Abstract
Regulation of muscle contraction via the myosin filaments occurs in vertebrate smooth and many invertebrate striated muscles. Studies of unphosphorylated vertebrate smooth muscle myosin suggest that activity is switched off through an intramolecular interaction between the actin-binding region of one head and the converter and essential light chains of the other, inhibiting ATPase activity and actin interaction. The same interaction (and additional interaction with the tail) is seen in three-dimensional reconstructions of relaxed, native myosin filaments from tarantula striated muscle, suggesting that such interactions are likely to underlie the off-state of myosin across a wide spectrum of the animal kingdom. We have tested this hypothesis by carrying out cryo-electron microscopy and 3D image reconstruction of myosin filaments from horseshoe crab (Limulus) muscle. The same head-head and head-tail interactions seen in tarantula are also seen in Limulus, supporting the hypothesis. Other data suggest that this motif may underlie the relaxed state of myosin II in all species (including myosin II in nonmuscle cells), with the possible exception of insect flight muscle.
The molecular organization of the myosin tails in the backbone of muscle thick filaments is unknown, and may differ between species. X-ray diffraction data support a general model for crustaceans in which tails associate together to form 4 nm diameter subfilaments, with these subfilaments assembling together to form the backbone. This model is supported by direct observation of 4 nm diameter, elongated strands in the tarantula reconstruction, suggesting that it might be a general structure across the arthropods. We observe a similar backbone organization in the Limulus reconstruction, supporting the general existence of such subfilaments.
Keywords: structure, cryo-electron microscopy, myosin filament, regulation, 3D reconstruction
Introduction
Contraction of muscle is regulated by Ca2+-sensitive molecular switches on the thick (myosin-containing) and thin (actin-containing) filaments.1–6 Myosin-linked regulation, found in almost all invertebrate striated muscles and in vertebrate smooth and nonmuscle cells, depends on phosphorylation of the myosin regulatory light chains or Ca2+ binding to the essential light chains, depending on species.2, 4, 7, 8 Recent studies have provided new insights into the mechanism by which regulated myosins are switched off, leading to muscle relaxation. Structural observations of purified vertebrate smooth muscle myosin in the dephosphorylated (off) state, either in 2D crystals or as single molecules, show that the two myosin heads interact with each other (Fig. 1).9, 10 This suggests a model for the myosin II off-state in which the actin-binding site of one head (the “blocked” head) is blocked by its binding to the other (“free”) head.9 Structural changes involved in ATP hydrolysis in the free head are inhibited, in turn, by the binding of the blocked head to the converter domain of the free head. Both heads would be switched off in different ways through this asymmetric interaction.
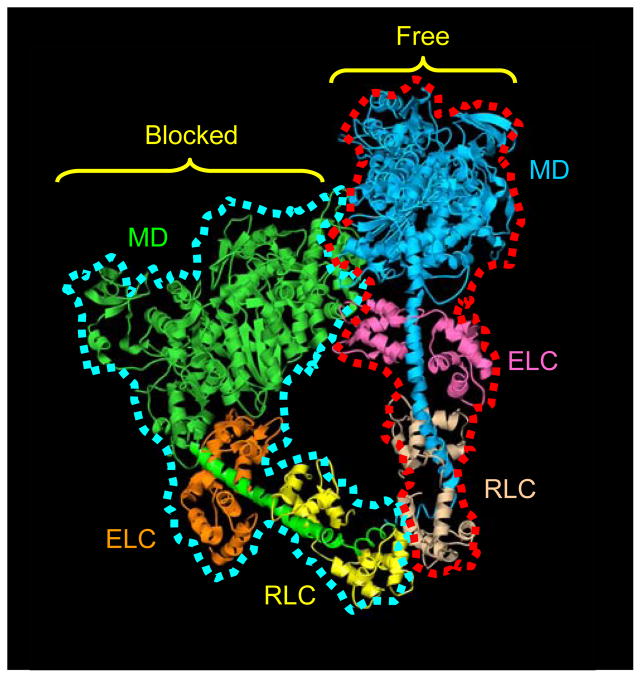
Figure 1. Atomic model of head-head interaction.
This ribbon model represents the best fit of the atomic model of vertebrate smooth muscle HMM (PDB1i84)9 into the tarantula 3D reconstruction.11 The actin-binding region of the blocked head, outlined in blue, binds to the converter region and essential light chain of the free head, outlined in red. Color scheme: motor domains (MD), blocked head, green; free head, blue; essential light chains (ELC), blocked head, orange; free head, pink; regulatory light chains (RLC), blocked head, yellow; free head, beige. Adapted from Ref. 11.
The key importance of this interaction in vivo has now been revealed by cryo-electron microscopic studies of myosin filaments from tarantula striated muscle.11 Myosin filaments are helical polymers of myosin II molecules.12–17 The α-helical myosin tails form the backbone of the filament, while the myosin heads lie at the surface in multi-start helical arrays, forming rotationally symmetric “crowns” every 14.5 nm.12, 13, 16, 17 Three-dimensional reconstructions of native tarantula filaments in the relaxed (off) state show that the two heads of each myosin molecule interact with each other11. Docking of the myosin head atomic structure with the reconstruction reveals that the interaction is essentially identical to that occurring in isolated vertebrate smooth muscle myosin molecules.9 The reconstruction also suggests interaction between the initial portion of the myosin tail and the blocked head, and this has recently been confirmed in single molecules.10 It further reveals interactions, not observed in the single molecule studies, between heads at different levels of the helix. These interactions would all contribute to the switching off of myosin molecules in the filament by a similar mechanism to that in isolated molecules.9, 11
The dramatic similarity between the head-head and head-tail interactions in vertebrate smooth muscle myosin molecules and in invertebrate striated muscle myosin filaments suggests that the interacting-head motif has been conserved for more than 600 million years,18 since before vertebrates and invertebrates diverged. It is therefore likely that most, if not all, regulated myosin II molecules are switched off by the same interactions.11 This proposal is supported by in vitro studies of isolated myosin molecules from several invertebrate and vertebrate species, from both muscle and nonmuscle sources.19, 20 Our goal here has been to directly test the hypothesis that head-head and head tail interactions similar to those in tarantula filaments are a general motif occurring in other myosin-regulated filaments. Here we study this question in thick filaments from Limulus muscle, which are regulated by phosphorylation7 and display helical ordering comparable in quality to tarantula, making them amenable to structural analysis.14, 21, 22 Using cryo-EM, single particle image reconstruction, and atomic fitting, we demonstrate intramolecular head-head interactions essentially identical to those in tarantula.
The backbone of myosin filaments consists of closely packed myosin tails (coiled-coil α-helices) running nearly parallel to each other and also to the long axis of the filament.12, 16 Because of their length, small diameter, and tight packing, the detailed molecular organization of the tails has not been defined experimentally. Theoretical models suggest that they may pack together as single molecules (molecular crystal model), or that they may associate into intermediate sized structures (subfilaments), which then assemble into the filament as a whole.23 X-ray diffraction data from intact muscles suggest that crustacean muscle filaments are built from subfilaments 4 nm in diameter and containing three myosin tails in cross-section.24 The tarantula filament reconstruction provides the first strong visual support for such a model,11 and extends it to the chelicerate subphylum. Reconstructions of other species of filament are required to test the generality of this model. We find evidence for a set of subfilaments in Limulus similar to those found in tarantula.
Results
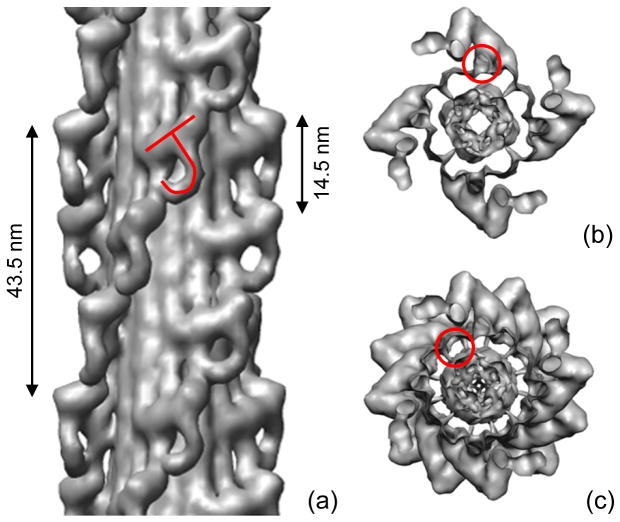
Frozen-hydrated Limulus thick filaments had a similar appearance to tarantula filaments11 (Fig. 2a, b). Characteristic arrowheads were observed pointing towards the central bare zone. These represent a projected view of the 4-fold symmetric arrangement of myosin heads, helically arranged on the filament surface, demonstrating good preservation of helical order, as found in previous studies of Limulus filaments.14, 21, 22 Averaged Fourier transforms of filaments showed layer lines indexing on a repeat of ~ 43.5 nm, with a meridional reflection at the third order, representing the axial distance of 14.5 nm between crowns of heads (Fig. 2c). The 3-fold relationship between the helical repeat and the distance between crowns, combined with the 4-fold rotational symmetry, means that neighboring crowns are rotated by 30° with respect to each other (Fig. 3a). Some filaments (especially those in which the heads were less well ordered) revealed that the filament backbone consisted of parallel elongated strands (Fig. 2b).
Figure 2. Cryo-electron microscopy of purified Limulus myosin filaments.
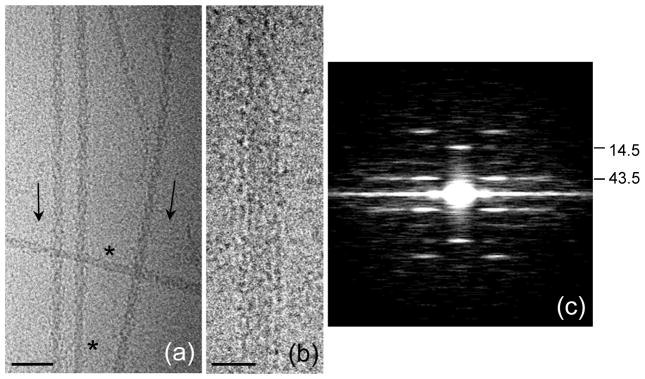
(a) Field of filaments in relaxing conditions. Arrowheads (direction shown by arrows) result from superposition of myosin helices on front and rear of filaments as seen in projection. Bare zones are indicated by *. (b) Filament in which heads are disordered, revealing parallel strands in backbone. (c) Average Fourier transform (intensities) of the 100 filament images used in the reconstruction, showing layer lines at orders of the 43.5 nm helical repeat, confirming order visualized in A. Scale bars: (a) 100 nm; (b) 20 nm.
Figure 3. Three-dimensional reconstruction of Limulus thick filament.
(a) Pairs of myosin heads, represented by tilted “J” motif, run in four parallel helices, with a repeat of 43.5 nm and an axial spacing between 4-fold rotationally symmetric ‘crowns’ of heads of 14.5 nm. (b) Transverse view of one 14.5 nm level of myosin heads, showing 4-fold rotational symmetric arrangement, viewed from bare zone. S2 is outlined. (c) Transverse view of one complete repeat (43.5 nm) of heads. In (b) and (c), four groups of three subfilaments appear as hollow tubes due to surface rendering (one has been outlined in (c)). The subfilaments in fact represent the highest density region of the map (Fig. 5a). In contrast, the center of the filament (interior to the tubes) appears solid but is in fact of low density (Fig. 5a).
A three-dimensional reconstruction was calculated from 100 of the best ordered filaments using single particle methods.11, 25 The resolution was 2.9 nm according to the Fourier shell correlation using a 0.5 threshold. The reconstruction showed two key features. At the surface of the filament was a helical arrangement of tilted, ‘J’-like motifs arranged in ‘crowns’ with 4-fold rotational symmetry, while at lower radius, the filament showed twelve elongated strands running parallel to the filament axis (Figs. 3, 5a). The similarity of these features to the tarantula filament reconstruction suggests that the J-motif represents a pair of interacting myosin heads, while the backbone strands represent subfilaments containing groups of myosin tails.
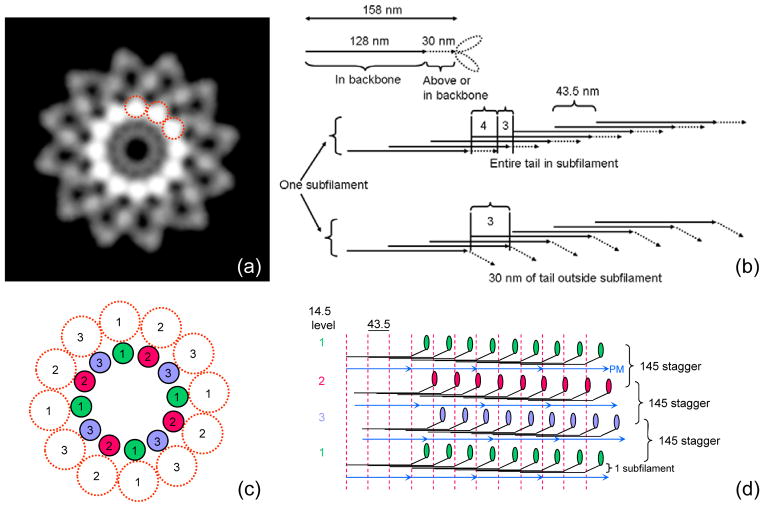
Figure 5. Backbone structure in Limulus thick filament.
(a) Density projection of one complete repeat of reconstruction (43.5 nm) along filament axis (protein white). The highest protein density is in a ring of twelve rods, about 4 nm in diameter (three have been circled), running parallel to the filament axis. Each represents one subfilament. Lower density at higher radius represents myosin heads. (b) Schematic diagram showing number of tails in cross-section of a subfilament, assuming a tail length of 158 nm (see text) and that molecules within a subfilament are staggered by 43.5 nm.24 Top: entire tail is in subfilament; subfilament thus has 4 tails in cross-section for ~ 2/3 of each repeat. Bottom: 30 nm of tail is outside subfilament; subfilament thus has 3 tails in cross-section along almost entire repeat. (c), (d) Hypothetical backbone structure showing how paramyosin molecules ~ 130 nm long might fit into the filament core, one associated with each subfilament every 3×43.5 nm. Numbers indicate relative axial levels of subfilaments and paramyosin (in increments of 14.5 nm). Molecules in each subfilament, and in subfilaments at the same level, have the same color. In this speculative model, paramyosins (assumed diameter 2 nm) are not quite close enough to interact. Such interaction, expected in any plausible model of the filament backbone, could occur, for example, if the coiled-coil diameter were slightly larger, or the filament dimensions slightly smaller than assumed. PM = paramyosin.
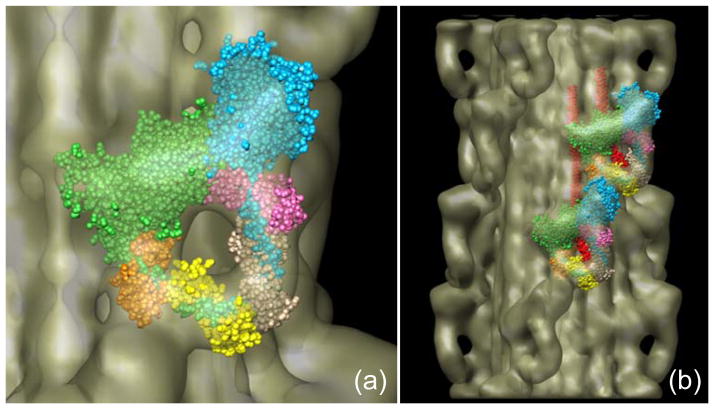
Detailed interpretation of the reconstruction was aided by manually docking the atomic model of the myosin head in the ADP.Pi state into the head features of the 3D map11 (Fig. 4a). The fitting was essentially identical to that obtained with the tarantula reconstruction. The actin-binding region of the blocked head abuts the converter domain and essential light chain of the free head (cf. Fig. 1). As is thought to be the case with vertebrate smooth muscle9 and tarantula striated muscle,11 this may be the structural basis for the switching off of myosin activity. In addition, a rod-like region of density was seen running from the heads towards the filament backbone. This was consistent with our observations for the tarantula density map where this region was assigned to the initial portion of myosin S2. As the S2 travels towards the filament backbone, it appears to come into close proximity with the actin-binding cleft region of the blocked head, which could help to inhibit blocked head activity.11
Figure 4. Docking of myosin head atomic model into J-motif of reconstruction.
(a) Fitting of myosin heads (PDB 1i84),9 each as two rigid bodies—the motor domain and the light chain domain—into the two heads of one J-motif, showing interaction of blocked head motor domain (green) with free head motor domain (blue) and essential light chain (pink). (b) Fitting of atomic model into heads at two adjacent axial levels, showing additional interactions between heads at different levels (cf.11).
When additional 14.5 nm levels of myosin heads were fitted, further putative interactions were revealed, this time between myosin molecules at different levels of the helix. On its course towards the filament backbone, S2 comes in contact with the SH3 and converter domains of the adjacent blocked head located closer to the bare zone. In addition, the free head actin-binding region appears to touch the essential light chain of the blocked head next closest to the bare zone. All of these interactions combined would add to the stability of the J-motif and may contribute to the maintenance of both heads in the off-state.
The backbone of the filament revealed an annular array of twelve continuous strands, ~ 4 nm in diameter, running parallel to the filament axis, centered at a radius of ~ 8 nm; the core of the filament was relatively empty (Fig. 5a). These strands (seen as apparently hollow tubes in surface renditions (Fig. 3b, c)) contain the highest protein density in the reconstruction when viewed in cross-section (see Fig. 5a), consistent with the presence of myosin tails running approximately lengthwise, oriented nearly parallel to the filament axis.
Discussion
Our reconstruction of frozen-hydrated, native, Limulus myosin filaments supports the hypothesis that head-head interaction is a motif common to regulated myosin filaments in the off-state.9, 11 In addition to tarantula and Limulus filaments, a similar motif is also seen in scallop striated muscle myosin filaments, which are regulated by direct Ca2+ binding rather than by light chain phosphorylation (unpublished data and26). This supports predictions of the widespread occurrence of the motif,11 based on the striking similarity of the two widely separated systems originally studied: isolated vertebrate smooth muscle myosin molecules and tarantula striated muscle myosin filaments. We conclude that a similar structural mechanism underlies regulation in all three myosin-regulated species so far studied. A similar motif is also seen at two of every three levels of heads in the MyBPC region of relaxed vertebrate striated muscle thick filaments,27 which are considered to be unregulated3 and thus always in the switched-on state. It seems likely that the motif exhibited by unregulated filaments may represent a weaker version of the interaction,20, 27 in this case possibly functioning to reduce interaction with actin filaments in the filament lattice (without switching myosin activity off), by keeping the heads in an ordered array close to the thick filament surface and away from actin.20, 27, 28 One type of filament in which this structure may be absent is in indirect insect flight muscle.29
The widespread occurrence of the motif demonstrated in native filament studies is further supported by observations of single myosin molecules in relaxing conditions. Vertebrate smooth,10, 19 scallop,19 and tarantula, Limulus, and vertebrate striated muscle myosin molecules20 all show similar interacting head-head and head-tail structures in the relaxed state.
Comparison with previous Limulus reconstructions
Previous reconstructions of Limulus filaments were based on negative stain images and Fourier-Bessel helical reconstruction methods.14, 22 As with similar reconstructions of tarantula15 and scorpion filaments,22 these studies lacked the resolution required for an unequivocal interpretation of head organization, and were carried out before myosin head atomic models were available for docking with the reconstruction. In all these cases it was concluded (based on 5 nm resolution data) that the heads of the myosin molecules were probably splayed apart so that heads from adjacent 14.5 nm levels interacted with each other.15, 22, 30 An alternative possibility considered was a parallel arrangement of heads pointing away from the bare zone22—the opposite of the direction that we find here. Cryo-EM images of unstained, frozen-hydrated filaments (Fig. 2a, b) reveal the entire structure of the filament (rather than just the surface features contrasted by negative stain), while single particle methods generate a higher resolution reconstruction by avoiding key limitations of helical image processing methods25 (compare11 with15). The ~ 2.9 nm resolution achieved here and with the earlier tarantula reconstruction enables unambiguous fitting of myosin head atomic models,11 revealing that the heads are not splayed. On the contrary, they interact with each other intramolecularly, and are both tilted back towards the myosin tail and thus towards the bare zone, as in isolated Limulus20 and other myosin molecules.10, 19, 20, 31
Previous studies of Limulus filaments showed that crosslinking of heads with an ATP dimer (bis22ATP) prevented dissolution of filaments by high salt.32 It was concluded that crosslinking must have been between heads at different 14.5 nm levels in order to maintain filament structure, and thus that the heads in Limulus filaments were splayed and not parallel, contrary to our findings. It is possible that the structure of the filaments following the experimental manipulations needed in these studies was not the same as the native structure revealed in our reconstruction.32
Intra- and intermolecular myosin interactions
Studies of single molecules of Limulus and other regulated myosins reveal two major intramolecular interactions—between the blocked head and both the converter domain and essential light chain of the free head.9, 10, 19, 20 These interactions would switch the molecule off by sterically interfering with actin binding to the blocked head, and with structural changes involved in ATP hydrolysis by the free head.9 An additional interaction revealed in isolated molecules10, 19, 20 and in this and earlier11 filament studies occurs between S2 and the blocked head, probably via a negatively charged patch on S2 and positively charged regions on the blocked head.33 This might, in fact, be one of the most important interactions in generating the off state, as regulation is lost when S2 is truncated so that the interaction is not possible,11, 34 and it has been noted that this interaction can still occur even when the head-head interaction is broken.10
When molecules with the interacting head structure are assembled into filaments, additional interactions and steric constraints are generated, which could further stabilize the off-state.11 Interaction of S2 with the converter and SH3 domains of the axially adjacent blocked head (Fig. 4b) may help to inhibit blocked head ATPase activity,11 thus adding to the inhibition of its actin-binding ability that already occurs from intramolecular interaction with the free head. While the actin-binding interface of the free head is ‘free’ in single molecules,9 when incorporated into the filament it interacts with the essential light chain of the next molecule along the helix, and also points towards the filament backbone. Both effects would add to the inhibition of its actin-binding activity by S2.11 We conclude that in filaments, the actin-binding and ATPase activities of both the blocked and the free heads are sterically inhibited.
Does phosphorylation regulate invertebrate myosin filaments in vivo?
Early studies of muscle regulation showed that most invertebrate muscles are dually regulated, via both the troponin-tropomyosin switch on the thin filaments and the myosin heads on the thick filaments.3 In many cases, it appears that the myosin switch is activated by phosphorylation of the regulatory light chains.7, 8, 35 How do such muscles respond rapidly to activation if one of the switches involves the slow (enzymatic) process of phosphorylation? In Limulus muscle, a high level of basal phosphorylation (~50%) is present even in the relaxed state, and force can be generated without change in the level of phosphorylation.36 Those myosin molecules that are basally phosphorylated may thus be poised for immediate interaction with actin, as soon as Ca2+ activates the thin filaments by binding to the troponin switch.5 In these muscles, troponin-tropomyosin would appear to be the dominant functional switch,36 and phosphorylation could modulate contraction (on a slower timescale), by activating additional myosin molecules during prolonged contractions. This mechanism would then be more akin to vertebrate striated muscle, in which phosphorylation is not required for myosin activity but nevertheless enhances longer term contractions.36, 37
Myosin filament backbone structure
The structure of the myosin filament backbone remains obscure. This is due to the tight packing of the narrow, elongated myosin tails within the backbone,23 making structural analysis difficult (in common with other coiled-coil filaments38). X-ray diffraction data have suggested a general model for different crustacean filaments constructed from 4 nm diameter subfilaments containing 3 tails in cross-section; these subfilaments are arranged in a ring, all at the same radius, creating a relatively hollow core.24 In this model, the number of subfilaments is equal to three times the rotational symmetry of the filament, and their slew angle is related to the helical repeat. When this repeat is an integral number of 14.5 nm repeats (the axial distance between crowns of myosin heads), the subfilaments are predicted to run parallel to the filament axis. Tarantula and Limulus filaments both have 4-fold rotational symmetry and a repeat that is 3 times the 14.5 nm axial rise of myosin.14, 15, 17, 21, 22, 39 In the subfilament model,24 this would imply twelve subfilaments running parallel to the filament axis, centered at a radius of ~ 8 nm. These are exactly the parameters we find for Limulus (as well as tarantula11) filaments (Fig. 5a). Our observations thus add support to the idea of 4 nm diameter subfilaments as basic building blocks in the chelicerate subphylum of arthropods (cf.11). Since the original X-ray data came from a different arthropod subphylum (crustacea24), it appears likely that all arthropods may have myosin subfilaments of this type. The same model may extend (with modifications) to other phyla.24
In Wray’s model,24 myosin tails within each subfilament coil around each other and are staggered by three times the 14.5 nm repeat. Each subfilament thus gives rise to a pair of myosin heads every 43.5 nm. The subfilaments themselves are displaced by 14.5 nm relative to their neighbors. This is exactly the arrangement we see in both Limulus (Fig. 2a) and tarantula11 reconstructions. Knowing the length of the myosin tail and of the portion of S2 that lies above the filament backbone, we can test the proposal that the subfilaments contain three tails in cross-section along most of their length.24 The measured length of the tail of vertebrate skeletal myosin is ~156 nm.40–42 Preliminary measurements of the tail length in rotary shadowed Limulus and tarantula molecules are similar (~157 and ~159 nm, respectively; H.S. Jung, personal communication). If the entire ~158 nm tail were incorporated into the subfilament, there would be four tails in cross-section for most of each 43.5 nm repeat, and three for only a short region (Fig. 5b). This would produce a subfilament closer to 5 nm in diameter, contrary to the X-ray data,24 and would center the subfilaments at a radius of ~ 9 nm, greater than the ~ 8 nm that we observe. Our reconstruction of tarantula filaments11 shows that ~30 nm of S2 lies above the filament surface, gradually descending from its junction with the two heads to the subfilament. While S2 is not as well resolved in the Limulus reconstruction, a similar S2 arrangement appears likely. With only ~128 nm of tail thus in the subfilament, there would be three molecules in cross-section along almost the entire repeat (~ 41 nm), with a short section (~ 3 nm) with only 2 molecules (Fig. 5b), consistent with X-ray data and the 4 nm diameter that we observe.
Invertebrate striated muscle myosin filaments also have small and varying amounts of paramyosin in their cores (~ 0.06–0.5 paramyosin:myosin molar ratio).39, 43 In Limulus and tarantula the ratio is ~ 0.3–0.5.39 Our Limulus and tarantula reconstructions do not appear to reveal paramyosin. This may reflect their limited resolution, or the possibility that paramyosin and myosin are not organized with identical symmetries. One possible model consistent with a 0.33 ratio would have one paramyosin molecule ~130 nm long44 associated with three myosin molecules in each subfilament every 3×43.5 nm, forming a layer ~ 2 nm thick (Fig. 5c,d). Azimuthally adjacent paramyosins would be staggered by 14.5 nm, and there would be head-to-tail contact at 130 nm intervals within each paramyosin strand.
Methods
Purification of Limulus muscle thick filaments
Horseshoe crabs (Limulus polyphemus) were obtained from the Marine Biological Laboratory, Woods Hole MA, and stored in a marine aquarium at 10°C. Preparative procedures were carried out at 0–4°C except where otherwise noted. Telson muscles were obtained by a modification of the method of Kensler and Levine.21 Muscles were quickly dissected from the animal and split into 1–2 mm bundles, which were tied to plastic plates. Their membranes were permeabilized by placing in 10 ml of mincing solution (0.1 M NaCl, 2 mM EGTA, 1 mM DTT, 5 mM MgCl2, 5 mM PIPES, 5 mM NaH2PO4, 1 mM NaN3, pH 7.2) with 1% saponin. After four hours rotation, the strips were transferred to 10 ml relaxing solution (0.1 M KCl, 5 mM EGTA, 1 mM DTT, 5 mM MgCl2, 5 mM PIPES, 5 mM MgATP, 5 mM NaH2PO4, 1 mM NaN3, pH 7.0) and rotated overnight. Filaments were purified, using this relaxing solution, according to Hidalgo et al.45 Briefly, 0.15 g of saponin-permeabilized muscle was finely chopped, homogenized, then briefly centrifuged to remove large debris. The supernatant was treated with bacterially expressed calcium-insensitive gelsolin46 to fragment the thin filaments, then centrifuged. The purified thick filaments (in the pellet) were resuspended in relaxing solution and used for electron microscopy.
Electron microscopy
Frozen-hydrated specimens were prepared at room temperature in a humidified chamber (R.H. ~70%). 6 μl of thick filament suspension was applied to a 400 mesh grid coated with a holey carbon film that had been rendered hydrophilic by glow discharge in n-amylamine vapor for three minutes. After allowing the filaments to absorb to the grid for 30 seconds, the grid was rinsed with relaxing rinse (100 mM NaAc, 3 mM MgCl2, 0.2 mM EGTA, 2 mM imidazole, 1 mM NaN3 and 1 mM MgATP, pH 7.0), blotted to a thin film using Whatman No. 42 filter paper, and immediately plunged under gravity into liquid ethane cooled by liquid nitrogen. Grids were examined using a Gatan (Pleasanton, CA) 626 DH cryo-holder at ~ −184°C in a Philips CM120 cryo-electron microscope (FEI, Hillsboro, OR) at 120 kV. Low dose (~600 e−/nm2) images of filaments suspended in vitreous ice over holes in the carbon film were recorded on a 2Kx2K CCD camera (F224HD, TVIPS, Gauting, Germany) with a nominal magnification of 28,000 × at the CCD (0.59 nm per pixel in the original specimen) and a defocus of ~ −1.9 μm.
Image processing
Filaments were aligned with the bare zone at the top to ensure the correct polarity in subsequent steps. Three-dimensional reconstruction was carried out by the Iterative Helical Real Space Reconstruction (IHRSR) single particle method,11, 25 using the SPIDER software package.47 The reconstruction was based on ~2500 segments, each 59 nm long with an overlap of 45 nm, from ~ 100 different filament halves. The total number of unique pairs of myosin heads that went into the final reconstruction was ~ 10,000. Initial reference models used for the reconstruction were helical reconstructions from negative stain data15 and from the current cryo data, and the tarantula cryo reconstruction.11 All gave the same final structure. The reconstruction was low pass filtered to 2.9 nm (the approximate resolution according to the Fourier shell correlation). Surface renderings were carried out with UCSF Chimera.48 Computational fitting of the atomic model of smooth muscle HMM (PDB 1i84) to the surface envelope of the reconstruction was carried out manually within Chimera as described previously.11
Acknowledgments
We thank Neil Nosworthy for help with bacterial expression of the N-terminal fragment of gelsolin, using cDNA kindly provided by Helen Yin. We are very grateful to HyunSuk Jung for his advice on various aspects of this work, for his preliminary measurements on the length of Limulus and tarantula myosin tails, and for comments on the manuscript. This work was supported by NIH grant AR34711 to RC. Electron microscopy was carried out in the Core Electron Microscopy Facility of the University of Massachusetts Medical School, supported in part by Diabetes Endocrinology Research Center grant DK32520. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081). This work has been published in preliminary form.26
References
- 1.Ebashi S, Endo M, Ohtsuki I. Control of muscle contraction. Quart Rev Biophys. 1969;2:351–384. doi: 10.1017/s0033583500001190. [DOI] [PubMed] [Google Scholar]
- 2.Kendrick-Jones J, Lehman W, Szent-Gyorgyi AG. Regulation in molluscan muscles. J Mol Biol. 1970;54:313–326. doi: 10.1016/0022-2836(70)90432-8. [DOI] [PubMed] [Google Scholar]
- 3.Lehman W, Szent-Gyorgyi AG. Regulation of muscular contraction. Distribution of actin control and myosin control in the animal kingdom. J Gen Physiol. 1975;66:1–30. doi: 10.1085/jgp.66.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szent-Gyorgyi AG, Kalabokis VN, Perreault-Micale CL. Regulation by molluscan myosins. Mol Cell Biochem. 1999;190:55–62. [PubMed] [Google Scholar]
- 5.Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol Rev. 2000;80:853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- 6.Perry SV. Activation of the contractile mechanism by calcium. In: Engel AG, Franzini-Armstrong C, editors. Myology. McGraw-Hill; New York, NY: 2004. pp. 281–306. [Google Scholar]
- 7.Sellers JR. Phosphorylation-dependent regulation of Limulus myosin. J Biol Chem. 1981;256:9274–9278. [PubMed] [Google Scholar]
- 8.Sellers JR. Myosins. Oxford University Press; New York: 1999. [Google Scholar]
- 9.Wendt T, Taylor D, Trybus KM, Taylor K. Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc Natl Acad Sci U S A. 2001;98:4361–4366. doi: 10.1073/pnas.071051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burgess SA, Yu S, Walker ML, Hawkins RJ, Chalovich JM, Knight PJ. Structures of smooth muscle myosin and heavy meromyosin in the folded, shutdown state. J Mol Biol. 2007;372:1165–1178. doi: 10.1016/j.jmb.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 11.Woodhead JL, Zhao FQ, Craig R, Egelman EH, Alamo L, Padron R. Atomic model of a myosin filament in the relaxed state. Nature. 2005;436:1195–1199. doi: 10.1038/nature03920. [DOI] [PubMed] [Google Scholar]
- 12.Huxley HE. Electron microscope studies on the structure of natural and synthetic protein filaments from striated muscle. J Mol Biol. 1963;7:281–308. doi: 10.1016/s0022-2836(63)80008-x. [DOI] [PubMed] [Google Scholar]
- 13.Huxley HE, Brown W. The low-angle x-ray diagram of vertebrate striated muscle and its behaviour during contraction and rigor. J Mol Biol. 1967;30:383–434. doi: 10.1016/s0022-2836(67)80046-9. [DOI] [PubMed] [Google Scholar]
- 14.Stewart M, Kensler RW, Levine RJ. Structure of Limulus telson muscle thick filaments. J Mol Biol. 1981;153:781–790. doi: 10.1016/0022-2836(81)90418-6. [DOI] [PubMed] [Google Scholar]
- 15.Crowther RA, Padron R, Craig R. Arrangement of the heads of myosin in relaxed thick filaments from tarantula muscle. J Mol Biol. 1985;184:429–439. doi: 10.1016/0022-2836(85)90292-x. [DOI] [PubMed] [Google Scholar]
- 16.Craig R, Woodhead JL. Structure and function of myosin filaments. Curr Opin Struct Biol. 2006;16:204–212. doi: 10.1016/j.sbi.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 17.Wray JS, Vibert PJ, Cohen C. Diversity of cross-bridge configurations in invertebrate muscles. Nature. 1975;257:561–564. doi: 10.1038/257561a0. [DOI] [PubMed] [Google Scholar]
- 18.Peterson KJ, Lyons JB, Nowak KS, Takacs CM, Wargo MJ, McPeek MA. Estimating metazoan divergence times with a molecular clock. Proc Natl Acad Sci U S A. 2004;101:6536–6541. doi: 10.1073/pnas.0401670101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jung HS, Burgess SA, Billington N, Colegrave M, Patel H, Chalovich JM, Chantler PD, Knight PJ. Conservation of the regulated structure of folded myosin 2 in species separated by at least 600 million years of independent evolution. Proc Natl Acad Sci U S A. 2008;105:6022–6026. doi: 10.1073/pnas.0707846105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung HS, Komatsu S, Ikebe M, Craig R. Head-head and head-tail interaction: a general mechanism for switching off myosin II activity in cells. Mol Biol Cell. 2008;19:3234–3242. doi: 10.1091/mbc.E08-02-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kensler RW, Levine RJ. An electron microscopic and optical diffraction analysis of the structure of Limulus telson muscle thick filaments. J Cell Biol. 1982;92:443–451. doi: 10.1083/jcb.92.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stewart M, Kensler RW, Levine RJ. Three-dimensional reconstruction of thick filaments from Limulus and scorpion muscle. J Cell Biol. 1985;101:402–411. doi: 10.1083/jcb.101.2.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Squire J. The Structural Basis of Muscular Contraction. Plenum Press; New York: 1981. [Google Scholar]
- 24.Wray JS. Structure of the backbone in myosin filaments of muscle. Nature. 1979;277:37–40. doi: 10.1038/277037a0. [DOI] [PubMed] [Google Scholar]
- 25.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 26.Zhao FQ, Woodhead JL, Craig R. Head-head interaction characterizes the relaxed state of scallop and Limulus muscle myosin filaments. 2008:630–Pos. doi: 10.1016/j.jmb.2008.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zoghbi ME, Woodhead JL, Moss RL, Craig R. Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proc Natl Acad Sci U S A. 2008;105:2386–2390. doi: 10.1073/pnas.0708912105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zoghbi ME, Woodhead JL, Craig R, Padron R. Helical order in tarantula thick filaments requires the “closed” conformation of the myosin head. J Mol Biol. 2004;342:1223–1236. doi: 10.1016/j.jmb.2004.07.037. [DOI] [PubMed] [Google Scholar]
- 29.al Khayat HA, Hudson L, Reedy MK, Irving TC, Squire JM. Myosin head configuration in relaxed insect flight muscle: x-ray modeled resting cross-bridges in a pre-powerstroke state are poised for actin binding. Biophys J. 2003;85:1063–1079. doi: 10.1016/S0006-3495(03)74545-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Offer G, Knight PJ, Burgess SA, Alamo L, Padron R. A new model for the surface arrangement of myosin molecules in tarantula thick filaments. J Mol Biol. 2000;298:239–260. doi: 10.1006/jmbi.2000.3664. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki H, Stafford WF, Slayter HS, Seidel JC. A conformational transition in gizzard heavy meromyosin involving the head-tail junction, resulting in changes in sedimentation coeffeicient, ATPase activity, and orientation of heads. J Biol Chem. 1985;260:14810–14817. [PubMed] [Google Scholar]
- 32.Levine RJ, Chantler PD, Kensler RW. Arrangement of myosin heads on Limulus thick filaments. J Cell Biol. 1988;107:1739–1747. doi: 10.1083/jcb.107.5.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blankenfeldt W, Thoma NH, Wray JS, Gautel M, Schlichting I. Crystal structures of human cardiac beta-myosin II S2-Delta provide insight into the functional role of the S2 subfragment. Proc Natl Acad Sci U S A. 2006;103:17713–17717. doi: 10.1073/pnas.0606741103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trybus KM, Freyzon Y, Faust LZ, Sweeney HL. Spare the rod, spoil the regulation: necessity for a myosin rod. Proc Natl Acad Sci U S A. 1997;94:48–52. doi: 10.1073/pnas.94.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Craig R, Padron R, Kendrick-Jones J. Structural changes accompanying phosphorylation of tarantula muscle myosin filaments. J Cell Biol. 1987;105:1319–1327. doi: 10.1083/jcb.105.3.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ritter O, Haase H, Morano I. Regulation of Limulus skeletal muscle contraction. FEBS Lett. 1999;446:233–235. doi: 10.1016/s0014-5793(99)00224-0. [DOI] [PubMed] [Google Scholar]
- 37.Sweeney HL, Bowman BF, Stull JT. Myosin light chain phosphorylation in vertebrate striated muscle: regulation and function. Am J Physiol. 1993;264:C1085–C1095. doi: 10.1152/ajpcell.1993.264.5.C1085. [DOI] [PubMed] [Google Scholar]
- 38.Parry DA, Strelkov SV, Burkhard P, Aebi U, Herrmann H. Towards a molecular description of intermediate filament structure and assembly. Exp Cell Res. 2007;313:2204–2216. doi: 10.1016/j.yexcr.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 39.Levine RJ, Kensler RW, Reedy MC, Hofmann W, King HA. Structure and paramyosin content of tarantula thick filaments. J Cell Biol. 1983;97:186–195. doi: 10.1083/jcb.97.1.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elliott A, Offer G. Shape and flexibility of the myosin molecule. J Mol Biol. 1978;123:505–519. doi: 10.1016/0022-2836(78)90204-8. [DOI] [PubMed] [Google Scholar]
- 41.Walker M, Knight P, Trinick J. Negative staining of myosin molecules. J Mol Biol. 1985;184:535–542. doi: 10.1016/0022-2836(85)90300-6. [DOI] [PubMed] [Google Scholar]
- 42.Knight PJ. Dynamic behaviour of the head-tail junction of myosin. J Mol Biol. 1996;255:269–274. doi: 10.1006/jmbi.1996.0022. [DOI] [PubMed] [Google Scholar]
- 43.Levine RJ, Elfvin M, Dewey MM, Walcott B. Paramyosin in invertebrate muscles. II Content in relation to structure and function. J Cell Biol. 1976;71:273–279. doi: 10.1083/jcb.71.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kendrick-Jones J, Cohen C, Szent-Gyorgyi AG, Longley W. Paramyosin: molecular length and assembly. Science. 1969;163:1196–1198. doi: 10.1126/science.163.3872.1196. [DOI] [PubMed] [Google Scholar]
- 45.Hidalgo C, Padron R, Horowitz R, Zhao FQ, Craig R. Purification of native myosin filaments from muscle. Biophys J. 2001;81:2817–2826. doi: 10.1016/S0006-3495(01)75923-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chaponnier C, Janmey PA, Yin HL. The actin filament-severing domain of plasma gelsolin. J Cell Biol. 1986;103:1473–1481. doi: 10.1083/jcb.103.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 48.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]