Abstract
Tea green leafhopper is one of the most damaging tea pests in main tea production regions of East Asia. For lack of recognized morphological characters, the dominant species of tea green leafhoppers in Mainland China, Taiwan and Japan have always been named as Empoasca vitis Göthe, Jacobiasca formosana Paoli and Empoasca onukii MATSUDA, respectively. Furthermore, nothing is known about the genetic relationships among them. In this study, we collected six populations from Mainland China, four populations from Japan and one population from Taiwan, and examined the genetic distances in the COI and 16sRNA regions of mtDNA among them. The results showed that the genetic distances based on single gene or the combined sequences among eleven leafhopper populations were 0.3–1.2%, which were all less than the species boundary of 2%. Moreover, there were at least two haplotypes shared by two distinct populations from different regions. The phylogenetic analysis based on single gene or combined sets also supported that tea green leafhoppers from Mainland China, Taiwan and Japan were closely related to each other, and there were at least two specimens from different regions clustered ahead of those from the same region. Therefore, we propose that the view of recognizing the dominant species of tea green leafhoppers in three adjacent tea production regions of East Asia as different species is unreliable or questionable and suggest that they are a single species.
Background
Tea, Camellia sinensis (L.) O. Kuntze, originated in China and is now grown in almost sixty countries. In East Asia, Mainland China, Japan and Taiwan are three main tea production regions. The tea plantation is a very stable ecosystem for plenty of insects to colonize. Globally, 1031 arthropod species are associated with tea plants, which are always the most damaging constraint, causing on average a 5% to 55% yield loss [1], [2]. The distribution of insect pests in tea plantations is related to the regional climate, planting history, tea varieties, cultivation management, pest management model, tea production characteristic, as well as other factors.
Tea green leafhopper is one of the most important pests in East Asia. It belongs to Hemiptera, Jassidioea, Cicadellidae, which is one of the most diverse families of terrestrial organisms [3], [4]. As the most dominant pest in Chinese tea plantations, tea green leafhopper has been attracting a lot of attention from both Chinese governments and farmers [5]. The adult tea green leafhopper is about 3 mm long and green with transparent colourless wings. There are four nymphal instars that look like the adults with no wings. Both adults and nymphs of tea leafhopper pierce and suck the sap of tender tea shoots, which results in leaf edge yellowing, leaf tine curling, vein reddening, tea shoot growing slowly or stagnating, and even leaf edge and tine hennaing and withering [6]. In most regions of Mainland and Taiwan, there are 9 to 15 overlapping generations of tea green leafhoppers per year, and it causes, on average 15–20% annual yield loss and deterioration of commercial tea quality [7]. In Japan, tea green leafhopper occurs not so frequently as China with 5 to 8 generations a year.
The leafhopper is an extraordinarily large group with a wide range of host plants. Over 25,000 species of Cicadellidae are described and new leafhopper species are being discovered almost every day, but few useful morphological characteristics are developed to understand the relationships between leafhopper species and ecological factors for diversification [4], [8]. Until now, it has been clear that there is just one dominant species of tea green leafhopper in these three regions, respectively [9]–[11]. However, for lack of recognized morphological characters and compared research communications, the dominant species of tea green leafhoppers in Mainland China, Taiwan and Japan are always named as different species; Empoasca vitis Göthe, Jacobiasca formosana Paoli and Empoasca onukii MATSUDA, respectively [5], [9], [11].
Mitochondrial DNA (mtDNA) is widely used in taxonomy and systematics to elucidate the phylogenetic relationships of insects [12], [13], especially among some sibling species and cryptic species which are difficult to distinguish solely by morphology [14], [15]. The mtDNA sequence was first employed to find relationships among species within a genus of leafhoppers by Dietrich in 1997 [4]. Since then, more and more mtDNA markers were developed to combine with morphological characters to diagnose the systematics of Cicadellidae and the co-evolution of leafhoppers and their bacterial symbionts. The mtDNA sequence information can be useful for detecting differences among putative insect species [16] or well-resolved biotypes [17]–[19]. Cytochrome Oxidase I (COI), is successfully used for leafhopper species identification, phylogeographical and phylogeny studies of sibling species, thus being well-suited for examining intra- and inter-specific variation at both the species and genus level [20]. The 16sRNA is already used for species-level phylogenetic studies of leafhoppers (Cicadellidae) and especially the closely related species with discrete informative morphological characters [4].
The tea plant spread from Mainland China to neighbouring regions such as Japan and Taiwan. Tea green leafhoppers, important insect pests in tea plantations, which have identical morphological characteristics and biological habits, are differently named in Mainland China, Taiwan and Japan. Furthermore, nothing is known about tea green leafhoppers evolution relationship in these regions. Here, we present the hypothesis that they might be a single species. In this study, we surveyed the mtDNA sequences of tea leafhoppers among eleven populations in Mainland China, Taiwan and Japan, to understand the genetic divergences, phylogenetic relationship, to prove the hypothesis that the tea green leafhopper species of E. vitis, J. formosana and E. onukii are a single species.
Materials and Methods
Ethics Statement
No specific permits were required for this study. The tea green leafhoppers are agricultural pests and are not endangered or protected species. All samples were collected in open tea plantations and not from any national parks or protected areas.
Sample collection and preparation
A relative number of collection sites were chosen from each area (Fig. 1). To mitigate the possibility of collecting other leafhopper species that migrate from neighbouring habitats into the tea plantations to survive during the cold season, we collected all the specimens in the seasons of tea leafhopper outbreak from 2005 to 2010. Tea leafhopper specimens were collected by sweep netting from six main tea producing provinces of Zhejiang, Anhui, Guangdong, Fujian, Hainan and Hunan in Mainland China, four main tea production areas of Shizuoka, Kyoto, Saitama and Kagoshima in Japan, and Taoyuan in Taiwan (Table 1). The peach green leafhopper Empoasca flavescens was as the out-group and collected at a riverside open park, which was absolutely isolated from tea plantations, in downtown Hangzhou of Zhejiang province. All insects were rinsed in 70% ethanol three times, and stored at −80°C in 100% ethanol until DNA was extracted. All tea green leafhoppers were first identified under a stereo microscope following the keysincluding head and thorax, forewing, wing, and genitalia characters were used for diagnosing specimens before being used [9]. The insects and DNA samples were deposited in the Tea Research Institute, Chinese Academy of Agricultural Sciences, CAAS, Hangzhou, China.
Figure 1. The distribution map of eleven tea green leafhopper populations.
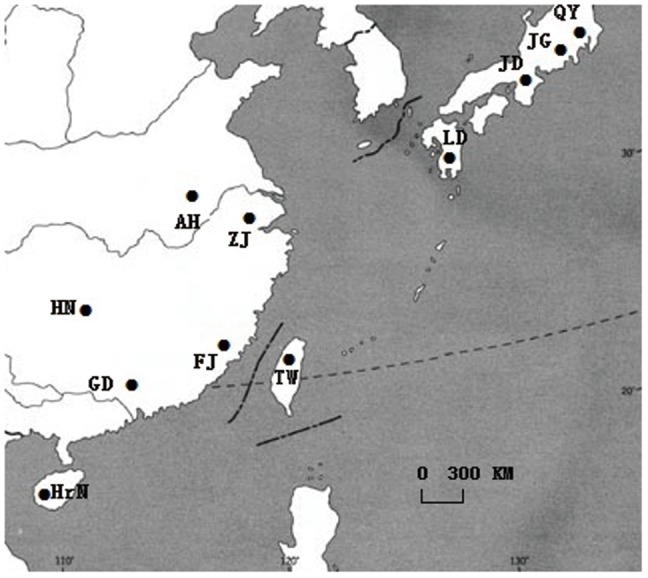
Six populations were sampled from Anhui (AH), Zhejiang (ZJ), Hunan (HN), Fujian (FJ), Guangdong (GD) and Hainan (HrN) in main tea production regions of Mainland China. One population was collected from Taiwan, and four populations were collected from Shizuoka (JG), Kyoto (JD), Saitama (QY) and Kagoshima (LD) in Japan.
Table 1. The collecting information and bioinformatics for leafhopper specimens.
| Leafhopper specimens | Collecting locality | Longitude°(E)/Latitude°(N) | Collecting date (M/Y) | Population number | Accession number/Haplotypes | |
| Ingroup Population | COI /73 | 16sRNA/67 | ||||
| Empoasca vitis (Göthe) | Mainland China, (Anhui, AH) | 119.08/30.58 | 6/2005 | 12 | KC172464–KC172527 | KC172397–KC172456, KC172463 |
| Empoasca vitis (Göthe) | Mainland China, (Fujian, FJ) | 119.13/26.05 | 8/2004 | 14 | ||
| Empoasca vitis (Göthe) | Mainland China, (Guangdong, GD) | 113.24/24.12 | 5/2008 | 13 | ||
| Empoasca vitis (Göthe) | Mainland China, (Hainan, HrN) | 109.29/19.30 | 9/2005 | 14 | ||
| Empoasca vitis (Göthe) | Mainland China, (Hunan, HN) | 113.26/28.27 | 6/2009 | 6 | ||
| Empoasca vitis (Göthe) | Mainland China, (Zhejiang, ZJ) | 120.10/30.16 | 7/2004 | 14 | ||
| Empoasca onukii MATSUDA | Japan, (Shizuoka, JG) | 138.38/34.98 | 9/2008 | 15 | ||
| Empoasca onukii MATSUDA | Japan, (Kyoto, JD) | 135.46/35.10 | 10/2008 | 12 | ||
| Empoasca onukii MATSUDA | Japan, (Saitama, QY) | 139.65/35.86 | 9/2008 | 14 | ||
| Empoasca onukii MATSUDA | Japan, (Kagoshima, LD) | 130.33/31.35 | 9/2008 | 14 | ||
| Jacobiasca formosana (Paoli) | Taiwan, (Taoyuan, TW) | 121.18/24.59 | 6/2006 | 13 | ||
| Outgroup species | ||||||
| Empoasca flavescens (Fabricius) | China, (Zhejiang, Hangzhou) | 120.12/30.16 | 7/2010 | 9 | KC191708–KC191716 | KC172457–KC172462 |
DNA extraction
Total DNA was extracted from a single insect body after it air-dried for 20 min. The specimen tissue was first ground with a flame-sealed pipette tip in a 1.5 ml microcentrifuge tube, and then incubated at 55°C in 0.5 ml lysis buffer containing 100 mM Tris-HCl, 5 mM EDTA, 200 mM NaCl, 0.5% SDS, and 100 µg/ml proteinase K for 5 hours. The lysis products were treated with a standard phenol-chloroform-isoamyl alcohol (25∶24∶1) method, and DNA was precipitated from the supernatant with two volumes of cold ethanol, centrifuged, washed, dried and dissolved in 100 µl deionized and sterilized water, and then stored at −80°C until use.
PCR amplification and sequencing
Polymerase chain reactions were carried out on an ABI Veriti thermocycler in 50 µl total volumes containing 5 ng DNA, 1.5 mM MgCl2, 0.25 mM dNTPs, 0.2 µM of each primer and 0.5 Units taq NEB DNA polymerase. We amplified the COI genes with primers C1-J-2195: 5′-TTGATTTTTTGGTCAYCCWGAAGT-3′ and TL2-N-3014: 5′-TTCATTGCACTAATCTGCCATACTA-3′ [21]. The 16sRNA fragments were obtained by primers LR-J-12887: 5′-CCGGTYTGAACTCARATCAWGT-3′ and LR-N-13398: 5′-CTGTTTAWCAAAAACATTTC-3′ [4]. The PCR program was denatured 4 min at 94°C, followed by 30 cycles of 94°C for 30 sec, 55°C for 30 sec, 72°C for 1 min and extension at 72°C for 10 min. The PCR products were detected by electrophoresis on a 1.2% agarose gel stained with ethidium bromide, purified with the Gel DNA extraction kit (Axygen), introduced into pGEM T-easy vector (Promega) and sequenced bidirectionally by an ABI 3730 automated sequencer with the universal primers M13F: 5′-CGCCAGGGTTTTCCCAGTCACGAC-3′ and M13R: 5′-AGCGGATAACAATTTCACACAGGA-3′. At least two copies of each clone were sequenced simultaneously.
Phylogenetic analysis and haplotype network
Both sense and antisense sequences were automatically aligned and assembled by DNAMAN 4.0 software, and the single consensus sequence was obtained by manual splicing. All these mtDNA sequences were further confirmed by checking if they could be appropriately translated into protein using invertebrate mitochondrial codes by MEGA 6.0 program [22]. The COI, 16sRNA and their combined sequences were aligned by Clustal Omega software online (http://www.ebi.ac.uk/Tools/msa/clustalo/), the alignments were downloaded and converted to compatible format by MEGA 6.0 for analysis below.
All these analyses were done based on both single gene and the combined sequences. The sequence polymorphism level including nucleotide diversity (Pi), haplotype diversity (Hd), and the number of segregating sites (S) was calculated by DnaSP 5.0. Genetic distances between gene pairs were calculated using the Kimura 2-parameter (K2-P) model via MEGA 6.0. Neutrality tests of Tajima's D, Fu and Li's D, Fu and Li's F were performed by DnaSP 5.0 [23]. The amount of variation within a population and among populations within a region was calculated by the hierarchical analysis of molecular variance (AMOVA) framework carried out using ARLEQUIN 3.11, and significant difference was tested by a nonparametric permutation procedure with 1000 permutations. The patterns of mitochondrial DNA polymorphism and evolution among E. vitis, J. formosana and E. onukii were also analyzed. Population differentiation was also quantified with non hierarchical analysis of molecular variance by estimating fixation index (Fst) of Mainland China-Japan, Japan-Taiwan, and Mainland-Taiwan populations.
The phylogenetic trees were constructed based on single gene and combined COI and 16sRNA by two methods of Maximum likelihood (ML) and Bayesian (BI). ML analysis was performed by RAxML online (http://www.trex.uqam.ca/index.php?action=raxml) and BI analysis was performed using GTR+I+G model via MrBayes 3.2.2. The BI trees were visualized and produced using FigTree software version 1.4.2, and only three BI trees for single gene and combined sets are presented here. The haplotypes of combined sequences were analyzed using the median-joining algorithm, and the network reconstruction were manual performed with Network 4.6.1.0 [24].
Results
Morphological examination
The E. onukii was nominated from leafhoppers in tea plantation by Matsuda in 1952 [25]. The dominant species of E. vitis and J. formosana in tea plantations of China Mainland and Taiwan were both named according to nominated species from other host plants. E. vitis was first named in 1875, and it was recognized as the dominant tea leafhopper species of China Mainland in 1988 [9], [26]; J. formosana was named in 1932, and it was identified as the dominant species in tea plantations of Taiwan [5], [27]. Utilizing the main, generally accepted, keys for tea green leafhopper [9], [25], we found no difference in these eleven population specimens.
Sequences analysis
A total of 149 partial COI and 16sRNA fragments were separately obtained and sequenced, of which 140 were from eleven leafhopper populations and nine from the out-group E. flavescens. The mtDNA COI fragment was 773 bp long with 208 polymorphic sites and 175 parsimony informative sites, and the A+T content was about 71%. In total, we obtained 64 haplotypes and the haplotype diversity index (Hd) was 0.99. As well, all nine COI genes from peach green leafhoppers are haplotypes. The 16sRNA gene was 535 bp long, which contained 129 polymorphic sites and 85 parsimony informative sites, and the A+T content was about 75%. We detected 61 haplotypes for 16sRNA in tea green leafhopper populations and the haplotype diversity was 0.95, and nine out-group sequences are all haplotypes. Sequences of 64 haplotypes for COI and 61 haplotypes for 16sRNA of tea green leafhoppers were finally deposited in GenBank (COI Accession Nos. KC172464-KC172527 and 16sRNA Accession Nos. KC172397-KC172456, KC172463), and the genes for out-group E. flavescens were also deposited respectively (Accession Nos. KC191708-KC191716 and KC172457-KC172462) (Table 1).
We also analyzed the combined mtDNA COI and 16sRNA genes simultaneously; 112 haplotypes were observed in 140 tea green leafhoppers specimens, and the haplotype diversity was high, more than 0.99. There were only eight haplotypes that contained more than two samples, of which FJ13 was the largest one and shared by nine samples of GD2, GD12, GD14, HrN6, HrN8, HrN12, TW4, TW10 and LD16 from all three regions of Mainland China, Taiwan and Japan. The haplotype of FJ2 was shared by five samples of FJ10, JG5, JG15, QY12 and QY17 from two regions of Mainland China and Japan. Three haplotypes of AH3, GD1 and AH11 were shared by two, two and seven samples from Mainland China respectively. The haplotypes of JD17, JG12 and QY8 were shared by two, three and three samples from Japan respectively. The other 104 haplotypes were all unique ones and the nine combined sets from E. flavescens were all unique haplotypes, too. The patterns of mitochondrial DNA polymorphism and evolution among the three leafhopper species are shown in Table 2. The values detected in E. vitis and E. onukii were much lower than those of J. formosana. Furthermore, the tested values for Tajima's D, Fu and Li's D and Fu and Li's F in E. vitis and E. onukii were all obviously less than those in J. formosana.
Table 2. Comparison of combined COI and 16sRNA sequences evolution of E. vitis, J. formosana and E. onukii.
| Species | n | h | Hd | S | Pi | D | Tajima's D | Fu and Li's D | Fu and Li's F |
| E. vitis | 74 | 59 | 0.983 | 94 | 0.00565 | −2.10751* | −2.16308** | −4.51381** | −4.25515** |
| E. onukii | 56 | 48 | 0.992 | 58 | 0.00368 | −2.19429*** | −2.71467*** | −4.19148** | −4.11439** |
| J. formosana | 10 | 9 | 0.978 | 24 | 0.00644 | −1.280251 | −0.22352 | 0.13177 | 0.04879 |
The letters n, h, Hd, S, Pi and D are the no. of tea green leafhopper samples, no. of mitochondrial DNA haplotype, no. of segregating sites, haplotype diversity, nucleotide diversity and genetic divergence, respectively. *: P<0.05; **: P<0.02; ***: P<0.01.
Genetic distance
The genetic divergences for COI, 16sRNA and combined sequences among 64, 61 and 112 leafhopper haplotypes of eleven populations were 0.3%–1.2%, 0.3%–0.6% and 0.3%–0.9%, and the average values were 0.7%, 0.8% and 0.6% respectively. All the genetic distances were at low levels and those for combined datasets are shown in Table 3. All the pairwise genetic distances between each two samples of tea green leafhopper and out-group E. flavescens were 31.3%–31.6%, which was 40-fold greater than those of in-group. The mean distances within each leafhopper population were also calculated and shown in Table 4 based on COI, 16sRNA and combined sets, which were also at much lower levels with the values of 0.2%–0.9%.
Table 3. Mean distance among tea green leafhopper populations based on combined datasets of COI and 16sRNA.
| Population | AH | FJ | GD | HN | HrN | ZJ | TW | JG | JD | QY | LD | PL |
| AH | ||||||||||||
| FJ | 0.005 | |||||||||||
| GD | 0.006 | 0.005 | ||||||||||
| HN | 0.005 | 0.005 | 0.005 | |||||||||
| HrN | 0.006 | 0.005 | 0.005 | 0.005 | ||||||||
| ZJ | 0.006 | 0.005 | 0.005 | 0.005 | 0.005 | |||||||
| TW | 0.008 | 0.007 | 0.007 | 0.007 | 0.007 | 0.007 | ||||||
| JG | 0.009 | 0.007 | 0.008 | 0.008 | 0.007 | 0.008 | 0.008 | |||||
| JD | 0.009 | 0.008 | 0.008 | 0.008 | 0.008 | 0.008 | 0.008 | 0.003 | ||||
| QY | 0.009 | 0.008 | 0.008 | 0.008 | 0.008 | 0.008 | 0.008 | 0.003 | 0.003 | |||
| LD | 0.008 | 0.007 | 0.007 | 0.007 | 0.007 | 0.007 | 0.008 | 0.004 | 0.005 | 0.004 | ||
| PL | 0.315 | 0.313 | 0.314 | 0.314 | 0.314 | 0.315 | 0.317 | 0.316 | 0.316 | 0.316 | 0.316 |
Table 4. Mean distance within tea green leafhopper populations based on COI, 16sRNA and combined datasets.
| Population | COI | 16sRNA | COI+16sRNA |
| AH | 0.0072 | 0.0054 | 0.0061 |
| FJ | 0.0085 | 0.0044 | 0.0048 |
| GD | 0.0059 | 0.0043 | 0.0054 |
| HN | 0.0061 | 0.0029 | 0.0044 |
| HrN | 0.0078 | 0.0026 | 0.0053 |
| ZJ | 0.0059 | 0.0048 | 0.0052 |
| JG | 0.0022 | 0.0057 | 0.0026 |
| JD | 0.0051 | 0.0046 | 0.0038 |
| QY | 0.0026 | 0.0059 | 0.0026 |
| LD | 0.0042 | 0.0058 | 0.0053 |
| TW | 0.0070 | 0.0059 | 0.0084 |
Phylogenetic analysis and haplotype network
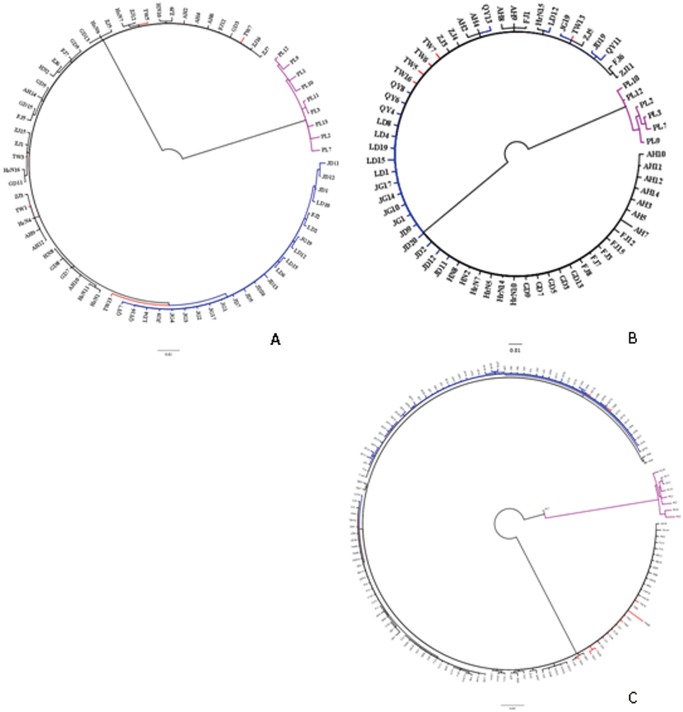
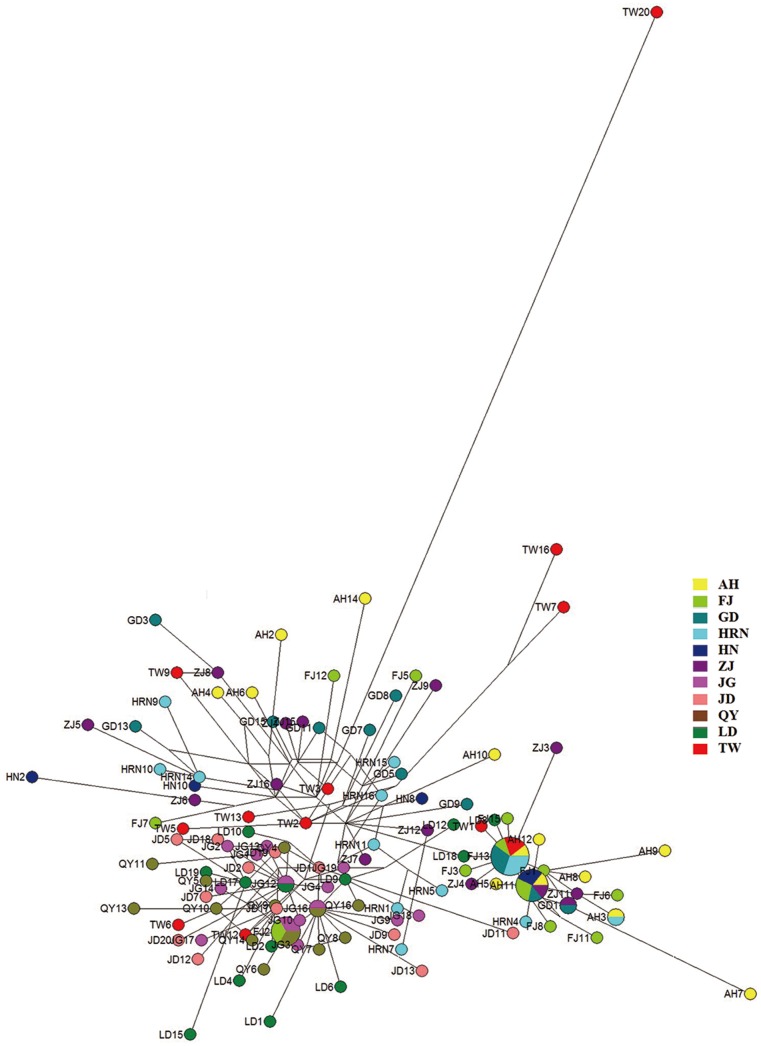
In the both BI and ML trees for COI gene, although most Japanese haplotypes clustered closely, the FJ2 haplotype from Mainland China first clustered with LD2 from Japan, and then clustered with other Japanese haplotypes together. Furthermore, the five Taiwan haplotypes always clustered first with haplotypes from at least three populations in Mainland China. To the both two trees for 16sRNA genes, no prior clustering pattern was found for haplotypes from the same regions. In the BI and ML trees for combined sequences, three samples LD8, LD16 and LD18 form Japan clustered prior with many samples from Mainland China, and two samples FJ2, FJ10 from Mainland China and two samples TW6, TW12 from Taiwan clustered prior with some samples from Japan. The BI trees for haplotypes of COI and 16sRNA, and the BI tree for combined sequences (S1, S2, S3 Figures) are only presented here (Fig. 2). Summarizing these phylogenetic trees, we also found that the leafhopper samples from Taiwan were relatively closer to Mainland China than those from Japan. The haplotype network that was reconstructed based on combined sequences showed no obvious divergent group (Fig. 3). The haplotypes FJ13, AH11 and FJ2 were three dominant haplotypes. FJ13 occurred in three populations of Mainland China, one population of Taiwan and one population of Japan. AH11 distributed in four populations from Mainland China, and FJ2 distributed in one population from Mainland China and two populations from Japan.
Figure 2. The BI phylogenetic trees for COI (A), 16sRNA (B) and combined sequences (C) of eleven leafhopper populations from Mainland China, Taiwan and Japan.
The samples from Mainland China, Japan and Taiwan were shown with black, blue and red colour, respectively; and the out-group of E. flavescens was shown with purple colour. These analyses involved 73, 67 and 149 nucleotide sequences including the out-group, respectively. Evolutionary analyses were conducted using MrBayes 3.2.2.
Figure 3. The haplotype network for combined COI and 16sRNA sequences of leafhoppers.
The populations were presented with different colours and the circle area showed the proportional of haplotype number.
Discussion
Tea plant spread from Mainland China to Japan and Taiwan along with the insect fauna. Tea green leafhoppers in the three regions have consistent morphological characters and biological habits [1], [6], [9], [11], but they are recognized as different species. Previously, we tested the 28S D2–D3 segments of genomic DNA among these leafhopper populations, and found the genetic divergences were much less than 0.1%. We had also tried some other mitochondrial genes to look for more helpful markers for genetic relationship studies; finally we found that only COI and 16sRNA genes could be stably amplified in all leafhopper DNA samples. Furthermore, we have investigated twelve populations of tea green leafhopper in China using the 16sRNA gene by another pair of primers and found the genetic distances among them were all less than 0.8% [28]. In a word, these two mitochondrial genes had been proved useful and accepted in leafhoppers diagnosis [21], [29], [30], and their sequences were successfully obtained, so the COI and 16sRNA fragments were selected in this study.
It has been proved that the genetic distance for insect species boundary was 2% [31]. All eleven populations from Mainland China, Taiwan and Japan showed such close relationships that the genetic distances for single gene and combined sequences were both less than 1.2%, and the mean values were less than 0.8%, which were below the criterion of 2% for insect species differentiation. These genetic relationship results support the hypothesis that E. vitis, J. formosana and E. onukii are a single tea green leafhopper species. On the whole, the genetic distances between each two populations from Mainland and Taiwan were 0.5–0.8%, while the genetic distances among populations from either Mainland or Taiwan with populations from Japan were 0.7–0.9%. It revealed that the genetic relationship between E. vitis and J. formosana was closer than either of them with E. onukii, and all of which were below species level.
Both the single gene and combined datasets showed high haplotype diversity (Hd) greater than 0.95, which implied a high diversity in tea green leafhoppers. In addition, at least one haplotype was shared together by leafhoppers from Mainland China, Taiwan and Japan. For the COI, 16sRNA and combined sequences, the haplotypes of FJ2 and AH3, QY14, LD6, FJ7 and FJ13, and FJ13 were shared by E. vitis, J. formosana and E. onukii, respectively. The ubiquitous shared haplotypes indicated that tea green leafhoppers from three regions were not distinct clades and they might be a single species at evolution level.
All the ML and BI trees based on the same genes were consistent and; therefore, only three BI trees were presented here. The trees, whether for single gene or combined sequences, showed that at least two samples from different regions clustered prior to some samples from one region. Indicating that the three tea green leafhopper species had no strict distinct boundary, therefore they might be a single species, according to their mutual phylogenetic relationships. Furthermore, all of the leafhopper samples from Taiwan showed a prior cluster with samples from Mainland China than those from Japan. This result indicated the obvious relationships below species level among tea green leafhoppers from East Asia, and J. formosana had a closer relationship with E. vitis than E. onukii. Although both Japan and Taiwan learned tea from Mainland China, Japan grew tea plants one thousand years earlier than Taiwan, which led to a relatively long-term isolation. Moreover, the climate of Taiwan was more coincident with Mainland compared with that of Japan. These might be the two most possible reasons for E. vitis being much closer to J. formosana than to E. onukii based upon genetic distances and phylogenetic trees.
In summary, we first conducted genetic distances and phylogenetic relationships analysis of the COI and 16sRNA regions of mtDNA for tea green leafhopper dominant species in East Asia. Both the results showed that within E. vitis, J. formosana and E. onukii there existed small genetic differences that were still below species level, which supported the hypothesis of a single species. However, further research is needed such as the creation of useful morphological keys, as well as cross-breeding test to understand genetic relationships of tea green leafhoppers on a large scale.
Supporting Information
The BI phylogenetic tree for COI of eleven leafhopper populations from Mainland China, Taiwan and Japan.
(EPS)
The BI phylogenetic tree for 16sRNA of eleven leafhopper populations from Mainland China, Taiwan and Japan.
(EPS)
The BI phylogenetic tree for combined sequences of eleven leafhopper populations from Mainland China, Taiwan and Japan.
(EPS)
Acknowledgments
The authors wish to thank all the reviewers for their comments that help improve the manuscript. Thank Dr Yasushi Sato for constructive advices and collecting leafhoppers from Japan. Thank Dr Andi Shau-mei Ou for collecting leafhoppers from Taiwan, and other people for collecting specimens.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Natural Science Foundation of China (31470693, 31100503), the public technology application research of Zhejiang Province (2013C32086), the Fundamental Research Funds of National Nonprofit Research Institute for Tea Research Institute, CAAS, (0032014014), the Natural Science Foundation of Zhejiang province, China (Y3080150), the Fundamental Research Funds of National Science and Technology (2013FY113200), and the Key Innovation Team of Zhejiang Province (Grant No. 2011R09027-13). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hazarika LK, Bhuyan M, Hazarika BN (2009) Insect pests of tea and their management. Ann Rev Entomol 54:267–284. [DOI] [PubMed] [Google Scholar]
- 2.Zhang HH, Tan JC (2004) Tea pests and non-pollution management in China. Anhui Science and Technology Publishing House: 3–9.
- 3.Oman P, Knight WJ, Nielson MW (1990) Leafhoppers (Cicadellidae): A Bibliography, Generic Check-list and Index to the World Literature 1956–1985: CAB International.
- 4. Dietrich CH, Whitcomb RF, Black WC (1997) Phylogeny of the grassland leafhopper genus Flexamia (Homoptera: Cicadellidae) based on mitochondrial DNA sequences. Mol Phylogenet Evol 8:139–149. [DOI] [PubMed] [Google Scholar]
- 5. Cho JY, Mizutani M, Shimizu B, Kinoshita T, Ogura M, et al. (2007) Chemical profiling and gene expression profiling during the manufacturing process of Taiwan oolong tea "Oriental Beauty". Biosci Biotechnol Biochem 71:1476–1486. [DOI] [PubMed] [Google Scholar]
- 6. Miao J, Han B (2007) Probing behavior of the tea green leafhopper on different tea plant cultivars. Acta Ecol Sin 27:3973–3982. [Google Scholar]
- 7. Mu D, Cui L, Ge J, Wang M-X, Liu L-F, et al. (2012) Behavioral responses for evaluating the attractiveness of specific tea shoot volatiles to the tea green leafhopper, Empoaca vitis . Insect Sci 19:229–238. [Google Scholar]
- 8. Hicks AL, Whitcomb RF (1996) Diversity of the leafhopper (Homoptera: Cicadellidae) fauna of northern Chihuahuan grasslands, with emphasis on gypsum grasslands and description of a new species of Athysanella (Cicadellidae: Deltocephalinae). P Entomol Soc Wash 98:145–157. [Google Scholar]
- 9. Zhao DX, Chen ZM, Cheng JA (2000) Belongings of tea leafhopper dominant species. J Tea Sci 20:101–104. [Google Scholar]
- 10. Fu JY, Han BY (2005) A Molecular Analysis on Genetic Relationships among Individuals of Tea leafhopper. Bull Sci Technol 5:550–556. [Google Scholar]
- 11. Mochizuki M, Ohtaishi M, Honma K (1994) Yellow sticky trap of flat type is useful for monitoring the occurrence of tea green leafhopper, Empoasca onukii Matsuda (Homoptera: Cicadellidae). Bull NRIVOPT Tea Ser B (Tea) 7:29–37. [Google Scholar]
- 12. Simon C, Frati F, Beckenbach A, Crespi B, Liu H, et al. (1994) Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am 87:651–701. [Google Scholar]
- 13. Boykin LM, Shatters RG, Hall DG, Burns RE, Franqui RA (2006) Analysis of host preference and geographical distribution of Anastrepha suspensa (Diptera: Tephritidae) using phylogenetic analyses of mitochondrial cytochrome oxidase I DNA sequence data. Bull Entomol Res 96:457–469. [PubMed] [Google Scholar]
- 14. Monti MM, Nappo AG, Giorgini M (2005) Molecular characterization of closely related species in the parasitic genus Encarsia (Hymenoptera: Aphelinidae) based on the mitochondrial cytochrome oxidase subunit I gene. Bull Entomol Res 95:401–408. [DOI] [PubMed] [Google Scholar]
- 15. Polaszek A, Manzari S, Quicke DLJ (2004) Morphological and molecular taxonomic analysis of the Encarsia meritoria species-complex (Hymenoptera, Aphelinidae), parasitoids of whiteflies (Hemiptera, Aleyrodidae) of economic importance. Zool Scr 33:403–421. [Google Scholar]
- 16. Smith MA, Wood DM, Janzen DH, Hallwachs W, Hebert PDN (2007) DNA barcodes affirm that 16 species of apparently generalist tropical parasitoid flies (Diptera, Tachinidae) are not all generalists. PNAS 104:4967–4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boykin LM, Shatters RG, Rosell RC, McKenzie CL, Bagnall RA, et al. (2007) Global relationships of Bemisia tabaci (Hemiptera: Aleyrodidae) revealed using Bayesian analysis of mitochondrial COI DNA sequences. Mol Phylogenet Evol 44:1306–1319. [DOI] [PubMed] [Google Scholar]
- 18. Lozier JD, Roderick GK, Mills NJ (2009) Molecular markers reveal strong geographic, but not host associated, genetic differentiation in Aphidius transcaspicus, a parasitoid of the aphid genus Hyalopterus . Bull Entomol Res 99:83–96. [DOI] [PubMed] [Google Scholar]
- 19. Smith PT (2005) Mitochondrial DNA variation among populations of the glassy-winged sharpshooter, Homalodisca coagulata . J Insect Sci 5:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Castalanelli MA, Baker AM, Munyard KA, Grimm M, Groth DM (2012) Molecular phylogeny supports the paraphyletic nature of the genus Trogoderma (Coleoptera: Dermestidae) collected in the Australasian ecozone. Bull Entomol Res 102:17–28. [DOI] [PubMed] [Google Scholar]
- 21. Takiya DM, Tran PL, Dietrich CH, Moran NA (2006) Co-cladogenesis spanning three phyla: leafhoppers (Insecta: Hemiptera: Cicadellidae) and their dual bacterial symbionts. Mol Ecol 15:4175–4191. [DOI] [PubMed] [Google Scholar]
- 22. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sun XJ, Xiao JH, Cook JM, Feng G, Huang DW (2011) Comparisons of host mitochondrial, nuclear and endosymbiont bacterial genes reveal cryptic fig wasp species and the effects of Wolbachia on host mtDNA evolution and diversity. BMC Evol Biol 11:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48. [DOI] [PubMed] [Google Scholar]
- 25. Matsuda R (1952) A new green Empoasca leaf hopper injurious to the tea shrub in Japan. Oyo-Kontyu Tokyo 8:19–21. [Google Scholar]
- 26.Ge ZLZ (1988) Research on the Cicadellidae species damaging Chinese tea shrub (I). J Tea Bus: 15–18.
- 27.Paoli G (1932) A new species distribution of Empoasca (Hemiptera, Homoptera). Memories for Italia Entomology Society: 109–122.
- 28. LI L, Fu JY, Xiao Q (2013) Sequence Analysis of mtDNA Gene and Genetic Differentiation in Geographic Population of Empoasca vitis (Gothe). Chin J Appl Entomol 50:675–685. [Google Scholar]
- 29. Fang Q, Black WC, Blocker HD, Whitcomb RF (1993) A phylogeny of New World Deltocephalus-like leafhopper genera based on mitochondrial 16S ribosomal DNA sequences. Mol Phylogenet Evol 2:119–131. [DOI] [PubMed] [Google Scholar]
- 30. Johnson DG, Carstens BC, Sheppard WS, Zack RS (2005) Phylogeny of leafhopper subgenus Errhomus (Erronus) (Hemiptera: Cicadellidae) based on mitochondrial DNA sequences. Ann Entomol Soc Am 98:165–172. [Google Scholar]
- 31. deWaard JR, Hebert PDN, Humble LM (2011) A Comprehensive DNA Barcode Library for the Looper Moths (Lepidoptera: Geometridae) of British Columbia, Canada. PLoS ONE 6:e18290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The BI phylogenetic tree for COI of eleven leafhopper populations from Mainland China, Taiwan and Japan.
(EPS)
The BI phylogenetic tree for 16sRNA of eleven leafhopper populations from Mainland China, Taiwan and Japan.
(EPS)
The BI phylogenetic tree for combined sequences of eleven leafhopper populations from Mainland China, Taiwan and Japan.
(EPS)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.