SUMMARY
Hematopoietic stem cells (HSC) reside within a specialized niche where interactions with vasculature, osteoblasts and stromal components regulate their self-renewal and differentiation. Little is known about bone marrow niche formation or the role of its cellular components in HSC development; therefore, we established the timing of murine fetal long bone vascularization and ossification relative to the onset of HSC activity. Adult-repopulating HSC emerged at E16.5, coincident with marrow vascularization, and were contained within the c-Kit+Sca-1+Lin− (KSL) population. We used Osterix-null (Osx−/−) mice that form vascularized marrow, but lack osteolineage cells to dissect the role(s) of these cellular components in HSC development. Osx−/− fetal bone marrow cells formed multi-lineage colonies in vitro, but were hyper-proliferative and failed to home to and/or engraft transplant recipients. Thus, in developing bone marrow, the vasculature can sustain multi-lineage progenitors, but interactions with osteolineage cells are needed to regulate LT-HSC proliferation and potential.
INTRODUCTION
Hematopoietic stem cells (HSC) provide a lifelong blood supply and this critical function relies on balanced regulation of self-renewal and differentiation provided within a specialized microenvironment (niche). Bone marrow serves as the main HSC niche in vertebrates after birth; however, during embryogenesis, HSC emerge and/or migrate through different tissues (reviewed in (Coskun and Hirschi, 2010)). In mice, multi-lineage hematopoietic progenitors first arise within the extraembryonic yolk sac at ~E8.25 (Palis et al., 1999). The placenta and embryonic aorta-gonad-mesonephros (AGM) region exhibit de novo HSC potential at ~E10 (Gekas et al., 2005; Medvinsky and Dzierzak, 1996; Ottersbach and Dzierzak, 2005) and at E11.0, HSC reside within fetal liver (Morrison et al., 1995). Finally, HSC home to and/or emerge within fetal bone marrow shortly before birth, where they reside postnatally.
One common feature of all hematopoietic sites is that multi-lineage hematopoietic stem/progenitor cells are closely associated with blood vessels; in fact, they arise from specialized (hemogenic) vascular endothelial cells in the yolk sac, AGM and placenta (reviewed in (Hirschi, 2012)). Even in adult bone marrow, HSC are closely associated with sinusoidal endothelial cells and perivascular cells in the trabecular regions of long bones (Ding et al., 2012; Kiel et al., 2005; Nombela-Arrieta et al., 2013), which regulate HSC survival and proliferation (Garrett and Emerson, 2009). Furthermore, mesenchymal cells around arterioles have been shown to regulate HSC quiescence (Kunisaki et al., 2013). HSC within adult bone marrow are also closely associated with osteoblasts (Calvi et al., 2003; Zhang et al., 2003), which are unique to the bone marrow microenvironment. The extent to which the vasculature and osteoblasts are developed within fetal bone at the onset of HSC activity, and their relative contribution to the regulation of HSC phenotype, proliferation and function are investigated herein.
We first established the time course of vascularization and ossification of murine fetal long bones (femur), relative to the onset of HSC activity, and assessed the phenotype of fetal bone marrow HSC. We found that adult repopulating HSC were first present at E16.5, coincident with vascular perfusion, and were contained within the c-Kit+Sca-1+Lin− (KSL) population. Collagen type I, alpha 1 (Col1a1)-expressing mature osteoblasts were also present within fetal bone at E16.5, although predominantly localized within the periosteum. To discern the roles of vasculature vs. osteolineage cells in the regulation of fetal bone marrow HSC function, we used Osx−/− mutants that form vascularized marrow but lack osteoblasts and osteolineage cells. Fetal bone marrow cells from Osx−/− mutants formed multi-lineage hematopoietic colonies in vitro. However, KSL cells isolated from Osx−/− fetal bone marrow exhibited hyper-proliferation and dysregulation of cell cycle genes, and failed to engraft transplant recipients. In contrast, fetal liver cells from Osx−/− mutants exhibited higher engraftment than wild type littermates. In addition, total blood cell counts in Osx−/− mutants were normal, suggesting that Osx−/− mutants do not exhibit a defect in definitive hematopoiesis, per se, but rather a defective fetal bone marrow microenvironment that cannot support the development or maintenance of LT-HSC. Further elucidating the molecular role of cellular niche components in HSC development should provide insights needed to modulate adult hematopoiesis, and enable recapitulation of a niche microenvironment ex vivo to sustain HSC for clinical therapies.
RESULTS
Time Course of Fetal Bone Marrow Niche Development
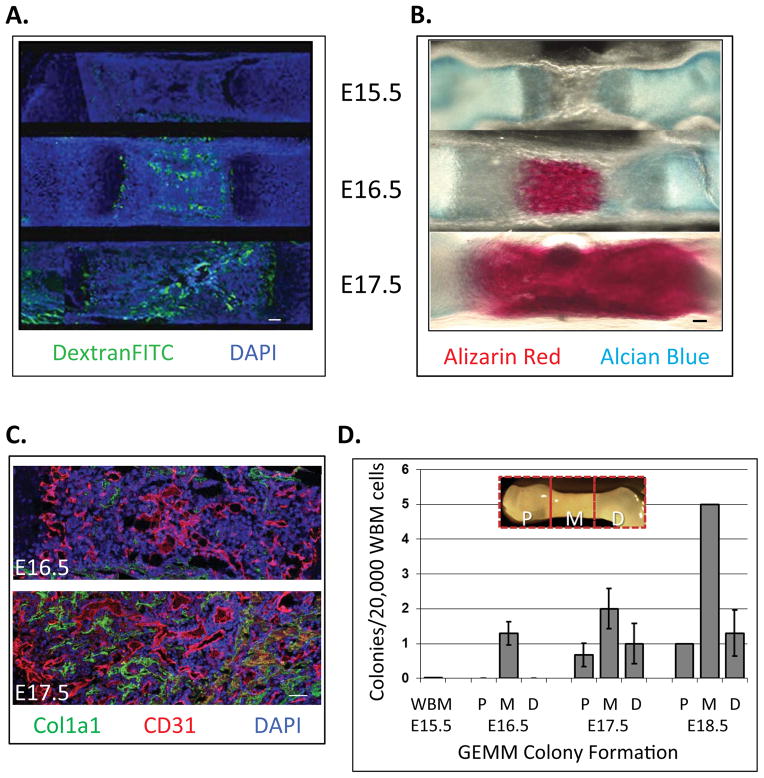
To define the formation of the fetal bone marrow niche and begin to dissect the effects of its cellular components on HSC phenotype and function, we first established the time course of vascularization of developing femurs of C57BL/6J mice at embryonic day (E) 15.5 through E17.5 via intravenous Dextran-FITC injection (Kienstra et al., 2007). We found that the cartilagenous bone templates were avascular at E15.5 (Figure 1A) and exhibited perfusion in the periosteum region and epiphyseal plate at E16 (Figure S1A). The middle, marrow region of fetal long bone was not perfused until E16.5 (Figure 1A). The functional vasculature then extended bidirectionally away from the marrow center, and at E17.5 most of the bone marrow cavity was vascularized.
Figure 1. Fetal bone is vascularized within the middle region at E16.5 where hematopoietic stem/progenitor cell (HSPC) activity is initially detected.
A) Mouse E15.5–17.5 fetuses were injected with 100 μL Dextran-FITC (70 kDa) via superficial temporal vein to reveal functional blood vessels. Fetal femurs were avascular at E15.5, but perfused by E16.5, specifically in the middle regions (Longitudional bone cross section, Green: Dextran-FITC, Blue: nuclear DAPI. Bar = 100 μm). B) Composition of fetal bone was revealed via alizarin red (mineralized tissue) and alcian blue (cartilage) staining. Calcification was apparent by E16.5 in the middle regions, and present throughout the tissue by E17.5. C) At E16.5, Col1a1-positive mature osteoblasts were detected at the periosteum, and throughout the bone at E17.5 (Bar = 100 μm). D) HSPC activity was first detected at E16.5 in the middle regions of bone. Multi-lineage colonies containing granulocyte, erythrocyte, monocyte, megakaryocyte (CFU-GEMM) were scored from 20,000 cells/well for each embryonic time points by Methocult® assay (Data represent mean ± SEM, N=3).
To establish the time course of calcification/osteogenesis of developing long bones, relative to vascular perfusion, we used alcian blue and alizarin red stains to distinguish cartilage and calcified bone, respectively (Inouye, 1976). At E15.5 and E16, fetal femurs were composed only of cartilage (Figure 1B and Figure S1B, respectively; light blue staining). Calcification was apparent by E16.5 in the middle regions (Figure 1B, dark red staining), and present throughout the tissue by E17.5. Thus, the pattern of calcification over time paralleled that of the forming vascular network. We next evaluated the presence of osteoblasts within the femurs, and localized them relative to the developing endothelium via co-immunofluorescence using antibodies against Collagen type I, alpha 1 (Col1a1, osteoblasts) and CD31 (endothelial cells). Osteoblasts were not present at E15.5 (not shown), but localized to the periosteum at E16.5 (Figure 1C), and throughout the bone marrow cavity and periosteum at E17.5 (Figure 1C).
HSC Activity Localizes to Vascularized Regions of Fetal Bone
We next defined the temporal and spatial emergence of HSC activity within fetal long bones. Femurs from E15.5–17.5 fetuses were dissected into three anatomical regions: proximal (P), middle (M) and distal (D) (Figure 1D, insert). Single cell suspensions obtained from each tissue were subjected to an in vitro hematopoietic (Methocult®) assay in which the formation of colonies containing multiple blood cell lineages including granulocytes, erythrocytes, monocytes and megakaryocytes (CFU-GEMM) indicates the presence of multi-lineage hematopoietic stem/progenitor cells (HSPC). The first fetal bone marrow HSPC activity was detected at E16.5 (Figure 1D), and restricted to the vascularized (middle) region of the developing bones (Figure 1A). At E17.5, CFU-GEMM colony-forming HSPC were detected throughout the bone tissue (Figure 1D). Thus, unlike adult HSC that reside mainly within trabecular (proximal/distal) regions of bone, fetal bone HSPC are localized to the central (middle), vascularized marrow.
Defining the Phenotype of Fetal Bone Marrow HSC
To define the phenotype of HSPC in fetal bone marrow, we used methods previously applied to adult mouse bone marrow: Hoechst dye exclusion to identify side population (SP) cells (Goodell et al., 1996); and isolation based on expression of c-Kit and Sca-1, and absence of blood lineage markers (KSL population) (Morrison and Weissman, 1994). Mac-1 antibodies were excluded from lineage cocktail since fetal HSPC express Mac-1 (Morrison et al., 1995). Unlike in adult bone marrow, SP cells were not detectable within fetal bone marrow at E16.5 and E17.5 but were present at E18.5 onward when evaluating the same number of whole bone marrow (WBM) cells at each time point (Figure S2A). In contrast, KSL cells were present within fetal bone marrow beginning at E16.5, which we found to be the onset of HSPC activity (Figure S2B, C). Furthermore, CFU-GEMM colony-forming activity was restricted to KSL cells at all stages of bone marrow development (Figure 2A). Consistent with this, cells co-expressing c-Kit and Sca-1 were localized to the central region of fetal bone marrow that is highly enriched for HSPC activity (Figure S2D). We further evaluated the phenotype of fetal bone marrow KSL cells at E17.5, and found that 19.63% ± 1.01% of KSL cells were CD150+CD48−, which is thought to be the phenotype of LT-HSC within adult bone marrow (Yilmaz et al., 2006).
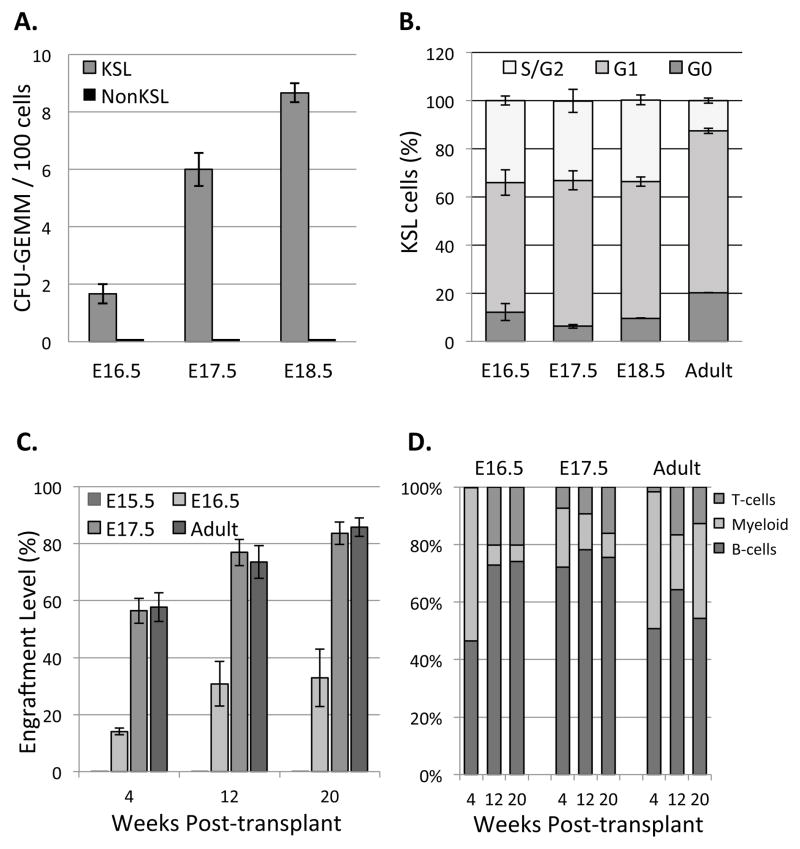
Figure 2. Fetal bone marrow hematopoietic activity is restricted to the KSL population, which is more actively proliferating than adult bone marrow KSL and gives rise to all blood lineages.
A) 100 KSL and 1000 NonKSL cells from fetal bone marrow were sorted and cultured in Methocult®; only KSL cells exhibited multi-lineage colony forming activity (Data represent mean ± SEM, N=3; p=0.007 (E16.5), p=0.0005 (E17.5) and p=0.00001 (E18.5)). B) Cell cycle distribution of the fetal and adult bone marrow KSL cells was revealed using PyroninY/Hoechst dyes and flow cytometry. KSL cells were more actively proliferating in fetal vs. adult bone marrow (Data represent mean ± SEM, N=3). C) Engraftment levels were determined at 4, 12 and 20 weeks post-transplantation by analyzing peripheral blood CD45.2 (donor) and CD45.1 (recipient) levels. LT-HSC were first present at E16.5 and showed engraftment levels up to 35% at 20 week post-transplant. By E17.5, engraftment level was comparable to adult bone marrow HSC. (Data represent mean ± SEM, N=3; Differences between E16.5 and either E17.5 or adult were significant; p=0.00085 (week 4), p=0.00326 (week 12) and p=0.00503 (week 20). D) Lineage analysis revealed fetal bone marrow cells (E16.5–17.5) gave rise to all blood lineages in vivo. (Data represent mean ± SEM, N=3).
Fetal Bone Marrow KSL Cells are Highly Proliferative
Although HSC within adult bone marrow are largely quiescent, HSC within fetal liver are more actively proliferating (Bowie et al., 2006). To compare the proliferative status of fetal and adult bone marrow HSC, we isolated KSL cells from fetal and adult femurs, stained them with Hoechst and Pyronin Y to label DNA and RNA, respectively (Shapiro, 1981), and used flow cytometry to quantify the proportion in each cell cycle phase based on relative DNA and RNA content. We found that ~35% of fetal bone marrow KSL cells were in S/G2/M phase vs. ~13% of adult KSL cells (Figure 2B). In contrast, ~20% adult KSL cells were in G0, relative to ~10% fetal KSL cells, indicating KSL cells are more actively proliferating in fetal vs. adult bone marrow.
Adult Repopulating HSC are Present Within Fetal Bone Marrow at E16.5
Although we detected multi-lineage hematopoietic potential in E16.5 fetal bone marrow cells in vitro, we performed bone marrow transplantation studies to assess engraftment and blood cell generation in vivo, to confirm the onset of long-term (LT)-HSC activity. Isolation of fetal bone marrow cells, pooled from multiple litters, typically yields a few million cells of which we expect only 0.01% to be LT-HSC (Challen et al., 2009). Thus, we used unfractionated WBM for these time course studies, which were isolated from E15.5–17.5 fetal hindlimbs of CD45.2 donor and transplanted into lethally irradiated adult CD45.1 recipients (1×106 donor WBM cells + 2×105 CD45.1 adult WBM helper cells/recipient; n=3 per group). In vivo engraftment was first detected at E16.5, with levels of 14.1%, 30.8% and 32.8% at 4, 12 and 20 week post-transplantation, respectively (Figure 2C and Figure S3). Our data showed HSPC activity present within fetal bone marrow at E16.5, which is a day earlier than previously described (Christensen et al., 2004) and consistent with our in vitro colony-forming assays. Lineage distribution analysis of the engrafted cells confirmed that E16.5 fetal bone marrow contains LT-HSC that give rise to all blood lineages in adult recipients (Figure 2D and Figure S4). By E17.5, fetal WBM engraftment was comparable to adult bone marrow HSC.
Fetal Bone Marrow Fails to Support LT-HSC Function in the Absence of Osteoblasts
To discern the roles of the vasculature and osteolineage cells in supporting LT-HSC within fetal bone marrow, we used C57BL/6J Osx−/− mice that lack osteoblasts and osteoblast lineage cells and die perinatally (Nakashima et al., 2002). Osx−/− fetuses exhibited abnormal skeletal development (Figure 3A, E17.5 hindlimbs shown), but had normal marrow that was vascularized similarly to wild type littermates (Figure 3A). Osx−/− fetal marrow also contained an equal proportion of KSL cells (Figure S5A), which exhibited similar apoptosis levels (Figure S5B), compared to wild type littermates.
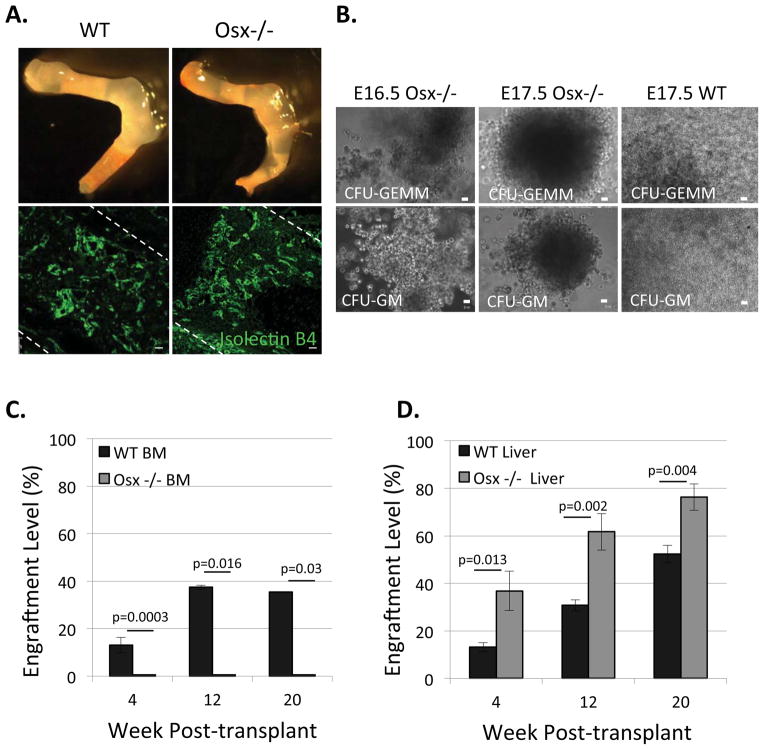
Figure 3. Osx−/− fetal bone marrow cells form multi-lineage CFU-GEMM colonies in vitro, but fail to repopulate transplanted recipients.
A) Osx−/− fetal long bones lack osteoblasts and have structural defects compared to wild type (WT), however, they have a normally vascularized marrow, as revealed by isolectin B4-FITC staining. Bar=20 μm B) Representative images of CFU-GEMM and CFU-GM colonies formed from E16.5 and E17.5 Osx−/− fetal bone marrow cells, similar to E17.5 WT fetal bone marrow cells. C) 400,000 WT and Osx−/− WBM cells at E17.5 were transplanted into sublethally irradiated neonates and engraftment levels were determined by analyzing recipients’ peripheral blood up to 20 week post-transplant; E17.5 Osx−/− WBM cells did not engraft. (Data represent mean ± SEM, N=3; p=0.0003 (week 4), p=0.016 (week12) and p=0.03 (week 20). D) WT and Osx−/− derived E17.5 fetal liver cells repopulated transplant recipients’ peripheral blood in vivo (Data represent mean ± SEM, N=3; p=0.013 (week 4), p=0.002 (week12) and p=0.004 (week 20).
We initially tested the hematopoietic function of E16.5 and E17.5 Osx−/− fetal marrow cells in vitro, and found that they formed multi-lineage CFU-GEMM colonies similar to wild type (Figure 3B). To test their LT-HSC activity in vivo, we first defined the minimum number of fetal bone marrow cells required to repopulate recipients at a reproducible engraftment level. We isolated E17.5 wild type (C57BL/6J) WBM cells, transplanted them into sublethally irradiated neonates (50,000–500,000 CD45.2 donor fetal cells + 200,000 CD45.1 adult WBM helper cells/recipient; n=4 per group). We used neonatal recipients in these studies because we could achieve more consistent engraftment with smaller numbers of donor cells, relative to adult recipients. We found that 100,000 E17.5 WT WBM cells/recipient yielded 5–10% engraftment at 4-week post-transplantation (Figure S6A). We were able to isolate and transplant ~400,000 WBM cells from each E17.5 Osx−/− fetus; thus, expecting >5–10% engraftment; however, Osx−/− WBM cells failed to repopulate recipients even 20-week post-transplantation (Figure 3C). In contrast, Osx−/− derived E17.5 fetal liver cells exhibited significantly higher engraftment levels than wild type fetal liver cells (Figure 3D). In addition, we did not observe differences in total blood cell counts E17.5 Osx−/− mutants compared to wild type littermates (Figure S6B, S6C). Thus, although Osx−/− mutants did not exhibit a global defect in definitive hematopoiesis, and Osx−/− fetal bone marrow cells exhibited HSPC activity in vitro, they failed to home to, or engraft within, transplant recipients.
Fetal Bone Marrow Cells Derived from Osx−/− Mutants Exhibit a Homing Defect
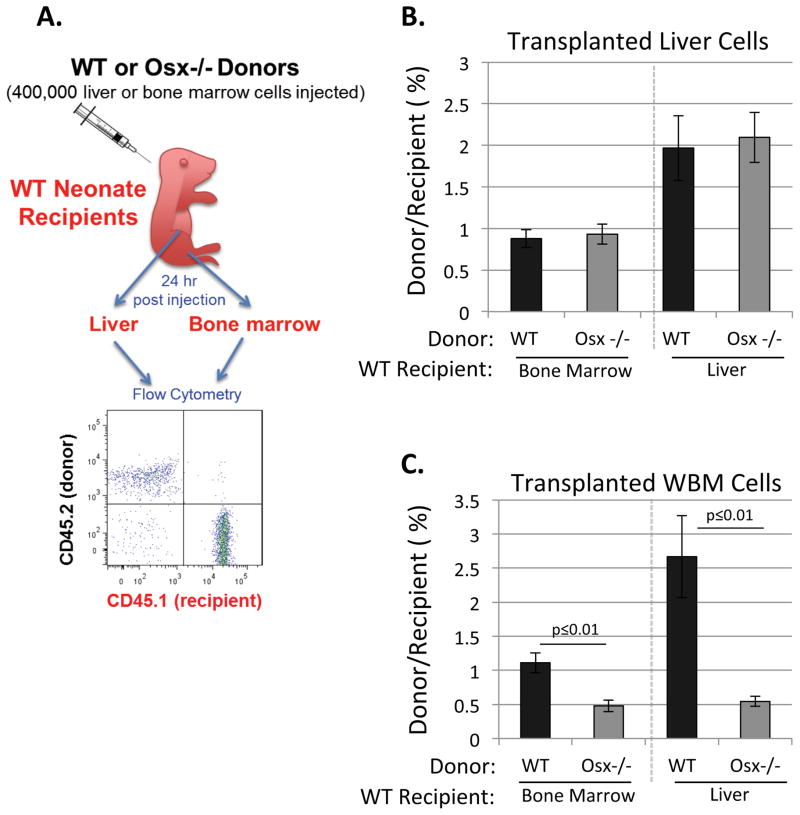
To evaluate the HSC homing ability of Osx−/− vs. wild type littermates, we isolated E17.5 fetal WBM and fetal liver cells from each group of embryos. As depicted in Figure 4A, 400,000 cells of each type (either WBM or liver cells from CD45.2 donor embryos) were then injected into P0-P2 CD45.1 neonatal recipients via the superficial temporal vein. Recipients were euthanized after 24 hr, then bone marrow and liver cells were collected immediately. All cells were incubated with CD45.1-FITC (recipient) and CD45.2-APC (donor) antibodies and analyzed by flow cytometry to measure the ratio of donor:recipient cells in bone marrow and liver. Cells derived from Osx−/− fetal liver showed normal homing to recipient liver and bone marrow (Figure 4B); however, Osx−/− fetal bone marrow cells exhibited significantly reduced homing ability (Figure 4C). Thus, osteoblasts or osteoblast lineage cells appear to be required within fetal bone marrow to maintain the homing ability of LT-HSC.
Figure 4. Fetal Bone Marrow Cells Derived from Osx−/− Mutants Exhibit a Homing Defect.
A) P0-P2 recipient neonates were euthanized 24 h post-transplantation and bone marrow and liver cells were analyzed with flow cytometry for donor/recipient cell ratios. B) Fetal liver cells from E17.5 WT and Osx−/− mutants exhibited similar homing to liver and bone marrow of WT recipients. C) In contrast, fetal WBM cells derived from E17.5 Osx−/− mutants showed significantly reduced homing compared to WT (Data represent mean ± SEM, N=3).
Dysregulation of the Fetal Bone Marrow Niche and HSPC in Osterix-null Mutants
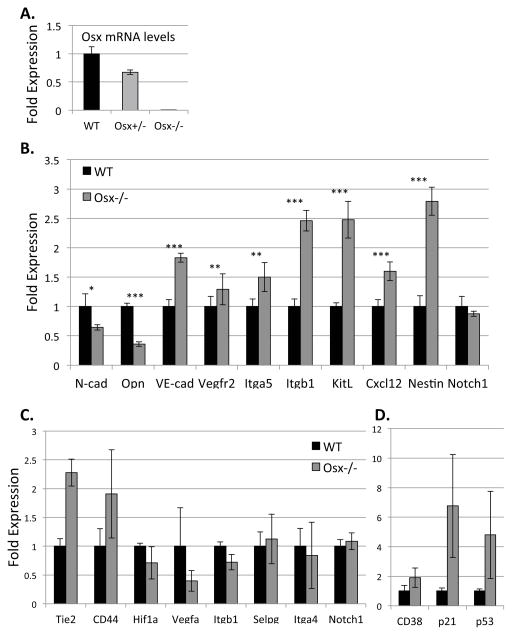
To investigate the impact of osteoblast deficiency on the fetal bone marrow microenvironment, we isolated total RNA from E17.5 Osx−/−, Osx+/− and wild type WBM cells and compared their relative expression of niche regulatory and cell type-specific genes (Table S1). As expected, Osx gene expression was ~40% lower in Osx+/− WBM and absent in Osx−/− WBM (Figure 5A); thus, validating our qPCR analysis. Osx−/− WBM also exhibited significantly decreased expression of osteoblast-derived osteopontin (opn) and N-cadherin (cdh2) (Figure 5B). In contrast, expression of endothelial-associated genes VE-cadherin (cdh5), vascular endothelial growth factor receptor 2 (vegfr2), and integrin subunits α5 (itga5) and β1 (itgb1) were significantly increased in Osx−/− WBM (Figure 5B). Nestin, which is expressed by proliferating endothelial cells (Suzuki et al., 2010), and endothelial-derived Cxcl12 (SDF-1) and Kit ligand (KitL or SCF), which regulate HSC migration and homing (Christensen et al., 2004), were also significantly increased in Osx−/− WBM, suggesting increased representation of vascular cells in osterix-null fetal bone marrow, which is consistent with flow cytometric analysis of the cellular composition of Osx−/− fetal bone marrow, relative to WT littermates (Figure S6D).
Figure 5. Gene expression studies revealed that genes specific to the bone marrow niche and HSPC are dysregulated in osterix mutants.
A) Gene expression analysis (via qPCR) was performed on E17.5 WT and Osx−/− WBM cells. Osx expression was ~40% decreased in Osx+/− fetuses and absent in Osx−/− mutants. B) N-cadherin (cdh2) and osteopontin (opn) were significantly downregulated in Osx−/− mutants; whereas, Cxcl12, kitL (SCF), nestin, vascular endothelial cell adhesion protein, VE-cadherin (cdh5), vascular endothelial growth factor receptor 2 (vegfr2), integrin subunits β1 (itgb1) and α5 (itga5) were significantly upregulated compared to WT (Asterix (*), (**), and (***) denotes p≤0.01, p≤0.001 and p≤0.0001 respectively). β-actin gene expression was used as an internal control. For each gene of interest, triplicates from four different cDNA samples were analyzed (Data represent mean ± SEM, N=4, triplicates for each sample). C–D) KSL cells were isolated from the hindlimbs of E17.5 WT and Osx−/− mutants and analyzed in 10-cell pools. In Osx−/− KSL cells, the expression of CD38, p21, p53, as well as Tie2 and CD44, trended toward higher than wild type littermates; whereas, expression of Hif1α and VegfA trended toward lower. β-actin served as an internal control. Statistical analyses were done using the two-tailed unpaired student t-test (Data represent mean ± SEM, N=4; p=0.30 (CD38), p=0.15 (p21), p=0.30 (p53), p=0.17 (Tie-2), p=0.28 (CD44), p=0.44 (Hif1a), p=0.33 (Vegfa), p=0.5 (itgb1), p=0.79 (Selpg), p=0.42 (itga4), p=0.29 (notch1).
To determine the effects of osterix-deficiency on the phenotype of HSPC, specifically, we sorted fetal bone marrow KSL cells from the hindlimbs and forelimbs of E17.5 Osx−/− and wild type littermates, and pooled them into 10-cell samples, the minimum number that could be reproducibly analyzed. Total RNA was isolated and analyzed via qPCR. We found that KSL cells isolated from Osx−/− fetal bone marrow exhibited a trend toward downregulation of β1 integrin (itgb1, Figure 5C), which plays a significant role in HSC retention in hematopoietic tissues (Potocnik et al., 2000). Hypoxia-activated gene Hif1α and one of its downstream targets Vegf-A, which is known to regulate HSC survival in adult bone marrow (Takubo et al., 2010), were also reduced in KSL cells isolated from Osx−/− fetal bone marrow (Figure 5C). In contrast, we found a trend toward upregulation of genes known to regulate HSC proliferation, including p21, p53 and CD38, in Osx−/− mutants (Figure 5D). These expression data are consistent with the observed hyper-proliferative phenotype of KSL cells isolated from Osx−/− fetal bone marrow that exhibited 30% less cells in G0 phase and 2.2-fold more cells in S/G2/M, compared to wild type littermates (Figure S5C). Collectively, our data suggest that changes in cellular composition and gene expression within the fetal bone marrow microenvironment in Osterix-null mutants leads to impaired ability to sustain LT-HSC.
DISCUSSION
The adult HSC niche has been extensively studied and specific cellular components, including osteoblasts (Calvi et al., 2003; Zhang et al., 2003), vascular/perivascular cells (Ding et al., 2012; Kiel et al., 2005; Kunisaki et al., 2013; Mendez-Ferrer et al., 2010; Sugiyama et al., 2006), nestin-expressing MSC (Mendez-Ferrer et al., 2010), adipocytes (DiMascio et al., 2007), and neurons of the sympathetic nervous system (Katayama et al., 2006), have been identified as important regulators of HSC maintenance and function. However, the formation of the bone marrow niche during fetal development has not been studied, and the impact of specific niche components on fetal bone marrow HSC phenotype, proliferation and function has not been defined.
We investigated these issues herein and found that fetal long bones were vascularized, and LT-HSC emerged, coincidentally at E16.5. Furthermore, fetal bone marrow HSC were contained within the KSL population, and localized specifically to the vascularized regions of fetal long bones. HSC within adult bone marrow are also closely associated with vascular endothelial cells (Kiel et al., 2005; Nombela-Arrieta et al., 2013), and the sinusoidal vascular niche is thought to support active proliferation of HSC (Winkler et al., 2012). This is consistent with our observations in fetal bone marrow; wherein, vascular-associated HSC were more proliferative than adult HSC. The subset of adult HSC that are more quiescent are thought to be associated with osteoblasts (Wilson et al., 2007); although the osteoblastic and vascular niches may not be entirely separate entities (Ellis et al., 2009). We found osteoblasts present within fetal bone at the onset of HSC activity (E16.5); thus, we aimed to dissect the differential roles of vascular and osteolineage niche components on HSC development using osterix-null mice that exhibit normal marrow vascularization but lack osteoblasts and osteolineage cells.
We observed that fetal bone marrow cells from Osx−/− mutants exhibited multi-lineage colony-forming activity in vitro, but were hyper-proliferative relative to wild type littermates, and failed to repopulate transplant recipients. In contrast, fetal liver cells isolated from Osx−/− successfully repopulated recipients, and blood cell counts were similar between Osx−/− and wild type littermates, demonstrating that there is not a global defect in HSC and definitive hematopoiesis in Osx−/− mutants, but rather a specific defect(s) in the bone marrow microenvironment that fails to support the generation and/or maintenance of LT-HSC. These results are consistent with previous studies of ectopic bone formation that suggest osteoblasts/progenitors are necessary, but not sufficient, for HSC niche formation (Chan et al., 2009).
To gain better understanding of the impact of osterix deficiency on the bone marrow microenvironment, we performed expression analysis of Osx−/− and wild type fetal WBM cells. We found that the Osx−/− mutants exhibited significantly decreased N-cadherin (cdh2) and increased VE-cadherin (cdh5) expression. Increased association of HSC with endothelial cells via VE-cadherin, and concomitant lack of interaction with osteoblasts would be consistent with the increased HSPC proliferation that we observed in the Osx−/− mutants, given that N-cadherin knockdown in HSC accelerates their division and reduces their engraftment (Hosokawa et al., 2010).
We also compared the gene expression of KSL cells, specifically, from Osx−/− and wild type littermates. Osx−/− KSL cells exhibited a trend toward upregulation of Tie2, CD38 and CD44 genes, all of which facilitate HSC adhesion/maintenance within their niche, and regulate their function (Arai et al., 2004; Higuchi et al., 2003; Williams et al., 2013). For example, CD38lo/− HSC engraft better than CD38+ HSC (Higuchi et al., 2003); thus, higher levels of CD38 in Osx−/− HSPC may contribute to their lack of engraftment. p21 and p53 levels were also somewhat higher in KSL cells isolated from Osx−/− fetal bone marrow, which may contribute to their disrupted cell cycle and lack of stem cell function. In fact, constitutive expression of p53 results in exhaustion of the adult HSC pool and premature aging (Tyner et al., 2002).
Collectively, our data demonstrate that osterix-deficiency significantly disrupts development of the fetal bone marrow microenvironment. Osterix-Cre labeled cells have been shown to give rise to osteoblasts and other stromal components in adult bone marrow (Chen et al., 2014; Liu et al., 2013). Other osterix-lineage tracing studies revealed that, although osterix-expressing cells predominantly give rise to osteoblasts in the adult, they contribute to the development of stromal components within bone marrow before and soon after birth (Mizoguchi et al., 2014). Therefore, in Osx−/− fetal bone marrow, critical stromal components, including nestin-expressing MSC (Mendez-Ferrer et al., 2010), CXCL12-abundant reticular cells (Sugiyama et al., 2006), adipocytes (DiMascio et al., 2007) and even neurons of the sympathetic nervous system (Katayama et al., 2006), may be poorly developed, contributing to the defects in HSC cell cycle control and engraftment potential that we observed. In fact, phenotypically identical HSC (Lin-CD41-Sca1+c-Kit+CD48−CD150+) isolated from the endosteal vs. central marrow regions of adult bone, where there are significant differences in stromal composition, exhibit different engraftment abilities (Grassinger et al., 2010; Guezguez et al., 2013; Oguro et al., 2013).
Further studies are clearly needed to define the molecular mechanisms by which specific niche components regulate the establishment and maintenance of LT-HSC during development. However, thus far, our data suggest that the vasculature maintains HSPC during the development of osteolineage cells, which supports the HSC-niche crosstalk model. That is, HSC are thought to promote the differentiation osteolineage cells (Jung et al., 2008), which then provide signals needed to induce quiescence and enable the development and maintenance of LT-HSC (Mendez-Ferrer et al., 2010).
Our study provides new insights into the establishment of the bone marrow niche during fetal development. Continuing to study this process in genetically malleable models, in parallel with studies of the regulation of hematopoiesis in the adult, will provide collective insights needed to optimize cell therapies, as well as generate and maintain HSC outside the body, for the treatment of hematopoietic disorders.
EXPERIMENTAL PROCEDURES
Fetal Bone Marrow Cell Isolation
Animal experiments were approved by the Institutional Animal Care and Use Committees (IACUC) of Baylor College of Medicine and Yale University. Osx+/− mice (Nakashima et al., 2002) were obtained from Dr. Benoit de Crombrugghe, University of Texas MD Anderson Cancer Center. Mouse breeding pairs (C57BL/6 and C57BL/6 Osx+/−) were monitored daily early in the morning for vaginal plugs; plugged mice were labeled as embryonic day (E) 0.5. Embryos from E14.5–18.5 and postnatal pups were extracted and dissected. Fetal liver and long bones were collected in cold HBSS+ medium (HBSS, 10mM HEPES, 2% FBS) and transferred into DMEM+ medium (DMEM, 10mM HEPES, 2% FBS). Fetal bone marrow and liver tissues were dissected into small pieces in DMEM+, and single cell suspensions were obtained via gentle passage through 18-G needles and filtration through 70-μm (bone marrow) or 40μm (liver) nylon strainers (BD Falcon).
Dextran-FITC Injections
Live embryos were injected with 100 μL Dextran-FITC (Sigma) via superficial temporal vein (Kienstra et al., 2007) and after 15 min, embryos were decapitated and fixed in 4% PFA overnight at 4°C. Following PBS washes; embryos were transferred to 30% sucrose, and then embedded in OCT (Sakura Finetek USA, Torrance CA). 10 μm thick tissue sections were obtained using a cryostat (Shandon). Confocal images were captured using a Zeiss LSM 510 META confocal inverted microscope.
Fetal Bone Histology
Fetal long bones (femurs) were dehydrated and incubated in staining solution (0.3% Alcian blue in 70% ethanol: 0.1% Alizarin red in 95% ethanol: Acetic acid: 70% Ethanol in 1:1:1:17 ratio) for 3 days at 37°C, as described (Inouye, 1976). Images were captured using a Zeiss Axiovert 200M microscope.
Immunohistochemistry
Tissues frozen in OCT were sectioned as described above. Immunohistochemistry was performed using primary mouse antibodies at 1:100 (rat anti–mouse c-Kit (eBiosciences), rat anti–mouse sca-1 (eBiosciences), goat anti-mouse CD31 (R&D System) and rabbit anti-mouse Col1A1 (Millipore antibodies), incubated overnight at 4°C. Sections were incubated for 1 hr at RT with secondary antibodies (Molecular Probes, Carlsbad, CA) diluted 1:1000 then mounted in VectaShield with DAPI (Vector Laboratories, Burlingame, CA) for imaging and analysis. To visualize bone marrow vasculature, cryosections were also stained by using isolectin B4-FITC staining solution (Sigma-Aldrich) diluted in PBLec Buffer (1mM CaCl2, 1mM MgCl2, 0.1mM MnCl2, 1% tritonX100 in PBS). Images were captured with a Zeiss AxioVert 200M microscope and AxioCamMRM camera and AxioVision software (Carl Zeiss MicroImaging, Thornwood, NY).
Fetal Bone Marrow Cell Phenotyping
For KSL analysis, freshly isolated fetal bone marrow cells were suspended in ice-cold DMEM+ at 107 cells/mL, and stained for 15 min on ice with the anti-c-Kit-PE, anti-Sca-1-FITC, and PE-Cy5 conjugated lineage marker antibodies (CD4, CD8, B220, TER119, and Gr-1) (eBiosciences). Samples were then washed with 10X volume of HBSS+ and centrifuged at 2000 rpm for 8 min at 4°C. Cell pellets were resuspended in DMEM+ with propidium iodide (PI: 2 μg/mL) and samples were immediately analyzed/sorted by flow cytometry (BD FACSAria).
For SP analysis, fetal bone marrow cells were suspended at 106 cells/mL in DMEM with 2% FBS/10 mM HEPES, then stained with 5μg/mL Hoechst 33342 (Sigma-Aldrich) for 90 min at 37°C, as described (Goodell, 2005). After centrifugation, the cell pellets were resuspended in cold HBSS+ (HBSS/2% FBS/10 mM HEPES) with PI (2 μg/mL). Analysis and collection of SP cells were performed on a triple-laser MoFlo instrument (Cytomation Inc., Fort Collins, CO). All phenotyping experiments were repeated at least three times; data represent mean +/− SEM of all data collected.
Methylcellulose Hematopoietic Colony-Forming Assay
For in vitro progenitor activity assay, 20,000 WBM cells were isolated from three distinct anatomical regions of the bone (proximal, middle, distal) from E15.5 to E18.5 fetal femurs and cultured on MethoCult® GFM3434 culture medium (StemCell Technologies, Vancouver, BC) in 35 mm petriplates. For KSL vs. Non KSL HSPC activity studies, 100 KSL cells or 1000 nonKSL cells were sorted directly into 200 μL of MethoCult® GFM3434 culture medium (StemCell Technologies, Vancouver, BC) in 48-well culture plates. Plates were incubated at 37°C with 5% CO2, and scored for colony formation at days 7 and 14. Representative colony images were taken, as described above.
Fetal Bone Marrow Transplantation
To evaluate fetal HSC function in vivo, we performed bone marrow transplantations into irradiated (lethal or sublethal) recipients (adult or P0-P3 neonates). Embryos were obtained from CD45.2 breeders. 1×106 WBM cells were isolated as described above and resuspended in in PBS with 2% FBS. Recipients received a double dose of 5 Gy irradiation with 2–4 hour interval (lethal dose) (XRAD Cs137 irradiator, J.L. Shepherd and Assoc, Inc). Adult recipients were injected with donor cells via tail vein. For neonatal injections, pups were sedated using mild hypothermia and donor-derived CD45.2 (~400.000 WBM) were injected in 100 μL PBS with 2% FBS via the superficial temporal vein. All transplantations were performed with adult WBM helper cells (200,000 CD45.1 cells). Engraftment levels were tested at 4, 12 and 20 weeks post-transplantation. All transplantation studies were repeated at least three times; data represent mean +/− SEM of all data collected.
Engraftment Analysis
Mice were sedated with Avertin® (Sigma Aldrich) via i.p. injection. Peripheral blood was withdrawn via retro-orbital bleeding and collected into tubes containing PBS with 2% Dextran T500, EDTA (3mg/mL) and heparin (1 unit/mL). After 20 min, the top phase was transferred to new tubes and centrifuged at 2000 rpm for 8 min at 4°C. The pellet was resuspended in PharM Lyse (Pharmingen) buffer diluted in H20 (1:10) for 5 min at RT for red blood cell lysis. After centrifugation, the cell pellets were washed in PBS and resuspended and stained anti-CD45.1 FITC, anti-CD45.2 APC, and lineage markers (PacB conjugated anti-CD4, anti-CD8, anti-CD45R and PE conjugated anti-Mac1, anti-Gr1, anti-CD45R, BD Pharmingen) diluted in PBS with 2% FBS (1:100) and incubated on ice for 15 min. After washing, samples were resuspended in HBSS+ with PI and analyzed with flow cytometry. Data represent the mean +/− SEM of triplicate samples collected at each time point in at least three independent experiments.
Cell Cycle Analysis
Fetal bone marrow cells were isolated as above and resuspended in 1×107 cell per mL in 10μg/mL Hoechst and 50μg/mL verapamil (Sigma-Aldrich). After 30 min, cells were stained with anti-c-Kit-APC-Cy7, anti-Sca-1-APC, and FITC conjugated lineage marker antibodies (CD4, CD8, B220, TER119, and Gr-1) (1:100 final concentration) and Pyronin Y at 1μg/mL for 10 min at 37°C. Following washing and resuspension, samples were kept on ice and immediately analyzed via flow cytometry. Data represent the mean ± SEM of triplicate samples.
Homing Assay
From E17.5 CD45.2 WT or Osx−/− fetuses, 400.000 WBM or fetal liver cells were isolated and injected into CD45.1 WT newborn (P0-P2) mice via the superficial temporal vein. 24 hr after cellular injections, the recipient neonates were euthanized, and their liver and bone marrow cells were collected immediately. Cells were incubated with anti-CD45.1-FITC and anti-CD45.2-APC antibodies (BD Pharmingen) diluted in PBS with 2% FBS (1:100) for 15 min on ice. Following washing and resuspension, samples were kept on ice and immediately analyzed via flow cytometry to determine the ratio of donor:recipient cells in bone marrow and liver tissues (Figure 4A).
Analysis of Gene Expression
Fetal bone marrow cells were isolated from the long bones of E17.5 Osx−/− and WT littermates. Total RNA was isolated using RNAeasy (Qiagen), and cDNA was synthesized using iScript cDNA synthesis kit (BioRad). qPCR reactions were performed using the Fast EVAGreen Supermix (BioRad) and analyzed using the CFX-96 Touch Real Time PCR Detection System (BioRad). Primers (Table S1) were designed using PrimerBank (Wang et al., 2012). Data represent the mean ± SEM of triplicate samples from at least three independent experiments.
Statistical Analysis
Data were evaluated using either the Student’s two-tail t-test or two-way ANOVA; p<0.05 was considered to be statistically significant.
Supplementary Material
Primers used for qPCR analysis. Gene accession number for each gene was obtained from NCBI database (http://www.ncbi.nlm.nih.gov/nuccore). Forward (F) and Reverse (R) primer sequences were obtained from PrimerBank database (http://pga.mgh.harvard.edu/primerbank).
Figure S1. A) Mouse E16 fetuses were injected with 100 μL Dextran-FITC (70 kDa) via the superficial temporal vein to reveal functional blood vessels within long bones. At E16, perfused vessels were detectable within periosteum and epiphyseal plate, but not within the middle marrow region of fetal long bone (Longitudinal femur cross section; dashed lines indicate bone edges. Green: Dextran-FITC, Blue: nuclear DAPI. Bar = 100 μm). B) At E16, fetal bone exhibited Alcian Blue (cartilage) staining, but no Alizarin Red (calcification) staining. (Light blue: Alcian Blue; Bar = 100 μm).
Figure S2. Related to Figure 1. A) Fetal bone marrow cells were stained with Hoechst dye to detect an SP population within fetal bone marrow; SP cells were not present in fetal bone marrow until E18.5. B) KSL cells were characterized by expression of c-Kit and Sca-1 proteins and lack of blood lineage protein expression (CD4, CD8, CD45R, Ter119, Gr-1 and Mac-1). Mac-1 was excluded from lineage cocktail for fetal bone marrow analysis since fetal HSC are positive for Mac-1. KSL cells were present in fetal bone marrow starting at E16.5, the onset of HSPC activity. C) Isotype controls were tested for each conjugated antibody. Single compensation controls and unstained controls were also performed. D) Femurs isolated from P0 embryos were fixed and stained with anti-c-Kit and anti-Sca-1 antibodies and with Alexa 594 and Alexa 488 conjugated secondary antibodies, respectively. Merged images show HSPC expressed both c-Kit and Sca-1 in the central bone marrow cavity. Bar=20 μm (insert: magnified view of a double positive HSPC). All flow cytometry studies were conducted at least three times (Data represent mean ± SEM).
Figure S3. Related to Figure 2. Fetal whole bone marrow cells isolated from femurs at E16.5 and onwards showed long term engraftment. Engraftment levels were determined at 4, 12 and 20 weeks after transplantation by analyzing peripheral blood donor/recipient ratio using anti-CD45.2 and CD45.1 antibodies respectively (Data represent mean ± SEM, N=3).
Figure S4. Related to Figure 2. Fetal whole bone marrow cells isolated from femurs at E16.5 and onward showed long term engraftment, and contribution to all three blood lineages in the recipients, similar to adult bone marrow cells. Donor derived blood cells (i.e. CD45.2 positive) were evaluated for their expression of lineage markers (CD4, CD8, B220, Mac1 and Gr1). (Data represent mean ± SEM, N =3).
Figure S5. Related to Figure 3. Properties of KSL cells in WT and Osx−/− fetal bone marrow at E17.5. A) The proportion of KSL cells within whole bone marrow of Osx−/− mutants was comparable to that of WT littermates. Absolute KSL cell numbers, in all long bones combined, from each E17.5 WT and Osx−/− fetus were 402.70 ± 43.59 and 426.86 ± 125.64, respectively. Data (mean ± SEM, N=3; p=0.86) were normalized based on the same number of events in each group. B) The AnnexinV/7AAD apoptosis assay revealed no significant difference (p=0.99) in the proportion of KSL cells undergoing apoptosis in E17.5 WT (6.98, 8.71 and 7.37% KSL cells; mean ± SEM = 7.69 ± 0.52%) and Osx−/− littermates (4.95, 15.50 and 2.70% KSL cells; mean ± SEM = 7.72 ± 3.94%). C) Osx−/− KSL cells exhibited a decreased proportion of cells in G0, and increased proportion of cells in G1 and S/G2/M phases, relative to WT KSL cells.
Figure S6. Related to Figure 3. A) Engraftment curve for fetal bone marrow cells. 50,000–500,000 WBM cells from E17.5 WT femurs were transplanted into sublethally irradiated neonate recipients. 100,000 WBM cells from E17.5 bone marrow were sufficient to detect the engraftment levels above 5% at 12 weeks post transplantation. (Data represent mean ± SEM, N =4). B–D) Cellular composition of bone marrow from E17.5 Osx−/− and WT littermates. There were no differences in total red blood cell (RBC; B) or total white blood cell (WBC; C) counts within bone marrow of E17.5 Osx−/− and WT littermates (Data represent normalized Mean ± SEM, N=4). Endothelial (CD31+CD45−; D) cell number was measured by flow cytometry and absolute percentage within WBM cells was compared (Data represent Mean ± SE, N=4). Total endothelial cell number tended to be higher in Osx−/− mutants compared to WT littermates.
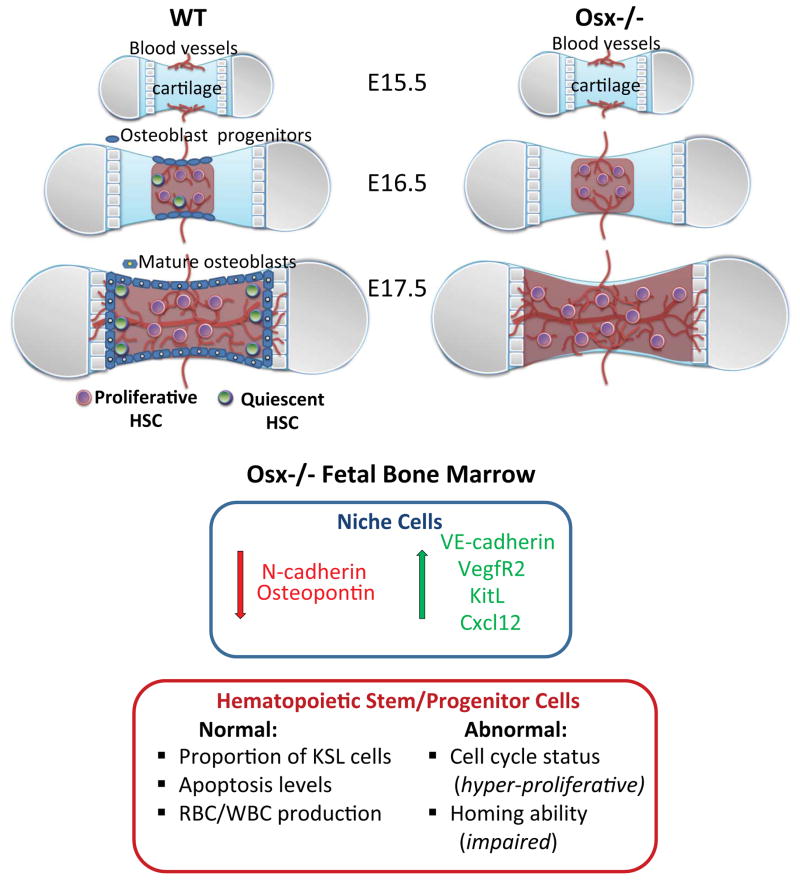
Figure 6. Working model.
In wild type (WT) fetal long bones (femur), adult repopulating HSC are first present at E16.5, coincident with vascular perfusion, localized to vascularized regions of bone, and are highly proliferative relative to adult HSC. Osteoblast progenitors are also present within fetal bone at this stage, but appear to be restricted to the periosteum. From E17.5 onward, HSC are also detected within the proximal and distal ends of bone that are fully vascularized and contain mature osteoblasts that are thought to regulate HSC quiescence. In Osx−/− fetal bone marrow, osteoblasts and osteolineage cells do not develop, and there is increased expression of vascular niche associated genes, including VE-cadherin, Vegfr2, KitL and cxcl12, as well as decreased expression of N-cadherin and osteopontin, which suggests increased representation of vascular endothelial cells in this microenvironment in osterix-null mutants. HSPC phenotype and function are also altered in the osteolineage cell-deficient microenvironment. Although KSL cells are present in equal numbers compared to WT, and exhibit similar apoptosis levels, KSL cells isolated from Osx−/− fetal bone marrow exhibited dysregulated cell cycle progression and defective homing ability. These data suggest that osteolineage cells in fetal bone marrow play a critical role in establishing and sustaining LT-HSC phenotype and function during development.
Acknowledgments
The authors thank Chris Threeton, Joel Sederstrom, and Amanda White for help with flow cytometry and Brittney Gibbons for suggestions during manuscript preparation. These studies were supported by NIH R01 grants HL077675, HL096360 and EB005173 to KKH.
Footnotes
Author Contributions
SC designed and performed the research, analyzed and interpreted the data, and wrote the manuscript. HC, HV, KH and NG assisted during in vivo transplant experiments, RT-PCR, cell sorting and neonate injections respectively. XZ and BD provided Osx+/− mice and advice on bone analysis. KKH provided intellectual input, funding and experimental oversight for all aspects of the project, as well as edited figures and manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arai F, Hirao A, Ohmura M, Sato H, Matsuoka S, Takubo K, Ito K, Koh GY, Suda T. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell. 2004;118:149–161. doi: 10.1016/j.cell.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Bowie MB, McKnight KD, Kent DG, McCaffrey L, Hoodless PA, Eaves CJ. Hematopoietic stem cells proliferate until after birth and show a reversible phase-specific engraftment defect. J Clin Invest. 2006;116:2808–2816. doi: 10.1172/JCI28310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, Martin RP, Schipani E, Divieti P, Bringhurst FR, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- Challen GA, Boles N, Lin KK, Goodell MA. Mouse hematopoietic stem cell identification and analysis. Cytometry A. 2009;75:14–24. doi: 10.1002/cyto.a.20674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Shi Y, Regan J, Karuppaiah K, Ornitz DM, Long F. Osx-Cre targets multiple cell types besides osteoblast lineage in postnatal mice. PLoS One. 2014;9:e85161. doi: 10.1371/journal.pone.0085161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen JL, Wright DE, Wagers AJ, Weissman IL. Circulation and chemotaxis of fetal hematopoietic stem cells. PLoS Biol. 2004;2:E75. doi: 10.1371/journal.pbio.0020075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coskun S, Hirschi KK. Establishment and regulation of the HSC niche: Roles of osteoblastic and vascular compartments. Birth Defects Res C Embryo Today. 2010;90:229–242. doi: 10.1002/bdrc.20194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMascio L, Voermans C, Uqoezwa M, Duncan A, Lu D, Wu J, Sankar U, Reya T. Identification of adiponectin as a novel hemopoietic stem cell growth factor. J Immunol. 2007;178:3511–3520. doi: 10.4049/jimmunol.178.6.3511. [DOI] [PubMed] [Google Scholar]
- Ding L, Saunders TL, Enikolopov G, Morrison SJ. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481:457–462. doi: 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis SL, Williams B, Asquith S, Bertoncello I, Nilsson SK. An innovative triple immunogold labeling method to investigate the hemopoietic stem cell niche in situ. Microsc Microanal. 2009;15:403–414. doi: 10.1017/S1431927609990924. [DOI] [PubMed] [Google Scholar]
- Garrett RW, Emerson SG. Bone and blood vessels: the hard and the soft of hematopoietic stem cell niches. Cell Stem Cell. 2009;4:503–506. doi: 10.1016/j.stem.2009.05.011. [DOI] [PubMed] [Google Scholar]
- Gekas C, Dieterlen-Lievre F, Orkin SH, Mikkola HK. The placenta is a niche for hematopoietic stem cells. Dev Cell. 2005;8:365–375. doi: 10.1016/j.devcel.2004.12.016. [DOI] [PubMed] [Google Scholar]
- Goodell MA. Stem cell identification and sorting using the Hoechst 33342 side population (SP) Curr Protoc Cytom. 2005;Chapter 9(Unit 9):18. doi: 10.1002/0471142956.cy0918s34. [DOI] [PubMed] [Google Scholar]
- Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassinger J, Haylock DN, Williams B, Olsen GH, Nilsson SK. Phenotypically identical hemopoietic stem cells isolated from different regions of bone marrow have different biologic potential. Blood. 2010;116:3185–3196. doi: 10.1182/blood-2009-12-260703. [DOI] [PubMed] [Google Scholar]
- Guezguez B, Campbell CJ, Boyd AL, Karanu F, Casado FL, Di Cresce C, Collins TJ, Shapovalova Z, Xenocostas A, Bhatia M. Regional localization within the bone marrow influences the functional capacity of human HSCs. Cell Stem Cell. 2013;13:175–189. doi: 10.1016/j.stem.2013.06.015. [DOI] [PubMed] [Google Scholar]
- Higuchi Y, Zeng H, Ogawa M. CD38 expression by hematopoietic stem cells of newborn and juvenile mice. Leukemia. 2003;17:171–174. doi: 10.1038/sj.leu.2402785. [DOI] [PubMed] [Google Scholar]
- Hirschi KK. Hemogenic endothelium during development and beyond. Blood. 2012;119:4823–4827. doi: 10.1182/blood-2011-12-353466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa K, Arai F, Yoshihara H, Iwasaki H, Nakamura Y, Gomei Y, Suda T. Knockdown of N-cadherin suppresses the long-term engraftment of hematopoietic stem cells. Blood. 2010;116:554–563. doi: 10.1182/blood-2009-05-224857. [DOI] [PubMed] [Google Scholar]
- Inouye M. Differential staining of cartilage and bone in fetal mouse skeleton by alcian blue and alizarin red. Congenital Anomalies. 1976;16:171–173. [Google Scholar]
- Jung Y, Song J, Shiozawa Y, Wang J, Wang Z, Williams B, Havens A, Schneider A, Ge C, Franceschi RT, et al. Hematopoietic stem cells regulate mesenchymal stromal cell induction into osteoblasts thereby participating in the formation of the stem cell niche. Stem Cells. 2008;26:2042–2051. doi: 10.1634/stemcells.2008-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama Y, Battista M, Kao WM, Hidalgo A, Peired AJ, Thomas SA, Frenette PS. Signals from the sympathetic nervous system regulate hematopoietic stem cell egress from bone marrow. Cell. 2006;124:407–421. doi: 10.1016/j.cell.2005.10.041. [DOI] [PubMed] [Google Scholar]
- Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Kienstra KA, Freysdottir D, Gonzales NM, Hirschi KK. Murine neonatal intravascular injections: modeling newborn disease. J Am Assoc Lab Anim Sci. 2007;46:50–54. [PubMed] [Google Scholar]
- Kunisaki Y, Bruns I, Scheiermann C, Ahmed J, Pinho S, Zhang D, Mizoguchi T, Wei Q, Lucas D, Ito K, et al. Arteriolar niches maintain haematopoietic stem cell quiescence. Nature. 2013;502:637–643. doi: 10.1038/nature12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Strecker S, Wang L, Kronenberg MS, Wang W, Rowe DW, Maye P. Osterix-cre labeled progenitor cells contribute to the formation and maintenance of the bone marrow stroma. PLoS One. 2013;8:e71318. doi: 10.1371/journal.pone.0071318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvinsky A, Dzierzak E. Definitive hematopoiesis is autonomously initiated by the AGM region. Cell. 1996;86:897–906. doi: 10.1016/s0092-8674(00)80165-8. [DOI] [PubMed] [Google Scholar]
- Mendez-Ferrer S, Michurina TV, Ferraro F, Mazloom AR, Macarthur BD, Lira SA, Scadden DT, Ma’ayan A, Enikolopov GN, Frenette PS. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Pinho S, Ahmed J, Kunisaki Y, Hanoun M, Mendelson A, Ono N, Kronenberg HM, Frenette PS. Osterix Marks Distinct Waves of Primitive and Definitive Stromal Progenitors during Bone Marrow Development. Dev Cell. 2014;29:340–349. doi: 10.1016/j.devcel.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Hemmati HD, Wandycz AM, Weissman IL. The purification and characterization of fetal liver hematopoietic stem cells. Proc Natl Acad Sci U S A. 1995;92:10302–10306. doi: 10.1073/pnas.92.22.10302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Weissman IL. The long-term repopulating subset of hematopoietic stem cells is deterministic and isolatable by phenotype. Immunity. 1994;1:661–673. doi: 10.1016/1074-7613(94)90037-x. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- Nombela-Arrieta C, Pivarnik G, Winkel B, Canty KJ, Harley B, Mahoney JE, Park SY, Lu J, Protopopov A, Silberstein LE. Quantitative imaging of haematopoietic stem and progenitor cell localization and hypoxic status in the bone marrow microenvironment. Nature cell biology. 2013;15:533–543. doi: 10.1038/ncb2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguro H, Ding L, Morrison SJ. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell. 2013;13:102–116. doi: 10.1016/j.stem.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottersbach K, Dzierzak E. The murine placenta contains hematopoietic stem cells within the vascular labyrinth region. Dev Cell. 2005;8:377–387. doi: 10.1016/j.devcel.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Palis J, Robertson S, Kennedy M, Wall C, Keller G. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development. 1999;126:5073–5084. doi: 10.1242/dev.126.22.5073. [DOI] [PubMed] [Google Scholar]
- Potocnik AJ, Brakebusch C, Fassler R. Fetal and adult hematopoietic stem cells require beta1 integrin function for colonizing fetal liver, spleen, and bone marrow. Immunity. 2000;12:653–663. doi: 10.1016/s1074-7613(00)80216-2. [DOI] [PubMed] [Google Scholar]
- Shapiro HM. Flow cytometric estimation of DNA and RNA content in intact cells stained with Hoechst 33342 and pyronin Y. Cytometry. 1981;2:143–150. doi: 10.1002/cyto.990020302. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Namiki J, Shibata S, Mastuzaki Y, Okano H. The neural stem/progenitor cell marker nestin is expressed in proliferative endothelial cells, but not in mature vasculature. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 2010;58:721–730. doi: 10.1369/jhc.2010.955609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takubo K, Goda N, Yamada W, Iriuchishima H, Ikeda E, Kubota Y, Shima H, Johnson RS, Hirao A, Suematsu M, et al. Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell. 2010;7:391–402. doi: 10.1016/j.stem.2010.06.020. [DOI] [PubMed] [Google Scholar]
- Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H, Lu X, Soron G, Cooper B, Brayton C, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- Wang X, Spandidos A, Wang H, Seed B. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic acids research. 2012;40:D1144–1149. doi: 10.1093/nar/gkr1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Motiani K, Giridhar PV, Kasper S. CD44 integrates signaling in normal stem cell, cancer stem cell and (pre)metastatic niches. Experimental biology and medicine (Maywood, NJ) 2013;238:324–338. doi: 10.1177/1535370213480714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A, Oser GM, Jaworski M, Blanco-Bose WE, Laurenti E, Adolphe C, Essers MA, Macdonald HR, Trumpp A. Dormant and self-renewing hematopoietic stem cells and their niches. Ann N Y Acad Sci. 2007;1106:64–75. doi: 10.1196/annals.1392.021. [DOI] [PubMed] [Google Scholar]
- Winkler IG, Barbier V, Nowlan B, Jacobsen RN, Forristal CE, Patton JT, Magnani JL, Levesque JP. Vascular niche E-selectin regulates hematopoietic stem cell dormancy, self renewal and chemoresistance. Nat Med. 2012;18:1651–1657. doi: 10.1038/nm.2969. [DOI] [PubMed] [Google Scholar]
- Yilmaz OH, Kiel MJ, Morrison SJ. SLAM family markers are conserved among hematopoietic stem cells from old and reconstituted mice and markedly increase their purity. Blood. 2006;107:924–930. doi: 10.1182/blood-2005-05-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Niu C, Ye L, Huang H, He X, Tong WG, Ross J, Haug J, Johnson T, Feng JQ, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425:836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used for qPCR analysis. Gene accession number for each gene was obtained from NCBI database (http://www.ncbi.nlm.nih.gov/nuccore). Forward (F) and Reverse (R) primer sequences were obtained from PrimerBank database (http://pga.mgh.harvard.edu/primerbank).
Figure S1. A) Mouse E16 fetuses were injected with 100 μL Dextran-FITC (70 kDa) via the superficial temporal vein to reveal functional blood vessels within long bones. At E16, perfused vessels were detectable within periosteum and epiphyseal plate, but not within the middle marrow region of fetal long bone (Longitudinal femur cross section; dashed lines indicate bone edges. Green: Dextran-FITC, Blue: nuclear DAPI. Bar = 100 μm). B) At E16, fetal bone exhibited Alcian Blue (cartilage) staining, but no Alizarin Red (calcification) staining. (Light blue: Alcian Blue; Bar = 100 μm).
Figure S2. Related to Figure 1. A) Fetal bone marrow cells were stained with Hoechst dye to detect an SP population within fetal bone marrow; SP cells were not present in fetal bone marrow until E18.5. B) KSL cells were characterized by expression of c-Kit and Sca-1 proteins and lack of blood lineage protein expression (CD4, CD8, CD45R, Ter119, Gr-1 and Mac-1). Mac-1 was excluded from lineage cocktail for fetal bone marrow analysis since fetal HSC are positive for Mac-1. KSL cells were present in fetal bone marrow starting at E16.5, the onset of HSPC activity. C) Isotype controls were tested for each conjugated antibody. Single compensation controls and unstained controls were also performed. D) Femurs isolated from P0 embryos were fixed and stained with anti-c-Kit and anti-Sca-1 antibodies and with Alexa 594 and Alexa 488 conjugated secondary antibodies, respectively. Merged images show HSPC expressed both c-Kit and Sca-1 in the central bone marrow cavity. Bar=20 μm (insert: magnified view of a double positive HSPC). All flow cytometry studies were conducted at least three times (Data represent mean ± SEM).
Figure S3. Related to Figure 2. Fetal whole bone marrow cells isolated from femurs at E16.5 and onwards showed long term engraftment. Engraftment levels were determined at 4, 12 and 20 weeks after transplantation by analyzing peripheral blood donor/recipient ratio using anti-CD45.2 and CD45.1 antibodies respectively (Data represent mean ± SEM, N=3).
Figure S4. Related to Figure 2. Fetal whole bone marrow cells isolated from femurs at E16.5 and onward showed long term engraftment, and contribution to all three blood lineages in the recipients, similar to adult bone marrow cells. Donor derived blood cells (i.e. CD45.2 positive) were evaluated for their expression of lineage markers (CD4, CD8, B220, Mac1 and Gr1). (Data represent mean ± SEM, N =3).
Figure S5. Related to Figure 3. Properties of KSL cells in WT and Osx−/− fetal bone marrow at E17.5. A) The proportion of KSL cells within whole bone marrow of Osx−/− mutants was comparable to that of WT littermates. Absolute KSL cell numbers, in all long bones combined, from each E17.5 WT and Osx−/− fetus were 402.70 ± 43.59 and 426.86 ± 125.64, respectively. Data (mean ± SEM, N=3; p=0.86) were normalized based on the same number of events in each group. B) The AnnexinV/7AAD apoptosis assay revealed no significant difference (p=0.99) in the proportion of KSL cells undergoing apoptosis in E17.5 WT (6.98, 8.71 and 7.37% KSL cells; mean ± SEM = 7.69 ± 0.52%) and Osx−/− littermates (4.95, 15.50 and 2.70% KSL cells; mean ± SEM = 7.72 ± 3.94%). C) Osx−/− KSL cells exhibited a decreased proportion of cells in G0, and increased proportion of cells in G1 and S/G2/M phases, relative to WT KSL cells.
Figure S6. Related to Figure 3. A) Engraftment curve for fetal bone marrow cells. 50,000–500,000 WBM cells from E17.5 WT femurs were transplanted into sublethally irradiated neonate recipients. 100,000 WBM cells from E17.5 bone marrow were sufficient to detect the engraftment levels above 5% at 12 weeks post transplantation. (Data represent mean ± SEM, N =4). B–D) Cellular composition of bone marrow from E17.5 Osx−/− and WT littermates. There were no differences in total red blood cell (RBC; B) or total white blood cell (WBC; C) counts within bone marrow of E17.5 Osx−/− and WT littermates (Data represent normalized Mean ± SEM, N=4). Endothelial (CD31+CD45−; D) cell number was measured by flow cytometry and absolute percentage within WBM cells was compared (Data represent Mean ± SE, N=4). Total endothelial cell number tended to be higher in Osx−/− mutants compared to WT littermates.