Abstract
Protein kinases are obvious drug targets against cancer due to their central role in cellular regulation. Since the discovery of Gleevec, a potent and specific inhibitor of Abl kinase, as a highly successful cancer therapeutic, the ability of this drug to distinguish between Abl and other tyrosine kinases like Src has been intensely investigated, but without much success. Using NMR and fast kinetics, we establish a novel model that solves this longstanding question of two tyrosine kinases adopting almost identical structures when bound to Gleevec, yet having vastly different affinities. In contrast to all other proposed models we show that the origin of Abl’s high affinity lies predominantly in a conformational change after binding. An energy landscape that provides tight affinity via an induced-fit and binding plasticity via conformational selection mechanism is likely to be general for many inhibitors.
INTRODUCTION
The fundamental importance of protein kinases is indisputable. Their central role in essential physiological processes have provoked extensive studies and resulted in a wealth of knowledge from biological signaling cascades to atomistic structural details1–3. Kinases are obvious attractive therapeutic drug targets, since different signaling cascades can be selectively regulated by inhibiting individual kinases4,5. However, all kinases share a great degree of similarity, making it difficult to design inhibitors that are specific for a particular kinase6–10. This complication has hampered progress in drug development and highlights the need for a deeper understanding of the biophysical principles that govern kinase-drug interactions11.
A prominent translational-research success story in treating chronic myeloid leukemia is the potent drug Gleevec (Imatinib) that specifically targets tyrosine kinase Abl. Its success is mainly due to the high specificity for the Abl subfamily of kinases as compared to its closest relative the Src subfamily. The kinase domain of Src shares 54% sequence identity with Abl, and its drug binding pocket with Gleevec bound is nearly identical to Abl in both sequence and structure, but surprisingly Src has about 3000 times weaker affinity for Gleevec12. The high clinical relevance and puzzling mismatch between structural similarity and different biochemical characteristics, has placed the selectivity of Gleevec for Abl under intense scrutiny for the last 20 years, but ultimately without decisive success12.
Early crystal structures showed that the highly conserved DFG-motif (Asp-Phe-Gly), in the activation loop of kinases, adopts two distinct conformations in Src and Abl. It was therefore proposed that the inactive conformation of Src prevents Gleevec binding due to direct steric clashes13–17. However a new structure solved later revealed that Src is in fact capable of adopting the Abl-like clash-free inactive conformation12. Moreover, it was also found that Abl is capable of adopting a Src-like inactive state18. With this initial hypothesis ruled out, two alternative explanations were put forward. According to the first one the difference in affinity is due to subtle changes in the drug binding pocket. Kuriyan and coworkers tested this idea by substituting residues in Src with the corresponding Abl residues12. This extensive mutagenesis screening showed that none of the substitutions (alone or in combinations) resulted in substantial increase in Gleevec affinity. This led to an alternative hypothesis in which both enzymes are capable of adopting a DFG-out conformation but they differ in the probability of occupying that conformation; thus binding of Gleevec is regulated via a conformational selection mechanism12,19–23. Monitoring the dynamics of the DFG-loop in kinases by NMR24,25 has not been successful because the corresponding peaks were missing in the apo spectra. Due to the lack of experimental results, several groups used molecular dynamics simulations to calculate different components of Gleevec binding free energy rationalizing the huge difference in affinity with controversial conclusions19–21,26. In summary, the question of why Gleevec is a potent inhibitor of Abl but not Src remains controversial and unresolved20. Here we set out to solve this open energetic question.
Extensive history in protein biochemistry shows that kinetic and energetic properties can rarely be inferred from high-resolution crystal structures alone. In this work we use a combination of pre-steady-state fluorescence kinetics and NMR spectroscopy to study directly the process of Gleevec binding to the catalytic domain of Abl and Src with millisecond time resolution and residue-specific precision. These data reveal a novel mechanism for Gleevec binding that quantitatively accounts for the difference in Gleevec affinity between Src and Abl.
RESULTS
NMR titration of Gleevec reveals an induced fit mechanism
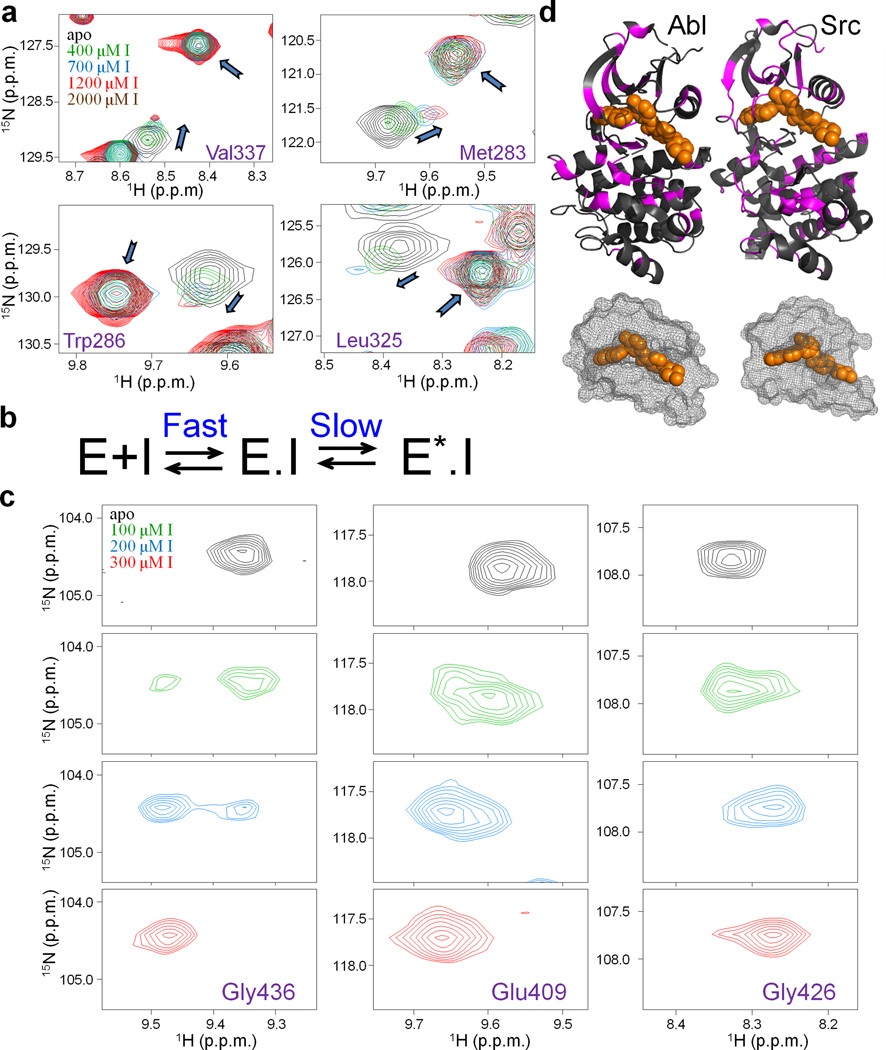
Binding of an inhibitor to its target protein is a dynamic process that cannot be understood solely based on structural data. NMR can provide information about structural changes within a protein during binding and detect timescales of these processes. To this end we titrated Gleevec into Src and Abl, and used [1H,15N]-HSQC spectra to monitor the binding. In the case of Src the pattern of peak movement was very unusual. Upon addition of increasing amounts of drug, peaks gradually shifted and simultaneously appeared at new positions (Fig. 1a). In general, peak shifting in a titration experiment indicates that the corresponding residue is in fast exchange between two states (>100 s−1 for typical chemical shift differences). This is in contrast to slow exchange (<1 s−1), wherein peaks disappear at one position and appear at another. In our case we observed both phenomena simultaneously, indicating that Gleevec binding is at least a two-step process, with one of the steps being fast and the other one slow. Residues that showed chemical shifts perturbations upon addition of Gleevec were not clustered around the Gleevec binding site or the DFG-loop. Instead they were distributed over a large fraction of the protein, suggestive of a global conformational change (Fig. 1b). In general, such conformational change can happen before or after binding. To differentiate between these two scenarios we simulated the spectroscopic behavior for all possible binding models27 (Supplementary Fig. 1a–d). The only scheme compatible with the observed pattern (Fig. 1a) is the one where Gleevec binding is fast, and is followed by a slow conformational change (Fig. 1b).
Figure 1.
Monitoring the Gleevec binding process to Src and Abl by NMR at 25 °C. (a) Zoom of [1H, 15N]-HSQC spectra showing an unusual pattern of chemical shifts perturbations (shifting of free peak position coinciding with the reappearance of Gleevec-bound peak) upon the titration of Src with Gleevec. (b) Biochemical scheme that can explain the observed titration patterns27 (see also Supplementary Fig. 1). E and E.I correspond to free and inhibitor bound kinase, E*.I corresponds to inhibitor bound kinase in a distinct conformational state. (c) Abl titration shows a simpler pattern with disappearance and reappearance of the peaks indicative of tight binding. (d) Residues with chemical shift changes upon Gleevec binding are plotted onto the crystal structures (pdb ids: 1OPJ, 2OIQ16,31) in magenta, Gleevec is shown in orange. The mesh representation of the Gleevec binding pocket illustrates how part of the drug is covered by the protein suggesting a conformational change after binding (step 2 in Fig. 1b).
This model of Gleevec binding is dramatically different from the models proposed earlier, in two distinct ways. First, our data uncover a previously undescribed step: a structural transition that follows Gleevec binding (E.I ⇔ E*.I, where E.I and E*.I are the two distinct conformations of the kinase with Gleevec bound) and stabilizes the Src.Gleevec complex. Second, in contrast to previous observations, this scheme implies that Gleevec binding is fast, and the rate-limiting step is, in fact, the structural transition.
NMR titration of Abl with Gleevec showed a simpler pattern (Fig. 1c) with peaks disappearing at one position and appearing at a second position. This is an indication that in Abl the E.I ⇔ E*.I equilibrium is far shifted towards the E*.I state (Fig. 1b), resulting in negligible population of the E.I complex at any concentration of the drug. Under such a scenario, the fast drug-binding step is undetectable (Supplementary Fig. 1e). This NMR spectroscopic pattern could naturally be explained by a simple 2-state mechanism (E+I⇔E.I, where E, I and E.I are apo kinase, Gleevec and kinase-Gleevec complex, respectively), however such model was ruled out by experiments described below. Such fundamentally distinct spectroscopic characteristics delivered the first sign that the difference in enzyme affinities is not due to the difference in the initial binding step but caused by a different conformational equilibrium after binding.
Real-time Gleevec binding kinetics
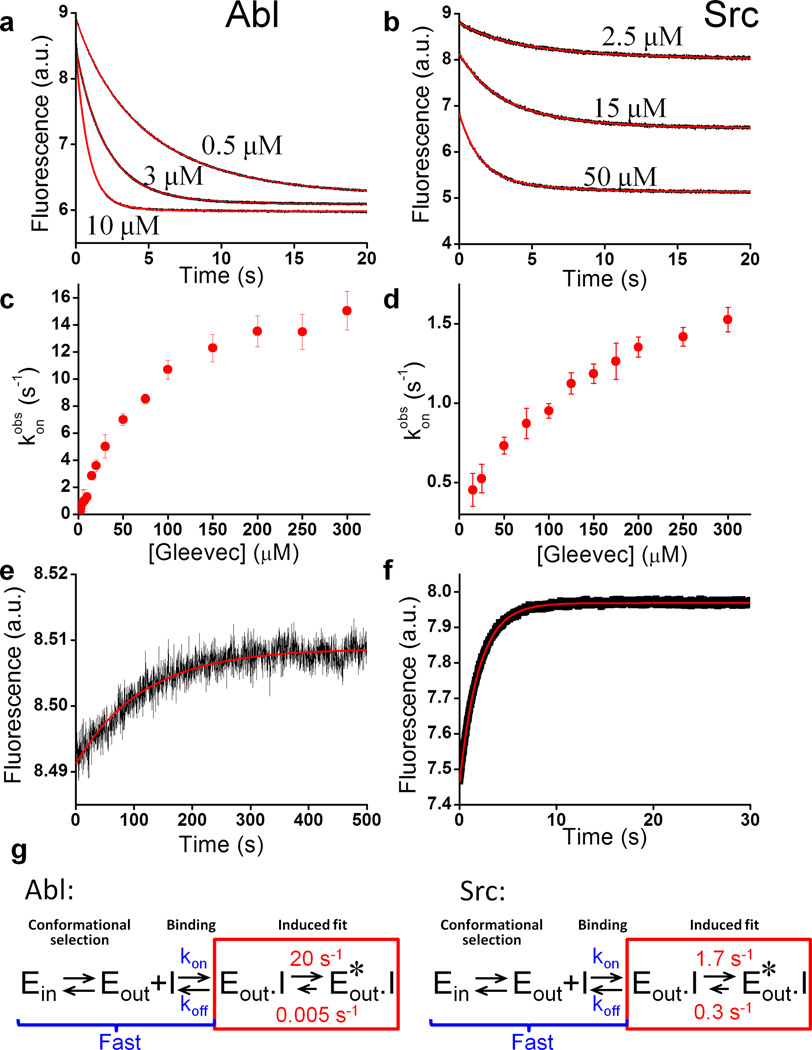
To buttress this hypothesis and to quantitatively characterize the energetics of Gleevec binding to Src and Abl, we performed a series of rapid-mixing experiments that allowed direct monitoring of inhibitor binding kinetics. Complex formation and conformational changes were detected using the enzymes’ intrinsic tryptophan fluorescence. For inhibitor binding to either enzyme, fluorescence kinetics at all Gleevec concentrations could be adequately fit by a single exponential at 25 °C (Fig. 2a,b). Interestingly, the dependence of the observed rate on drug concentration is non-linear with an apparent plateau at approximately 20 s−1 and 2 s−1 for Abl and Src, respectively (Fig. 2c,d).
Figure 2.
Kinetics of Gleevec binding to Abl and Src at 25 °C. Trp fluorescence change after mixing of Abl (a) or Src (b) with increasing amounts of Gleevec. Both Abl and Src kinetics are monoexponential. (c,d) The observed rates (konobs) for Gleevec binding to Abl (c) and Src (d) do not show the expected linear dependence on the Gleevec concentration, but a curvature approaching a plateau in agreement with the proposed binding scheme in Fig. 1b. (n=3 experiments, mean ± s. e. m.) (e,f) Dissociation kinetics of Gleevec from Abl and Src measured by dilution of Abl.Gleevec and Src.Gleevec complexes. (g) Binding scheme highlighting a 1000-fold tighter affinity for Abl caused by a newly identified conformational step after binding (red). Ein and Eout defines apo kinase with the DFG-loop in “in” and “out” position respectively, Eout.I and Eout*.I is kinase with DFG-loop in the “out” position and Gleevec bound, and the relevant rate constants are defined.
How could these data be interpreted? Non-linear concentration dependence of the observed binding rates indicates that it is not a simple pseudo-first-order binding, but rather protein conformational transitions are rate-limiting, in agreement with the scheme in Fig. 1b. At the same time, this scheme implies a two-step process that should be characterized by double-exponential kinetics, which is not observed. To resolve this apparent contradiction, we repeated the experiments at 5 °C with the idea to slow down possibly unresolved processes. At this temperature Gleevec binding to Abl is indeed clearly double-exponential (Fig. 3a). The fast phase (kobsbind) follows a linear dependence on Gleevec concentration (Fig. 3e) corresponding to the binding step, while the slower step (kobsconf) shows the curvature (Fig. 3f) seen before in Fig. 2c and corresponds to the conformational transition after binding (induced fit) (Fig. 2g, Table 1). Fits of these data demonstrate that rate constants for the binding step are fast, 1.5 µM−1s−1 and 25 s−1 respectively, and that the conformational transition is substantially slower (see “analysis of kinetic data” in online methods and Supplementary Fig. 6 for more details) This implies that the rates determined at 25 °C reflect the conformational transitions, and that the faster phase is not observed because at the higher temperature binding is finished within the dead time of the instrument (5 ms). With this insight Fig. 2a–d has the following interpretation: binding of the drug to both enzymes is fast, and the observed kinetics reflects the conformational transition induced by Gleevec. The fact that the observed rate of a conformational transition depends on ligand concentration might be counterintuitive, but at increasing drug concentration the transient concentration of the E.I complexes increases, which leads to an increase of the apparent rate (Fig. 1b, 2c,d) (see “analysis of kinetic data” section in online methods and Supplementary Fig. 6). We note that the binding step could not be directly observed for Src due to a too small population of the binding competent state as discussed below (Supplementary Fig. 2).
Figure 3.
Kinetics of Gleevec binding measured at 5 °C allows dissection of energetics of individual steps. (a) Gleevec binding to Abl at 5 °C is biphasic. Double-exponential fit (red) gives an excellent fit, but not a mono-exponential fit (cyan). (b) Gleevec binding to Src at 5 °C is monophasic. Mono-exponential fit (cyan) is as good as double-exponential fit (red). (c,d) Dissociation of Gleevec from Abl and Src. (e,f) Concentration dependence of the fast (e) and slow (f) phases of Gleevec binding to Abl identifies them as the binding and conformational step, respectively. (n=3 experiments, mean ± s. e. m.) (g) Gleevec’s overall observed Kdobs to Abl at 5 °C determined independently via steady-state measurement of Trp fluorescence quench by Gleevec. (n=3 experiments, mean ± s. e. m.)
Table 1.
Rate constants of binding (kon and koff) and conformational change steps (kconf+ and kconf−) and corresponding dissociation constants (Kd) and free energy changes (ΔG) in the Abl.Gleevec binding scheme obtained from data in Fig. 3.
| Binding | Conformational change |
|---|---|
| koff = 25±6 s−1 kon = 1.5±0.1 µM−1s−1 |
kconf−= (7±1) •10−4 s−1 kconf+=1.5±0.2 s−1 |
| Kdbind =17±4 µM (ΔG=−6.5±0.1 kcal/mol) |
Kdconf=(4.7±0.9)•10−4 (ΔG= −4.5±0.1 kcal/mol) |
| Kdkin = Kdbind•Kdconf /(1+Kdconf) = 8±3 nM (ΔG=−11±0.3 kcal/mol) | |
Determining factor for dramatic difference in Gleevec’s affinity
Where do these kinetic experiments take us towards understanding the underlying mechanism for the 3000-fold difference in Gleevec affinity? A final experiment completes the puzzle, measuring the kinetics of dissociation. Enzyme incubated with Gleevec was rapidly diluted 11-fold to initiate dissociation. The observed rate of fluorescent change is remarkably slow, especially in Abl (Fig. 2e, 3c). Considering that even at 5 °C the actual “off” rate constant from the E.I state was 25 s−1 (Fig. 3e, Table 1), the observed slow process must be attributed to slow conformational change from E*.I to E.I. Thus we can conclude that the rate constants 0.3 s−1 and 0.005 s−1 obtained in the dilution experiments correspond to the E*.I →E.I transition in Src and Abl, respectively (Fig. 2g).
Strikingly, the measured difference in the conformational equilibrium between two drug-bound states accounts for an approximately 1000-fold difference in Gleevec affinity between Abl and Src: the forward rate constants differ 10-fold, and the difference in the reverse direction is 100-fold (Fig. 2g). These two effects account for all but 3-fold of the overall Gleevec binding affinity. This remaining difference probably arises from differences in the Ein/Eout equilibrium in the apo protein (corresponding to the “in” and “out” position of the DFG loop) and/or the binding step (Fig. 2g).
Energetic dissection of individual binding steps
The fact that both phases can be observed for Abl at 5 °C enables us to calculate the Kd for Gleevec binding purely based on the measured kinetics (Kdkin), and then to compare it with the global Kdobs from thermodynamic experiments (Fig. 3g). These two independently determined Kd’s are remarkably close (Table 1, Fig. 3g), substantiating our new proposed mechanism underlying Gleevec specificity. As a second test of our model a global fit of all fluorescence kinetics data was performed using numerical simulations (Fig. 4)28. First, all fitted microscopic rate constants are in excellent agreement with the values from the analysis described in Fig. 3, and second, possible alternative models fail to fit the data (Supplementary Fig. 3 and Supplementary Fig. 4).
Figure 4.
Global fit of Abl binding and dissociation kinetics data measured by Trp fluorescence at 5 °C. Black circles – experimental data, red lines - global numerical simulation of the whole dataset using Kintek Explorer software (Kintek Corp)28,32. The induced fit model (E⇔E.I⇔E*.I) was used and the global fitting results (kon=1.3 ± 0.3 µM−1s−1, koff = 23 ± 5 s−1, kconf+ = 1.3 ± 0.2 s−1, kconf− = (6±2)•10−4 s−1) were in excellent agreement with individual fits (Table 1) validating the robustness of the model.
Importantly, the kinetic experiments deliver a quantitative dissection of the binding free energy contributions from each microscopic step, highlighting that about half of the favorable free energy comes from the conformational transition after binding (Table 1). It is exactly this step that accounts for Abl’s ability to tightly bind Gleevec in contrast to Src, with a difference of −4.6 kcal/mol between them, leaving only a minor energy contribution from the widely proposed DFGin/DFGout flip12,19–21.
Direct detection of the higher energy E.I state in Src
Since our mechanism differs drastically from the current paradigm in the field12,19–21, we felt the need of designing yet one additional experiment that could directly detect this critical conformational transition of the kinase.Gleevec complex. Close examination of the rate constants in the scheme (Fig. 2g) shows that in Src, the E.I⇔E*.I equilibrium is slow on the NMR timescale and is only moderately skewed (~15% in the E.I state), suggesting that peaks corresponding to this state should be directly visible in the HSQC spectrum of Src saturated with Gleevec. Since we did not observe this in our original titration (Fig. 1a), and such a spectroscopic feature was not seen in previous NMR data published on the Src.Gleevec complex29, we recorded spectra for 16 h to increase the signal-to-noise ratio. This maneuver indeed revealed the presence of minor peaks at positions corresponding to the E.I state (Fig. 5, Supplementary Fig. 5) with relative populations of the E.I and E*.I states being in agreement with the kinetics data (Fig. 2g). The same experiment performed on Abl-Gleevec complex showed no minor peaks, in agreement with our conclusion that for Abl, the E.I⇔E*.I equilibrium lies far towards the Abl*.I state (Fig. 2g). Our data establish a new model that resolves the controversy of how Src and Abl have drastically different affinities to the clinically relevant drug Gleevec while adopting almost identical structures when bound to it. A relatively large number of residues sensing conformational changes induced by Gleevec binding is in agreement with suggestions by Kuriyan and coworkers based on mutagenesis efforts and a recent NMR study of full-length Abl30.
Figure 5.
Direct observation of the conformational exchange step in the Src.Gleevec complex by NMR. [1H,15N]-HSQC spectrum of Src at saturating Gleevec concentrations recorded for 16 h shows presence of major and minor peaks corresponding to the Src*.I and Src.I complexes respectively. The minor peaks appear exactly where predicted from the Gleevec titrations (Fig. 1a, Supplementary Fig. 5).
DISCUSSION
These results enable us to envision the free energy landscape of Gleevec binding and to quantitatively rationalize Gleevec selectivity. Multiple lines of evidence, including direct detection of Gleevec binding by NMR and stopped-flow fluorescence, identify an induced-fit conformational change in the kinase. Gleevec complex as the origin of the observed high affinity of Gleevec to Abl. For Gleevec, the physical binding step results in only micromolar affinities for both Abl and Src, and only the induced-fit step brings Abl’s affinity into the clinically relevant nanomolar range. On the other hand, the ability of Abl and Src to sample the binding-competent DFG-out state (Eout) enables binding of Gleevec. Such sampling of multiple conformations by the apo enzyme (i.e. conformational selection) by definition weakens affinity by the fraction of the enzyme in the binding-incompetent state, but is required for more selective drugs that only bind to a unique enzyme conformation. This is in contrast to non-selective drugs that easily bind to multiple conformations and hence multiple targets1,11,21. The effect of increasing and decreasing affinities via induced fit versus conformational selection (in the case of Abl and Src it is Eout.I⇔Eout*.I and Ein⇔Eout steps) is, of course, based on simple thermodynamics of coupled equilibria. The free energy landscape of Gleevec binding to two important drug targets, characterized here, illustrates how nature solved the dual challenge of having high selectivity while maintaining strong affinity. Such principles may be general for many other tight-binding inhibitors. While the detailed path to rational drug design, encompassing the points above, is long, the first step is to characterize a quantitative model for binding as performed here, followed by an identification of the key residues underlying the energetics of binding (C.W., R.V.A., D.K., unpublished). Computation undoubtedly plays a key role in the energy calculations and design part, but the correct binding model with quantitative experimental data is crucial as best illustrated by the history of understanding Gleevec selectivity by computation19–21. By analogy with the advances in predicting protein structures using the large set of structural data combined with energy functions, we anticipate that availability of extensive quantitative data on the detailed energetics of drug binding as delivered here may have a big impact on rational drug design.
ONLINE METHODS
Protein expression and purification
Genes corresponding to kinase domains of Abl and Src were subcloned into an expression vector containing a His (for Src) or His/MBP (for Abl) tags for growth in BL21 (DE3) cell line. Cells were co-transformed with the YOPH phosphatase to ensure de-phosphorylated state of expressed protein and to lower toxicity due to overexpression. Culture was grown to OD of 0.8 at 37 °C degrees then switched to 20 °C and induced with 200µM of IPTG overnight. Protein was purified using a combination of ion affinity, anion exchange, and size-exclusion columns. Affinity-tags were removed by overnight incubation with TEV protease. Purity of the protein was confirmed by gel electrophoresis, and lack of tyrosin phosphorylation was confirmed by mass spectrometry. For NMR experiments [U-15N]- and U-[2H,15N]-labeled proteins were expressed in M9 minimal medium prepared using H2O or D2O, respectively. In both cases, the M9 medium contained 1 g/L 15NH4Cl as the sole nitrogen source and 3 g/L unlabeled D-glucose.
NMR experiments
[1H-15N]-TROSY-HSQC experiments were performed at 25 °C on an Agilent DD2 600 MHz four-channel spectrometer, equipped with a triple-resonance cryogenically cooled probehead. . To increase stability of the samples buffer composition were optimized using thermofluor-based high-throughput stability assay33. Solutions of Sypro Orange were mixed with 5 µL of protein at 100 µM and 15 µL of buffer of interest and added to a 96-well PCR plate. A control sample contained buffer and Sypro Orange only. The plates where sealed with optical sealing tap and heated in a real-time PCR machine from 20 to 100 °C with increments of 0.2 °C. Fluorescence was measured by exciting at 490 nm and measuring at 575 nm. Optimized NMR samples contained between 250–500 µM protein and 10 (v/v)% D2O in either 50 mM TRIS, 500 mM NaCl, 1 mM MgCl2, 1 mM TCEP (pH 8.0) or 10 mM EPPS, 100 mM NaCl, 1 mM TCEP (pH 8.0). NMR data were processed with the NMRPipe/NMRDraw software package34 and analyzed using the program CcpNmr35.
Pre-steady-state kinetics experiments
Stopped-flow experiments were performed with the Applied Photophysics SX-20 instrument equipped with a temperature control unit. Gleevec binding kinetics was monitored via changes in intrinsic tryptophan fluorescence. Samples were excited at 295 nm (9 nm bandwidth) and emission was detected with a long-pass 320 nm cut-off filter. To overcome Gleevec solubility issues, non-equal mixing ratio was utilized. Protein was loaded into a 0.5 mL syringe and the drug was loaded into a 5 ml syringe, resulting in a 1 to 10 mixing volume ratio. Final protein concentration was typically 0.1 µM, and Gleevec concentration was varied. To study dissociation kinetics, protein (at 0.1–1 µM) was preincubated with 0.1–100 µM of Gleevec for 10 minutes, placed into the 0.5 mL syringe and then diluted with buffer loaded into the 5 mL syringe. This setup allowed for an 11-fold dilution. All experiments were performed at 5 °C and 25 °C in a buffer containing 50 mM TRIS, 500 mM NaCl, 1 mM MgCl2, 1 mM TCEP and 5% DMSO (pH 8.0).
Data were analyzed using Applied Photophysics or Origin (OriginLab Corporation) software. Individual fluorescent transients were fitted to single or multi-exponential curves. To account for photobleaching, an additional exponential term was added to the fitting function. This rate was fixed to the value determined in control experiments where protein was mixed with buffer in the absence of Gleevec.
Association and dissociation of Gleevec with Abl at 5 °C (when both binding and conformational transitions could be clearly resolved) were also simulated and fitted globally using numerical algorithms with KinTek explorer program28,32. In these simulations the same set of kinetic rate constants was used to fit all fifteen datasets corresponding to different Gleevec concentrations.
Measurements of Gleevec affinity
Gleevec affinity to Abl at 5° C was measured with a Hitachi F-2500 fluorimeter. 10 nM of Abl was mixed with 2 nM to 75 nM of Gleevec. Binding was monitored via changes in Trp fluorescence. Tryptophanes were excited at 295 nm, and fluorescence was detected at 350 nm. Extracted intensities were fitted to a generalized binding equation:
where [Et] is total enzyme concentration, [I] concentration of Gleevec, F0 and A are background fluorescent and a scaling factor respectively.
Analysis of kinetic data
The following naming convention is used throughout the text:
Different states of enzyme without or with bound inhibitor are called E, E.I and E*.I respectively,
In cases where conformation of the DFG-loop is specified, subscripts “in” or “out” are added (e.g. Ein, Eout, Eout.I, etc).
Rates describing the time dependence of experimentally observed changes in fluorescence are called “observed rates” or “apparent rates”,
kon, koff, kconf+ and kconf− are called “rate constants” and correspond to individual microscopic steps in the reaction schemes. They are specified in Fig, 2g.
F denotes the amplitude of the observed fluorescent signal and is generated by combined fluorescence from all enzyme species.
1. Two-state model (pseudo-first order reaction) for binding
In the case of a simple pseudo-first-order reaction, the time-dependence of fluorescent signal is mono-exponential (Supplementary Fig. 6a). Binding and dissociation (kon and koff respectively) rate constants can be determined from the plot of observed rate as a function of inhibitor concentration (Supplementary Fig. 6b), where the slope is equal to kon and the intercept corresponds to koff.
2. Three-state model (binding followed by a conformational change)
In a more complicated case of a two-step reaction E+I⇔E.I⇔E*.I, a double-exponential behavior is observed (Supplementary Fig. 6c). If the rates corresponding to these steps are substantially different, each step can be treated separately, and two observed rates (kobsbind and kobsconf) can be extracted from the fits of Fluorescence vs. Time graph (Supplementary Fig. 6c,e).
The first step (i.e. binding) is defined by the observed binding rate kobsbind and follows the same laws as described above. In particular, the kobsbind vs. [I] plot will be linear, the slope will define kon while the intercept will define koff (Supplementary Fig. 6e). The second phase corresponds to the conformational transition. kobsconf vs. [I] plot is non-linear and plateaus at the value corresponding to the sum of kconf+ and kconf− rate constants (Supplementary Fig. 6e).
The fact that the observed rate of conformational transition depends on inhibitor concentration is counterintuitive, but can be rationalized in the following way. The rate of E*.I formation is determined by the differential equation:
meaning that it depends on the concentration of E.I. At higher inhibitor concentrations more inhibitor will be bound, effectively increasing the transient concentration of the E.I state (Supplementary Fig. 6d), consequently the observed rate of fluorescent changes will increase. At very high concentrations of I, the equilibrium of E+I⇔E.I reaction is heavily skewed to the right, at which point the concentration of E.I state is no longer [I]-dependent and the kobsconf vs. [I] graph reaches a plateau.
If the dissociation of I is fast, then kconf− can be reliably determined in a dilution experiment, when first the enzyme is saturated with I, and then diluted, causing I to dissociate. If the dissociation step (rate constant koff) is much faster than kconf−, then the observed fluorescent change will be monoexponential and the rate will be limited by the kconf− rate constant (remaking E.I from E*.I) (Supplementary Fig. 6f).
In summary, double exponential fit of Fluorescence vs Time graph, can be used to generate kobsbind vs [I] and kobsconf vs. [I] plots. The first one is linear and defines kon and koff, the second one is non-linear and defines the sum kconf++kconf−. If kconf− is independently determined in the dilution experiment, then both kconf+ and kconf− can be calculated. Thereby the system is fully determined.
Supplementary Material
Acknowledgement
We are grateful to M.A. Morando and F. Gervasio (Spanish National Cancer Research Center) for providing plasmids for the Src kinase domain and sharing expression and purification protocols. This work was supported by the Howard Hughes Medical Institute (HHMI) the Office of Basic Energy Sciences, Catalysis Science Program, US Department of Energy, award DE-FG02-05ER15699 and the US National Institute of Health GM100966-01. R.O. is an HHMI Fellow of the Damon Runyon Cancer Research Foundation, DRG-2114-12.
Footnotes
Author contributions: R.V.A. and C.W. contributed equally to this work. R.V.A, C.W. and D.K. designed and analyzed all experiments, R.V.A. and C.W. performed experiments, R.O. assisted with NMR data acquisition and analysis, V.B. assisted with MS experiments, R.V.A, C.W. and D.K. wrote the manuscript with contributions from all authors.
References
- 1.Jura N, et al. Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms. Mol Cell. 2011;42:9–22. doi: 10.1016/j.molcel.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kornev AP, Taylor SS. Defining the conserved internal architecture of a protein kinase. Biochim Biophys Acta. 2010;1804:440–444. doi: 10.1016/j.bbapap.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor SS, Keshwani MM, Steichen JM, Kornev AP. Evolution of the eukaryotic protein kinases as dynamic molecular switches. Philos Trans R Soc Lond B Biol Sci. 2012;367:2517–2528. doi: 10.1098/rstb.2012.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hunter T. Tyrosine phosphorylation: thirty years and counting. Curr Opin Cell Biol. 2009;21:140–146. doi: 10.1016/j.ceb.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen P. Protein kinases--the major drug targets of the twenty-first century? Nat Rev Drug Discov. 2002;1:309–315. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- 6.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 7.Nagar B. c-Abl tyrosine kinase and inhibition by the cancer drug imatinib (Gleevec/STI-571) J Nutr. 2007;137:1518S–1523S. doi: 10.1093/jn/137.6.1518S. discussion 1548S. [DOI] [PubMed] [Google Scholar]
- 8.Harrison SC. Variation on an Src-like theme. Cell. 2003;112:737–740. doi: 10.1016/s0092-8674(03)00196-x. [DOI] [PubMed] [Google Scholar]
- 9.Azam M, et al. Activity of dual SRC-ABL inhibitors highlights the role of BCR/ABL kinase dynamics in drug resistance. Proc Natl Acad Sci U S A. 2006;103:9244–9249. doi: 10.1073/pnas.0600001103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winter GE, et al. Systems-pharmacology dissection of a drug synergy in imatinib-resistant CML. Nat Chem Biol. 2012;8:905–912. doi: 10.1038/nchembio.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dar AC, Shokat KM. The evolution of protein kinase inhibitors from antagonists to agonists of cellular signaling. Annu Rev Biochem. 2011;80:769–795. doi: 10.1146/annurev-biochem-090308-173656. [DOI] [PubMed] [Google Scholar]
- 12.Seeliger MA, et al. c-Src binds to the cancer drug imatinib with an inactive Abl/c-Kit conformation and a distributed thermodynamic penalty. Structure. 2007;15:299–311. doi: 10.1016/j.str.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 13.Nagar B, et al. Crystal structures of the kinase domain of c-Abl in complex with the small molecule inhibitors PD173955 and imatinib (STI-571) Cancer Res. 2002;62:4236–4243. [PubMed] [Google Scholar]
- 14.Schindler T, et al. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 15.Williams JC, et al. The 2.35 A crystal structure of the inactivated form of chicken Src: a dynamic molecule with multiple regulatory interactions. J Mol Biol. 1997;274:757–775. doi: 10.1006/jmbi.1997.1426. [DOI] [PubMed] [Google Scholar]
- 16.Xu W, Doshi A, Lei M, Eck MJ, Harrison SC. Crystal structures of c-Src reveal features of its autoinhibitory mechanism. Mol Cell. 1999;3:629–638. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- 17.Xu W, Harrison SC, Eck MJ. Three-dimensional structure of the tyrosine kinase c-Src. Nature. 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 18.Levinson NM, et al. A Src-like inactive conformation in the abl tyrosine kinase domain. PLoS Biol. 2006;4:e144. doi: 10.1371/journal.pbio.0040144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aleksandrov A, Simonson T. Molecular dynamics simulations show that conformational selection governs the binding preferences of imatinib for several tyrosine kinases. J Biol Chem. 2010;285:13807–13815. doi: 10.1074/jbc.M110.109660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin YL, Meng Y, Jiang W, Roux B. Explaining why Gleevec is a specific and potent inhibitor of Abl kinase. Proc Natl Acad Sci U S A. 2013;110:1664–1669. doi: 10.1073/pnas.1214330110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lovera S, et al. The different flexibility of c-Src and c-Abl kinases regulates the accessibility of a druggable inactive conformation. J Am Chem Soc. 2012;134:2496–2499. doi: 10.1021/ja210751t. [DOI] [PubMed] [Google Scholar]
- 22.Cowan-Jacob SW, et al. The crystal structure of a c-Src complex in an active conformation suggests possible steps in c-Src activation. Structure. 2005;13:861–871. doi: 10.1016/j.str.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 23.Dar AC, Lopez MS, Shokat KM. Small molecule recognition of c-Src via the Imatinib-binding conformation. Chem Biol. 2008;15:1015–1022. doi: 10.1016/j.chembiol.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vajpai N, et al. Solution conformations and dynamics of ABL kinase-inhibitor complexes determined by NMR substantiate the different binding modes of imatinib/nilotinib and dasatinib. J Biol Chem. 2008;283:18292–18302. doi: 10.1074/jbc.M801337200. [DOI] [PubMed] [Google Scholar]
- 25.Vogtherr M, et al. NMR characterization of kinase p38 dynamics in free and ligand-bound forms. Angew Chem Int Ed Engl. 2006;45:993–997. doi: 10.1002/anie.200502770. [DOI] [PubMed] [Google Scholar]
- 26.Shan Y, et al. A conserved protonation-dependent switch controls drug binding in the Abl kinase. Proc Natl Acad Sci U S A. 2009;106:139–144. doi: 10.1073/pnas.0811223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kovrigin EL. NMR line shapes and multi-state binding equilibria. J Biomol NMR. 2012;53:257–270. doi: 10.1007/s10858-012-9636-3. [DOI] [PubMed] [Google Scholar]
- 28.Johnson KA. Fitting enzyme kinetic data with KinTek Global Kinetic Explorer. Methods Enzymol. 2009;467:601–626. doi: 10.1016/S0076-6879(09)67023-3. [DOI] [PubMed] [Google Scholar]
- 29.Campos-Olivas R, Marenchino M, Scapozza L, Gervasio FL. Backbone assignment of the tyrosine kinase Src catalytic domain in complex with imatinib. Biomol NMR Assign. 2011;5:221–224. doi: 10.1007/s12104-011-9304-7. [DOI] [PubMed] [Google Scholar]
- 30.Skora L, Mestan J, Fabbro D, Jahnke W, Grzesiek S. NMR reveals the allosteric opening and closing of Abelson tyrosine kinase by ATP-site and myristoyl pocket inhibitors. Proc Natl Acad Sci U S A. 2013;110:E4437–E4445. doi: 10.1073/pnas.1314712110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagar B, et al. Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell. 2003;112:859–871. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 32.Johnson KA, Simpson ZB, Blom T. FitSpace explorer: an algorithm to evaluate multidimensional parameter space in fitting kinetic data. Anal Biochem. 2009;387:30–41. doi: 10.1016/j.ab.2008.12.025. [DOI] [PubMed] [Google Scholar]
ONLINE METHODS REFERENCES
- 33.Ericsson UB, Hallberg BM, Detitta GT, Dekker N, Nordlund P. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal Biochem. 2006;357:289–298. doi: 10.1016/j.ab.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 34.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 35.Vranken WF, et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.