Abstract
Gene set analysis (GSA) is a promising tool for uncovering the polygenic effects associated with complex diseases. However, the available techniques reflect a wide variety of hypotheses about how genetic effects interact to contribute to disease susceptibility. The lack of consensus about the best way to perform GSA has led to confusion in the field and has made it difficult to compare results across methods. A clear understanding of the various choices made during GSA—such as how gene sets are defined, how SNPs are assigned to genes, and how individual SNP-level effects are aggregated to produce gene- or pathway-level effects—will improve the interpretability and comparability of results across methods and studies. In this review, we provide an overview of the various data sources used to construct gene sets and the statistical methods used to test for gene set association, as well as provide guidelines for ensuring the comparability of results.
Keywords: Gene Set Analysis, GWAS, Polygenic Effects, Complex Traits
Moving from genes to gene sets to understand complex disease
Genome-wide association studies (GWAS) have substantially improved our knowledge of the genes involved in complex diseases [1]. For most diseases, however, GWAS studies have revealed that strong single-gene effects are the exception, not the rule. It is now clear that genetic risk for most complex diseases involves the cumulative small effects of many genes, perhaps in concert with a few genes of moderate effect. This fact mandates a move from individual genetic associations to hypotheses about how interactions between the effects of multiple genes contribute to disease susceptibility and expression [2].
Gene set (or pathway) analyses, which test for association between curated collections of genes and a phenotype, are a primary approach for exploring the aforementioned polygenic effects. It is expected that this approach will allow researchers to extend the knowledge gained from GWAS, including those already conducted. As these methods test the cumulative effect across multiple genes, it is possible to detect effects at the gene set level, which may be the result of heterogeneous effects at the gene or variant level. Furthermore, gene set analyses improve the power to detect statistically significant associations both because collapsing individual SNPs into gene sets results in fewer statistical tests performed [3,4,5,6], and because individual weak effects, not detectable in a standard GWAS, can combined to produce a strong association signal.
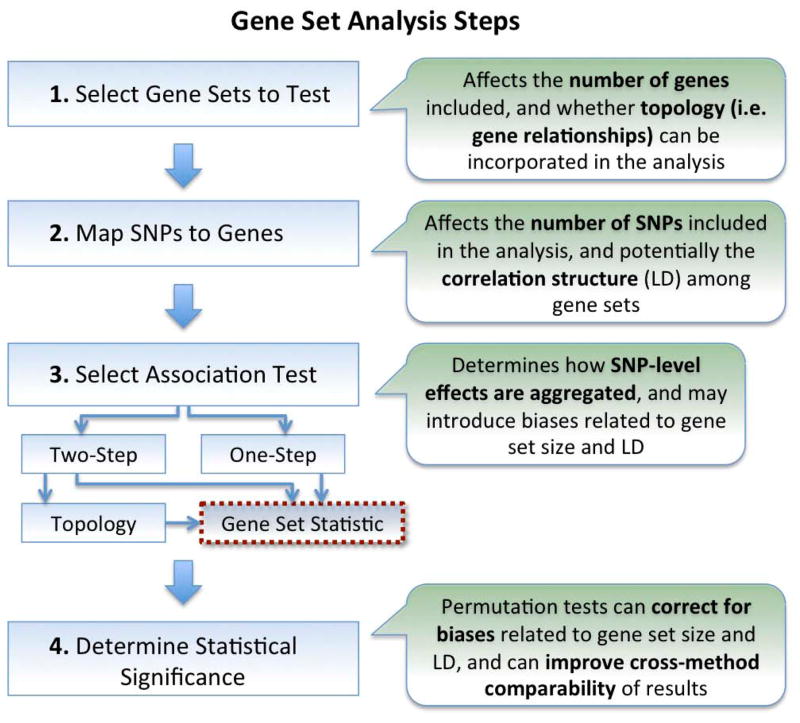
In order to realize the potential of gene set analyses to extend the knowledge gained from GWAS, the field requires a clear understanding of the different methods for defining gene sets, the different approaches to utilizing gene sets, and the statistical methods used to test gene sets. The available techniques encompass a wide variety of hypotheses about how genetic effects combine to contribute to disease susceptibility, which meaningfully influence the results obtained, and therefore must be recognized in order to use these methods effectively. The basic steps performed in a gene set analysis are illustrated in Figure 1.
Figure 1.
An overview of the steps performed in a gene set analysis. The effects of decisions made at each step are highlighted.
In this review, we aim to provide an overview of the methods available for gene set analysis, with special attention paid to the hypotheses and assumptions behind the analysis, the interpretation of results, and the comparability of results from different methods. A theme that will underlie much of our discussion here is the importance of context. The context relevant for gene set analyses can be conceptualized in two ways. Genomic context refers to the inherent structure of the genotype data and the statistical or mathematical methods used to transform SNP effects into pathway effects. Functional context refers to the details of how genes are hypothesized to interact biologically to produce a specific function. The definition of a gene set, including information about the relative importance of genes within the set and any interactions with the environment in which the gene set acts, is important for understanding the functional context of a gene set.
Having a clear understanding of both genomic and functional context will improve interpretation of results and will allow for comparison of results from different studies—something that is often difficult at present. Such an understanding will, we hope, contribute to the establishment of a unified perspective of gene set analysis, in turn advancing the genetic study of complex traits.
Gene Set Definitions
The concept of a gene set loosely refers to a curated collection of genes based on knowledge of genes and biological processes. Numerous databases now provide publicly available gene sets that can be used for gene set analyses, for example: the Pathway Commons database [7], the Kyoto Encyclopedia of Genes and Genomes (KEGG) [8,9], Reactome [10], the Gene Ontology [11], Metacore [12], Biocarta [13], Molecular Signatures Database (MSigDB) [14, 15], Pathway Interaction Database (PID) [16], Ingenuity [17], and ConsensusPathDB [18], among others. Researchers must be aware of the different models and corresponding definitions used to construct these gene sets, so as not to provide misleading interpretations of the results [19]. Here we will provide an overview of four of the most commonly used gene set definitions: biological pathways, networks, gene ontologies, and biomarkers.
Biological Pathways
The strictest definition of a gene set is the biological pathway. Jarvik and Botstein [20] originally described a biological pathway when referring to developmental events as “parts of pathways in which intermediates are processed in a defined sequence.” The definition has evolved to describe a pathway as a set of interacting genes (or their products) that together perform a specific biological function. Still, the idea of concerted action towards a specific endpoint remains a key element of a biological pathway [5, 21]. Therefore, a pathway must include information about the molecular entities involved, as well as information about how those entities interact.
More recently, four types of pathways were proposed to attempt to describe the heterogeneity of currently available pathway definitions: molecular, cellular, disease, and intervention pathways [5]. With the premise that biological pathways are driven from a specific starting point to a specific outcome, molecular pathways characterize biochemical actions on a molecule or compound (e.g. folate metabolism). By contrast, cellular pathways model the regulation of more global cellular processes, such as cell division or apoptosis. It was also suggested that pathways can be defined at higher levels, such as the organ or system level. An example of this type of high-level pathway would be the circadian rhythm pathway in KEGG [8, 9].
In addition, pathway models can describe disease processes or responses to interventions (e.g. drug response). An example of this type of pathways is the Alzheimer’s Disease pathway in KEGG, which contains key Alzheimer’s-related genes, such as APOE, APP and PSEN1. It was noted, however, that disease and intervention pathways may simply be collections of genes previously associated with a phenotype, rather than being based on knowledge of precise biological mechanisms [5]. In that case, they may not qualify as pathways at all based on the definitions proposed herein. Therefore, depending on the amount of information about the interactions between biological entities incorporated in disease and intervention pathways, it may be more appropriate to think of these types of gene sets as “biomarkers” (discussed below).
Currently available pathway databases illustrate two issues important for the interpretation of results from gene set analyses. The first issue is that pathways represent models of hypothesized biological processes. There are no standardized rules for constructing pathway models [21], although there are frameworks for representing them once they are constructed (such as BioPAX [22]). Although experimental data is often used to derive these models, the level of evidence supporting pathway models varies among databases and among pathways. Thus pathway models may differ among data sources. These differences can have consequences for the interpretation, confidence, and comparability of results from gene set analyses [19].
The second issue concerns the hierarchical nature of biological pathways, meaning that some pathways can represent subsets of a larger pathway. It was shown that “cross-talk” effects (the influence of one pathway on another) are largely due to the shared genes (overlap) between pathways. Using a simulated data set, the authors found a strong correlation between pathway similarity and p-value coupling, confirming that significantly overlapping pathways will have similar p-values [23], violating the independence assumptions of statistical tests. Researchers should be aware that gene overlap between pathways can lead to a statistically significant association for a pathway that is not necessarily biologically meaningful (e.g. not relevant in the specific biological context of the disease being studied).
Networks
Biological networks, such as protein-protein interaction (PPI) networks, can also be used to define gene sets. Unlike pathways, networks do not explicitly describe a specific biological function or process carried out in a specific biological context. Rather, networks simply aim to describe biological relationships (observed or predicted interactions) between multiple genes or gene products. In general, the evidence used to build publicly available PPI networks, such as BIND, DIP, IntAct, MINT, HPRD, and STRING [24, 25, 26, 27, 28, 29], is heterogeneous. For example, evidence for protein interaction can come from multiple types of experiments (e.g., Co-IP, ChiP-chip, gene expression, text-mining), which are performed in various tissue types and under various conditions [30]. In addition, the data used to infer an interaction can be of varying quality, and information about the confidence of interactions is not always available [31]. Therefore, it can be difficult to extract from these databases a high-confidence sub-network that is relevant to a specific biological context (e.g., disease process). Nevertheless, networks can be used to derive sets of related genes that can be tested for association with a phenotype. Manual curation of network models can provide disease-specific biological context, and can improve the ability to detect gene set associations. An example is the neurodevelopmental network identified in [32]. Other methods for extracting gene sets (sub-networks) from a genome-scale PPI network include community detection algorithms, which use topological measures to identify tightly clustered nodes [33, 34], and heuristic search algorithms that can identify active subnetworks [35, 36, 37]. It is also possible to simply overlay already defined gene sets (such as gene ontology categories) onto a PPI network to obtain interactions between the genes. See Box 1 for more information about extracting context-specific gene sets from PPI networks.
Box 1. Biological Context for Gene Sets.
The function of biological pathways often depends on a specific biological context (e.g., cell or tissue type). For example, there is evidence that protein interactions can change based on the cellular context in which those proteins are being observed (e.g., different stimuli or different tissues) [38, 39]. Methods used to test for association between a pathway and a phenotype should consider the effects of biological context [19].
Although manual curation can help refine a pathway definition related to a specific phenotype [32], bioinformatics techniques that integrate information from multiple sources can also be used to create context-specific gene sets. For instance, a method has been described that uses gene expression data and functional annotations to derive Cell Context-Specific Gene Sets (CSGS) from molecular interaction data [40]. The hypothesis underlying this method is that many gene set analyses that use predefined gene sets are limited because of the lack of biological context associated with each gene set and the biases in the literature used to define gene sets (e.g., many gene sets and pathways are based on studies of a few high-prevalence diseases).
In a gene set analysis of cardiomyopathy [40], CSGS were more robust than the predefined gene sets from the MSigDB [14, 15]. Specifically, CSGS were more frequently observed in the list of most significant gene sets when the analysis was repeated on multiple subsets of the data.
Another group created a framework for assigning context-sensitive weights to protein-protein interaction data [41]. Their method is based on the idea that proteins have preferred interaction partners depending on the cellular context. Applying this framework to identify pathways activated during influenza infection, they found that high-scoring context-sensitive paths in a PPI network were more likely to include biologically relevant proteins (those shown to have a significant effect on viral propagation and interferon production when silenced in an siRNA experiment) [41].
Although these types of context-sensitive methods are in their infancy, evidence suggests that accounting for cellular context is an important issue that should be addressed when designing studies to identify networks or pathways associated with complex diseases.
Gene Ontologies
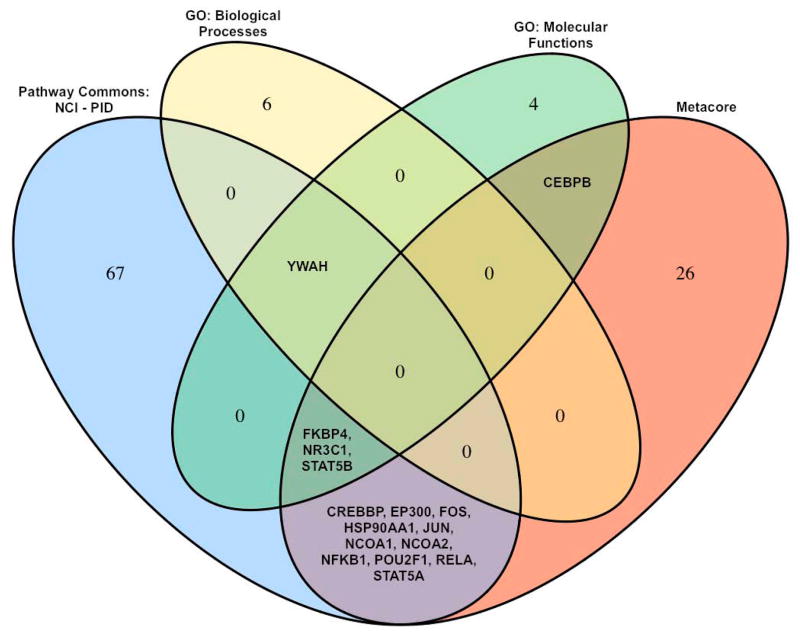
A popular source for gene set definitions is the Gene Ontology (GO) database, which attempts to describe gene functions using three hierarchical biological categories: molecular functions, biological processes, and cellular components. For gene set analyses it is common practice to define gene sets by grouping all genes associated with a particular molecular function or biological process. Examples of gene ontology biological processes and molecular functions are utilized in generating Figure 3.
Figure 3.
Gene sets related to glucocorticoid receptor (GCR) processes retrieved from four data sources. A single pathway in the Pathway Commons database (GCR regulatory network) and one in Metacore (Development_GCR signaling), along with two GO biological processes (GCR signaling pathway, Negative regulation of GCR signaling pathway) and two GO molecular functions (GCR binding, GCR activity) were identified. The Venn diagram shows the limited overlap among gene sets from the four data sources, highlighting the differences in membership among gene sets from different sources.
However, although genes assigned to a particular GO category may be associated with similar functions, this grouping does not indicate known relationships or interactions between these genes. Additionally, like pathways, the hierarchical nature of GO can produce significant overlap between categories, which has implications for both analysis and interpretation of results [23]. It is also important to examine evidence codes for GO assignments, which can range from experimentally determined to computationally inferred.
Disease Biomarkers
Genes that have previously been associated with a particular disease can be grouped to form another type of gene set, a disease biomarker. The difference between a disease biomarker and a GO category is that the genes that constitute a biomarker do not necessarily share functions. Also, unlike the concept of a disease pathway mentioned above, biomarker genes do not necessarily interact with one another.
The hypothesis behind multi-gene biomarkers is simply that the gene set is more strongly associated with the disease than any individual gene, but for many biomarkers this hypothesis is not necessarily confirmed. Indeed, because the evidence supporting individual biomarker genes may come from studies done on a diverse range of populations, the biomarker set may capture genetic heterogeneity of a disease rather than a unified marker. In practice, many multi-gene biomarkers that show statistical association at the population level are not able to accurately predict individual susceptibility for purposes of clinical usage [42]. From the strict definition of a clinical biomarker, therefore, the label “correlated gene set” is often more appropriate. An example of such a putative biomarker, which includes 14 genes associated with bone mineral density and fracture risk at the population level, is described in [43].
Implications of Gene Set Annotation Source
In addition to the underlying models, the data sources used to construct gene sets can influence the hypotheses tested in a gene set analysis. First, similar biological concepts can be described by different sets of genes. Second, the relationships between genes within a gene set can differ between annotation repositories (e.g., GO does not specify gene interactions). Finally, the total number of genes that are members of any gene set (genomic coverage) can vary significantly across data sources, which means that some genes may not be assigned to any gene sets and the effects of those genes will be ignored in the analysis. This last point is an important consideration as more and more candidate genes are produced from large consortium GWAS efforts. It is important to examine how well a particular collection of gene sets covers those genes hypothesized to affect the trait of interest in order to ensure the gene set analysis produces meaningful results.
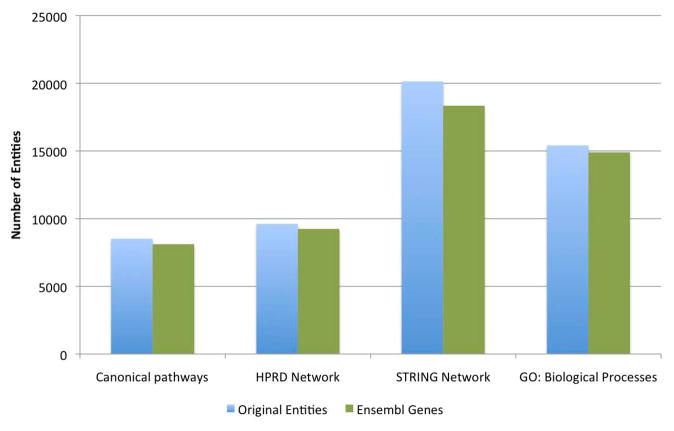
To demonstrate differences among the various gene set annotations, we compared the genomic coverage of four popular sources of gene sets: canonical pathways from the Pathway Commons database (version 4, all sources) [7], PPI networks from the Human Protein Reference Database (HPRD) (release 9) [28] and STRING (version 9.05) [29, 30], and the GO database (submission date: Dec. 11, 2013) [11]. Both HPRD and STRING were included to illustrate the difference between manually curated interaction data (HPRD) and interaction data that includes computational predictions (STRING). Figure 2 shows a significant difference in the level of genomic coverage between potential data sources. For instance, GO biological processes contain nearly twice the number of genes contained in all Pathway Commons pathways.
Figure 2.
The number of genomic entities (genes or proteins) contained in various data sources used for gene set analysis. The proportion of the genome covered by different gene set data sources varies significantly. “Original entities” refers to the specific type of identifier used for gene set members in each data set. These entities were then mapped to Ensembl Gene identifiers for comparison across data sources. “Canonical pathways” includes all unique human pathways in the Pathway Commons database. The Human Protein Reference Database (HPRD) is a manually curated PPI network. The STRING PPI network contains evidence of interaction from multiple sources, including interactions inferred from text mining. For the Gene Ontology data, only Biological Processes were included here, as this category most closely resembles pathways. Including GO Molecular Functions and Cellular Components would increase the genomic coverage of GO by ~20 %.
Related to the differences in genomic coverage are differences in membership among gene sets from different sources (Figure 3). As an illustration, we chose a specific biological concept, glucocorticoid receptors (GCR), and identified gene sets related to this concept from three different data sources: Pathway Commons, GO, and the proprietary pathway database Metacore. There was very little overlap in gene membership between the gene sets representing the same biological concept. For instance, the number of genes related to GCR ranged from 7 to 82 across the data sources, and the uniqueness of the gene sets (the proportion of genes related to GCR in only one data source) ranged from 44 % for GO Molecular Functions to 86 % for GO Biological Processes.
The above examples demonstrate that the choice of data source for gene sets can have significant implications for the biological hypotheses tested. It should be recognized that the use of different gene set sources may limit our ability to replicate specific findings.
In addition, genomic coverage is an important part of the overall genomic context in which a gene set analysis is performed. In particular, genomic coverage will affect the number of SNPs included in the analysis and may have an impact on the level of correlation between gene sets. For GWAS data, the correlation between gene sets is due to linkage disequilibrium among SNPs. This is fundamentally different from the correlation between genes in gene expression data (e.g. due to co-regulation of expression), but it results in similar challenges for data analysis [44]. See Box 2 and Box 3 for details about the influence of genomic context (including the influence of linkage disequilibrium and correlated genes) in gene set analyses.
Box 2. Genomic Context: Assigning SNPs to Genes.
Because gene set analyses are concerned with the effects of groups of genes, it is necessary to map data acquired from different sources (e.g. SNPs) to genes. The simplest methods for mapping SNPs to genes are based solely on SNP location. For instance a SNP is mapped to any gene within a specified distance, or simply to the nearest gene. These methods, although straightforward, do not take into account linkage disequilibrium (LD) patterns or the possibility of regulatory sequences distant from a gene.
LD patterns can be used to provide a more realistic way to map SNPs to genes. One such approach describes the use of an LD-weighted scoring method for assigning SNPs to various genomic features (3′-UTRs, exons, introns, etc.) [45]. SNPs are first annotated with genomic features based on position. Scores for each SNP are then calculated as the sum of correlation coefficients between the SNP and all other SNPs mapped to a particular feature. If the SNP’s score is above a chosen threshold, the SNP is assigned to the corresponding feature. To address the issue of correlated association measures, another group proposed the use of LD blocks, rather than genes, as features in a gene set analysis [46].
Functional annotations (i.e. the predicted functional consequences of SNPs) have also been used to prioritize and map SNPs to genes. This approach can be particularly useful in regions where genes on different strands overlap [35, 47].
Yet another approach addresses three important biases associated with mapping SNPs to genes [48]. First, using GWAS data from a study of HDL levels, the authors found that gene sets created with the largest genes (those that had the most SNPs assigned) were significantly enriched for a number of GO terms, demonstrating a bias in favor of gene sets containing large genes.
Second, they found that gene sets comprised of genes positioned closely together on the same chromosome had significant and widespread enrichment of GO terms when compared to random gene sets. To address this they suggest cleaning gene sets to remove genes in high LD with others in the same set.
Finally, they found that using imputed data increased the coverage of short genes, and that these short genes were less likely than longer genes to contain significant SNPs. This last issue will need to be addressed when comparing the results of gene set analyses performed with datasets containing different numbers of SNPs.
Box 3. Statistical Significance and the Impact of Pathway Size.
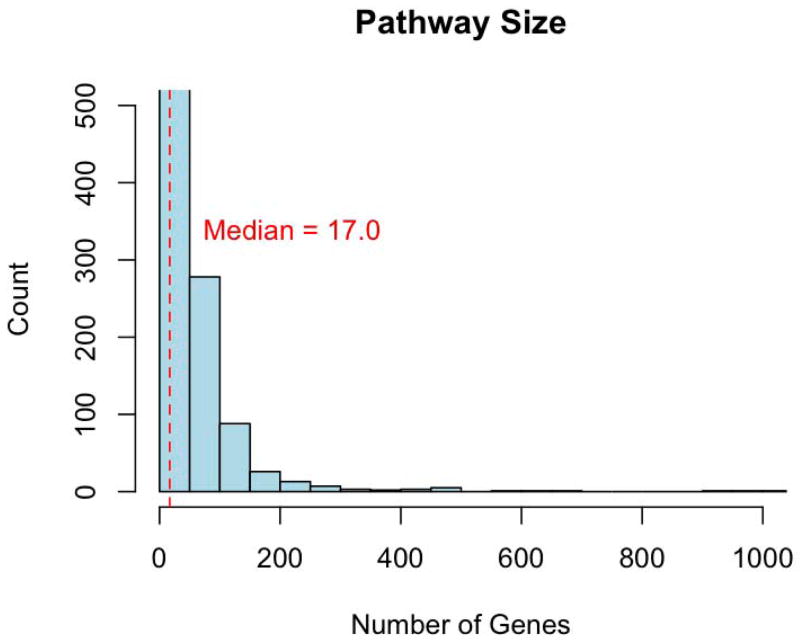
For all analysis methods, pathway size is a potential source of bias [4, 5]. For example, large gene sets are more likely to contain association signals simply by chance. On the other hand, very small gene sets may exhibit false-positive associations due to a strong single-SNP effect [3]. To address this, many studies limit the size of the gene sets tested. Although the limits are arbitrary, pathways used in gene set analyses are commonly limited to those containing between 10 and 200 genes. Figure I shows the distribution of sizes for all pathways in the Pathway Commons database (after removing any pathways containing only a single gene and mapping pathway members to Ensembl gene IDs). There are 732 pathways with fewer than 10 genes and 43 pathways with greater than 200 genes, illustrating that while constraining the size of pathways tested may reduce the potential for biased results, it may also lead to a loss of information.
In addition, there is no consensus on the best way to adjust significance measures for the size of a gene set (number of SNPs mapped to the set). It has been suggested that one simply multiply the gene set p-value by the number of LD-adjusted SNPs in the gene set [48]. However, this may not be adequate for all gene set statistical methods, and a number of permutation or randomization procedures for creating a null distribution of gene set p-values have also been proposed. The most widely used method is to repeatedly permute the phenotype (case/control status) to create a null distribution of gene set statistics. This method preserves the LD between SNPs. Another technique is to create random gene sets (by randomly sampling genes or SNPs or LD blocks) of the same size as the target gene set in order to create a null distribution [46, 62]. A weighted gene resampling procedure based on gene length has also been suggested [63], as has an innovative method called “circular genomic permutation” that aims to preserve LD structure among the permuted SNP sets [64]. A hybrid method, known as “restandardization” [65], combines both phenotype permutation and gene randomization. Because the level of LD among SNPs will vary among datasets, and because different statistical tests vary in their susceptibility to gene set size bias, the correlation between gene set size and gene set significance should always be examined, both before and after a correction procedure has been applied.
Figure I.
Histogram of pathway sizes for 2256 pathways from Pathway Commons. Both axes have been trimmed (Mean = 38, Max = 1757). There are 775 pathways (34 %) with fewer than 10 genes or greater than 200 genes.
Analysis Methods
A variety of gene set analysis methods have been developed which are particularly well suited for secondary analysis of GWAS data (Table 1). In the following sections we describe general methodological categories, which can be useful for classifying and comparing the numerous approaches. However, it is important to note that not all available methods fit neatly into these categories.
Table 1.
Gene Set Analysis Methods and Software Tools: (Focused on GWAS data)
| Method: | Test type: | Input: | References: |
|---|---|---|---|
| Gamma method | 2s, SC | Genotypes | [51] |
| Adaptive rank truncated product method | 2s, SC | Genotypes or P-values | [52] |
| VEGAS | 2s*, SC | P-values | [53] |
| BGSAsnp | 2s, SC | Genotypes | [54] |
| GRASS | 2s, SC | Genotypes | [55] |
| HYST | 2s, SC | P-values, R2 coef. | [66, 67] |
| PRP | 2s, SC, Top | P-values | [68] |
| GSA-SNP | 2s, C, SC | P-values | [69] |
| gamGWAS | 2s, C | P-values | [63] |
| Pathway-PDT (family-based tests) | 2s, C | Genotypes | [70] |
| WebGestalt | 2s, C | Genes | [71] |
| i-GSEA4GWAS | 2s, C | P-values | [72] |
| ICSNPathway | 2s, C | P-values | [73] |
| GenGen | 2s, C | Genotypes | [74] |
| ALIGATOR | 2s, C | P-values | [62] |
| GeSBAP | 2s, C | P-values | [75] |
| GSEA | 2s, C | Genotypes | [76] |
| GSEA-P | 2s, C | Ranked gene list | [77] |
| MAGENTA | 2s, C | P-values | [78] |
| Modified Fisher’s method | 1s, 2s, SC | P-values | [49] |
| Modified Fisher’s method | 1s, 2s, SC | P-values | [50] |
| Plink set-based test | 1s, SC | Genotypes | [79] |
| GSEA-SNP | 1s, SC | P-values | [80] |
| GLOSSI | 1s, SC | P-values | [81] |
| Supervised PC | 1s, SC | Genotypes | [56] |
| SNP ratio test | 1s, SC | Genotypes | [82] |
| PAGWAS (hierarchical model) | 1s, SC | Genotypes | [57] |
| GLMM | 1s, SC | P-values | [58] |
| BMApathway | 1s, SC | Genotypes | [59] |
| Synthetic Feature Random Forest | 1s | Genotypes | [60] |
| SGL-BCGD | 1s, SC | Genotypes | [61] |
| INRICH | 1s, C | Genomic regions | [83] |
| GREAT | C | Genomic regions | [84] |
| SSEA | 1s, SC | P-values | [85] |
| PARIS | 1s, C | P-values | [46] |
| Gowinda | 1s, C | SNPs | [86] |
| GeneSetDB (hypergeometric) | C | P-values | [87] |
| Gene set bagging | C | Genotypes | [88] |
| PoDA | 1s, C | Genotypes | [89] |
| ProxyGeneLD | NA | [48] | |
| GWAS Pathway Identifier | NA | [90] | |
| ConsensusPathDB | NA | [18] | |
| Markov Random Field Model | Top | Genes | [91] |
| EnrichNet | Top | Genes | [92] |
| TopoGSA | Top | Genes | [93] |
| CePa | Top | Genes | [94, 95] |
| THINK-Back-DS | C, Top | P-values, Genes | [96] |
| PINA v2.0 | C, Top | Genes | [34] |
| PANOGA | Top | P-values | [35] |
| dmGWAS | Top | P-values | [36] |
| HotNet2 | Top | P-values | [37] |
| Metacore | Top | Genes | [12] |
| DAPPLE | Top | Genes or genomic regions | [97] |
| GRAIL | Top, TM | SNPs or genomic regions | [98] |
| Chilibot | Top, TM | Genes | [99] |
1S = one-step, 2S = two-step, C = competitive; SC = self-contained; Top = topology-based; TM = text mining;
VEGAS is a gene-based test and can be used as the first step in a two-step GSA.
Two-step and One-Step Methods
Gene set analysis methods have been previously described as falling into two categories: two-step or one-step [6]. In two-step methods, gene-level statistics are calculated first. Next, the gene-level statistics are aggregated to calculate a single statistic for the entire gene set. By contrast, one-step methods use all SNPs in the gene set to calculate the final test statistic, disregarding any gene-level effects. These methods have complementary advantages and disadvantages.
The simplest method for calculating gene-level effects for the two-step approach is to calculate all SNP-level effects, and then for each gene select the minimum p-value from among the SNPs mapped to that gene. Although straightforward and computationally efficient, using a single SNP to summarize a gene-level effect is not always the most appropriate choice. Genomic context and the method for assigning SNPs to genes can have a non-trivial effect on the calculation of gene-level effects (Box 2 and Box 3). For instance, the greater the number of SNPs assigned to a gene, the greater the probability that one of those SNPs, and hence the gene, will be statistically significant.
Furthermore, the power to detect a gene set association may be reduced when the method used to create gene-level statistics does not take into account multiple associated SNPs within the same gene. For example, combining multiple SNPs with moderate association signals may be as significant as a single SNP with a strong association signal.
Within the two-step approaches, multiple SNPs within the same gene can be used to calculate a gene-level effect in two ways: (i) combining individual SNP p-values, or (ii) multi-SNP modeling. For the first, all SNPs within a gene are individually tested for association and then these test statistics are combined to produce a gene-level measure of association. Fisher’s method for combining p-values is a common approach, and numerous modifications have been proposed [49, 50]. Other closely related methods, such as the Gamma method and the adaptive rank truncated product method have also been proposed [51, 52]. Another method, VEGAS, calculates a gene-level statistic by summing the chi-squared statistics from each SNP in the gene and produces an empirical p-value via simulations derived from the multivariate normal distribution [53].
Alternatively, a gene-level effect can be calculated by jointly modeling multiple SNPs within the same gene using multiple regression approaches [54]. To account for LD among SNPs assigned to the same gene, variable selection methods, shrinkage methods (penalized regression), and dimension reduction methods (e.g., principal component analysis and kernel methods) are promising approaches [55, 56, 51]. In the case of principal component analysis, for example, rather than modeling all SNPs within a gene, the top components (e.g., those explaining 80% of variation) are used as variables in a regression model.
Methods for modeling multiple SNPs may also be adapted to “one-step” approaches, where all SNPs in the gene set are modeled to produce a measure of association for the entire gene set [57, 58]. However, for gene sets that contain many SNPs (i.e., more SNPs than samples) not all approaches will be appropriate. In those cases, variable selection methods, penalized regression methods, or dimension reduction methods may be viable options [59, 60, 61].
There is no consensus regarding the relative merit of one-step methods versus two-step methods [52]. However, an advantage of two-step methods is that individual gene contributions to a gene set association can be identified because gene-level effects are known. This may be particularly useful when interpreting results from sets containing a large number of genes, as not all genes in a gene set contribute equally to the association. As was pointed out, the power to detect gene set associations ultimately depends on the signal-to-noise ratio within the unit of analysis (e.g., SNPs vs. genes) and the amount of LD within a gene [52]. This signal-to-noise ratio will depend, in part, on the method of defining gene sets and the method of assigning SNPs to genes (Box 2).
It is common practice to perform gene set analysis as a secondary exploration of GWAS data (often by looking for enrichment of individual SNP associations). However, in order to truly address the issues of small effect size and statistical power inherent in GWAS, it may be useful for researchers to think of pathways (or gene sets) as the unit of analysis in the early stages of study design.
Competitive and Self-Contained Tests
Tests to determine the statistical significance of a gene set can be classified as either competitive or self-contained, based on the definition of the null hypothesis. Competitive methods, such as enrichment tests, compare the association signal among the genes in a gene set to the association signal among all genes outside the set. The null hypothesis is that the extent of association signal within the gene set is equal to the extent of association signal outside the gene set.
On the other hand, self-contained methods consider only the effects within a gene set of interest. These tests simply ask whether the gene set is associated with the trait of interest, the null hypothesis being no association. Self-contained methods do not require data from genes outside the gene set being tested. It has been suggested that two-step methods may be more powerful when self-contained statistical tests are used [6, 51]. Given the striking difference between the null hypotheses for these two categories of tests, it is critical that the method chosen reflect the question of interest.
Topology-based Methods
For gene sets derived from data sources that describe interactions between genes (e.g., pathways or networks), there is a growing collection of methods that incorporate graph topology into the procedure for scoring the gene set ([21] and references therein). Although most work done in this area has focused on gene expression data, some of the methods are adaptable for use with GWAS data, as they only require a gene list or p-value for each gene (see Table 1). HotNet2 and dmGWAS are examples of network-based methods designed specifically for DNA variant data [36, 37], and both can be used to search PPI networks for sub-networks significantly associated with a trait of interest.
Topology-based methods take into account not only individual gene effects, but also the relationship between genes and the relative “importance” of individual genes within the entire gene set. The hypotheses tested by topology-based methods are fundamentally different from other gene set analysis methods because not all genes in the gene set are treated equally. Topology-based methods are more concerned with the overall process performed by the gene set, rather than the cumulative effects of multiple independent genes. This is because genes hypothesized to play a more crucial role in the process are weighted more heavily when testing for association. These gene weights are often based on graph theory measures of centrality.
Because of the use of gene weights, it may not be possible to directly compare results from topology-based analyses with results from other gene set analyses. When interpreting results from topology-based analyses, it is important to examine the relative contribution (the combination of gene-level effects and gene weights) of individual genes in the overall association measure.
A potential source of bias for topology-based methods is the data used to calculate connectedness between genes. This issue is demonstrated by the difference between the HPRD (~38,000 interactions) and STRING (over 2,000,000 interactions) PPI networks (see Figure 2). When comparing results from different topology-based analyses it is important to account for the overall level of connectivity in the datasets used.
Concluding remarks
Gene set analyses have great potential to extend the knowledge gained from GWAS. However, the wide variety of available techniques for testing gene set associations makes it difficult to compare results across studies. Discrepant results are often difficult to reconcile without detailed knowledge of the statistical procedures used. Given the growing popularity of gene set analyses and the rapid development of techniques, it is crucial that steps are taken to improve the interpretability and comparability of results. To aid in this effort we have provided an overview of issues important for understanding both the genomic context and functional context in which gene set analyses are conducted. We have also provided guidelines for reporting the results of gene set analyses (Box 4), which we believe will enable researchers to more readily compare results across methods, and consequently to generate meaningful hypotheses about the genetic mechanisms involved in complex diseases.
Box 4. Unanswered Questions & Suggested Guidelines.
Despite the rapid development of methods for gene set analysis, several questions remain regarding how to appropriately represent and evaluate multi-gene associations. First, consensus is lacking about the best way to summarize SNP-level effects at the gene set-level. Second, it is unclear how best to evaluate the statistical significance of a pathway-level effect. This is complicated by known biases, such as gene or pathway size, and the complex correlation structure of the data both within an individual pathway and between different pathways.
Definitive answers to these questions are hindered by the fact that there is no gold standard by which to evaluate gene set analysis methods. Additionally, our lack of knowledge about the overall genetic architecture responsible for complex disease prevents the creation of realistically complex simulated datasets.
One strategy for building consensus in the field is to compare the ability of different methods to identify narrowly defined polygenic effects, such as the additive effect of many weakly associated SNPs or multiple interaction effects. Because it is unlikely that a single method will be sensitive to all types of polygenic effects, knowing which methods perform best in different situations will be of great value. Furthermore, because the cumulative genetic risk for complex disease is likely a combination of multiple types of polygenic effects, using an ensemble of analysis methods will probably be necessary to uncover all risk factors. This type of ensemble approach has previously been suggested for gene set analysis of gene expression data [100].
Another area for future research relates to the question of how best to map SNPs to genes. Mapping all intergenic SNPs to the nearest gene has the potential to add a significant amount of noise to the data (e.g., SNPs mapped to genes when they have no effect on that particular gene). Yet, ignoring all intergenic SNPs results in a significant loss of information. Development of new methods for mapping intergenic SNPs to genes, possibly through mappings to known regulatory elements, is an interesting area for future work.
Knowledge of how different analysis choices affect gene set analysis results will be crucial for our ability to compare studies and to judge the utility of gene set associations for predicting genetic risk or for generating biological hypotheses. To aid in the interpretation and comparison of results from different studies, we suggest the following guidelines for reporting the results of gene set analyses:
Provide the Source of Gene Set Definitions: Report the definitions of the gene sets tested, including database versions. List the genes belonging to any significantly associated gene sets.
Report Statistical Methods & Potential Biases: Provide a detailed description of the methods used to score pathways and determine statistical significance, including the methods used to “roll-up” effects from the SNP level to the pathway level, and any statistical adjustments made. An examination of the correlation between gene set size and significance should also be performed.
Report SNP- and Gene-level Effects: Provide a reference for the original GWAS results, or information about the distribution of individual SNP-level effects. Also, report the individual gene contributions to a significant gene set association.
Report Gene Set Overlap: Examine the possibility of cross-talk effects for significant gene sets.
Demonstrate Biological Context: Provide biological context and rationale for the joint effect of the genes in the significantly associated gene sets.
Highlights.
Gene set analyses (GSA) will extend the knowledge gained from GWAS.
There is no consensus about the best way to perform GSA.
The wide variety of analysis methods makes it difficult to compare GSA results.
Steps must be taken to improve the interpretability and comparability of GSA.
Acknowledgments
The authors would like to thank the two anonymous reviewers for their thoughtful comments and suggestions to improve the manuscript. Funding for this work was provided by the following grants: NIMH (1R01MH099064), NIH/NCI (5P30CA069533), NIH/NCATS (5UL1RR024140).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hindorff LA, MacArthur J, Morales J, Junkins HA, Hall PN, Klemm AK, Manolio TA European Bioinformatics Institute. [Accessed February 19, 2014];A Catalog of Published Genome-Wide Association Studies. Available at: www.genome.gov/gwastudies.
- 2.Hirschhorn JN. Genomewide association studies--illuminating biologic pathways. N Engl J Med. 2009 Apr 23;360(17):1699–701. doi: 10.1056/NEJMp0808934. [DOI] [PubMed] [Google Scholar]
- 3.Holmans P. Statistical methods for pathway analysis of genome-wide data for association with complex genetic traits. Adv Genet. 2010;72:141–79. doi: 10.1016/B978-0-12-380862-2.00007-2. [DOI] [PubMed] [Google Scholar]
- 4.Wang K, Li M, Hakonarson H. Analysing biological pathways in genome-wide association studies. Nat Rev Genet. 2010 Dec;11(12):843–54. doi: 10.1038/nrg2884. [DOI] [PubMed] [Google Scholar]
- 5.Ramanan VK, Shen L, Moore JH, Saykin AJ. Pathway analysis of genomic data: concepts, methods, and prospects for future development. Trends Genet. 2012 Jul;28(7):323–32. doi: 10.1016/j.tig.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fridley BL, Biernacka JM. Gene set analysis of SNP data: benefits, challenges, and future directions. Eur J Hum Genet. 2011 Aug;19(8):837–43. doi: 10.1038/ejhg.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerami EG, Gross BE, Demir E, Rodchenkov I, Babur O, Anwar N, Schultz N, Bader GD, Sander C. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011 Jan;39(Database issue):D685–90. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000 Jan 1;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014 Jan 1;42(1):D199–205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, Caudy M, Garapati P, Gillespie M, Kamdar MR, Jassal B, Jupe S, Matthews L, May B, Palatnik S, Rothfels K, Shamovsky V, Song H, Williams M, Birney E, Hermjakob H, Stein L, D’Eustachio P. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014 Jan 1;42(1):D472–7. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000 May;25(1):25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Metacore. thomsonreuters.com/metacore.
- 13.Biocarta. www.biocarta.com.
- 14.MSigDB. www.broadinstitute.org/gsea/msigdb/index.jsp.
- 15.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005 Oct 25;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009 Jan;37(Database issue):D674–9. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ingenuity. www.ingenuity.com.
- 18.Kamburov A, Pentchev K, Galicka H, Wierling C, Lehrach H, Herwig R. ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Res. 2011 Jan;39(Database issue):D712–7. doi: 10.1093/nar/gkq1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khatri P, Sirota M, Butte AJ. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput Biol. 2012;8(2):e1002375. doi: 10.1371/journal.pcbi.1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarvik J, Botstein D. A genetic method for determining the order of events in a biological pathway. Proc Natl Acad Sci U S A. 1973 Jul;70(7):2046–50. doi: 10.1073/pnas.70.7.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitrea C, Taghavi Z, Bokanizad B, Hanoudi S, Tagett R, Donato M, Voichiţa C, Drăghici S. Methods and approaches in the topology-based analysis of biological pathways. Front Physiol. 2013 Oct 10;4:278. doi: 10.3389/fphys.2013.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Demir E, Cary MP, Paley S, Fukuda K, Lemer C, Vastrik I, Wu G, D’Eustachio P, Schaefer C, Luciano J, Schacherer F, Martinez-Flores I, Hu Z, Jimenez-Jacinto V, Joshi-Tope G, Kandasamy K, Lopez-Fuentes AC, Mi H, Pichler E, Rodchenkov I, Splendiani A, Tkachev S, Zucker J, Gopinath G, Rajasimha H, Ramakrishnan R, Shah I, Syed M, Anwar N, Babur O, Blinov M, Brauner E, Corwin D, Donaldson S, Gibbons F, Goldberg R, Hornbeck P, Luna A, Murray-Rust P, Neumann E, Ruebenacker O, Samwald M, van Iersel M, Wimalaratne S, Allen K, Braun B, Whirl-Carrillo M, Cheung KH, Dahlquist K, Finney A, Gillespie M, Glass E, Gong L, Haw R, Honig M, Hubaut O, Kane D, Krupa S, Kutmon M, Leonard J, Marks D, Merberg D, Petri V, Pico A, Ravenscroft D, Ren L, Shah N, Sunshine M, Tang R, Whaley R, Letovksy S, Buetow KH, Rzhetsky A, Schachter V, Sobral BS, Dogrusoz U, McWeeney S, Aladjem M, Birney E, Collado-Vides J, Goto S, Hucka M, Le Novère N, Maltsev N, Pandey A, Thomas P, Wingender E, Karp PD, Sander C, Bader GD. The BioPAX community standard for pathway data sharing. Nat Biotechnol. 2010 Sep;28(9):935–42. doi: 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donato M, Xu Z, Tomoiaga A, Granneman JG, Mackenzie RG, Bao R, Than NG, Westfall PH, Romero R, Drăghici S. Analysis and correction of crosstalk effects in pathway analysis. Genome Res. 2013 Nov;23(11):1885–93. doi: 10.1101/gr.153551.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bader GD, Betel D, Hogue CW. BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res. 2003 Jan 1;31(1):248–50. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004 Jan 1;32(Database issue):D449–51. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerrien S, Aranda B, Breuza L, Bridge A, Broackes-Carter F, Chen C, Duesbury M, Dumousseau M, Feuermann M, Hinz U, Jandrasits C, Jimenez RC, Khadake J, Mahadevan U, Masson P, Pedruzzi I, Pfeiffenberger E, Porras P, Raghunath A, Roechert B, Orchard S, Hermjakob H. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012 Jan;40(Database issue):D841–6. doi: 10.1093/nar/gkr1088. Epub 2011 Nov 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Licata L, Briganti L, Peluso D, Perfetto L, Iannuccelli M, Galeota E, Sacco F, Palma A, Nardozza AP, Santonico E, Castagnoli L, Cesareni G. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 2012 Jan;40(Database issue):D857–61. doi: 10.1093/nar/gkr930. Epub 2011 Nov 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, Balakrishnan L, Marimuthu A, Banerjee S, Somanathan DS, Sebastian A, Rani S, Ray S, Harrys Kishore CJ, Kanth S, Ahmed M, Kashyap MK, Mohmood R, Ramachandra YL, Krishna V, Rahiman BA, Mohan S, Ranganathan P, Ramabadran S, Chaerkady R, Pandey A. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009 Jan;37(Database issue):D767–72. doi: 10.1093/nar/gkn892. Epub 2008 Nov 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009 Jan;37(Database issue):D412–6. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013 Jan;41(Database issue):D808–15. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orchard S, Salwinski L, Kerrien S, Montecchi-Palazzi L, Oesterheld M, Stümpflen V, Ceol A, Chatr-aryamontri A, Armstrong J, Woollard P, Salama JJ, Moore S, Wojcik J, Bader GD, Vidal M, Cusick ME, Gerstein M, Gavin AC, Superti-Furga G, Greenblatt J, Bader J, Uetz P, Tyers M, Legrain P, Fields S, Mulder N, Gilson M, Niepmann M, Burgoon L, De Las Rivas J, Prieto C, Perreau VM, Hogue C, Mewes HW, Apweiler R, Xenarios I, Eisenberg D, Cesareni G, Hermjakob H. The minimum information required for reporting a molecular interaction experiment (MIMIx) Nat Biotechnol. 2007 Aug;25(8):894–8. doi: 10.1038/nbt1324. [DOI] [PubMed] [Google Scholar]
- 32.Poelmans G, Pauls DL, Buitelaar JK, Franke B. Integrated genome-wide association study findings: identification of a neurodevelopmental network for attention deficit hyperactivity disorder. Am J Psychiatry. 2011 Apr;168(4):365–77. doi: 10.1176/appi.ajp.2010.10070948. [DOI] [PubMed] [Google Scholar]
- 33.Xu G, Bennett L, Papageorgiou LG, Tsoka S. Module detection in complex networks using integer optimisation. Algorithms Mol Biol. 2010 Nov 12;5:36. doi: 10.1186/1748-7188-5-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cowley MJ, Pinese M, Kassahn KS, Waddell N, Pearson JV, Grimmond SM, Biankin AV, Hautaniemi S, Wu J. PINA v2.0: mining interactome modules. Nucleic Acids Res. 2012 Jan;40(Database issue):D862–5. doi: 10.1093/nar/gkr967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bakir-Gungor B, Sezerman OU. A new methodology to associate SNPs with human diseases according to their pathway related context. PLoS One. 2011;6(10):e26277. doi: 10.1371/journal.pone.0026277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jia P, Zheng S, Long J, Zheng W, Zhao Z. dmGWAS: dense module searching for genome-wide association studies in protein-protein interaction networks. Bioinformatics. 2011 Jan 1;27(1):95–102. doi: 10.1093/bioinformatics/btq615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vandin F, Upfal E, Raphael BJ. Algorithms for detecting significantly mutated pathways in cancer. J Comput Biol. 2011 Mar;18(3):507–22. doi: 10.1089/cmb.2010.0265. [DOI] [PubMed] [Google Scholar]
- 38.de Las Heras JI, Meinke P, Batrakou DG, Srsen V, Zuleger N, Kerr AR, Schirmer EC. Tissue specificity in the nuclear envelope supports its functional complexity. Nucleus. 2013 Nov 1;4(6):460–477. doi: 10.4161/nucl.26872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bisson N, James DA, Ivosev G, Tate SA, Bonner R, Taylor L, Pawson T. Selected reaction monitoring mass spectrometry reveals the dynamics of signaling through the GRB2 adaptor. Nat Biotechnol. 2011 Jun 26;29(7):653–8. doi: 10.1038/nbt.1905. [DOI] [PubMed] [Google Scholar]
- 40.Liu M, King V, Lim WK. Assembling cell context-specific gene sets: a case in cardiomyopathy. J Integr Bioinform. 2013 Dec 13;10(1):234. doi: 10.2390/biecoll-jib-2013-234. [DOI] [PubMed] [Google Scholar]
- 41.Lan A, Ziv-Ukelson M, Yeger-Lotem E. A context-sensitive framework for the analysis of human signalling pathways in molecular interaction networks. Bioinformatics. 2013 Jul 1;29(13):i210–6. doi: 10.1093/bioinformatics/btt240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.National Biomarker Development Alliance. Organization to promote biomarker development. J Nucl Med. 2014 Mar;55(3):11N. [PubMed] [Google Scholar]
- 43.Estrada K, Styrkarsdottir U, Evangelou E, Hsu YH, Duncan EL, Ntzani EE, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet. 2012 Apr 15;44(5):491–501. doi: 10.1038/ng.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu D, Smyth GK. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 2012 Sep 1;40(17):e133. doi: 10.1093/nar/gks461. Epub 2012 May 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schork AJ, Thompson WK, Pham P, Torkamani A, Roddey JC, Sullivan PF, Kelsoe JR, O’Donovan MC, Furberg H, Schork NJ, Andreassen OA, Dale AM Tobacco and Genetics Consortium; Bipolar Disorder Psychiatric Genomics Consortium; Schizophrenia Psychiatric Genomics Consortium. All SNPs are not created equal: genome-wide association studies reveal a consistent pattern of enrichment among functionally annotated SNPs. PLoS Genet. 2013 Apr;9(4):e1003449. doi: 10.1371/journal.pgen.1003449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yaspan BL, Bush WS, Torstenson ES, Ma D, Pericak-Vance MA, Ritchie MD, Sutcliffe JS, Haines JL. Genetic analysis of biological pathway data through genomic randomization. Hum Genet. 2011 May;129(5):563–71. doi: 10.1007/s00439-011-0956-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saccone SF, Bolze R, Thomas P, Quan J, Mehta G, Deelman E, Tischfield JA, Rice JP. SPOT: a web-based tool for using biological databases to prioritize SNPs after a genome-wide association study. Nucleic Acids Res. 2010 Jul;38(Web Server issue):W201–9. doi: 10.1093/nar/gkq513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hong MG, Pawitan Y, Magnusson PK, Prince JA. Strategies and issues in the detection of pathway enrichment in genome-wide association studies. Hum Genet. 2009 Aug;126(2):289–301. doi: 10.1007/s00439-009-0676-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De la Cruz O, Wen X, Ke B, Song M, Nicolae DL. Gene, region and pathway level analyses in whole-genome studies. Genet Epidemiol. 2010 Apr;34(3):222–31. doi: 10.1002/gepi.20452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo L, Peng G, Zhu Y, Dong H, Amos CI, Xiong M. Genome-wide gene and pathway analysis. Eur J Hum Genet. 2010 Sep;18(9):1045–53. doi: 10.1038/ejhg.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Biernacka JM, Jenkins GD, Wang L, Moyer AM, Fridley BL. Use of the gamma method for self-contained gene-set analysis of SNP data. Eur J Hum Genet. 2012 May;20(5):565–71. doi: 10.1038/ejhg.2011.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu K, Li Q, Bergen AW, Pfeiffer RM, Rosenberg PS, Caporaso N, Kraft P, Chatterjee N. Pathway analysis by adaptive combination of P-values. Genet Epidemiol. 2009 Dec;33(8):700–9. doi: 10.1002/gepi.20422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, Hayward NK, Montgomery GW, Visscher PM, Martin NG, Macgregor S AMFS Investigators. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010 Jul 9;87(1):139–45. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shahbaba B, Shachaf CM, Yu Z. A pathway analysis method for genome-wide association studies. Stat Med. 2012 May 10;31(10):988–1000. doi: 10.1002/sim.4477. [DOI] [PubMed] [Google Scholar]
- 55.Chen LS, Hutter CM, Potter JD, Liu Y, Prentice RL, Peters U, Hsu L. Insights into colon cancer etiology via a regularized approach to gene set analysis of GWAS data. Am J Hum Genet. 2010 Jun 11;86(6):860–71. doi: 10.1016/j.ajhg.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen X, Wang L, Hu B, Guo M, Barnard J, Zhu X. Pathway-based analysis for genome-wide association studies using supervised principal components. Genet Epidemiol. 2010 Nov;34(7):716–24. doi: 10.1002/gepi.20532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Evangelou M, Dudbridge F, Wernisch L. Two novel pathway analysis methods based on a hierarchical model. Bioinformatics. 2014 Mar 1;30(5):690–7. doi: 10.1093/bioinformatics/btt583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang L, Jia P, Wolfinger RD, Chen X, Grayson BL, Aune TM, Zhao Z. An efficient hierarchical generalized linear mixed model for pathway analysis of genome-wide association studies. Bioinformatics. 2011 Mar 1;27(5):686–92. doi: 10.1093/bioinformatics/btq728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carbonetto P, Stephens M. Integrated enrichment analysis of variants and pathways in genome-wide association studies indicates central role for IL-2 signaling genes in type 1 diabetes, and cytokine signaling genes in Crohn’s disease. PLoS Genet. 2013;9(10):e1003770. doi: 10.1371/journal.pgen.1003770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pan Q, Hu T, Malley JD, Andrew AS, Karagas MR, Moore JH. A system-level pathway-phenotype association analysis using synthetic feature random forest. Genet Epidemiol. 2014 Apr;38(3):209–19. doi: 10.1002/gepi.21794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Silver M, Chen P, Li R, Cheng CY, Wong TY, Tai ES, Teo YY, Montana G. Pathways-driven sparse regression identifies pathways and genes associated with high-density lipoprotein cholesterol in two Asian cohorts. PLoS Genet. 2013 Nov;9(11):e1003939. doi: 10.1371/journal.pgen.1003939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Holmans P, Green EK, Pahwa JS, Ferreira MA, Purcell SM, Sklar P, Owen MJ, O’Donovan MC, Craddock N Wellcome Trust Case-Control Consortium. Gene ontology analysis of GWA study data sets provides insights into the biology of bipolar disorder. Am J Hum Genet. 2009 Jul;85(1):13–24. doi: 10.1016/j.ajhg.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jia P, Wang L, Fanous AH, Chen X, Kendler KS, Zhao Z International Schizophrenia Consortium. A bias-reducing pathway enrichment analysis of genome-wide association data confirmed association of the MHC region with schizophrenia. J Med Genet. 2012 Feb;49(2):96–103. doi: 10.1136/jmedgenet-2011-100397. [DOI] [PubMed] [Google Scholar]
- 64.Cabrera CP, Navarro P, Huffman JE, Wright AF, Hayward C, Campbell H, Wilson JF, Rudan I, Hastie ND, Vitart V, Haley CS. Uncovering networks from genome-wide association studies via circular genomic permutation. G3 (Bethesda) 2012 Sep;2(9):1067–75. doi: 10.1534/g3.112.002618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Efron B, Tibshirani R. On testing the significance of sets of genes. Ann Appl Stat. 2007;1:107. [Google Scholar]
- 66.Li MX, Kwan JS, Sham PC. HYST: a hybrid set-based test for genome-wide association studies, with application to protein-protein interaction-based association analysis. Am J Hum Genet. 2012 Sep 7;91(3):478–88. doi: 10.1016/j.ajhg.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li MX, Gui HS, Kwan JS, Sham PC. GATES: a rapid and powerful gene-based association test using extended Simes procedure. Am J Hum Genet. 2011 Mar 11;88(3):283–93. doi: 10.1016/j.ajhg.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen L, Zhang L, Zhao Y, Xu L, Shang Y, Wang Q, Li W, Wang H, Li X. Prioritizing risk pathways: a novel association approach to searching for disease pathways fusing SNPs and pathways. Bioinformatics. 2009 Jan 15;25(2):237–42. doi: 10.1093/bioinformatics/btn613. [DOI] [PubMed] [Google Scholar]
- 69.Nam D, Kim J, Kim SY, Kim S. GSA-SNP: a general approach for gene set analysis of polymorphisms. Nucleic Acids Res. 2010 Jul;38(Web Server issue):W749–54. doi: 10.1093/nar/gkq428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park YS, Schmidt M, Martin ER, Pericak-Vance MA, Chung RH. Pathway-PDT: a flexible pathway analysis tool for nuclear families. BMC Bioinformatics. 2013 Sep 4;14:267. doi: 10.1186/1471-2105-14-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013 Jul;41(Web Server issue):W77–83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang K, Cui S, Chang S, Zhang L, Wang J. i-GSEA4GWAS: a web server for identification of pathways/gene sets associated with traits by applying an improved gene set enrichment analysis to genome-wide association study. Nucleic Acids Res. 2010 Jul;38(Web Server issue):W90–5. doi: 10.1093/nar/gkq324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang K, Chang S, Cui S, Guo L, Zhang L, Wang J. ICSNPathway: identify candidate causal SNPs and pathways from genome-wide association study by one analytical framework. Nucleic Acids Res. 2011 Jul;39(Web Server issue):W437–43. doi: 10.1093/nar/gkr391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang K, Zhang H, Kugathasan S, Annese V, Bradfield JP, Russell RK, Sleiman PM, Imielinski M, Glessner J, Hou C, Wilson DC, Walters T, Kim C, Frackelton EC, Lionetti P, Barabino A, Van Limbergen J, Guthery S, Denson L, Piccoli D, Li M, Dubinsky M, Silverberg M, Griffiths A, Grant SF, Satsangi J, Baldassano R, Hakonarson H. Diverse genome-wide association studies associate the IL12/IL23 pathway with Crohn Disease. Am J Hum Genet. 2009 Mar;84(3):399–405. doi: 10.1016/j.ajhg.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Medina I, Montaner D, Bonifaci N, Pujana MA, Carbonell J, Tarraga J, Al-Shahrour F, Dopazo J. Gene set-based analysis of polymorphisms: finding pathways or biological processes associated to traits in genome-wide association studies. Nucleic Acids Res. 2009 Jul;37(Web Server issue):W340–4. doi: 10.1093/nar/gkp481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang K, Li M, Bucan M. Pathway-based approaches for analysis of genomewide association studies. Am J Hum Genet. 2007 Dec;81(6):1278–83. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005 Oct 25;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Segrè AV, Groop L, Mootha VK, Daly MJ, Altshuler D DIAGRAM Consortium; MAGIC investigators. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010 Aug;12(6):8. doi: 10.1371/journal.pgen.1001058. pii: e1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007 Sep;81(3):559–75. doi: 10.1086/519795. Epub 2007 Jul 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Holden M, Deng S, Wojnowski L, Kulle B. GSEA-SNP: applying gene set enrichment analysis to SNP data from genome-wide association studies. Bioinformatics. 2008 Dec 1;24(23):2784–5. doi: 10.1093/bioinformatics/btn516. [DOI] [PubMed] [Google Scholar]
- 81.Chai HS, Sicotte H, Bailey KR, Turner ST, Asmann YW, Kocher JP. GLOSSI: a method to assess the association of genetic loci-sets with complex diseases. BMC Bioinformatics. 2009 Apr 3;10:102. doi: 10.1186/1471-2105-10-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.O’Dushlaine C, Kenny E, Heron EA, Segurado R, Gill M, Morris DW, Corvin A. The SNP ratio test: pathway analysis of genome-wide association datasets. Bioinformatics. 2009 Oct 15;25(20):2762–3. doi: 10.1093/bioinformatics/btp448. [DOI] [PubMed] [Google Scholar]
- 83.Lee PH, O’Dushlaine C, Thomas B, Purcell SM. INRICH: interval-based enrichment analysis for genome-wide association studies. Bioinformatics. 2012 Jul 1;28(13):1797–9. doi: 10.1093/bioinformatics/bts191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010 May;28(5):495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weng L, Macciardi F, Subramanian A, Guffanti G, Potkin SG, Yu Z, Xie X. SNP-based pathway enrichment analysis for genome-wide association studies. BMC Bioinformatics. 2011 Apr 15;12:99. doi: 10.1186/1471-2105-12-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kofler R, Schlötterer C. Gowinda: unbiased analysis of gene set enrichment for genome-wide association studies. Bioinformatics. 2012 Aug 1;28(15):2084–5. doi: 10.1093/bioinformatics/bts315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Araki H, Knapp C, Tsai P, Print C. GeneSetDB: A comprehensive meta-database, statistical and visualisation framework for gene set analysis. FEBS Open Bio. 2012 Apr 17;2:76–82. doi: 10.1016/j.fob.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jaffe AE, Storey JD, Ji H, Leek JT. Gene set bagging for estimating the probability a statistically significant result will replicate. BMC Bioinformatics. 2013 Dec 12;14(1):360. doi: 10.1186/1471-2105-14-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Braun R, Buetow K. Pathways of distinction analysis: a new technique for multi-SNP analysis of GWAS data. PLoS Genet. 2011 Jun;7(6):e1002101. doi: 10.1371/journal.pgen.1002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Büchel F, Mittag F, Wrzodek C, Zell A, Gasser T, Sharma M. Integrative pathway-based approach for genome-wide association studies: identification of new pathways for rheumatoid arthritis and type 1 diabetes. PLoS One. 2013 Oct 25;8(10):e78577. doi: 10.1371/journal.pone.0078577. eCollection 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen M, Cho J, Zhao H. Incorporating biological pathways via a Markov random field model in genome-wide association studies. PLoS Genet. 2011 Apr;7(4):e1001353. doi: 10.1371/journal.pgen.1001353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Glaab E, Baudot A, Krasnogor N, Schneider R, Valencia A. EnrichNet: network-based gene set enrichment analysis. Bioinformatics. 2012 Sep 15;28(18):i451–i457. doi: 10.1093/bioinformatics/bts389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Glaab E, Baudot A, Krasnogor N, Valencia A. TopoGSA: network topological gene set analysis. Bioinformatics. 2010 May 1;26(9):1271–2. doi: 10.1093/bioinformatics/btq131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gu Z, Liu J, Cao K, Zhang J, Wang J. Centrality-based pathway enrichment: a systematic approach for finding significant pathways dominated by key genes. BMC Syst Biol. 2012 Jun 6;6:56. doi: 10.1186/1752-0509-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gu Z, Wang J. CePa: an R package for finding significant pathways weighted by multiple network centralities. Bioinformatics. 2013 Mar 1;29(5):658–60. doi: 10.1093/bioinformatics/btt008. [DOI] [PubMed] [Google Scholar]
- 96.Farfán F, Ma J, Sartor MA, Michailidis G, Jagadish HV. THINK Back: KNowledge-based Interpretation of High Throughput data. BMC Bioinformatics. 2012 Mar 13;13( Suppl 2):S4. doi: 10.1186/1471-2105-13-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rossin EJ, Lage K, Raychaudhuri S, Xavier RJ, Tatar D, Benita Y, Cotsapas C, Daly MJ International Inflammatory Bowel Disease Genetics Constortium. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011 Jan 13;7(1):e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Raychaudhuri S, Plenge RM, Rossin EJ, Ng AC, Purcell SM, Sklar P, Scolnick EM, Xavier RJ, Altshuler D, Daly MJ International Schizophrenia Consortium. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009 Jun;5(6):e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen H, Sharp BM. Content-rich biological network constructed by mining PubMed abstracts. BMC Bioinformatics. 2004 Oct 8;5:147. doi: 10.1186/1471-2105-5-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Väremo L, Nielsen J, Nookaew I. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 2013 Apr;41(8):4378–91. doi: 10.1093/nar/gkt111. [DOI] [PMC free article] [PubMed] [Google Scholar]