Abstract
West Nile virus (WNV) is a zoonotic virus, which is transmitted by mosquitoes. It is the causative agent of the disease syndrome called West Nile fever. In some human cases, a WNV infection can be associated with severe neurological symptoms. The immune response to WNV is multifactorial and includes both humoral and cellular immunity. T-cell epitope mapping of the WNV envelope (E) protein has been performed in C57BL/6 mice, but not in BALB/c mice. Therefore, we performed in BALB/c mice a T-cell epitope mapping using a series of peptides spanning the WNV envelope (E) protein. To this end, the WNV-E specific T cell repertoire was first expanded by vaccinating BALB/c mice with a DNA vaccine that generates subviral particles that resemble West Nile virus. Furthermore, the WNV structural protein was expressed in Escherichia coli as a series of overlapping 20-mer peptides fused to a carrier-protein. Cytokine-based ELISPOT assays using these purified peptides revealed positive WNV-specific T cell responses to peptides within the different domains of the E-protein.
Introduction
West Nile virus (WNV) is a small enveloped single-stranded positive sense RNA-containing virus that belongs to the family Flaviviridae [1]. The virus is maintained in an enzootic cycle where it is transmitted between ornithophilic mosquitoes of the Culex genus and birds. Equine and humans are considered dead-end hosts since they do not mount high enough viremia for mosquitoes to become infected following feeding [2]. Human infection with the virus leads to a wide range of diseases from mildly febrile to severe neurologic complications and death, but asymptomatic infections occur most frequently [3]. Humoral immunity is considered an essential aspect of protective immunity since it limits WNV dissemination into the nervous system. This was demonstrated in mice lacking B cells which developed high-grade viremia, early dissemination into the brain and uniform mortality [4]. The envelope E glycoprotein is the principal antigen that elicits neutralizing antibodies and as such is a primary target for vaccine development [5]. Studies in animal models have also demonstrated that T lymphocytes are an essential component of protection against WNV. Mice deficient in CD8+ T cells develop persistent WNV infections in the brain [6], [7]. Studies in mice have also shown that CD4+ T cells control WNV infection by priming B cell and antibody responses, and by sustaining CD8+ T cell activity [8]. Mapping of antigenic peptide sequences from proteins of relevant pathogens recognized by T helper (Th) and by cytolytic T lymphocytes (CTL) may help to understand virus immunity and pathogenesis. The majority of T cells recognize peptide epitopes bound to major histocompatibility complex (MHC)-encoded glycoproteins on the surface of professional antigen-presenting cells APC [9]. Most T cells are specific for peptide epitopes in association with either classical MHC class I molecules (H2-K, D, and L in mice) in the case of CD8+ T cells, or class II molecules (H2-A and E in mice) for CD4+ T cells [10]. These peptide antigens are subsequently detected by the T cell receptor of T cells, which proliferate, secrete cytokines and differentiate into antigen-specific effector cells [11], [12]. Nearly all of the epitopes associated with protective responses against WNV using mice models are non-linear and conformational B-cell epitopes and most of these are specific to the envelope (E) protein. The majority of these B-cell epitopes have been defined in BALB/c mice [13] whereas C57BL/6 mice have been used mostly to identify T-cell epitopes [13]–[16]. Both mouse strains are equally susceptible to WNV infection [17], [18] but C57BL/6 mice demonstrate elevated blood-brain barrier permeability [17]. In addition, as C57BL/6 mice predominantly show Th1-dependent immune responses upon infection, whereas BALB/c mice tend to favor Th2-responses, T-cell epitopes identified in one strain might not be directly transferable to the other.
Bioinformatics experts have developed computer-driven algorithms methods to predict T cell epitopes which have significantly decreased the experimental burden that is associated with epitope identification [11], [19]–[21]. To predict in BALB/c mice potential T-cell epitopes of the E-protein of the WNV, we used the Immune Epitope Database and Analysis Resource (IEDB, [22]). To confirm these predictions with in vivo immunogenicity, we vaccinated BALB/c mice with a plasmid expressing the membrane protein M (prM) and E protein. Subsequently, we measured the CD8+ and CD4+ T cell cytokine responses using a series of peptides derived from WNV E-protein. As the DNA vaccine leads to the expression of virus-like-particles resembling correctly folded E and M proteins, we can assume that the identified epitopes will also exist during the course of a WNV infection [23].
Findings
The sequence spanning the E protein of the lineage 1 WNV strain “Ita09” (NCBI Acc#.210 GU011992) was separated into 26 clones, each coding for 30 amino acid long peptides with an overlap of 10 amino acids on both sides. The DNA fragments encoding these peptides were cloned in the bacterial expression vector pEXP1 by ATG:biosynthetics (Freiburg, Germany) after the sequence of glutathione-S-transferase (GST) from Schistosoma japonicum. The sequence of the different bacterial expression vectors were sequenced to ensure that they contained the right base pair sequences and hence encoded the desired peptides. After transformation of these vectors into the E. coli BL21 strain, selected colonies were grown in Luria Bertani broth containing kanamycine and induced overnight with isopropylthio-β-galactoside (IPTG, 1 mM final concentration, Sigma–Aldrich (Diegem, Belgium)). The bacteria were collected by centrifugation (20 min at 4,000 g at 4°C) and the cell pellet was resuspended in lysis buffer (phosphate buffered saline (PBS, Invitrogen (Merelbeke, Belgium)) containing protease inhibitors cocktail (Roche, Mannheim, Germany), 1 mM DTT, 1 mM EDTA, 1% Triton X-100 and Lysozyme (1 mg/ml)). Lysates were obtained after sonication and were subsequently clarified by centrifugation (30 min at 15,000×g at 4°C). Recombinant peptides derived from the E-protein were purified by glutathione affinity purification and dialyzed against sterile PBS before use. The purified proteins were analyzed in denaturing protein gels and the size difference compared to GST alone was visualized as previously described by Chabierski et al. [24]. The expression levels were very high to low (Table 1 and Fig. 1).
Table 1. An overview of used E-protein derived peptides and their characteristics.
| Peptide n° | E-proteinderivedpeptides | MHCclass I | MHCclass II | Expression | Domain withinWNV E protein | Sequences ofknown humanT cell epitopewithin our peptides | References |
| 15 | M66-75/E1-20 | − | I | ||||
| 16 | E11-40 | ++ | I | ||||
| 17 | E31-60 | x | − | I/II | |||
| 18 | E51-80 | x | x | + | II | ||
| 19 | E71-100 | ++ | II | ||||
| 20 | E91-120 | x | ++ | II | RGWGNGCGLFGKGSI | [30], [31] | |
| 21 | E111-140 | x | ++ | II/I | |||
| 22 | E131-160 | x | − | I | |||
| 23 | E151-180 | x | x | ++ | I | ||
| 24 | E171-200 | ++ | I | ||||
| 25 | E191-220 | x | + | I/II | |||
| 26 | E211-240 | x | + | II | TFLVHREWFMDLNLPW | [13], [32] | |
| 27 | E231-260 | x | ++ | II | |||
| 28 | E251-280 | x | x | + | II | ||
| 29 | E271-300 | x | − | II/I | |||
| 30 | E291-320 | x | + | I/III | EKLQLKGTTYGVCSKAFK | [30] | |
| 31 | E311-340 | x | + | III | |||
| 32 | E331-360 | x | − | III | |||
| 33 | E351-380 | +/− | III | ||||
| 34 | E371-400 | x | + | III | KVLIELEPPFGDSYIVV | [31] | |
| 35 | E391-420 | x | + | III | HKSGSSIGKAFTTTLKGA | [31] | |
| 36 | E411-440 | x | x | − | III | SVGGVFTSV | [33], [34], [35] |
| WDFGSVGGVFTSVGKAVH | [13] | ||||||
| 37 | E431-460 | x | + | III | FRSLFGGMSWITQGLLGA | [30] | |
| 38 | E451-480 | x | + | III | FRSLFGGMSWITQGLLGA | [30], [31] | |
| 39 | E471-500 | x | x | +/− | III | ||
| 40 | E491-501/NS11-18 | + | III |
Amino acid sequences of the E-protein derived peptides used in this study, their expression level in E. coli, and the presence of predicted MHC class I or class II epitopes in the different domains of the E-protein compared to known human T-cell epitopes. The sequence of the peptides can be found in Chabierski et al. [24].
Abbreviations. E: WNV envelope protein, M: WNV membrane protein, NS: WNV non-structural protein ++: very high expression, +: high expression, +/−: moderate expression and –: very low expression.
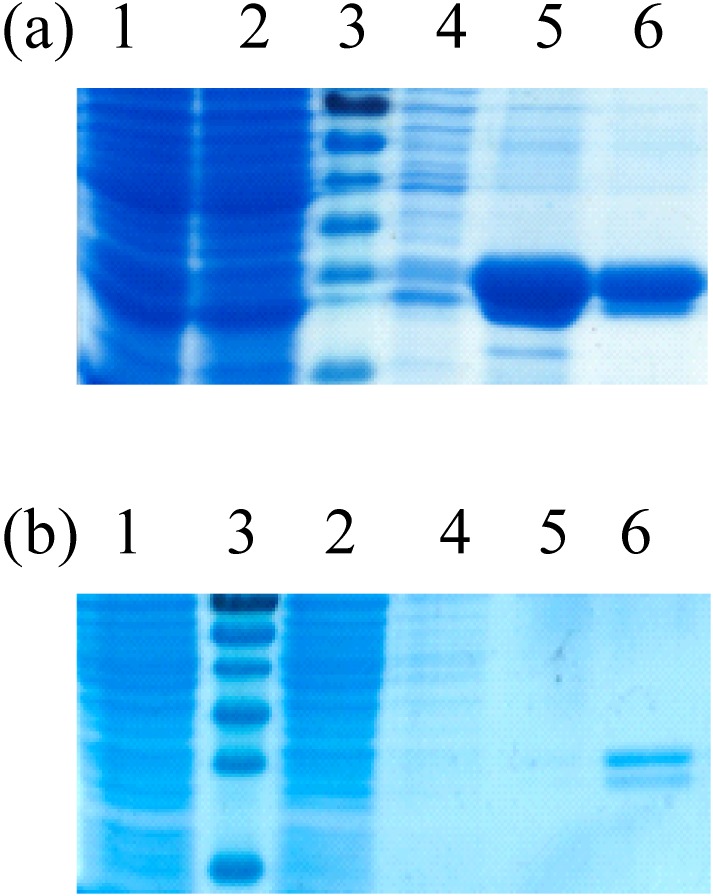
Figure 1. Expression and purification of recombinant GST tagged peptides.
SDS-PAGE showing crude lysates of protein-expressing bacteria and purification steps of peptide E471 showing a moderate expression (a) and peptide E131 showing very low expression (b). Lane 1, crude lysate; lane 2, lysate supernatant after centrifugation; lane 3, size marker; lanes 4–5, respectively elution fraction 1 and 2 of recombinant peptide after glutathione affinity purification.
From six clones the amount of protein obtained was not sufficient to be used as stimulant in ELISPOT assays. To expand the WNV-E specific T-cell repertoire, five BALB/c mice were intramuscularly immunized (two vaccinations with four-weeks interval) followed by in vivo electroporation (BTX ECM 830, Harvard apparatus, Holliston, MA, USA) of a plasmid encoding the prM/E expression cassette [23]. This plasmid was prepared by cloning the prM/E expression cassette from pCBWN (kindly provided by Mike Diamond, Washington University) into the pVAX1 (Invitrogen) backbone. Five untreated BALB/c mice served as naive control mice. The ethics committee of the faculty of Veterinary Medicine of Ghent University authorized the animal experiments (permit EC2012/124). Two weeks after the boost, blood was collected and splenocytes were isolated. Splenocyte suspensions were depleted of red blood cells using ammonium chloride hypotonic lysis and passed through a m cell strainer. To obtain enough splenocytes for CD4+ and CD8+ isolation, we pooled the splenocytes of the vaccinated as well as the naive control mice, and subsequently divided the pooled cells into two cell preparations. In the first preparation CD8+ T cells were depleted with magnetic CD8 Dynabeads (Life Technologies, Merelbeke, Belgium) and in the other preparation CD4+ cells were depleted with magnetic CD4 Dynabeads (Life Technologies, Merelbeke, Belgium) in accordance with the manufacturer’s recommended protocol. To confirm the depletion, spleen cells were stained with PerCP-anti-CD4 (Biolegend, Uithoorn, The Netherlands) or PerCP anti-CD8 (Biolegend, Uithoorn, The Netherlands) and analyzed by flow cytometry (Accuri C6, BD Biosciences, Erembodegem, Belgium). The extent of depletion obtained was 97% and 93% for respectively CD4− and CD8-depletion. Next, ELISPOT assays were performed to measure the IFN-γ and IL-4 production by CD4 and CD8 depleted splenocytes. In detail, sterile 96-well Maxisorp immuno-plates were coated with anti-IL-4 (Biolegend, Uithoorn, The Netherlands) or anti-IFN-γ monoclonal antibodies (Biolegend, Uithoorn, The Netherlands) and blocked with sterile PBS containing 1% BSA for 1 h at 37°C. Next, 3×105 CD8− or CD4− depleted splenocytes were plated in 100 µl of culture medium and stimulated during 16h with culture medium with GST (negative control), g/mL Concanavalin A (ConA, positive control, Sigma–Aldrich (Diegem, Belgium)), 2 µg/ml purified E-protein [25] or 2 µg/ml of purified recombinant GST-peptides. After stimulation the plates were washed two times with PBS and four times with PBS containing 0,05% Tween-20. IL-4 or IFN-γ trapped on the plates was detected by a biotinylated monoclonal anti-IL-4 (Biolegend, Uithoorn, The Netherlands) or anti-IFN-γ antibody (Biolegend, Uithoorn, The Netherlands). Subsequent incubation with GABA-conjugated streptavidin (U-Cytech Biosciences, Utrecht, The Netherlands) was used to develop silver spots at places where immune cells secreted IL-4 or IFN-γ during stimulation. Splenocytes from the vaccinated and non-vaccinated mice were analyzed in triplicate and the spots were counted using the Bioreader 5000 (Bio-sys, Karben, Germany). IgG antibody titers were determined as described by Oliphant et al. [25]. Briefly, two weeks after the boost, blood samples were collected by cardiac puncture. Blood was allowed to clot for 60 min at 37°C, and serum was obtained by combining the supernatant from two successive centrifugations. The titers of E-specific IgG1 and IgG2a antibodies in the serum were determined by ELISA in 96-well Maxisorp immuno-plates (Nalge Nunc, Rochester, USA) coated overnight with recombinant E-protein (1 µg/ml in carbonate buffer, 100 µl/well, 4°C). After coating, the plates were washed three times with PBS containing 0.1% Tween-20 and blocked with 3% skim milk in PBS. Next, three-fold serial dilutions of mouse serum, starting with a 1/100 dilution, were incubated for 1 h while shaking at room temperature. After washing goat-derived anti-mouse serum conjugated with horseradish peroxidase specific for mouse isotypes IgG1 or IgG2a (Southern Biotechnology Associates, Birmingham, USA) and tetramethylbenzidine substrate (BD Biosciences) were used to determine specific antibody titers. Antibody titers are defined as the reciprocal of the highest dilution with an OD450 that is at least double the OD450 of pre-immune serum samples.
To predict MHC class I and II T cell epitopes, we used the immune epitope database (IEDB) analysis tool (http://www.iedb.org) [22]. The binding affinity of peptides (15-mers) to MHC class II (H2-IEd and H2-IAd) was predicted. In this case, the SMM-align method was employed to find out good MHC class II candidate binders. The top scoring peptides were selected by setting cut-off values of IC50 for the predicted binders at 250 nM. The MHC class I binding predictions were made using the IEDB analysis resource Consensus tool, which combines predictions from ANN aka NetMHC (3.4), SMM and Comblib. All the available MHC alleles were selected and the peptide lengths were set at respectively 9, 10 and 12 for H2-Kd, H2-Dd and H2-Ld before making prediction. Strong binder to selected MHC I alleles were classified based on the binding affinity thresholds ≤50 nM. Overall, this computer driven algorithm predicted that 14 and 11 of the E-protein derived peptides contained epitopes that were specific for respectively MHC class I and II (Table 1). Interestingly, we found a clear difference of the epitope distribution within the different structural domains of the E-protein. Most of the epitopes located in domain III were peptides binding to MHC class I molecules, whereas the majority of the epitopes located in domain II are peptides that preferentially bind to MHC class II molecules (Table 1). Next, we wanted to confirm that the predicted epitopes were indeed recognized by CD8+ or CD4+ T cells. Therefore, we stimulated the CD4− or CD8-depleted splenocytes with the purified E-protein derived peptides and measured the IFN-γ and IL-4secretion via ELISPOT assays. Twenty recombinant polypeptides were tested for their ability to stimulate CD8+ or CD4+ spleen-derived T cells from BALB/c mice immunized against WNV E-protein. As a negative control, splenocytes preparations from naïve mice were used. Out of the 14 predicted MHC class I epitopes we could purify 12 peptides and, 11 were confirmed to be able to induce IFN-γ secretion by CD8+ T cells. However, peptide E391 that was also predicted to stimulate CD8+ cells did not stimulate IFN-γ production. Interestingly, a new epitope (present in peptide E211), for which the MHC class I restriction was not ascertained by the IEDB analysis tool, was identified (Fig. 2a).
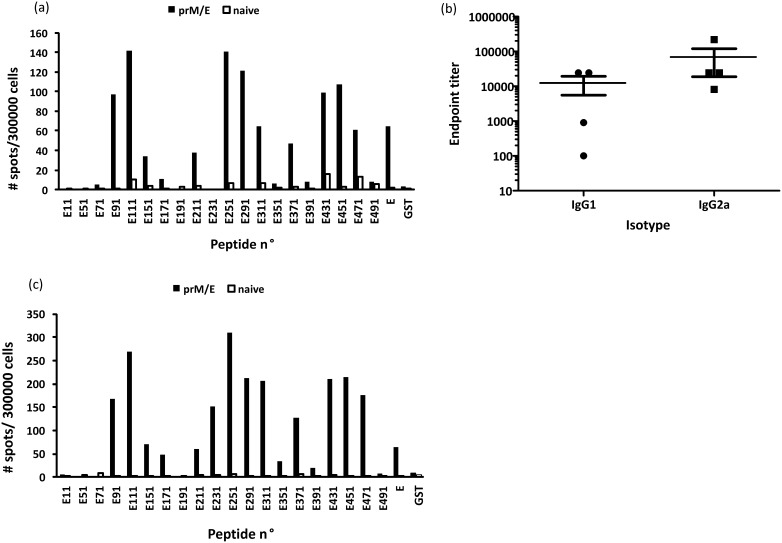
Figure 2. Detection of cellular and humoral immune response following pDNA-based vaccination.
IFN-γ production by (a) CD4-depleted and (c) CD8-depleted splenocytes after stimulation with purified recombinant GST tagged E-protein derived peptides. The WNV E-protein specific T-cell repertoire in BALB/c mice was expanded by two DNA vaccinations. Splenocytes obtained two weeks after the boost were stimulated with different recombinant GST tagged E-protein derived peptides and the numbers of cells producing IFN-γ were determined via ELISPOT. (b) Detection of serum IgG1 and IgG2a titers to the WNV E-protein two weeks after the boost via ELISA.
Peptides with atypical anchoring residues for a related MHC-I allele always display very low binding affinity for MHC-I molecule and probably therefore epitope E211 was not picked up by the IEDB analysis tool. However, some of these peptides may possess potent antigenicity to induce robust and specific T cell responses. The absence of an IFN-γ response to the different peptides in naïve mice demonstrates the specificity of the anti-peptide responses observed. Brien et al. identified six CD8 T cell epitopes in C57BL/6 mice by using overlapping WNV 15-mer peptides [14]. We identified more positive peptides most likely due to the fact that we used 30-mer peptides and certain epitopes are present in two peptides like e.g. H2-Dd epitope RSLFGGMSWI that is present in both peptides E431 and E451. Fig. 3 depicts the location of our peptide sequences in the WNV E protein.
Figure 3. Location of the peptide sequences in the E protein that, based on our in vivo experiments, contain strong CD4+ (underlined) and CD8+ (bold) T cell epitopes.
The shown amino acid sequence is that of the E protein of lineage 1 WNV strain Ita09. Sequences that are in bold and underlined contain strong CD4+ as well as CD8+ T cell epitopes.
Some of our detected peptides match with T-cell epitopes that were identified in WNV-infected humans (e.g. E91, E211, E291 (see also Table 1)). Additionally, some of the peptides that elicited a strong IFN-γ response are known epitopes for neutralizing antibodies or belong to regions of the WNV E-protein that are very important for the virus’s functionality. For example, peptide E91 is part of the fusion loop domain [26], E291 and E311 are located in lateral ridge of domain III, which has been shown to elicit virus-specific neutralizing antibodies [27], and E431 is part of the stem-anchor region that is important in virus-cell membrane fusion [28].
Surprisingly, we could not detect IL-4 responses in the CD4+ splenocytes with any of the twenty purified peptides. Additionally, even when the full-length E protein was used to stimulate the CD4+ cells we did not observe IL-4 production (data not shown). On the other hand, immunized mice were able to raise antibodies to the E-protein and a Th1-type response was noticed since higher levels of IgG2a were obtained compared to the IgG1 levels (Fig. 2b). However, with thirteen of the purified peptides we could detect a robust IFN-γ response after ex vivo stimulation of the CD4+ splenocytes (Fig. 2c). Only five of them were predicted by the IEDB analysis tools. Two of the predicted peptides (18 and 25) did not stimulate IFN-γ secretion, whereas several new epitopes were identified in the domain III of the E protein (peptides E291, E311, E371, E431). Since still 7% of the CD8+ cells were not depleted in the CD4 splenocytes preparation it is conceivable that some of the peptides detected may be restricted to MHC class I molecules. However, the number of spots were reasonably higher compared to the number of spots obtained in the CD8+ assays. On the other hand, since the peptides are 30 amino acids long it is possible that they contain both CD4 and CD8 epitopes. Indeed, Hughes et al. identified in C57/BL/6J mice a strong CD4+ epitope in WNV E between amino acids 466–495 [16] that had previously been shown by Brien et al. [14] to contain a potent CD8+ T cell epitope with protective cytotoxic capabilities. Peptide E231 specifically induced IFN-γ production in CD4+ splenocytes but not in CD8+ splenocytes.
One may notice that the number of spots obtained after stimulation of the CD4+ and CD8+ T cells with WNV E protein is lower than after stimulation with the purified GST fusion peptides (Fig. 2a and 2c). This can be explained as follows. For the stimulation of the CD4+ and CD8+ T cells we used an equal “mass/volume” concentration of the E-protein and the purified recombinant GST-peptides. However, the molecular weight of the E-protein (the ectodomain was used in this study) is about twice that of the GST-peptides. Consequently, the molar concentration of the GST-peptides was about twice the molar concentration of the E-protein. Thus, T cells stimulated with the GST-peptides were exposed to a two-fold higher number of epitopes. This explains why the number of spots after stimulation with the E-protein was lower.
To conclude, to our knowledge this is the first study that describes a detailed analysis of WNV E determinants recognized by CD4+ or CD8+ T cells in BALB/c mice. We identified several CD8+ and CD4+ T cell epitopes in BALB/c mice immunized against WNV E-protein by employing overlapping peptides spanning the entire E-protein sequence. The DNA vaccine leads to the expression of subviral particles that resemble WNV. Therefore, we can assume that the identified epitopes will also exist during the course of a WNV infection. Computer-assisted prediction of MHC peptide binding is a useful approach to identify possible CD8+ epitopes, although we demonstrated that the accuracy of these predictions must be verified empirically. Regarding the identification of CD4+ T cell epitopes, the less strict binding requirements and thus the limited predictive value of MHC class II motifs [29] makes this approach less suitable. Only epitopes which are proven to stimulate CD4+ T cells in vitro or in vivo can be used as potential subunit components for the design of vaccines against WNV. Both the results of the cytokine profile produced by WNV-specific CD4+ T cells and the IgG isotype suggest that the CD4 immune response after DNA vaccination shows a Th1 bias.
Acknowledgments
We thank Michael Diamond (Washington University, USA) for providing us the E-protein and the prM/E expression cassette.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.
Funding Statement
This work was supported by the European Union under FP7 (MDF, SC, SU, NNS), Project 261426 (WINGS-West Nile Integrated Shield Project), and by the Research Foundation - Flanders (OA, NNS) (http://www.fwo.be/en). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Heinz CM, Purcell RH, Gould EA, Howard CR, Houghton M, et al. (2000) Virus Taxonomy. 7th Report of the Interntional Committee for the Taxonomy of Virusses.
- 2. Hayes EB, Gubler DJ (2006) West Nile virus: epidemiology and clinical features of an emerging epidemic in the United States. Annu Rev Med 57:181–194. [DOI] [PubMed] [Google Scholar]
- 3. Hayes EB, Sejvar JJ, Zaki SR, Lanciotti RS, Bode AV, et al. (2005) Virology, pathology, and clinical manifestations of West Nile virus disease. Emerg Infect Dis 11:1174–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Diamond MS, Shrestha B, Marri A, Mahan D, Engle M (2003) B cells and antibody play critical roles in the immediate defense of disseminated infection by West Nile encephalitis virus. J Virol 77:2578–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roehrig JT (2003) Antigenic structure of flavivirus proteins. Adv Virus Res 59:141–175. [DOI] [PubMed] [Google Scholar]
- 6. Shrestha B, Diamond MS (2004) Role of CD8+ T cells in control of West Nile virus infection. J Virol 78:8312–8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang Y, Lobigs M, Lee E, Mullbacher A (2003) CD8+ T cells mediate recovery and immunopathology in West Nile virus encephalitis. J Virol 77:13323–13334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sitati EM, Diamond MS (2006) CD4+ T-cell responses are required for clearance of West Nile virus from the central nervous system. J Virol 80:12060–12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yewdell JW, Haeryfar SM (2005) Understanding presentation of viral antigens to CD8+ T cells in vivo: the key to rational vaccine design. Annu Rev Immunol 23:651–682. [DOI] [PubMed] [Google Scholar]
- 10. Guermonprez P, Valladeau J, Zitvogel L, Thery C, Amigorena S (2002) Antigen presentation and T cell stimulation by dendritic cells. Annu Rev Immunol 20:621–667. [DOI] [PubMed] [Google Scholar]
- 11. Bevan MJ (2004) Helping the CD8(+) T-cell response. Nat Rev Immunol 4:595–602. [DOI] [PubMed] [Google Scholar]
- 12. Harty JT, Tvinnereim AR, White DW (2000) CD8+ T cell effector mechanisms in resistance to infection. Annu Rev Immunol 18:275–308. [DOI] [PubMed] [Google Scholar]
- 13. Vaughan K, Greenbaum J, Blythe M, Peters B, Sette A (2010) Meta-analysis of all immune epitope data in the Flavivirus genus: inventory of current immune epitope data status in the context of virus immunity and immunopathology. Viral Immunol 23:259–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brien JD, Uhrlaub JL, Nikolich-Zugich J (2007) Protective capacity and epitope specificity of CD8(+) T cells responding to lethal West Nile virus infection. Eur J Immunol 37:1855–1863. [DOI] [PubMed] [Google Scholar]
- 15. Brien JD, Uhrlaub JL, Nikolich-Zugich J (2008) West Nile virus-specific CD4 T cells exhibit direct antiviral cytokine secretion and cytotoxicity and are sufficient for antiviral protection. J Immunol 181:8568–8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hughes HR, Crill WD, Davis BS, Chang GJ (2012) A West Nile virus CD4 T cell epitope improves the immunogenicity of dengue virus serotype 2 vaccines. Virology 424:129–137. [DOI] [PubMed] [Google Scholar]
- 17. Morrey JD, Olsen AL, Siddharthan V, Motter NE, Wang H, et al. (2008) Increased blood-brain barrier permeability is not a primary determinant for lethality of West Nile virus infection in rodents. J Gen Virol 89:467–473. [DOI] [PubMed] [Google Scholar]
- 18. Pinto AK, Richner JM, Poore EA, Patil PP, Amanna IJ, et al. (2013) A hydrogen peroxide-inactivated virus vaccine elicits humoral and cellular immunity and protects against lethal West Nile virus infection in aged mice. J Virol 87:1926–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nielsen M, Lundegaard C, Worning P, Hvid CS, Lamberth K, et al. (2004) Improved prediction of MHC class I and class II epitopes using a novel Gibbs sampling approach. Bioinformatics 20:1388–1397. [DOI] [PubMed] [Google Scholar]
- 20. Nielsen M, Lundegaard C, Worning P, Lauemoller SL, Lamberth K, et al. (2003) Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci 12:1007–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reche PA, Glutting JP, Zhang H, Reinherz EL (2004) Enhancement to the RANKPEP resource for the prediction of peptide binding to MHC molecules using profiles. Immunogenetics 56:405–419. [DOI] [PubMed] [Google Scholar]
- 22. Vita R, Zarebski L, Greenbaum JA, Emami H, Hoof I, et al. (2010) The immune epitope database 2.0. Nucleic Acids Res 38:D854–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Davis BS, Chang GJ, Cropp B, Roehrig JT, Martin DA, et al. (2001) West Nile virus recombinant DNA vaccine protects mouse and horse from virus challenge and expresses in vitro a noninfectious recombinant antigen that can be used in enzyme-linked immunosorbent assays. J Virol 75:4040–4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chabierski S, Makert GR, Kerzhner A, Barzon L, Fiebig P, et al. (2013) Antibody responses in humans infected with newly emerging strains of West Nile Virus in Europe. PLoS One 8:e66507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Oliphant T, Nybakken GE, Austin SK, Xu Q, Bramson J, et al. (2007) Induction of epitope-specific neutralizing antibodies against West Nile virus. J Virol 81:11828–11839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chabierski S, Barzon L, Papa A, Niedrig M, Bramson JL, et al. (2014) Distinguishing West Nile virus infection using a recombinant envelope protein with mutations in the conserved fusion-loop. BMC Infect Dis 14:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Diamond MS, Pierson TC, Fremont DH (2008) The structural immunology of antibody protection against West Nile virus. Immunol Rev 225:212–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schmidt AG, Yang PL, Harrison SC (2010) Peptide inhibitors of dengue-virus entry target a late-stage fusion intermediate. PLoS Pathog 6:e1000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sinigaglia F, Hammer J (1994) Defining rules for the peptide-MHC class II interaction. Curr Opin Immunol 6:52–56. [DOI] [PubMed] [Google Scholar]
- 30. Koo QY, Khan AM, Jung KO, Ramdas S, Miotto O, et al. (2009) Conservation and variability of West Nile virus proteins. PLoS One 4:e5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jung KO, Khan AM, Tan BY, Hu Y, Simon GG, et al. (2012) West Nile virus T-cell ligand sequences shared with other flaviviruses: a multitude of variant sequences as potential altered peptide ligands. J Virol 86:7616–7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lanteri MC, Heitman JW, Owen RE, Busch T, Gefter N, et al. (2008) Comprehensive analysis of west nile virus-specific T cell responses in humans. J Infect Dis 197:1296–1306. [DOI] [PubMed] [Google Scholar]
- 33. Parsons R, Lelic A, Hayes L, Carter A, Marshall L, et al. (2008) The memory T cell response to West Nile virus in symptomatic humans following natural infection is not influenced by age and is dominated by a restricted set of CD8+ T cell epitopes. J Immunol 181:1563–1572. [DOI] [PubMed] [Google Scholar]
- 34. Kim S, Li L, McMurtrey CP, Hildebrand WH, Weidanz JA, et al. (2010) Single-chain HLA-A2 MHC trimers that incorporate an immundominant peptide elicit protective T cell immunity against lethal West Nile virus infection. J Immunol 184:4423–4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Piazza P, McMurtrey CP, Lelic A, Cook RL, Hess R, et al. (2010) Surface phenotype and functionality of WNV specific T cells differ with age and disease severity. PLoS One 5:e15343. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.