Abstract
During development, epigenetic programs are “installed” on the genome that direct differentiation and normal tissue and organ function in adulthood. Consequently, development is also a period of susceptibility to reprogramming of the epigenome. Developmental reprogramming occurs when an adverse stimulus or insult interrupts the proper “install” of epigenetic programs during development, reprogramming normal physiological responses in such a way as to promote disease later in life. Some of the best examples of developmental reprogramming involve the reproductive tract, where early life exposures to environmental estrogens can increase susceptibility to benign and malignant tumors in adulthood including leiomyoma (fibroids), endometrial and prostate cancer. Although specific mechanism(s) by which environmental estrogens reprogram the developing epigenome were unknown, both DNA and histone methylation were considered likely targets for epigenetic reprogramming. We have now identified a mechanism by which developmental exposures to environmental estrogens reprogram the epigenome by inducing inappropriate activation of nongenomic estrogen receptor (ER) signaling. Activation of non-genomic ER signaling via the PI3K pathway activates the kinase AKT/PKB in the developing reproductive tract, which phosphorylates the histone lysine methyltransferase (HKMT) EZH2, the key “installer” of epigenetic histone H3 lysine 27 trimethylation (H3K27me3). AKT phosphorylation inactivates EZH2, decreasing levels of H3K27 methylation, a repressive mark that inhibits gene expression, in the developing uterus. As a result of this developmental reprogramming, many estrogen-responsive genes become hypersensitive to estrogen in adulthood, exhibiting elevated expression throughout the estrus cycle, and resulting in a “hyper-estrogenized” phenotype in the adult uterus that promotes development of hormone dependent tumors.
Keywords: Epigenetics, Leiomyoma, Uterus, Prostate, reproductive tract, environmental exposures, estrogen, histone methyltransferase
Developmental Reprogramming by Early Life Environmental Exposures
Early life exposures to environmental agents can have profound effects on adult disease outcome via an epigenetic process termed developmental reprogramming (Gluckman and Hanson, 2004; Jirtle and Skinner, 2007). The developmental reprogramming hypothesis proposes that exposure of developing tissues to an adverse stimulus or insult can permanently reprogram normal physiological responses, and so give rise to metabolic and hormonal disorders later in life (Barker, 2002; Couzin, 2002; Frankel and others, 1996; Hattersley and Tooke, 1999). Developmental reprogramming can also be induced by early life exposures to environmental agents, such as xenoestrogens that mimic the female hormone estrogen and act as estrogen receptor (ER) agonists. There are now many examples of developmental reprogramming by environmental estrogens increasing susceptibility to adult diseases such as obesity and cancer.
From the 1940-70s, the xenoestrogen diethylstilbestrol (DES) was extensively prescribed to pregnant women at risk for miscarriage. Women exposed to DES in utero during critical periods of reproductive tract development developed several types of reproductive tract abnormalities, as well as an increased incidence of cervical-vaginal cancer later in life (Herbst and others, 1971). Animal studies that simulate the human DES experience have since shown that exposure of the developing reproductive tract of CD-1 mice to DES imparts a permanent estrogen imprint that alters reproductive tract morphology, induces persistent expression of the lactoferrin and c-fos genes and induces a high incidence of uterine adenocarcinoma (Li and others, 2003; Newbold and others, 1990; Newbold and others, 1997). DES exposure also induces changes in the expression of several uterine genes involved in tissue patterning, such as Wnt7a, Hoxa9, Hoxa10 and Hoxa11, contributing to changes in tissue architecture and morphology (Block and others, 2000; Ma and others, 1998; Miller and others, 1998). DES-induced developmental programming has been demonstrated to require estrogen receptor α (ERα) (Couse and others, 2001), suggesting that signaling through this receptor is crucial for establishing the imprint. These initial observations with DES firmly established the developmental period as a window of susceptibility during which an inappropriate xenoestrogen exposure can induce developmental programming and increase risk for diseases, including cancer, later in life.
We have recently shown that developmental reprogramming induced by environmental estrogens can increase susceptibility to uterine tumorigenesis by increasing the penetrance of a tumor suppressor gene defect in adulthood (Cook and others, 2005). Utilizing rats carrying a germline defect in the tuberous sclerosis complex 2 (Tsc-2) tumor suppressor gene that are predisposed to uterine leiomyomas, we found that an early life exposure to diethylstilbestrol (DES) during development of the uterus increased tumor suppressor gene penetrance from 65% to >90%, increased tumor multiplicity and size in genetically predisposed animals, but failed to induce tumors in wild-type rats. Importantly, we found that DES exposure had imparted a hormonal “imprint” on the developing uterine myometrium that was independent of the genetic defect (i.e. occurred in both wild-type and carrier females), causing an increase in expression of estrogen-responsive genes prior to the onset of tumors. Like many other benign and malignant reproductive tract tumors, uterine leiomyoma are hormone dependent (Walker and Stewart, 2005). Transcriptional profiling of tumors compared to age- and stage-matched myometrium identified 171 genes differentially expressed in leiomyomas relative to normal myometrium (Greathouse and others, 2008). Of the estrogen-responsive genes analyzed, over half were reprogrammed in the adult myometrium as a result of developmental exposure to DES. Reprogramming caused these estrogen-responsive genes to become hyper-responsive to hormone, resulting in increased expression during the estrous cycle when estrogen was high, and sustained expression even when estrogen levels were low. Importantly, this reprogramming was observed in the adult myometrium 8-10 months prior to the onset of tumors. When this developmental reprogramming of estrogen-responsive genes occurred in the presence of the Tsc-2 tumor suppressor gene defect, the reprogrammed “hyperestrogenized” phenotype of the myometrium promoted the development of leiomyomas in genetically susceptible animals.
Ho and colleagues have reported similar data for the developing male reproductive tract. They found that neonatal exposure of male rats to the environmental xenoestrogen bisphenol A (BPA) predisposed them to develop prostate cancer (PCa) or high-grade prostatic intraepithelial neoplasia (HG-PIN) later in life (Ho and others, 2006). They went on to demonstrate that this early life exposure could induce aberrant DNA methylation of CpG islands in the promoter region of specific genes such as Pde4d4, leading to inappropriate gene expression in the adult prostate. These data indicate that in both males and females, environmental exposures during development can permanently reprogram the developing reproductive tract and lead to increased susceptibility to develop tumors in adulthood.
Histone Methylation: An Epigenetic Mechanism for Developmental Reprogramming
Epigenetics refers to heritable changes in gene expression not mediated by changes in DNA sequence. Feinberg (Feinberg and Tycko, 2004) has classified epigenetic information into three categories: cytosine methylation at CpG sites in the DNA, genomic imprinting (parent-of-origin-specific allele silencing which may also occur via cytosine methylation) and histone modifications. Importantly, although epigenetic alterations are heritable and stably maintained, they are potentially reversible pharmacologically (e.g. by treatment with 5-azacytadine which reduces cytosine methylation) or nutritionally (for example with folate supplementation which can increase levels of 5-methyl cytosine). Therefore, elucidating the epigenetic alterations induced by environmental agents not only broadens our understanding of the mechanisms of action of these agents, but potentially opens new avenues to reverse or prevent their adverse health effects.
Until recently, epigenetic analyses for developmental reprogramming have focused on changes in DNA methylation. However, our group has recently demonstrated that epigenetic histone modifications are direct targets for developmental reprogramming by environmental estrogens. Histones are the architectural proteins that make up nucleosomes, the basic repeat unit of chromatin. The nucleosome is composed of DNA packaged in units of 146 base-pairs wrapped around a histone octamer composed of two units each of histones H2A, H2B, H3, and H4 (Luger and Richmond, 1998). Nucleosomal DNA is further compacted by linker histone H1 and other, non-histone proteins into higher order chromatin. Post-translational modification of histone proteins alters chromatin conformation and regulates gene expression (Cheung and Lau, 2005; Sims and others, 2003; Zhang and Reinberg, 2001). These post-translational modifications include acetylation, phosphorylation, methylation, ADP ribosylation, glycosylation, SUMOylation and ubiqutination. Covalent modifications of histones affect the structural dynamics of the nucleosome and modulate DNA accessibility.
Different covalent modifications, alone or in combination, occur at multiple sites on different histones, generating tremendous diversity in histone/nucleosome structure. Several histone methyltransferase (HMT) enzymes have been identified, each of which covalently modifies specific lysine (HKMTs) or arginine (PRMT) residues on core histones. HMTs catalyze the mono- and di-methylation of arginine and mono-, di-, and trimethylation of lysine residues on histone proteins H1, H2A, H3, and H4 (Bannister and others, 2002; Jenuwein, 2006; Lachner and others, 2003; Zhang and Reinberg, 2001), with histone H3 and H4 containing the majority of characterized histone methylation sites. While acetylation and phosphorylation are reversible and generally associated with inducible gene expression, histone methylation can be stably inherited by daughter cells after cell division and is an epigenetic regulator of gene expression.
In contrast to DNA methylation, histone methylation can result in either activation (eg H3K4 and H3R17) or silencing (eg H3K9 and H3K27) of transcription (Berger, 2007; Lee and others, 2005). Site-specific methylation of histones generates binding sites for various chromatin remodeling complexes as well as other epigenetic effectors, such as DNA methyltransferases (Bannister and others, 2001; Lachner and others, 2001; Smallwood and others, 2007; Vire and others, 2006; Zhao and others, 2009). Different combinations of histone modifications are believed to comprise a “histone code” that directs cellular processes by the recruitment of specific chromatin associated proteins, resulting in distinct gene expression outcomes (Jenuwein and Allis, 2001; Rice and Allis, 2001; Strahl and Allis, 2000; Turner, 2000). Since epigenetic reprogramming occurs during development, it is evident that it must also be possible to reset histone methylation patterns. Three classes of enzymes that can remove arginine- and lysine-methylation (PAD4, LSD1 and JmjC) have recently been identified (Metzger and others, 2005; Tsukada and others, 2005; Wang and others, 2004), however, the role of these enzymes in epigenetic reprogramming is not well understood at this time.
Although it is clear that histone modifications are heritable, the precise mechanism(s) responsible for inheritance of these modifications are still being elucidated. Lysine-methylation is stable and persists through cell division, where parental nucleosomes are recycled and deposited onto daughter strands, potentially transferring modified histones from generation to generation (Jackson and Chalkley, 1985; Sogo and others, 1986). Alternatively, the parental octamer may divide in a semi-conservative manner and segregate onto the daughter strands, with nucleosome assembly complexes adding newly-synthesized histones to complete the octamer (Tagami and others, 2004). In both these scenarios, it is still necessary to faithfully “copy” the histone modifications, either onto the newly assembled nucleosomes that fill in the gaps once intact octamers are distributed onto the daughter strands or to the “new half” of the nucleosome from the preexisting “old half”. In contrast to the well-understood process for faithfully copying DNA methylation patterns from hemi-methylated DNA, we still know little about the process by which histone modifications are replicated.
To date, the sequence of molecular events that establish and maintain heritable chromatin states has not been fully characterized. Recent studies indicate that histone methylation precedes or occurs concurrently with DNA methylation (Martin and Zhang, 2007). Enhancer of Zeste homolog 2 (EZH2) is the histone methyltransferase (HMT) found in the Polycomb repressive complex 2 (PRC2), which methylates histone H3 lysine 27 (H3K27). Importantly, H3K27 methylation appears to play a role in de novo DNA methylation in cancer. Schlesinger et al (Schlesinger and others, 2007) demonstrated that H3K27 methylation profiles in stem cells direct the silencing of specific gene targets by DNA methylation during carcinogenesis, suggesting that histone methylation patterns established during fetal development precede changes in DNA methylation and can have functional consequences much later in life and/or during pathogenesis.
Nongenomic Estrogen Receptor Signaling by Environmental Estrogens
The classical effects of steroid hormones are mediated by nuclear hormone receptors functioning as ligand-activated transcription factors. However, it now appreciated that not all the effects of steroid hormones are manifested by their activity as transcription factors (Bjornstrom and Sjoberg, 2005; Castoria and others, 1999; Cato and others, 2002; Cheskis, 2004; Edwards, 2005; Levin, 2005; Losel and Wehling, 2003). The effects of steroid hormones that occur independent from gene transcription have been termed nongenomic to distinguish them from the direct, or genomic, effects of their receptors as transcription factors in the nucleus. The nongenomic effects of steroid hormones can be manifested by both a subpopulation of classical receptors that associate with signaling complexes in the cytoplasm or plasma cell membrane.
For estrogens, nongenomic receptor interactions occur more rapidly than genomic responses (minutes to hours), are ligand-mediated, blocked by the pure antiestrogen ICI and can occur in association with an ER lacking a NLS. Stimulation of PI3K signaling is a well characterized example of a nongenomic function of the ER. PI3K signaling occurs downstream of many receptor tyrosine kinases such as the insulin and EGF receptors, and is involved in key cellular processes including survival, proliferation, growth and motility (Sulis and Parsons, 2003; Vivanco and Sawyers, 2002). ERs have been shown to directly associate with the p85 regulatory subunit of PI3K (Simoncini and others, 2000) and caveoli have been identified as sites for assembly of ER:PI3K complexes that activate AKT (Losel and others, 2003). In this cellular compartment, caveolin mediates retention of ERs in caveoli, where they associate with and activate PI3K (Kim and others, 1999; Razandi and others, 2002; Schlegel and others, 1999). In addition to activating other signaling molecules downstream from AKT such as mTOR, AKT mediates rapid phosphorylation of eNOS at the plasma membrane, resulting in production of nitric oxide (Chambliss and others, 2000; Chen and others, 1999; Dimmeler and others, 1999; Haynes and others, 2000). Caveoli are abundant in smooth muscle and endothelial cells, and PI3K/AKT phosphorylation of eNOS is thought to mediate many of the protective effects of estrogen in the cardiovascular system such as acute vasodilatation. Ligand-activated ER also associates with and activates the insulin-like growth factor receptor (IGF-1R) (Kahlert and others, 2000) and can directly bind to Src and Shc (Castoria and others, 2001; Kousteni and others, 2001; Migliaccio and others, 2000; Migliaccio and others, 1996; Song and others, 2004; Song and others, 2002a; Song and others, 2002b; Song and others, 2005; Wong and others, 2002), leading to activation of PI3K and MAPK signaling.
Developmental Reprogramming of Histone Methylation in the Developing Uterus
What are the targets for rapid, nongenomic ER signaling? In some cases it is the steroid hormone receptors themselves, as their classical genomic action as transcription factors is potentiated when phosphorylated at specific amino acids (Kato and others, 1995). Importantly, it now appears that these same signal transduction pathways can phosphorylate chromatin remodeling proteins and induce alterations in epigenetic histone modifications.
The best data to date involve PI3K signaling via AKT and phosphorylation of EZH2. Although AKT is activated at the membrane, this kinase subsequently translocates to several sub-cellular compartments, including the nucleus, to phosphorylate target substrates (Ahmed and others, 1993; Meier and others, 1997; Neri and others, 2002). As mentioned above, EZH2 is a member of the polycomb family of HKMTs, which methylates lysine 27 (K27) of histone H3 to epigenetically silence genes such as HOXA9 (Cao and Zhang, 2004). A specific AKT phosphorylation site at serine 21 in EZH2 is phosphorylated by this kinase, inactivating this HKMT and reducing K27 trimethylation of H3 (Cha and others, 2005). As a result, levels of H3 trimethylated K27 are depressed in cells with activated AKT, which correlated with derepression of silenced genes including HOXA9. This study demonstrated that the HKMT EZH2 was regulated by AKT and that phosphorylation of EZH2 by AKT suppressed histone H3 methylation, disrupting epigenetic gene regulation (Cha and others, 2005). Importantly, these data demonstrate that activation of cell signaling, and in particular AKT activation, can result in phosphorylation of chromatin remodeling proteins in the nucleus, leading to changes in histone modifications, chromatin conformation and changes in gene expression.
We recently identified a novel role for this non-genomic (or more appropriately pre-genomic) signaling by xenoestrogens via activation of membrane-associated ER and regulation of EZH2 to reprogram the developing uterus (Bredfeldt and others, 2010). In response to both 17-b estradiol (E2) and the xenoestrogen diethylstilbestrol (DES), ER signaling via PI3K/AKT phosphorylates EZH2 at S21, reducing H3K27Me3 levels in hormone-responsive cells. During windows of uterine development that are susceptible to developmental reprogramming, activation of this ER signaling pathway by DES results in phosphorylation of EZH2 and reduced H3K27Me3 levels in chromatin of the developing uterus. In response to this inappropriate activation of pre-genomic signaling, the expression profile of estrogen-responsive genes in uterine myometrial cells is reprogrammed, suggesting this as a potential mechanism for developmental reprogramming caused by early life exposure to xenoestrogens. Thus, pre-genomic, rapid ER signaling provides the first direct linkage between xenoestrogen-induced nuclear hormone receptor signaling and modulation of the epigenetic machinery during tissue development to induce developmental reprogramming.
Summary
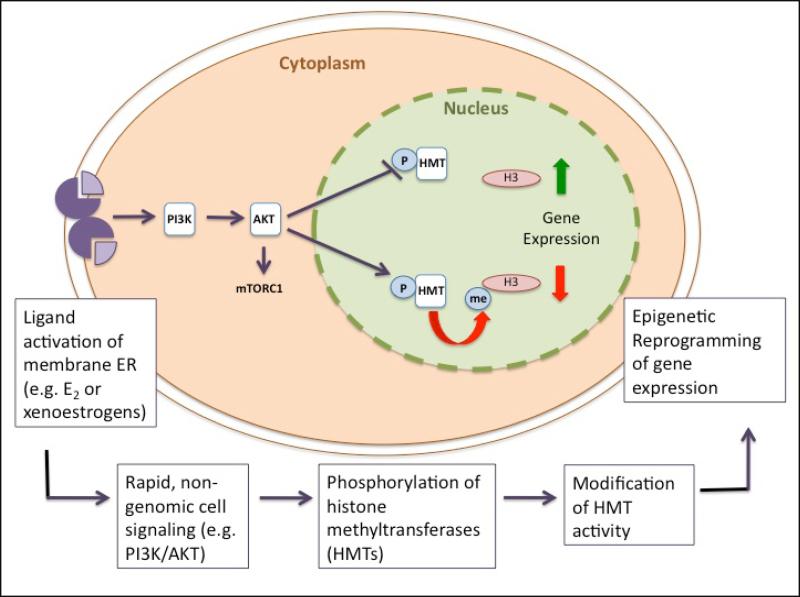
The emerging picture for epigenetic reprogramming of the developing reproductive tract involves inappropriate activation of cell signaling via non-genomic ER signaling, post-translational modification of histone methyltransferases, alteration of histone methylation patterns and effects on gene expression as shown in Figure 1. Activation of non-genomic signaling by environmental estrogens to regulate histone methyltransferase activity provides the first direct pathway linking environmental exposures to the cell’s epigenetic machinery and developmental reprogramming. While current data support activation of PI3K signaling and AKT to modulate histone methyltransferase activity, it is clearly possible that activation of other signaling pathways such as MAPK signaling and ERK, could also be involved, as these pathways are activated in response to non-genomic signaling as well. Furthermore, while in the case of EZH2, histone methyltransferase activity is inhibited by phosphorylation, it is likely that phosphorylation could increase the activity of other histone methyltransferases, or modulate the activity of histone demethylases to reprogram the developing epigenome.
Figure 1.
Non-genomic Signaling Links Xenoestrogen Exposure to Epigenetic Reprogramming. Exposure to xenoestrogens inappropriately actuates cell signaling pathways such as P13K. This non-genomic ER signaling can phosphorylate and modify the activity of epigenetic “writers” such as histone methyltransferases. As a result, epigenetic programs are not properly “installed”, resulting in developmental reprogramming and enhanced disease susceptibility.
Acknowledgments
Grants: This research is supported in part by the National Institutes of Health through MD Anderson's Cancer Center Support Grant P30CA016672, and National Institute of Environmental Health Sciences Grants RC2ES018789, R01ES008263, and P30ES007784.
Literature Cited
- Ahmed NN, Franke TF, Bellacosa A, Datta K, Gonzalez-Portal ME, Taguchi T, Testa JR, Tsichlis PN. The proteins encoded by c-akt and v-akt differ in post-translational modification, subcellular localization and oncogenic potential. Oncogene. 1993;8(7):1957–1963. [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Kouzarides T. Histone methylation: dynamic or static? Cell. 2002;109(7):801–806. doi: 10.1016/s0092-8674(02)00798-5. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410(6824):120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Barker DJ. Fetal programming of coronary heart disease. Trends Endocrinol Metab. 2002;13(9):364–368. doi: 10.1016/s1043-2760(02)00689-6. [DOI] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447(7143):407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Bjornstrom L, Sjoberg M. Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol Endocrinol. 2005;19(4):833–842. doi: 10.1210/me.2004-0486. [DOI] [PubMed] [Google Scholar]
- Block K, Kardana A, Igarashi P, Taylor HS. In utero diethylstilbestrol (DES) exposure alters Hox gene expression in the developing mullerian system. FASEB J. 2000;14(9):1101–1108. doi: 10.1096/fasebj.14.9.1101. [DOI] [PubMed] [Google Scholar]
- Bredfeldt TG, Greathouse KL, Safe SH, Hung MC, Bedford MT, Walker CL. Xenoestrogen-induced regulation of EZH2 and histone methylation via estrogen receptor signaling to PI3K/AKT. Mol Endocrinol. 2010;24(5):993–1006. doi: 10.1210/me.2009-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15(1):57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Castoria G, Barone MV, Di Domenico M, Bilancio A, Ametrano D, Migliaccio A, Auricchio F. Non transcriptional action of oestradiol and progestin triggers DNA synthesis. EMBO J. 1999;18(9):2500–2510. doi: 10.1093/emboj/18.9.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoria G, Migliaccio A, Bilancio A, Di Domenico M, de Falco A, Lombardi M, Fiorentino R, Varricchio L, Barone MV, Auricchio F. PI3-kinase in concert with Src promotes the S-phase entry of oestradiol-stimulated MCF-7 cells. EMBO J. 2001;20(21):6050–6059. doi: 10.1093/emboj/20.21.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cato AC, Nestl A, Mink S. Rapid actions of steroid receptors in cellular signaling pathways. Sci STKE. 2002;2002(138):RE9. doi: 10.1126/stke.2002.138.re9. [DOI] [PubMed] [Google Scholar]
- Cha TL, Zhou BP, Xia W, Wu Y, Yang CC, Chen CT, Ping B, Otte AP, Hung MC. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science. 2005;310(5746):306–310. doi: 10.1126/science.1118947. [DOI] [PubMed] [Google Scholar]
- Chambliss KL, Yuhanna IS, Mineo C, Liu P, German Z, Sherman TS, Mendelsohn ME, Anderson RG, Shaul PW. Estrogen receptor alpha and endothelial nitric oxide synthase are organized into a functional signaling module in caveolae. Circ Res. 2000;87(11):E44–52. doi: 10.1161/01.res.87.11.e44. [DOI] [PubMed] [Google Scholar]
- Chen Z, Yuhanna IS, Galcheva-Gargova Z, Karas RH, Mendelsohn ME, Shaul PW. Estrogen receptor alpha mediates the nongenomic activation of endothelial nitric oxide synthase by estrogen. J Clin Invest. 1999;103(3):401–406. doi: 10.1172/JCI5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheskis BJ. Regulation of cell signalling cascades by steroid hormones. J Cell Biochem. 2004;93(1):20–27. doi: 10.1002/jcb.20180. [DOI] [PubMed] [Google Scholar]
- Cheung P, Lau P. Epigenetic regulation by histone methylation and histone variants. Mol Endocrinol. 2005;19(3):563–573. doi: 10.1210/me.2004-0496. [DOI] [PubMed] [Google Scholar]
- Cook JD, Davis BJ, Cai SL, Barrett JC, Conti CJ, Walker CL. Interaction between genetic susceptibility and early-life environmental exposure determines tumor-suppressor-gene penetrance. Proc Natl Acad Sci U S A. 2005;102(24):8644–8649. doi: 10.1073/pnas.0503218102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couse JF, Dixon D, Yates M, Moore AB, Ma L, Maas R, Korach KS. Estrogen receptor-alpha knockout mice exhibit resistance to the developmental effects of neonatal diethylstilbestrol exposure on the female reproductive tract. Dev Biol. 2001;238(2):224–238. doi: 10.1006/dbio.2001.0413. [DOI] [PubMed] [Google Scholar]
- Couzin J. Quirks of fetal environment felt decades later. Science. 2002;296(5576):2167–2169. doi: 10.1126/science.296.5576.2167. [DOI] [PubMed] [Google Scholar]
- Dimmeler S, Fleming I, Fisslthaler B, Hermann C, Busse R, Zeiher AM. Activation of nitric oxide synthase in endothelial cells by Akt-dependent phosphorylation. Nature. 1999;399(6736):601–605. doi: 10.1038/21224. [DOI] [PubMed] [Google Scholar]
- Edwards DP. Regulation of signal transduction pathways by estrogen and progesterone. Annu Rev Physiol. 2005;67:335–376. doi: 10.1146/annurev.physiol.67.040403.120151. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4(2):143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- Frankel S, Elwood P, Sweetnam P, Yarnell J, Smith GD. Birthweight, body-mass index in middle age, and incident coronary heart disease. Lancet. 1996;348(9040):1478–1480. doi: 10.1016/S0140-6736(96)03482-4. [DOI] [PubMed] [Google Scholar]
- Gluckman PD, Hanson MA. Living with the past: evolution, development, and patterns of disease. Science. 2004;305(5691):1733–1736. doi: 10.1126/science.1095292. [DOI] [PubMed] [Google Scholar]
- Hattersley AT, Tooke JE. The fetal insulin hypothesis: an alternative explanation of the association of low birthweight with diabetes and vascular disease. Lancet. 1999;353(9166):1789–1792. doi: 10.1016/S0140-6736(98)07546-1. [DOI] [PubMed] [Google Scholar]
- Haynes MP, Sinha D, Russell KS, Collinge M, Fulton D, Morales-Ruiz M, Sessa WC, Bender JR. Membrane estrogen receptor engagement activates endothelial nitric oxide synthase via the PI3-kinase-Akt pathway in human endothelial cells. Circ Res. 2000;87(8):677–682. doi: 10.1161/01.res.87.8.677. [DOI] [PubMed] [Google Scholar]
- Herbst AL, Ulfelder H, Poskanzer DC. Adenocarcinoma of the vagina. Association of maternal stilbestrol therapy with tumor appearance in young women. N Engl J Med. 1971;284(15):878–881. doi: 10.1056/NEJM197104222841604. [DOI] [PubMed] [Google Scholar]
- Ho SM, Tang WY, Belmonte de Frausto J, Prins GS. Developmental exposure to estradiol and bisphenol A increases susceptibility to prostate carcinogenesis and epigenetically regulates phosphodiesterase type 4 variant 4. Cancer Res. 2006;66(11):5624–5632. doi: 10.1158/0008-5472.CAN-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson V, Chalkley R. Histone segregation on replicating chromatin. Biochem. 1985;24(24):6930–6938. doi: 10.1021/bi00345a027. [DOI] [PubMed] [Google Scholar]
- Jenuwein T. The epigenetic magic of histone lysine methylation. FEBS J. 2006;273(14):3121–3135. doi: 10.1111/j.1742-4658.2006.05343.x. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8(4):253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlert S, Nuedling S, van Eickels M, Vetter H, Meyer R, Grohe C. Estrogen receptor alpha rapidly activates the IGF-1 receptor pathway. J Biol Chem. 2000;275(24):18447–18453. doi: 10.1074/jbc.M910345199. [DOI] [PubMed] [Google Scholar]
- Kato S, Endoh H, Masuhiro Y, Kitamoto T, Uchiyama S, Sasaki H, Masushige S, Gotoh Y, Nishida E, Kawashima H, et al. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 1995;270(5241):1491–1494. doi: 10.1126/science.270.5241.1491. [DOI] [PubMed] [Google Scholar]
- Kim HP, Lee JY, Jeong JK, Bae SW, Lee HK, Jo I. Nongenomic stimulation of nitric oxide release by estrogen is mediated by estrogen receptor alpha localized in caveolae. Biochem Biophys Res Commun. 1999;263(1):257–262. doi: 10.1006/bbrc.1999.1348. [DOI] [PubMed] [Google Scholar]
- Kousteni S, Bellido T, Plotkin LI, O'Brien CA, Bodenner DL, Han L, Han K, DiGregorio GB, Katzenellenbogen JA, Katzenellenbogen BS, Roberson PK, Weinstein RS, Jilka RL, Manolagas SC. Nongenotropic, sex-nonspecific signaling through the estrogen or androgen receptors: dissociation from transcriptional activity. Cell. 2001;104(5):719–730. [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410(6824):116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Sullivan RJ, Jenuwein T. An epigenetic road map for histone lysine methylation. J Cell Sci. 2003;116(Pt 11):2117–2124. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- Lee DY, Teyssier C, Strahl BD, Stallcup MR. Role of protein methylation in regulation of transcription. Endocr Rev. 2005;26(2):147–170. doi: 10.1210/er.2004-0008. [DOI] [PubMed] [Google Scholar]
- Levin ER. Integration of the extranuclear and nuclear actions of estrogen. Mol Endocrinol. 2005;19(8):1951–1959. doi: 10.1210/me.2004-0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Hansman R, Newbold R, Davis B, McLachlan JA, Barrett JC. Neonatal diethylstilbestrol exposure induces persistent elevation of c-fos expression and hypomethylation in its exon-4 in mouse uterus. Mol Carcinog. 2003;38(2):78–84. doi: 10.1002/mc.10147. [DOI] [PubMed] [Google Scholar]
- Losel R, Wehling M. Nongenomic actions of steroid hormones. Nat Rev Mol Cell Biol. 2003;4(1):46–56. doi: 10.1038/nrm1009. [DOI] [PubMed] [Google Scholar]
- Losel RM, Falkenstein E, Feuring M, Schultz A, Tillmann HC, Rossol-Haseroth K, Wehling M. Nongenomic steroid action: controversies, questions, and answers. Physiol Rev. 2003;83(3):965–1016. doi: 10.1152/physrev.00003.2003. [DOI] [PubMed] [Google Scholar]
- Luger K, Richmond TJ. DNA binding within the nucleosome core. Curr Opin Struct Biol. 1998;8(1):33–40. doi: 10.1016/s0959-440x(98)80007-9. [DOI] [PubMed] [Google Scholar]
- Ma L, Benson GV, Lim H, Dey SK, Maas RL. Abdominal B (AbdB) Hoxa genes: regulation in adult uterus by estrogen and progesterone and repression in mullerian duct by the synthetic estrogen diethylstilbestrol (DES). Dev Biol. 1998;197(2):141–154. doi: 10.1006/dbio.1998.8907. [DOI] [PubMed] [Google Scholar]
- Martin C, Zhang Y. Mechanisms of epigenetic inheritance. Curr Opin Cell Biol. 2007;19(3):266–272. doi: 10.1016/j.ceb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Meier R, Alessi DR, Cron P, Andjelkovic M, Hemmings BA. Mitogenic activation, phosphorylation, and nuclear translocation of protein kinase Bbeta. J Biol Chem. 1997;272(48):30491–30497. doi: 10.1074/jbc.272.48.30491. [DOI] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437(7057):436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- Migliaccio A, Castoria G, Di Domenico M, de Falco A, Bilancio A, Lombardi M, Barone MV, Ametrano D, Zannini MS, Abbondanza C, Auricchio F. Steroid-induced androgen receptor-oestradiol receptor beta-Src complex triggers prostate cancer cell proliferation. EMBO J. 2000;19(20):5406–5417. doi: 10.1093/emboj/19.20.5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliaccio A, Di Domenico M, Castoria G, de Falco A, Bontempo P, Nola E, Auricchio F. Tyrosine kinase/p21ras/MAP-kinase pathway activation by estradiol-receptor complex in MCF-7 cells. EMBO J. 1996;15(6):1292–1300. [PMC free article] [PubMed] [Google Scholar]
- Miller C, Degenhardt K, Sassoon DA. Fetal exposure to DES results in de-regulation of Wnt7a during uterine morphogenesis. Nat Genet. 1998;20(3):228–230. doi: 10.1038/3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri LM, Borgatti P, Capitani S, Martelli AM. The nuclear phosphoinositide 3-kinase/AKT pathway: a new second messenger system. Biochim Biophys Acta. 2002;1584(2-3):73–80. doi: 10.1016/s1388-1981(02)00300-1. [DOI] [PubMed] [Google Scholar]
- Newbold RR, Bullock BC, McLachlan JA. Uterine adenocarcinoma in mice following developmental treatment with estrogens: a model for hormonal carcinogenesis. Cancer Res. 1990;50(23):7677–7681. [PubMed] [Google Scholar]
- Newbold RR, Hanson RB, Jefferson WN. Ontogeny of lactoferrin in the developing mouse uterus: a marker of early hormone response. Biol Reprod. 1997;56(5):1147–1157. doi: 10.1095/biolreprod56.5.1147. [DOI] [PubMed] [Google Scholar]
- Razandi M, Oh P, Pedram A, Schnitzer J, Levin ER. ERs associate with and regulate the production of caveolin: implications for signaling and cellular actions. Mol Endocrinol. 2002;16(1):100–115. doi: 10.1210/mend.16.1.0757. [DOI] [PubMed] [Google Scholar]
- Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr Opin Cell Biol. 2001;13(3):263–273. doi: 10.1016/s0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- Schlegel A, Wang C, Katzenellenbogen BS, Pestell RG, Lisanti MP. Caveolin-1 potentiates estrogen receptor alpha (ERalpha) signaling. caveolin-1 drives ligand-independent nuclear translocation and activation of ERalpha. J Biol Chem. 1999;274(47):33551–33556. doi: 10.1074/jbc.274.47.33551. [DOI] [PubMed] [Google Scholar]
- Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, Bergman Y, Simon I, Cedar H. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39(2):232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- Simoncini T, Hafezi-Moghadam A, Brazil DP, Ley K, Chin WW, Liao JK. Interaction of oestrogen receptor with the regulatory subunit of phosphatidylinositol-3-OH kinase. Nature. 2000;407(6803):538–541. doi: 10.1038/35035131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Nishioka K, Reinberg D. Histone lysine methylation: a signature for chromatin function. Trends Genet. 2003;19(11):629–639. doi: 10.1016/j.tig.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Smallwood A, Esteve PO, Pradhan S, Carey M. Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev. 2007;21(10):1169–1178. doi: 10.1101/gad.1536807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo JM, Stahl H, Koller T, Knippers R. Structure of replicating simian virus 40 minichromosomes. The replication fork, core histone segregation and terminal structures. J Mol Biol. 1986;189(1):189–204. doi: 10.1016/0022-2836(86)90390-6. [DOI] [PubMed] [Google Scholar]
- Song RX, Barnes CJ, Zhang Z, Bao Y, Kumar R, Santen RJ. The role of Shc and insulin-like growth factor 1 receptor in mediating the translocation of estrogen receptor alpha to the plasma membrane. Proc Natl Acad Sci U S A. 2004;101(7):2076–2081. doi: 10.1073/pnas.0308334100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song RX, McPherson RA, Adam L, Bao Y, Shupnik M, Kumar R, Santen RJ. Linkage of rapid estrogen action to MAPK activation by ERalpha-Shc association and Shc pathway activation. Mol Endocrinol. 2002a;16(1):116–127. doi: 10.1210/mend.16.1.0748. [DOI] [PubMed] [Google Scholar]
- Song RX, Santen RJ, Kumar R, Adam L, Jeng MH, Masamura S, Yue W. Adaptive mechanisms induced by long-term estrogen deprivation in breast cancer cells. Mol Cell Endocrinol. 2002b;193(1-2):29–42. doi: 10.1016/s0303-7207(02)00093-x. [DOI] [PubMed] [Google Scholar]
- Song RX, Zhang Z, Santen RJ. Estrogen rapid action via protein complex formation involving ERalpha and Src. Trends Endocrinol Metab. 2005;16(8):347–353. doi: 10.1016/j.tem.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Sulis ML, Parsons R. PTEN: from pathology to biology. Trends Cell Biol. 2003;13(9):478–483. doi: 10.1016/s0962-8924(03)00175-2. [DOI] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116(1):51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- Tsukada YI, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2005 doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22(9):836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, Bollen M, Esteller M, Di Croce L, de Launoit Y, Fuks F. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439(7078):871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2(7):489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y, Roeder RG, Clarke S, Stallcup MR, Allis CD, Coonrod SA. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306(5694):279–283. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- Wong CW, McNally C, Nickbarg E, Komm BS, Cheskis BJ. Estrogen receptor-interacting protein that modulates its nongenomic activity-crosstalk with Src/Erk phosphorylation cascade. Proc Natl Acad Sci U S A. 2002;99(23):14783–14788. doi: 10.1073/pnas.192569699. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15(18):2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Rank G, Tan YT, Li H, Moritz RL, Simpson RJ, Cerruti L, Curtis DJ, Patel DJ, Allis CD, Cunningham JM, Jane SM. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat Struct Mol Biol. 2009;16(3):304–311. doi: 10.1038/nsmb.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]