Abstract
Pancreatic ductal adenocarcinoma (PDAC) accounts for approximately 90-95% exocrine malignant tumors of the pancreas. The high prevalence of metastasis and the difficulty of early diagnosis lead to a dismal prognosis. MicroRNAs (miRNAs) play a critical role in extensive biological processes. The purpose of this study was to evaluate the feasibility of stool miRNAs as novel biomarker for PDAC screening. MiRNAs were extracted from clinical specimens which included cancer and matched adjacent benign pancreatic tissues of 30 PDAC patients, pancreatic juice of 20 from the 30 PDAC patients and 10 chronic pancreatitis (CP) patients, stool samples of the 30 PDAC patients, the 10 CP patients and 15 healthy volunteers. Relative expression of a panel of 5 dysregulated miRNAs (miR-21, miR-155, miR-196a, miR-216 and miR-217) was analyzed with qRT-PCR. Receiver operating characteristic curve (ROC) analysis was performed to assess the diagnosing value of stool miRNAs in PDAC patients. The study showed that our methods of extracting and detecting miRNAs from pancreatic juice and stool specimens had high reproducibility. Compared to matched adjacent benign pancreatic tissues and pancreatic juice of CP patients, the expression of miR-21 (P = 0.0021 and P = 0.0027) as well as miR-155 (P = 0.0087 and P = 0.0067) was significantly higher and the expression of miR-216 (P < 0.0001 and P = 0.0044) was significantly lower in primary tumor tissues and pancreatic juice of PDAC patients. PDAC patients had a significantly higher stool miR-21 and miR-155 (P = 0.0049 and P = 0.0112) and lower miR-216 level (P = 0.0002) compared to normal controls. The same results were obtained in the expression levels of stool miR-21, miR-155 and miR-216 between PDAC and CP patients (P = 0.0337, P = 0.0388 and P = 0.0117, respectively). Receiver operating characteristic (ROC) analysis by using stool miRNAs expression indicated that combination of miR-21 and miR-155 had best sensitivity of 93.33% while the combination of miR-21, miR-155 and miR-216 would be best for detecting and screening PDAC with area under the curve (AUC) of 0.8667 (95% CI: 0.7722-0.9612) and a better balance of sensitivity and specificity (83.33% vs. 83.33%). Our data indicate that miRNAs could be extracted and detected from pancreatic juice and stool efficiently and reproducibly. MiR-21, miR-155 and miR-216 in stool have the potential of becoming biomarkers for screening PDAC.
Keywords: PDAC, stool, microRNA, pancreatic juice, diagnosis
Introduction
Pancreatic cancer is the sixth leading cause of cancer-related mortality in China and the fourth in the United States, a total of 90-95% pancreatic cancers are exocrine tumors, known as pancreatic ductal adenocarcinoma (PDAC) [1,2]. Although chemoradiotherapy has been shown to prolong survival time in recent years, the prognosis of PDAC remains dismally with a median survival time of less than 2 years [3,4]. In addition to its aggressive malignant behavior, the extremely poor prognosis also due to absence of clinically useful screening and detecting measures of early PDAC.
In the past decade, lots of body fluid biomarker researches have provided an insight into new areas of screening and detecting PDAC [1,5-7]. However, most of these biomarkers discovered by new technologies suffered from significant limitations such as lack of methodological standardization and quality control, lower or no correlation with tumor stage and tumor invasiveness as well as minimal utility to assess prognosis or predict recurrence. Therefore, a stable, noninvasive and high sensitivity/specificity screening test is urgently needed.
MicroRNAs (miRNAs) are small noncoding transcripts of 22 nucleotides, which have recently been demonstrated as a new class of cellular molecules with important diagnostic, prognostic, and therapeutic implications [8,9]. Because of their small size and exosomal presence, miRNAs are notably well protected from endogenous and exogenous RNase activity [10-12]. Recent reports have proposed the possibility of miRNAs as biomarkers to screen and detect PDAC in blood and pancreatic juice while both of approaches are still invasive [13,14]. It has been suggested that gastrointestinal cancer-related genetic and epigenetic alterations may also noninvasively be detected in stools [7,15,16]. Considering the high volume of pancreatic juice excreted into the bowel, it has been assumed that pancreatic cancer-related miRNAs expression profile changes may be detected in stools of PDAC patients [7,16].
Option of candidate miRNAs in our study was performed according to following criteria: a) previously reported as potential PDAC development-related miRNAs (miR-21, miR-155, miR-196a, miR-216 miR-217) [17,18]; b) differential expression of miRNAs has been reported in blood (miR-21, miR-155, miR-196a) [12,13] and/or pancreatic juice (miR-21, miR-155) of pancreatic carcinoma patients [18,19].
Materials and methods
All clinical samples were collected from patients treated in Biliary-Pancreatic Surgery Department of Renji Hospital, School of Medicine, Shanghai Jiao tong University from January 2012 to December 2013. The diagnosis of PDAC and chronic pancreatitis (CP) was confirmed by the pathological results of resected specimens. All patients and volunteers including 30 PDAC patients, 10 CP patients and 15 healthy volunteers provided written informed consent and the study was approved by the Institutional Review Board of Renji Hospital, School of Medicine, Shanghai Jiao tong University. Clinical and demographical information of the patients are presented in Table 1.
Table 1.
Clinic-pathological Characteristics of patients included to the study
| PDAC | CP | Controls | |
|---|---|---|---|
| Number | 30 | 10 | 15 |
| Sex | |||
| Women | 18 | 6 | 8 |
| Men | 12 | 4 | 7 |
| Age (mean ± SD) | 66.9 ± 11 | 57.3 ± 8.6 | 30.4 ± 9.6 |
| Diagnosis | |||
| PDAC | 30 | ||
| CP | 10 | ||
| Tumor stage | |||
| I | 7 | ||
| II | 11 | ||
| III | 12 | ||
| Location | |||
| Head | 20 | ||
| Corpus | 4 | ||
| Tail | 6 | ||
| CA19-9 (U/ml) (mean ± SD) | 569.6 ± 803 | ||
| Tumor diameter (cm) | 2.9 ± 1.0 |
Tissue and pancreatic juice samples
Cancer tissues (0.5 cm in diameter from center of cancer tissues) and matched adjacent benign pancreatic tissues (same size at least 2 cm apart from the tumor) samples were collected from 30 PDAC patients during surgical resection. At least 2 ml pancreatic juice was extracted from ectatic pancreatic duct under the guidance of intraoperative B-mode ultrasound (LOGIQ 5; GE Healthcare, Milwaukee, WI, USA) with a 12 MHz probe. Finally, we obtained 30 pancreatic juice samples including 20 of the PDAC patients and 10 of the CP patients. All samples were snap-frozen in liquid nitrogen and stored at -80°C subsequently.
Stool samples
A total of 55 stool specimens, obtaining from the 30 PDAC patients, 10 CP patients and 15 healthy volunteers were included in the study. All stool samples were collected in the morning with a 40 ml aseptic specimen cup. Prior to reaching the laboratory, the stool samples were kept at -20°C freezer for short-term and then transferred to -80°C freezer within 24 h for long-term storage. Four categories of stool consistency were defined: ‘firm’, ‘soft’, ‘loose’ and ‘watery’. Only ‘firm’, ‘soft’, ‘loose’ stools were collected and analyzed in the study [20].
Isolation of miRNAs from tissue, pancreatic juice and stool samples
Total miRNA of tissue samples was isolated from 20-30 mg tissue using E.Z.N.ATM miRNA Kit according to the instruction of the manufacturer. E.Z.N.ATM miRNA Kit and E.Z.N.ATM stool RNA Kit (OMEGA, GA, USA) were used jointly to isolate total miRNA of pancreatic juice and stool samples with some modifications of the manufacturer’s guidelines. Briefly, an equal volume of acidic phenol, RPL buffer solution and chloroform was added to 200 ul pancreatic juice or 200 mg stool samples, and the aqueous phase was loaded onto the HiBind RNA membrane after the addition of the buffer RB and absolute ethanol. After removal of large RNAs (> 200 nt), apply the flow through from the HiBind RNA column into the MicroElute RNA column with 0.9 volume of absolute ethanol. Finally, miRNA was eluted in 20 µl DEPC water. The concentrations of all miRNA samples were quantified by the NanoDrop 2000 (NanoDrop, Wilmington, DE, USA).
MiRNA quantification by real-time PCR
After miRNA extraction, the samples were reverse-transcribed with All-in-One™ First-Strand cDNA Synthesis Kit (GeneCopoeia, MD, USA). 25 ul reverse transcriptase reactions contained 1 ul 2.5 U/ul PolyA Polymerase, 1 ul RTase Mix, 5 ul 5* PAP/RT Buffer, the volume of 100 ng miRNA and corresponding nuclease-free H2O. The reaction program was at 37°C for 60 min, followed by an incubation step at 85°C for 5 min. After reverse transcription, samples were run using Applied Biosystems step one plus RT-PCR detection system. Quantitative miRNA expression analyses were performed using SYBR green method (SYBR® Premix Ex TaqTM -Tli RNaseH Plus, TaKaRa, Japan) according to the manufacturers’ instructions. The 10 ul PCR reaction included 2.8 ul RT production (1:3 diluton), 1 ul universal reverse primer, 1 ul of specific sense primer, 5 ul SYBR green and 0.2 ul ROX Reference Dye II. To avoid an inter-plate bias, miRNA expression analyses were performed using a 96-well plate in triplicate with 40 cycles of 95°C for 5 s and 60°C for 30 s after 95°C for 30 s. Differences between the groups are presented as ΔCt, representing the difference of the Ct value between the interesting miRNA and the normalizer miRNA. We selected RNU6B as the normalizer. Primer sequences for the RT-PCR assays are listed in the Supplementary Table 1.
Statistical analysis
Data analyses were performed with GraphPad Prism 5.0 software (San Diego, CA, USA) and SPSS 17.0 (SPSS, Chicago, IL, USA). The differences between two groups were analyzed using Student’s t-test. Correlation analyses were performed using Pearson’s test. The reproducibility of miRNAs extraction and detection was analyzed by linear correlation. Receiver operating characteristic (ROC) curve analysis was used to determine the highest assay sensitivity and specificity. Two sided P < 0.05 was regarded as significant.
Results
Detectability of miRNAs in tissue, pancreatic juice and stool samples
As expected, we detected the expression of 5 miRNAs in all samples (Figure 1A-D). The expression levels of 5 miRNAs were high in cancer and benign pancreatic tissues. The expression levels of miRNAs in pancreatic juice were similar to those in stool but were lower than those in cancer and benign pancreatic tissues. The mean Ct values of the 5 miRNAs ranged from 13.11 to 25.53 in cancer and benign pancreatic tissues and from 30.99 to 35.45 in pancreatic juice and stool.
Figure 1.
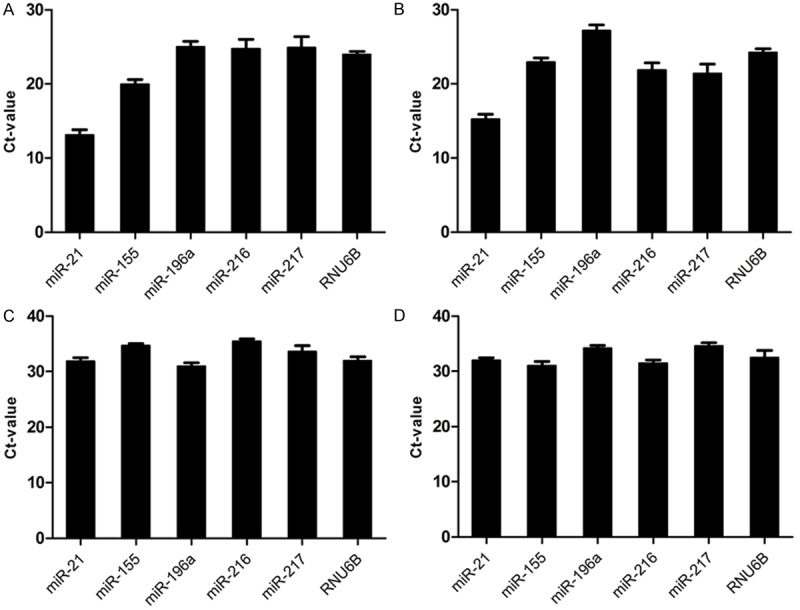
Detectability of miRNAs and RNU6B. To evaluate the detectability of selected miRNAs (miR-21, miR-155, miR-196a, miR-216, miR-217 and RNU6B), we analyze all Ct value of miRNAs and RNU6B. A. Shows the expression of all miRNAs and RNU6B in all primary cancer tissue of PDAC patients. B. Hows the expression of all miRNAs and RNU6B in all matched adjacent benign pancreatic tissues of PDAC patients. C. Hows the expression of all miRNAs and RNU6B in all pancreatic juice of PDAC patients. D. Hows the expression of all miRNAs and RNU6B in all stools of PDAC patients. The Figure shows the detectability miRNAs and RNU6B in tissue, pancreatic juice and stool.
Normalization of miRNA levels
For accurate quantization of miRNA levels with RT-PCR, normalization is critical but no consensus yet. RNU6B was commonly used as an endogenous control in miRNA studies. In this study, RNU6B could be detected in all samples. Furthermore, the expression of RNU6B in all samples is similar to that of miR-155 which is overexpressed miRNA relating to PDAC (Figure 2). So we selected RNU6B as our normalizer for subsequent analyses.
Figure 2.
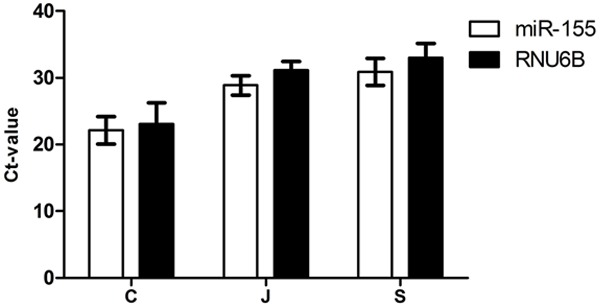
Expression levels of miR-155 and RNU6B in cancer tissue (C), pancreatic juice (J) and stool (S) of all PDAC patients.
Reproducibility of miRNAs extraction and detection in pancreatic juice and stool samples
To assess the reproducibility of our method for miRNAs extraction and detection, we performed twice independent extraction and detection. As shown in Figure 3, there was a high liner correlation in miRNA concentration of the pancreatic juice (R2 = 0.9609, P < 0.0001, Figure 3A) and the stool (R2 = 0.9882, P < 0.0001, Figure 3B). The same results were obtained in RNU6B detection of the pancreatic juice (R2 = 0.9357, P < 0.0001, Figure 3C) and the stool (R2 = 0.9068, P < 0.0001, Figure 3D).
Figure 3.
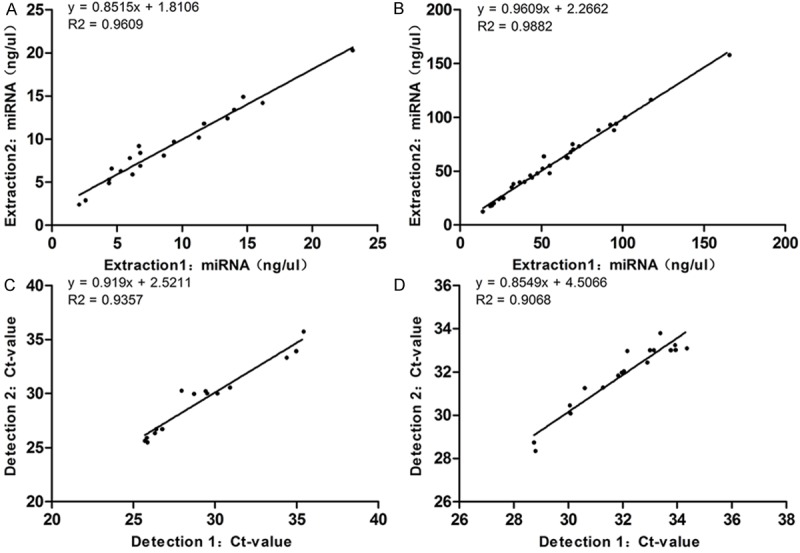
Reproducibility of miRNAs extraction and detection in pancreatic juice and stool samples. A. Reproducibility of our methodology for extraction of miRNAs in pancreatic juice shows a high linear correlation (R2 = 0.9609, P < 0.0001). B. Reproducibility of our methodology for extraction of miRNAs in stool shows a high linear correlation (R2 = 0.9882, P < 0.0001). C. Reproducibility of miRNAs detection in pancreatic juice shows a high linear correlation (R2 = 0.9357, P < 0.0001). D. Reproducibility of miRNAs detection in stool shows a high linear correlation (R2 = 0.9068, P < 0.0001).
Differential expression of miRNAs in tissue, pancreatic juice and stool samples
Figure 4 shows that the relative expression levels of 3 miRNAs (miR-21: P = 0.0021, miR-155: P = 0.0087, miR-196a: P = 0.0042) were significantly higher, while the levels of the other 2 (miR-216: P < 0.0001, miR-217: P = 0.0004) were significantly lower in primary cancer tissue as compared to matched adjacent benign pancreatic tissues.
Figure 4.
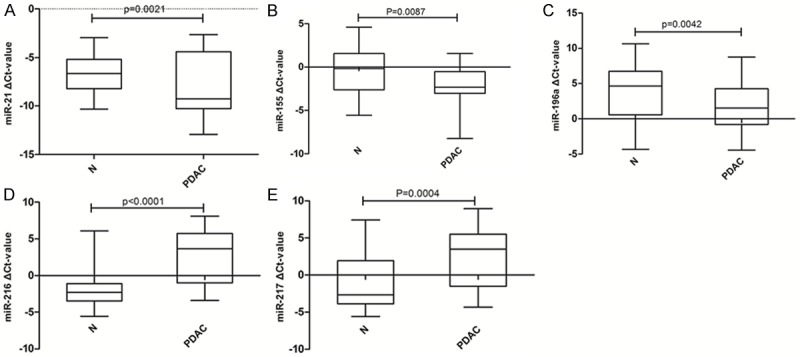
MiRNA expression patterns in primary cancer tissue and matched adjacent benign pancreatic tissues. A-E. Represent the difference expression of different miRNAs between primary cancer tissue and matched adjacent benign pancreatic tissues. Abbreviations: N- adjacent benign pancreatic tissues, PDAC- primary cancer tissue. The data are present as box-and-whiskers plot: the upper and lower limits of the boxes indicate the 75th and 25th percentiles, the lines inside the boxes-the medians, and the upper and lower horizontal bars denote the 90th and 10th percentiles, respectively.
Figure 5 shows that the relative expression levels of 2 miRNAs (miR-21: P = 0.0027, miR-155: P = 0.0067) were significantly higher, while the levels of other 2 (miR-196a: P = 0.0026, miR-216: P = 0.0044) were significantly lower in pancreatic juice of PDAC patients as compared to those of CP patients. There was no significantly difference of miR-217 between PDAC and CP pancreatic juice.
Figure 5.
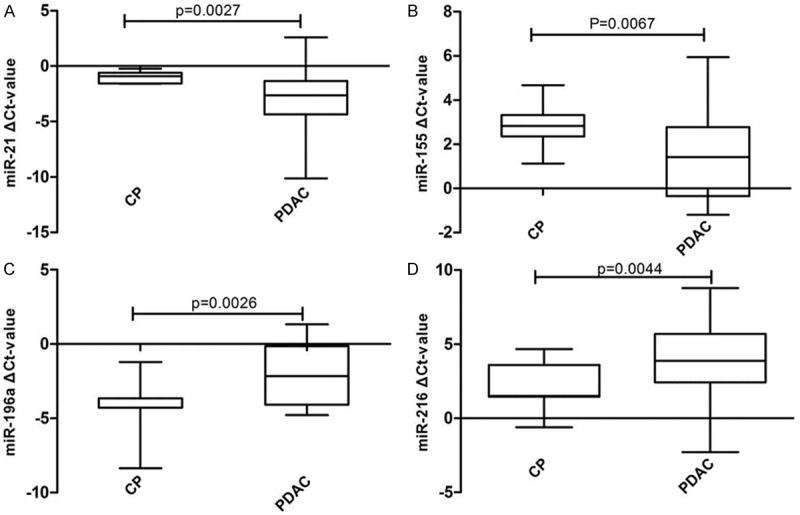
MiRNA expression patterns in pancreatic juice of CP and pancreatic juice of PDAC. A-D. Represent the difference expression of different miRNAs between pancreatic juice of CP and pancreatic juice of PDAC. Abbreviations: CP- chronic pancreatitis, PDAC- pancreatic ductal adenocarcinoma.
Figure 6 shows that the relative expression levels of 2 miRNAs (miR-21: P = 0.0049, miR-155: P = 0.0112) were significantly higher, while the level of miR-216 (P = 0.0044) was significantly lower in stool of PDAC patients as compared to those of normal controls. There was no significantly difference of miR-196a and miR-217 between PDAC and normal controls stools. The same results were obtained between PDAC and CP patients (miR-21: P = 0.0337, miR-155: P = 0.0388, miR-216: P = 0.0117, miR-196a: P = 0.1812, miR-217: P = 0.1093).
Figure 6.
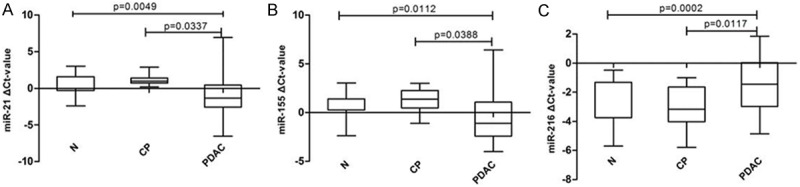
MiRNA expression patterns in stool of normal controls and stool of PDAC. A-C. Represent the difference expression of different miRNAs between stool of normal controls and stool of PDAC. Abbreviations: N- normal volunteers, PDAC- pancreatic ductal adenocarcinoma.
Our data show that the signature of miR-21, miR-155 and miR-216 was fully consistent in PDAC cancer tissues, pancreatic juice and stool. Thus, the 3 miRNAs were analyzed in our study.
Diagnosis performance of stool miRNAs
For diagnostic purpose, the actual clinical utility of miRNAs was analyzed by comparing the AUC of ROC curves and sensitivity at given specificities. For differentiating PDAC from normal controls, the AUC-ROC of miR-21, miR-155 and miR-216 were 0.7989 (95% CI: 0.6779-0.9198), 0.7189 (95% CI: 0.5795-0.8588) and 0.7344 (95% CI: 0.6049-0.8640). The corresponding sensitivity and specificity of miR-21, miR-155 and miR-216 were 90% vs. 66.67%, 76.67% vs. 73.33% and 86.67% vs. 60% (Figure 7). As showed in Figure 8 and Table 2, the diagnostic capacity increased by combining the two or three miRNAs. Combination of miR-21, miR-155 and miR-216 would have a good balance sensitivity and specificity of 83.33% and 83.33% with better AUC up to 0.8667 (95% CI: 0.7722-0.9612).
Figure 7.
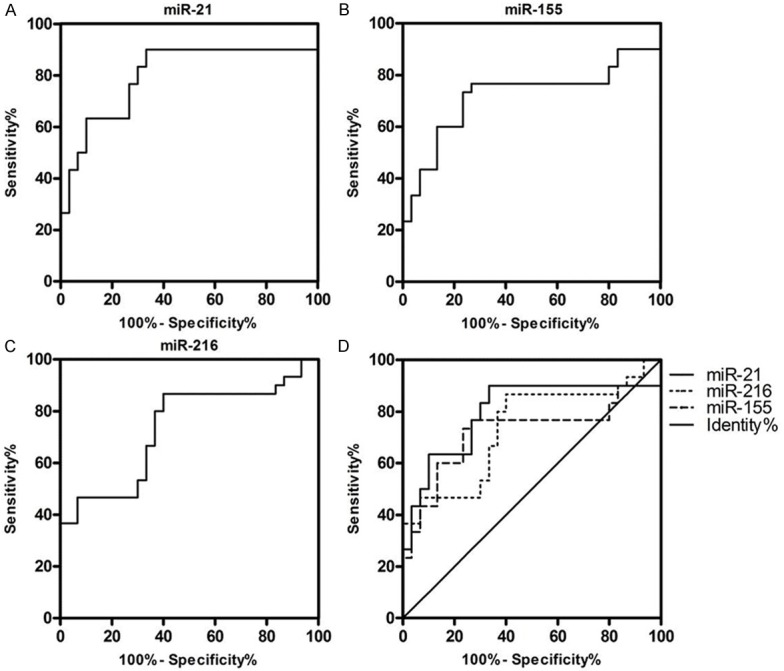
Diagnostic performance of the expression data of fecal miR-21, miR-155 and miR-216. A-C. Represents the AUC-ROC of different single fecal miRNAs of PDAC patients from controls. D. Shows the difference among the AUC-ROC of miR-21, miR-155 and miR-216. ROC = receiver operating characteristics.
Figure 8.
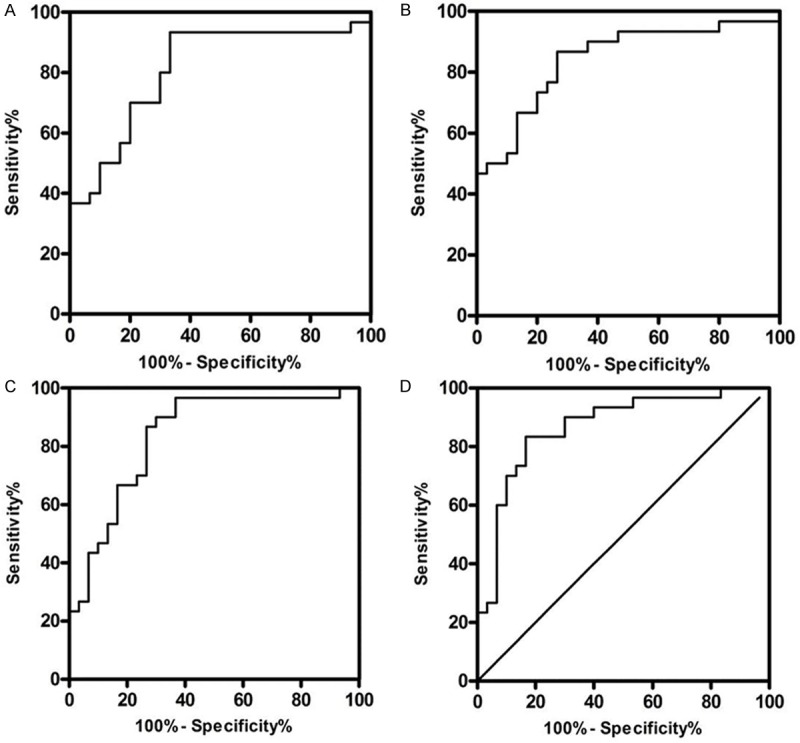
Diagnostic performance of combination of two or three miRNAs (miR-21, miR-155 and miR-216). A-D. Represents the AUC-ROC of combination of miR-21 and miR-155, combination of miR-21 and miR-216, combination of miR-155 and miR-216 and combination of miR-21, miR-155 and miR-216 of PDAC from controls. The data of combination of miRNAs obtained through processing the original expression data of miR-21, miR-155 and miR-216 by logistic regression analysis. ROC = receiver operating characteristics.
Table 2.
ROC analysis of the ΔCt of fecal miR-21, miR-155, miR-216 and the combination of two or three miRNAs
| Sensitivity (%) | Specificity (%) | AUC | 95% AUC | P-value | |
|---|---|---|---|---|---|
| miR-21 | 90% | 66.67% | 0.7989 | 0.6779-0.9198 | < 0.0001 |
| miR-155 | 76.67% | 73.33% | 0.7189 | 0.5795-0.8583 | 0.0036 |
| miR-216 | 86.67% | 60% | 0.7344 | 0.6049-0.8640 | 0.0018 |
| miR-21 & miR-155 | 93.33% | 66.67% | 0.8111 | 0.6981-0.9242 | < 0.0001 |
| miR-21 & miR-216 | 86.67% | 73.33% | 0.8422 | 0.7396-0.9448 | < 0.0001 |
| miR-155 & miR-216 | 90% | 70% | 0.8356 | 0.7308-0.9403 | < 0.0001 |
| miR-21 & miR-155 & miR-216 | 83.33% | 83.33% | 0.8667 | 0.7722-0.9612 | < 0.0001 |
Discussion
In this proof-of-principle study, we evaluated the feasibility of stool-based miRNAs as potential biomarkers for screening PDAC. Using a subset of miRNAs dysregulated frequently in PDAC, we found that miR-21 and miR-155 were overexpressed and miR-216 was down expressed in PDAC tissues, pancreatic juice and stool specimens compared to their controls. In addition, we validated good reproducibility in miRNAs extracting and detecting from stool and pancreatic juice. Lastly, we indicated that a combination of a subset of miRNAs may enhance the diagnostic value with a higher sensitivity and specificity for screening PDAC.
At present, only CA19-9 which has some markedly limitations including poor specificity, lack of expression in Lewis negative phenotype and higher false-positive elevation in the presence of obstructive jaundice is regarded as a useful clinical biomarker in the early detection of PDAC patients [21,22]. Thus, there is a clear need for alternative biomarkers for screening and detecting of PDAC. It is considered that the biomarkers in pancreatic juice are more related and specific with PDAC because of the same histological origin, but they are not suitable for screening purpose mainly due to the invasive collecting measures [1,23,24]. Although blood-based biomarker test may be practical and tremendous potential for screening, it is highly likely that the earliest and the most specific biomarker changes can be detected in stools rather than in blood basing on constant considerable shedding of exfoliated cells and production of pancreatic fluid [7,15,25]. So we suppose that the tissue-related miRNAs in stool may be ideal biomarkers for PDAC screening.
Basing on existing knowledge, we selected miR-21, miR-155, miR-196a, miR-216, and miR-217 as candidate miRNAs in our study [12,14,16-19,26]. However, It is noted in the present study that the microRNAs signature in different types of samples was not fully consistent [17,26]. Our data showed that the stool miR-196a and miR-217 signature was not concordant with those of tissues and/or pancreatic juice. Two reasons might result in the discrepancy: a) The degradation of free miRNAs and partial exosome miRNAs in stools [27]; b) the accidents in the process with which miRNAs from the tumor-derived exosomes in blood are secreted into the digestive tract [28]. Finally, only miR-21, miR-155 and miR-216 were analyzed in the study.
Stool-based miRNA extraction and detection was high reproducible (R2 = 0.9882, P < 0.0001 and R2 = 0.9068, P < 0.0001) in our study. From recent researches, we could know the stability of miRNAs in stool stemmed from the protection of exfoliative cellular membrane and exosome [27,29,30]. Pancreatic juice including exfoliative cells and tumor exosomes are released continuously and mixed homogenously with the stool. So, repeated sampling from the same stool samples results in similar test outcome [20,28]. These features favour the use of stool-based miRNA as screening biomarkers.
Several independent studies have demonstrated that the overexpressing miRNAs in gastrointestinal cancer tissues also shown higher expression levels in stool samples because of the same origin of miRNAs from exfoliative cells and the exosomes [15,20,31]. At present, Sadakari et al has demonstrated that the expression of miR-21 and miR-155 were significantly higher in PDAC tumor tissues and pancreatic juice [14]. Link A et al has found that miR-21, miR-155 and miR-216 had significantly high expression levels in stool samples of pancreatic cancer patients and the same result was obtained in tissues of pancreatic cancer patients by Bloomston et al [16,17]. But there was few researches focusing on miRNAs expression of paired cancer tissues, pancreatic juice and stool samples of PDAC patients. In this research, we detected the pancreas-related miRNAs in PDAC tissues, matched pancreatic juice and corresponding stool samples and found the tendency of differential expression of miR-21, miR-155 and miR-216 was consistent among PDAC tissues, pancreatic juice and stools. Basing on description above, we have reasons to believe that the stable pancreas-related miRNAs in stools mainly come from exfoliative cells and the exosomes which are excreted to intestine via pancreatic juice from PDAC tumor tissues. More important, we proved the feasibility of stool-based miRNAs for screening PDAC patients.
Stool miR-21, miR-155 and miR-216 were identified to have significant power in differentiating PDAC patients from CP patients and normal subjects in the study. With regard to pancreatic cancer, varying expression profiles of tissues microRNAs distinguishing malignant lesions from normal pancreatic samples and CP have also been reported [17,32]. However, few of these varying expression profiles of microRNAs have been detected in stool samples. Our data showed stool miR-21, miR-155 and miR-216 could differentiate PDAC from normal pancreas or CP, but could not differentiate CP from normal pancreas.
The potential of stool miRNAs as biomarkers for screening PDAC was assessed by ROC in our research. For single miRNA, miR-21 had better sensitivity of 90% and AUC of 0.7989 (95% CI: 0.6779-0.9198). Joint detection of two or three miRNAs can get a better result. Combination of miR-21 and miR-155 had best sensitivity of 93.33% while the combination of the three miRNAs would be best for detecting and screening PDAC with AUC of 0.8667 (95% CI: 0.7722-0.9612) and a better balance of sensitivity and specificity (83.33% and 83.33%). Comparing with the low sensitivity and specificity (68% and 70%) of CA19-9 which are used extensively in clinic [33], our stool miRNAs group is obviously more significant for PDAC detecting and screening.
Basing on the markedly features of stability, high sensitivity/specificity and non-invasion, stool miRNAs testing for screening PDAC in clinic is promising. So far, there are no reports about stool miRNAs in PDAC, so we believe that our significant finding will attract more attention to this area. Because of the retrospective feature of the research, we were unable to obtain more samples such as stools of precancerous lesions to validate whether the stool miRNAs can distinguish the precancerous lesion from PDAC. Additionally, systematic analyses of stools using microarray or ultra-high throughput sequencing will help in uncovering novel stool miRNAs and allow a methodological selection of potential biomarkers. In light of these facts, further intensive investigations prior to clinical application are needed.
In summary, the method for miRNAs extraction and detection was efficiently and reproducibly in pancreatic juice and stools. The consistent expression of miR-21, miR-155 and miR-216 among PDAC cancer tissues, pancreatic juice and stools demonstrated the feasibility of using stool miRNAs as noninvasive tools for screening PDAC. Moreover, combination of two or three stool miRNAs (miR-21, miR-155 and miR-216) could provide acceptable capacity for screening PDAC.
Acknowledgements
This work is supported by funds from Shanghai Health Bureau (20114213).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Ballehaninna UK, Chamberlain RS. Biomarkers for pancreatic cancer: promising new markers and options beyond CA 19-9. Tumour Biol. 2013;34:3279–3292. doi: 10.1007/s13277-013-1033-3. [DOI] [PubMed] [Google Scholar]
- 2.Taira N, Arai M, Ikeda M, Iwasaki M, Okamura H, Takamatsu K, Yamamoto S, Ohsumi S, Mukai H. The Japanese Breast Cancer Society clinical practice guideline for epidemiology and prevention of breast cancer. Breast Cancer. 2014 doi: 10.1007/s12282-014-0555-x. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 3.Hirata K, Egawa S, Kimura Y, Nobuoka T, Oshima H, Katsuramaki T, Mizuguchi T, Furuhata T. Current status of surgery for pancreatic cancer. Dig Surg. 2007;24:137–147. doi: 10.1159/000102067. [DOI] [PubMed] [Google Scholar]
- 4.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 5.Harsha HC, Kandasamy K, Ranganathan P, Rani S, Ramabadran S, Gollapudi S, Balakrishnan L, Dwivedi SB, Telikicherla D, Selvan LD, Goel R, Mathivanan S, Marimuthu A, Kashyap M, Vizza RF, Mayer RJ, Decaprio JA, Srivastava S, Hanash SM, Hruban RH, Pandey A. A compendium of potential biomarkers of pancreatic cancer. PLoS Med. 2009;6:e1000046. doi: 10.1371/journal.pmed.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L, Farrell JJ, Zhou H, Elashoff D, Akin D, Park NH, Chia D, Wong DT. Salivary transcriptomic biomarkers for detection of resectable pancreatic cancer. Gastroenterology. 2010;138:949–957. doi: 10.1053/j.gastro.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haug U, Wente MN, Seiler CM, Jesenofsky R, Brenner H. Stool testing for the early detection of pancreatic cancer: rationale and current evidence. Expert Rev Mol Diagn. 2008;8:753–759. doi: 10.1586/14737159.8.6.753. [DOI] [PubMed] [Google Scholar]
- 8.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 9.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hui AB, Shi W, Boutros PC, Miller N, Pintilie M, Fyles T, McCready D, Wong D, Gerster K, Waldron L, Jurisica I, Penn LZ, Liu FF. Robust global micro-RNA profiling with formalin-fixed paraffin-embedded breast cancer tissues. Lab Invest. 2009;89:597–606. doi: 10.1038/labinvest.2009.12. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Chen J, Chang P, LeBlanc A, Li D, Abbruzzesse JL, Frazier ML, Killary AM, Sen S. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Phila) 2009;2:807–813. doi: 10.1158/1940-6207.CAPR-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong X, Du Y, Wang G, Gao J, Gong Y, Li L, Zhang Z, Zhu J, Jing Q, Qin Y, Li Z. Detection of differentially expressed microRNAs in serum of pancreatic ductal adenocarcinoma patients: miR-196a could be a potential marker for poor prognosis. Dig Dis Sci. 2011;56:602–609. doi: 10.1007/s10620-010-1285-3. [DOI] [PubMed] [Google Scholar]
- 14.Sadakari Y, Ohtsuka T, Ohuchida K, Tsutsumi K, Takahata S, Nakamura M, Mizumoto K, Tanaka M. MicroRNA expression analyses in preoperative pancreatic juice samples of pancreatic ductal adenocarcinoma. JOP. 2010;11:587–592. [PubMed] [Google Scholar]
- 15.Link A, Balaguer F, Shen Y, Nagasaka T, Lozano JJ, Boland CR, Goel A. Fecal MicroRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiol Biomarkers Prev. 2010;19:1766–1774. doi: 10.1158/1055-9965.EPI-10-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Link A, Becker V, Goel A, Wex T, Malfertheiner P. Feasibility of fecal microRNAs as novel biomarkers for pancreatic cancer. PLoS One. 2012;7:e42933. doi: 10.1371/journal.pone.0042933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 18.Szafranska AE, Davison TS, John J, Cannon T, Sipos B, Maghnouj A, Labourier E, Hahn SA. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442–4452. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 19.Habbe N, Koorstra JB, Mendell JT, Offerhaus GJ, Ryu JK, Feldmann G, Mullendore ME, Goggins MG, Hong SM, Maitra A. MicroRNA miR-155 is a biomarker of early pancreatic neoplasia. Cancer Biol Ther. 2009;8:340–346. doi: 10.4161/cbt.8.4.7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu CW, Ng SS, Dong YJ, Ng SC, Leung WW, Lee CW, Wong YN, Chan FK, Yu J, Sung JJ. Detection of miR-92a and miR-21 in stool samples as potential screening biomarkers for colorectal cancer and polyps. Gut. 2012;61:739–745. doi: 10.1136/gut.2011.239236. [DOI] [PubMed] [Google Scholar]
- 21.Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605–1617. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 22.Ballehaninna UK, Chamberlain RS. The clinical utility of serum CA 19-9 in the diagnosis, prognosis and management of pancreatic adenocarcinoma: An evidence based appraisal. J Gastrointest Oncol. 2012;3:105–119. doi: 10.3978/j.issn.2078-6891.2011.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin SH, Kim SC, Hong SM, Kim YH, Song KB, Park KM, Lee YJ. Genetic alterations of K-ras, p53, c-erbB-2, and DPC4 in pancreatic ductal adenocarcinoma and their correlation with patient survival. Pancreas. 2013;42:216–222. doi: 10.1097/MPA.0b013e31825b6ab0. [DOI] [PubMed] [Google Scholar]
- 24.Teich N, Mossner J. Molecular analysis of pancreatic juice: a helpful tool to differentiate benign and malignant pancreatic tumors? Dig Dis. 2004;22:235–238. doi: 10.1159/000082794. [DOI] [PubMed] [Google Scholar]
- 25.Nagasaka T, Tanaka N, Cullings HM, Sun DS, Sasamoto H, Uchida T, Koi M, Nishida N, Naomoto Y, Boland CR, Matsubara N, Goel A. Analysis of fecal DNA methylation to detect gastrointestinal neoplasia. J Natl Cancer Inst. 2009;101:1244–1258. doi: 10.1093/jnci/djp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Li M, Wang H, Fisher WE, Lin PH, Yao Q, Chen C. Profiling of 95 microRNAs in pancreatic cancer cell lines and surgical specimens by real-time PCR analysis. World J Surg. 2009;33:698–709. doi: 10.1007/s00268-008-9833-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koga Y, Yasunaga M, Moriya Y, Akasu T, Fujita S, Yamamoto S, Matsumura Y. Exosome can prevent RNase from degrading microRNA in feces. J Gastrointest Oncol. 2011;2:215–222. doi: 10.3978/j.issn.2078-6891.2011.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren Y, Gao J, Liu JQ, Wang XW, Gu JJ, Huang HJ, Gong YF, Li ZS. Differential signature of fecal microRNAs in patients with pancreatic cancer. Mol Med Rep. 2012;6:201–209. doi: 10.3892/mmr.2012.862. [DOI] [PubMed] [Google Scholar]
- 29.Gusachenko ON, Zenkova MA, Vlassov VV. Nucleic acids in exosomes: disease markers and intercellular communication molecules. Biochemistry (Mosc) 2013;78:1–7. doi: 10.1134/S000629791301001X. [DOI] [PubMed] [Google Scholar]
- 30.Zoller M. Pancreatic cancer diagnosis by free and exosomal miRNA. World J Gastrointest Pathophysiol. 2013;4:74–90. doi: 10.4291/wjgp.v4.i4.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalimutho M, Del VBG, Di Cecilia S, Sileri P, Cretella M, Pallone F, Federici G, Bernardini S. Differential expression of miR-144* as a novel fecal-based diagnostic marker for colorectal cancer. J Gastroenterol. 2011;46:1391–1402. doi: 10.1007/s00535-011-0456-0. [DOI] [PubMed] [Google Scholar]
- 32.Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, Calin GA, Volinia S, Liu CG, Scarpa A, Croce CM. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J. Clin. Oncol. 2006;24:4677–4684. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- 33.Bedi MM, Gandhi MD, Jacob G, Lekha V, Venugopal A, Ramesh H. CA 19-9 to differentiate benign and malignant masses in chronic pancreatitis: is there any benefit? Indian J Gastroenterol. 2009;28:24–27. doi: 10.1007/s12664-009-0005-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


