J Biol Chem. 1994 Jan 28;269:2380. [PubMed] [Google Scholar]
- 38.Brakmann S, Grzeszik S. An error-prone T7 RNA polymerase mutant generated by directed evolution. Chembiochem. 2001 Mar 2;2:212. doi: 10.1002/1439-7633(20010302)2:3<212::AID-CBIC212>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 39.Roberts JD, Bebenek K, Kunkel TA. The accuracy of reverse transcriptase from HIV-1. Science. 1988 Nov 25;242:1171. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- 40.Bebenek K, Abbotts J, Wilson SH, Kunkel TA. Error-prone polymerization by HIV-1 reverse transcriptase. Contribution of template-primer misalignment, miscoding, and termination probability to mutational hot spots. J Biol Chem. 1993 May 15;268:10324. [PubMed] [Google Scholar]
- 41.Esvelt KM, Carlson JC, Liu DR. A system for the continuous directed evolution of biomolecules. Nature. 2011 Apr 28;472:499. doi: 10.1038/nature09929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amir Y, et al. Universal computing by DNA origami robots in a living animal. Nat Nanotechnol. 2014 May;9:353. doi: 10.1038/nnano.2014.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qian L, Winfree E, Bruck J. Neural network computation with DNA strand displacement cascades. Nature. 2011 Jul 21;475:368. doi: 10.1038/nature10262. [DOI] [PubMed] [Google Scholar]
- 44.Seelig G, Soloveichik D, Zhang DY, Winfree E. Enzyme-free nucleic acid logic circuits. Science. 2006 Dec 8;314:1585. doi: 10.1126/science.1132493. [DOI] [PubMed] [Google Scholar]
- 45.Rothemund PW. Folding DNA to create nanoscale shapes and patterns. Nature. 2006 Mar 16;440:297. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 46.Douglas SM, et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 2009 May 21;459:414. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Douglas SM, Bachelet I, Church GM. A logic-gated nanorobot for targeted transport of molecular payloads. Science. 2012 Feb 17;335:831. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- 48.Chirieleison SM, Allen PB, Simpson ZB, Ellington AD, Chen X. Pattern transformation with DNA circuits. Nat Chem. 2013 Dec;5:1000. doi: 10.1038/nchem.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brosey CA, et al. A new structural framework for integrating replication protein A into DNA processing machinery. Nucleic Acids Res. 2013 Feb 1;41:2313. doi: 10.1093/nar/gks1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kosuri S, Church GM. Large-scale de novo DNA synthesis: technologies and applications. Nat Methods. 2014 May;11:499. doi: 10.1038/nmeth.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012 Jul;9:676. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.