Abstract
While androgen and androgen receptor (AR) activity have been strongly implicated in prostate cancer development and therapy, the influence of the CAG repeat, which is found within the first exon of the AR gene, on prostate carcinogenesis is still unclear. We investigated the differences in the length of the CAG repeat between prostate cancer patients and controls in the Chinese population as well as between TMPRSS2:ERG fusion positive and negative samples. A general association between prostate cancer and either longer or shorter AR CAG repeat length was not observed in the Chinese population. However, our data suggest that certain CAG repeat lengths may increase or decrease prostate cancer risk. Shorter CAG repeat length was also not shown to be associated with a higher induction rate of TMPRSS2 and ERG proximity, an essential step for TMPRSS2:ERG fusion formation. However, samples with a CAG repeat of 17 were found more frequently in the TMPRSS2:ERG fusion positive than negative prostate cancer cases and mediated a higher rate of androgen-induced TMPRSS2 and ERG co-localisation than AR with longer (24) and shorter (15) CAG repeats. This suggests that 17 CAG repeats may be associated with TMPRSS2:ERG fusion positive prostate cancer, but may have a preventive role for prostate cancer in the Chinese population, which has a low TMPRSS2:ERG fusion frequency. This study suggests that different mechanisms for the association of CAG repeat length polymorphism and prostate cancer exist in different ethnic populations.
Keywords: CAG repeat, length polymorphism, androgen receptor gene, prostate cancer
Introduction
Epidemiological studies demonstrate wide variations in the clinical incidence and mortality rates of prostate cancer in different countries. While prostate cancer is the most common male malignancy in Northern Europe, the USA and Canada, low rates are seen in the Far East [1,2]. The incidence and mortality of prostate cancer in Western countries is ten times more than that in China [1,3]. In the USA, there is a significantly higher mortality rate among African Americans than in Caucasians [1,2]. It is generally accepted that genetic, environmental and dietary factors contribute to these population differences in cancer incidence and mortality, but many of the individual factors are yet to be identified or confirmed [4].
It is well-established that androgen and androgen receptor (AR) play important roles in prostate cancer development and progression [1]. Androgen stimulates prostate cell growth through AR. In exon 1 of the AR gene there is a CAG repeat encoding a poly-glutamine track in the N-terminal of AR protein [5,6]. Polymorphism of the CAG repeat length exists and in vitro analysis has demonstrated that AR with a shorter poly-glutamine track (coded by CAG repeats) has greater AR transcription activity compared with longer poly-glutamine tracks [5,6]. It has been consistently observed in previous studies that there are differences in the CAG repeat length in different populations, and that this is inversely correlated with prostate cancer incidence and mortality rates in those populations [7]. The population reported as having the longest average CAG repeat length is the Hispanic population (23-25). The Chinese population has a longer average CAG repeat (average between 22-23) than that of the Caucasian population (average between 21-22), and the black population has the shortest average CAG repeats (average between 19-20) [8-19]. These observations suggest that shorter CAG repeat lengths, which are known to be associated with higher AR activity, may be associated with higher prostate cancer risk. However, separate studies using cases and controls from the same population showed contradictory results [20,21]. While several studies have reported that shorter CAG repeats are associated with higher prostate cancer risk [7,17,22-31], many other studies have failed to observe the same association [7,12,15,18,32-40]. In a few studies, mainly in the East Asian population, the average CAG repeat length in cancer samples was found to be longer than in normal controls [10,26,41], indicating that AR CAG repeat length may affect prostate carcinogenesis through molecular pathways that are different in the East Asian and Western populations.
In our recent prostate cancer genomic study, in which we compared Western and Chinese cancer samples, we showed that the Chinese cancer samples lack certain somatic genomic changes that are commonly found in Western prostate cancers, including the 21q22 deletion, which is associated with the TMPRSS2:ERG fusion gene, and the 10q23 deletion, which results in PTEN inactivation [42]. Further investigation into the mechanisms underlying these genomic differences revealed that high dose androgen treatment of prostate epithelial cells can induce TMPRSS2 and ERG gene proximity and the TMPRSS2:ERG fusion [43]. This observation has been supported by several subsequent studies from other research groups [44-46]. These studies suggest that androgen and AR activity may increase prostate cancer risk through the induction of the TMPRSS2:ERG fusion and other genomic alterations, and that differences in CAG repeat length might have a different impact on TMPRSS2:ERG fusion induction and consequently prostate cancer risk in Chinese and Western populations. We therefore investigated the CAG repeat length difference between prostate cancer patient and controls in the Chinese population and between TMPRSS2:ERG fusion positive and negative samples. The influence of AR with certain CAG repeat lengths in the induction of TMPRSS2 and ERG proximity was also investigated in vitro. We found that prostate cancer risk in the Chinese population and TMPRSS2:ERG fusion induction were not generally associated with shorter or longer AR CAG repeat length. However, specific CAG repeat lengths may be associated with prostate cancer risk and this risk may attribute to different mechanisms, including the induction of the TMPRSS2:ERG gene fusion.
Materials and methods
Subjects
A population based case control cohort (186 cases and 163 control) was used for the Chinese study. Samples were collected from the Urological Departments in Shanghai and Chongqing hospitals from patients treated with pathologically diagnosed prostate cancer. Controls samples were collected from age-matched (within±5 years) non-cancer male patients attending those hospitals. Whole blood samples were collected with patient consent and ethical approval from the ethical committees of Second Military Medical University and Chongqing Medical University. In addition, 38 fresh-frozen prostate surgical remaining samples were collected from cancer patients with ethical approval from the ethical committee of Second Military Medical University. All patients and controls are native Chinese residences. UK prostate cancer patient samples were obtained with patient consents from Barts and The London Hospital and Whipps Cross Hospital, either as prostate surgical remaining (49 fresh-frozen and 92 paraffin-embedded samples) or whole blood samples (n=42). The study was approved by the East London & the City Research Ethics Committee. TMPRSS2:ERG fusion status has been previously obtained from these fresh-frozen samples by single nucleotide polymorphism (SNP) array or reverse transcription PCR (RT-PCR) analyses [42,43,47,48] and for the paraffin-embedded tissue samples by fluorescent in situ hybridization (FISH) analysis [42].
DNA extraction
For blood samples, white blood cells were isolated using Ficoll and then using a DNA extraction kit (Tiangen Biotechnology, Beijing, China) for Chinese samples and phenol/chloroform method for UK samples) to extract whole genomic DNA. For tissue samples, tissue was cut into 10 μM sections. Fresh-frozen sections were directly digested with proteinase K and DNA was extracted using the standard phenol/chloroform method. Paraffin-embedded tissue sections were de-waxed using xylene and then digested with proteinase K over two nights, adding fresh proteinase K every 24 hours. Following digestion, DNA was extracted using the standard phenol/chloroform method.
Microsatellite analysis of AR CAG repeat length
AR CAG repeat length was determined by microsatellite analysis. The AR CAG repeat region was amplified by PCR from 50 ng template DNA using primers flanking the AR CAG repeat in exon 1 (AR_FAM_F: 5’FAM-ACCCAGAGGCCGCGAGCGCAG and AR_R: 5’-TTGCTGTTCCTCATCCAGGA). The HotStarTaq DNA polymerase kit (includes 10x PCR buffer, Q solution and HotStarTaq DNA polymerase) was used and run for 40 cycles with an annealing temperature of 58°C. PCR product was diluted as appropriate (ranging from 1:200 to 1:2000) and denatured before running on 3730 xl ABI sequencer against ROX-400/LIZ 600 (Genescan 400 HD or 600 LIZ size standards, Life Technologies, Paisley, UK). Samples were analyzed by Genemapper v 4.0 (Life Technologies).
DNA sequencing analysis
To confirm the CAG repeat length detected by microsatellite analysis, direct sequencing of PCR product from selected samples was performed using the 3730 xl ABI sequencer and primers AR_F (5’-ACCCAGAGGCCGCGAGCGCAG) and AR_R (5’-TTGCTGTTCCTCATCCAGGA).
Construction of AR plasmid and cell transfection
A three kb full length AR cDNA was PCR amplified from a commercial plasmid pRR-AR-5Z (Addgene, Cambridge, MA), using forward primer ACGGATGCTAGCATGGAAGTTCAATTGGGTTTG and reverse primer TTGACTTCTAGATCACTGGGTGTGGAAATAGATG, where NheI restriction enzyme cutting point was included. This AR cDNA was then ligated with a T vector, pCR2.1-TOPO (Life Technologies), and amplified in E Coli bacteria. Finally, the AR cDNA was subcloned as an NheI/EcoRV fragment into the pcDNA3.1+ plasmid (Life Technologies). AR DNA fragments containing the AR CAG repeat sequence (about 500 bp) were PCR amplified from patient samples known to have different CAG repeat lengths. The CAG repeat fragments were cloned into the AR-pcDNA3.1+ construct at NheI/AflII sites using a similar approach as described above. After confirming by sequencing analysis, the AR-pcDNA3.1+ constructs (with different CAG repeats) were transfected into AR negative DU145 prostate cancer cells using lipofectamine 2000 (Life Technologies) according to the manufacturer’s instruction. Stable transfected cell lines were generated by selection with 50 μg/ml G418 (Life Technologies).
Real-time quantitative RT-PCR
RNA, extracted using Trizol, was reverse transcribed using the M-MLV Reverse Transcriptase, RNase H Minus, Point Mutant (Promega) according to the manufacturer’s instruction. Relative mRNA levels were determined using predesigned TaqMan gene expression assays targeting AR (Hs00907244_m1) and GAPDH (Hs99999905_m1) (Life Technologies) and quantified by quantitative RT-PCR analysis using the ABI Prism 7700 Sequence detector (Life Technologies) and the standard real time PCR programme.
FISH analysis
DU145 cells cultured in medium with charcoal striped serum were treated with or without 10 nM dihydrotestosterone (DHT) and then harvested for FISH analysis as previously described [43]. TMPRSS2 and ERG co-localization analysis was performed using standard FISH protocol with two bacterial artificial chromosomes (BACs) RP11-535-H11 (TMPRSS2) and RP11-476D17 (ERG) as probes. RP11-535H11 was labeled with Fluorescein-12-dUTP (Green) and RP11-476D17 was labeled with Tetramethyl-rhodamine-5-dUTP (red) using the BioPrime labeling kit (Invitrogen). A minimum of 300 nuclei were counted per sample. Induced proximity was quantified and represented as the percentage of co-localized signal pairs.
Statistical analysis
Continuous data was analyzed using t test and category data was analyzed using Chi-squared test. All statistics was done in two-tails. P values of less than 0.05 were considered statistically significant.
Results
Firstly, we analyzed the average length of CAG repeats in the 224 cancer and 163 control samples from Chinese individuals to see if there is an association between overall shorter or longer CAG repeat length and Chinese prostate cancer, an approach which has been generally used in previous AR CAG repeat studies. We found that, in the Chinese population, CAG repeat length is not significantly different between cancer patients and normal individuals with average CAG repeat length of 22.48 and 22.62 respectively (Table 1). We also used more or less than 18, 20 and 22 repeats as cut off to see if there are any differences in the cancer and normal control groups, but none of these are significant (P=0.361, 0.685, and 0.9 respectively). A close examination of the frequency for each length of CAG repeat found that CAG repeats of 20, 24, and 27 are found more frequently in the cancer patients with borderline significance (P=0.077, 0.094 and 0.056 respectively, comparing the groups with and without each of those repeat length) and 17 repeats is observed significantly less frequently compared to the controls (P=0.037) (Figure 1).
Table 1.
AR CAG repeat length distribution in Chinese prostate cancer cases, controls and UK cancer cases
| CAG repeat size | China Tumour (n=224) | China Normal (n=163) | UK Tumour (n=183) |
|---|---|---|---|
| 11 | 1 | 1 | 0 |
| 12 | 1 | 0 | 0 |
| 13 | 0 | 0 | 1 |
| 14 | 0 | 0 | 1 |
| 15 | 5 | 2 | 1 |
| 16 | 1 | 2 | 2 |
| 17 | 3 | 8 | 11 |
| 18 | 10 | 7 | 23 |
| 19 | 11 | 8 | 19 |
| 20 | 20 | 7 | 21 |
| 21 | 30 | 19 | 25 |
| 22 | 40 | 33 | 14 |
| 23 | 19 | 21 | 17 |
| 24 | 29 | 13 | 19 |
| 25 | 18 | 19 | 9 |
| 26 | 11 | 11 | 15 |
| 27 | 15 | 4 | 2 |
| 28 | 7 | 3 | 1 |
| 29 | 1 | 2 | 2 |
| 30 | 0 | 2 | 0 |
| 31 | 0 | 1 | 0 |
| 32 | 0 | 0 | 0 |
| 33 | 0 | 0 | 0 |
| 34 | 1 | 0 | 0 |
| 35 | 1 | 0 | 0 |
| Average size | 22.48 | 22.62 | 21.31 |
Figure 1.
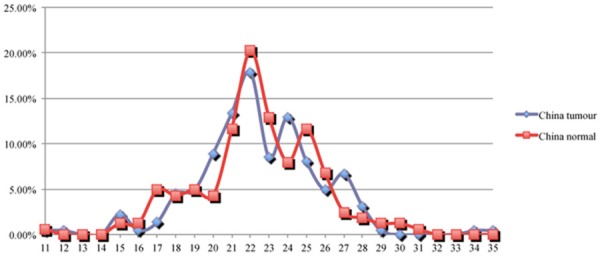
The distribution (in percentage, Y axis) of Chinese prostate cancer cases and controls at each AR CAG repeat length (X axis).
We then compared the CAG repeat length in the UK cancer patients with Chinese cancer patients. The average length of UK cancer patients is 21.31, which is much shorter than those detected in the Chinese cancer patients. Both t test analysis, using all the CAG repeat length distribution data (Table 1), and Chi squared analysis comparing these shorter than 21 to equal or longer than 21 repeats showed significantly shorter repeats in the UK cancer group than those detected in the Chinese cancer patients (P=0.0001 and < 0.0001 respectively). Taking these data together suggests that the number of CAG repeats in the AR gene may not generally affect prostate cancer risk and the difference in CAG repeat length in cancer patients from UK and Chinese samples may just reflect the difference of the baseline CAG repeat lengths in these two populations.
We further investigated AR CAG repeat length in TMPRSS2:ERG fusion positive and negative cases to determine if CAG repeat length is associated with fusion gene status. TMPRSS2:ERG fusion status has been previously determined in the fresh frozen samples by SNP array or RT-PCR analyses and in the paraffin-embedded samples by FISH [42,43,47,48]. In total, there were 55 fusion positive and 79 fusion negative UK cases. The CAG repeat length distribution is presented in Table 2 and Figure 2. The average CAG repeat length in the fusion negative group (21.09) is similar to the fusion positive group (21.07). However, if 23 CAG repeats was used as a cut off to categorize CAG repeat length into two groups (< 23 vs ≥ 23 repeats), a modest association between longer CAG repeat lengths and fusion positive cancer cases (P=0.319) was observed. TMPRSS2:ERG fusion status was available in 38 Chinese fresh frozen samples; however, as TMPRSS2:ERG is rare in the Chinese population, only three samples were fusion positive [42]. These three TMPRSS2:ERG fusion positive Chinese cases had shorter CAG repeat lengths (19, 21 and 22) when compared to the average CAG repeat length of either the 35 fusion negative cases (22.46) and all the 224 Chinese cancer cases (22.48) (Tables 1 and 3). When individual CAG repeat length was assessed in UK cases where the TMPRSS2:ERG fusion status is known, we observed a significantly higher frequency of cases detected with 24 CAG repeats in the fusion negative cases as compared to the fusion positive group (P=0.079 by comparing the cases with and without 24 repeats) and a trend that fusion positive cases had higher frequency of CAG repeat length of 17 and 22 than the fusion negative cases (P=0.205, for both) ( Figure 2).
Table 2.
AR CAG repeat length distribution in UK TMPRSS2:ERG fusion positive and negative prostate cancer cases
| CAG repeat size | UK fusion positive (n=55) | UK fusion negative (n=79) |
|---|---|---|
| 13 | 1 | 0 |
| 14 | 0 | 1 |
| 15 | 0 | 0 |
| 16 | 0 | 2 |
| 17 | 6 | 4 |
| 18 | 5 | 10 |
| 19 | 5 | 8 |
| 20 | 7 | 12 |
| 21 | 10 | 10 |
| 22 | 6 | 4 |
| 23 | 4 | 7 |
| 24 | 3 | 12 |
| 25 | 1 | 3 |
| 26 | 5 | 5 |
| 27 | 1 | 1 |
| 28 | 0 | 0 |
| 29 | 1 | 0 |
| Average size | 21.07 | 21.09 |
Figure 2.
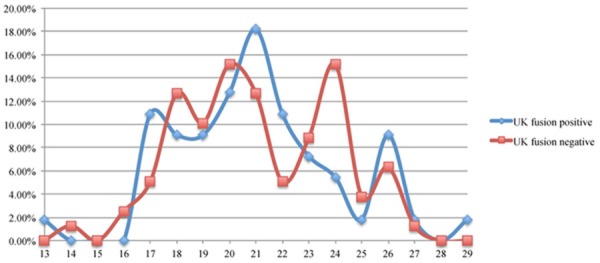
The distribution (in percentage, Y axis) of TMPRSS2:ERG fusion positive and negative cases at each AR CAG repeat length (X axis).
Table 3.
AR CAG repeat length distribution in Chinese TMPRSS2:ERG fusion positive and negative prostate cancer cases
| CAG repeat size | China fusion positive (n=3) | China fusion negative (n=35) |
|---|---|---|
| 11 | 0 | 1 |
| 12 | 0 | 0 |
| 13 | 0 | 0 |
| 14 | 0 | 0 |
| 15 | 0 | 0 |
| 16 | 0 | 1 |
| 17 | 0 | 0 |
| 18 | 0 | 2 |
| 19 | 1 | 1 |
| 20 | 0 | 4 |
| 21 | 1 | 5 |
| 22 | 1 | 4 |
| 23 | 0 | 2 |
| 24 | 0 | 5 |
| 25 | 0 | 2 |
| 26 | 0 | 3 |
| 27 | 0 | 3 |
| 28 | 0 | 2 |
| Average size | 20.67 | 22.46 |
To further investigate the relationship between AR CAG repeat length and TMPRSS2:ERG fusion gene, AR-negative prostate cancer cell line DU145 was transfected with constructs expressing AR with 15, 17 and 24 CAG repeats. Following transfection a significant increase (668-944 fold) in AR expression level was detected (Figure 3). Surprisingly, lower AR expression was detected in those cells transfected with constructs containing 17 CAG than 15 and 24 CAG repeats (Figure 3). However, this may not reflect the endogenous AR expression with those CAG repeat lengths. We further analyzed TMPRSS2 and ERG co-localization by FISH in untransfected DU145 cells and DU145 cells transfected with AR containing 15, 17 and 24 CAG repeats and found that the percentage of co-localized TMPRSS2 and ERG signals was increased in DU145 cells transfected with AR of all the three CAG repeat length compared to DU145 cells without AR transfection (Figure 4). Interestingly, the rate of TMPRSS2 and ERG co-localization in DU145 cells transfected with 17 CAG repeats is significant higher than in those cells transfected with 15 (P < 0.001) and 24 (P < 0.001) CAG repeats (Figure 4).
Figure 3.
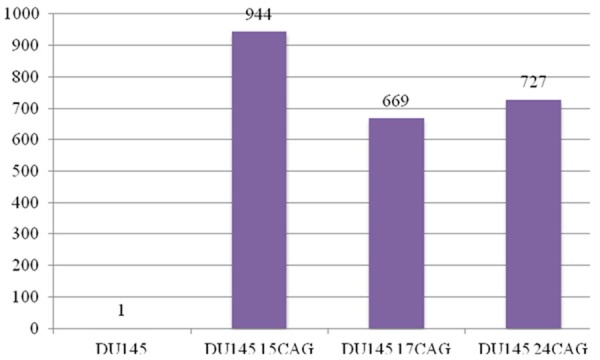
AR expression level detected in DU145 cells by Q-RT-PCR with and without the transfection of AR with 15 (15 CAG), 17 (17 CAG) and 24 (24 CAG) CAG repeats.
Figure 4.
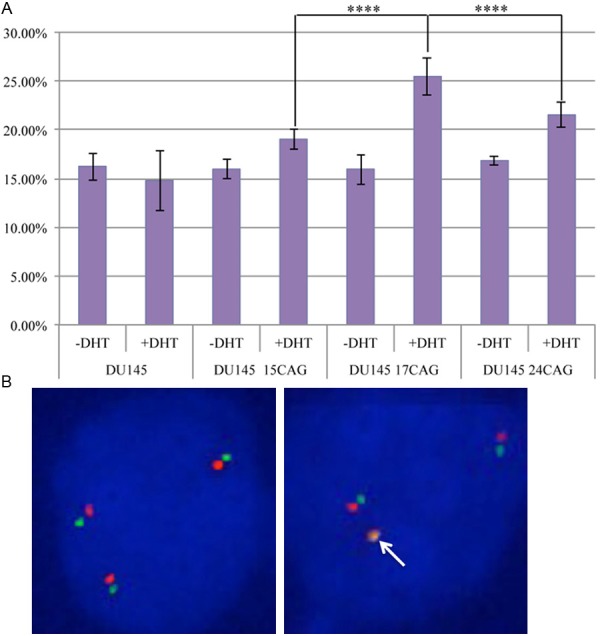
The influence of AR CAG repeat length in androgen-induced TMPRSS2 and ERG gene co-localization. A. TMPRSS2 (green) and ERG (red) co-localization rates in DU145 cells with and without the transfection of AR with 15 (15 CAG), 17 (17 CAG) and 24 (24 CAG) CAG repeats were assessed by FISH with and without DHT treatment. -DHT: cells cultured without androgen using charcoal striped serum; +DHT: cells cultured with charcoal striped serum and additional DHT at 100 nM; ****: P < 0.0001; B. Representative FISH images to show cells without TMPRSS2 and ERG co-localization (17 CAG repeats AR transfected cells without DHT treatment) and with a pair of TMPRSS2 and ERG co-localized (17 CAG repeats AR transfected cells with DHT treatment).
Discussion
While differences in AR CAG repeat length in different ethnic populations is well established, the association between the CAG repeat length polymorphism and prostate cancer risk is still debatable. Although extensive studies have attempted to address the association between AR CAG repeat length and prostate cancer, the results are conflicting [7,10,12,15,17,18,22-39,41]. A meta-analysis showed that overall, cancer patients have slightly shorter CAG repeats than controls, but this difference was less than one repeat [21]. It has been argued whether this CAG repeat difference can contribute a significant difference to prostate cancer risk. The number of case control studies that have investigated the association between AR CAG repeat length and prostate cancer risk in the Chinese population is limited and each of these studies has investigated a small number of cases [10,17,41]. Both shorter and longer AR CAG repeat length has been reported in cancer cases compared to controls. In this study we have shown, used a considerably larger cohort of Chinese patients and controls, that in the Chinese population prostate cancer risk is not associated with overall shorter or longer AR CAG repeat length. This suggests that the previously conflicting results may be caused by random sampling. If shorter CAG repeats, which are also associated with greater AR activity, are associated with a higher prostate cancer risk in the Chinese population, we may also expect a subtle difference of CAG repeat lengths between the Chinese and UK cancer patients. However, the difference in these two groups was significant, similar to the population difference, which further supports the lack of a general association between the shorter CAG repeat length and prostate cancer risk.
We have previously reported specific genetic differences between Western and Chinese prostate cancer cells, including different frequencies of TMPRSS2:ERG fusion and BRAF copy number changes [42,49]. These differences suggest that different mechanisms/pathways and etiological factors may be associated with different types of prostate cancers. The different frequency of TMPRSS2:ERG fusion in East Asian and Western populations may be associated with the relatively low androgen level and longer AR CAG repeats found in East Asian men compared with Western men. We and other researchers recently have shown that a high-dose of androgen can induce TMPRSS2 and ERG gene proximity, which facilitates TMPRSS2:ERG fusion [43-46]. As shorter CAG repeats are associated with higher AR transactivity [5,6], shorter CAG repeats may increase TMPRSS2:ERG fusion rate and consequently prostate carcinogenesis. We previously observed a trend that TMPRSS2:ERG fusion positive prostate cancer cases had an overall shorter AR CAG repeat length than fusion negative prostate cancer cases, but this difference was not statistically significant [43]. However, by including additional samples, we found that the trend for shorter AR CAG repeat lengths in TMPRSS2:ERG fusion positive UK prostate cancer cases disappeared. In a recent U.S. study that analyzed a greater number of prostate cancer samples with known TMPRSS2:ERG fusion status, a borderline association (P=0.06) was detected between shorter CAG repeats and the TMPRSS2:ERG fusion positive cases. More interestingly, TMPRSS2:ERG fusion positive cases have a significantly overall shorter AR CAG repeat length than the non-cancer controls, which may be explained by the increased statistical power gained from the large number of controls [40]. Although the TMPRSS2:ERG fusion positive cases in our Chinese samples have a shorter average AR CAG repeat length than the fusion negative cases as well as non-cancer control cases, the limited number of fusion positive cases makes it difficult to establish a reliable association. While studies using much larger cohort of samples are required, these current data suggest that overall shorter AR CAG repeat length may have a limited impact on TMPRSS2:ERG positive prostate cancer.
Although no clear association was observed between shorter CAG repeats with prostate cancer or TMPRSS2:ERG fusion positive cases, our data suggests that TMPRSS2:ERG fusion positive and negative prostate cancer may be differentially associated with certain CAG repeat lengths in AR. AR with 17 repeats was potentially associated with fusion positive cases, while AR with 24 repeats may be associated with fusion negative cases. In the Chinese population, where TMPRSS2:ERG fusion positive cases are far less frequent than the Western population, men with 20, 24 and 27 repeats AR may have higher risk of prostate cancer. The hypothesis of the association between a shorter CAG repeat length and prostate cancer risk was prompted by observations that populations with lower incidence of prostate cancer have longer CAG repeats and that with shorter CAG repeats are associated with a more active AR. However, functional studies of AR with different CAG repeats also showed that, although generally AR with more CAG repeats are less functionally active than these with less CAG repeats, the correlation of AR CAG repeat length either to AR expression level or AR activity is not linear [5,6]. Actually, AR expression level varies for those with CAG repeats range from 15 to 24 and many of those with shorter CAG repeats have less activity and/or lower expression than some with longer repeats [5] and the overall activity difference between the shorter ones and longer ones within this range is not dramatic [5,6]. Therefore, the association between AR CAG repeat length and prostate cancer risk may be specific for certain individual CAG repeat length rather than a generally shorter repeat length, which is supported by our data. As the number of samples in our study is limited, most of the associations of individual CAG repeat lengths with prostate cancer were borderline significant and should be validated in further studies using larger sample cohorts. Nevertheless our cell transfection study using AR with different CAG repeats demonstrated that AR with 17 CAG repeats may increases the risk of prostate cancer by more efficiently mediating androgen-induced TMPRSS2 and ERG proximity than AR with shorter (15) or longer (24) CAG repeats.
It is unclear why 17 CAG repeats is associated with a potential preventive role, but 20, 24 and 27 CAG repeats are more frequently associated with Chinese prostate cancer. It is also not clear why 24 CAG repeats is associated with TMPRSS2:ERG fusion negative prostate cancer. These observations warrant further investigation. The association between a repeat length of 24 with fusion negative cases is supported by both the comparison between fusion positive and negative cancer cases and our case control study of the Chinese prostate cancers, which are mainly comprised of fusion negative cases. While the number of cases in this study is small, particularly for the cases with TMPRSS2:ERG fusion status, the observation of the association between 24 CAG repeats with fusion negative prostate cancer in these two separate comparisons indicates that this repeat length is very likely to play a role in increasing the risk of prostate cancer without TMPRSS2:ERG fusion; however, the mechanism behind this is not yet clear.
Although morphologically the majority of prostate cancer falls only into one histopathological type, it is well recognized that prostate cancer is very heterogeneous, both in terms of clinical behavior and genetic alterations [50]. Our data suggest that certain AR CAG repeat lengths may be associated with TMPRSS2:ERG fusion positive and some associated with fusion negative cancers. If only certain subgroups of prostate cancer are associated with AR CAG repeat length polymorphism and each of such cancer sub-groups is associated with different AR CAG repeat lengths, analysis of a mixed population of all subgroups may not be able to detect any of the associations for those particular subgroups. Therefore, studies of the association of AR CAG repeat length with subgroups of prostate cancer in large cohorts of samples by consortiums are required to illustrate the real contribution of AR CAG repeat length to prostate carcinogenesis.
In summary, by comparing the Chinese patients and controls as well as the UK and Chinese cancer cases with the consideration of TMPRSS2:ERG fusion status, we have shown that both prostate cancer risk in Chinese population and TMPRSS2:ERG fusion in UK cases are not associated with overall differences in AR CAG repeat length. However, AR with certain CAG repeats may be associated with prostate cancer risk in the Chinese population, as well as the TMPRSS2:ERG fusion even. We speculate that AR with different CAG repeats may increase prostate cancer risk through different mechanisms. Subgroup analysis is strongly recommended for future studies investigating the association of specific CAG repeat lengths with prostate cancer. This study also demonstrates that the differentially presented factors in Western and Asian countries, here AR CAG repeat length, may interact with certain environmental factors and contribute differentially to somatic genomic alterations, such as the TMPRSS2:ERG fusion.
Acknowledgements
This work is supported by the grant from Orchidand National Natural Science foundation of China (Grant No: 30671793). We also thank Dr. N. Coll-Bastus, M Li and Y Yu for technical support and sample collection.
Disclosure of conflict of interest
None to declare.
References
- 1.Gronberg H. Prostate cancer epidemiology. Lancet. 2003;361:859–864. doi: 10.1016/S0140-6736(03)12713-4. [DOI] [PubMed] [Google Scholar]
- 2.Oishi K, Yoshida O, Schroeder FH. The geography of prostate cancer and its treatment in Japan. Cancer Surv. 1995;23:267–280. [PubMed] [Google Scholar]
- 3.Sim HG, Cheng CW. Changing demography of prostate cancer in Asia. Eur J Cancer. 2005;41:834–845. doi: 10.1016/j.ejca.2004.12.033. [DOI] [PubMed] [Google Scholar]
- 4.Williams H, Powell IJ. Epidemiology, pathology, and genetics of prostate cancer among African Americans compared with other ethnicities. Methods Mol Biol. 2009;472:439–453. doi: 10.1007/978-1-60327-492-0_21. [DOI] [PubMed] [Google Scholar]
- 5.Ding D, Xu L, Menon M, Reddy GP, Barrack ER. Effect of a short CAG (glutamine) repeat on human androgen receptor function. Prostate. 2004;58:23–32. doi: 10.1002/pros.10316. [DOI] [PubMed] [Google Scholar]
- 6.Irvine RA, Ma H, Yu MC, Ross RK, Stallcup MR, Coetzee GA. Inhibition of p160-mediated coactivation with increasing androgen receptor polyglutamine length. Hum Mol Genet. 2000;9:267–274. doi: 10.1093/hmg/9.2.267. [DOI] [PubMed] [Google Scholar]
- 7.Li J, Mercer E, Gou X, Lu YJ. Ethnical disparities of prostate cancer predisposition: genetic polymorphisms in androgen-related genes. Am J Cancer Res. 2013;3:127–151. [PMC free article] [PubMed] [Google Scholar]
- 8.Balic I, Graham ST, Troyer DA, Higgins BA, Pollock BH, Johnson-Pais TL, Thompson IM, Leach RJ. Androgen receptor length polymorphism associated with prostate cancer risk in Hispanic men. J Urol. 2002;168:2245–2248. doi: 10.1016/S0022-5347(05)64364-9. [DOI] [PubMed] [Google Scholar]
- 9.Giwercman C, Giwercman A, Pedersen HS, Toft G, Lundin K, Bonde JP, Lundberg Giwercman Y. Polymorphisms in genes regulating androgen activity among prostate cancer low-risk Inuit men and high-risk Scandinavians. Int J Androl. 2008;31:25–30. doi: 10.1111/j.1365-2605.2007.00750.x. [DOI] [PubMed] [Google Scholar]
- 10.Huang SP, Chou YH, Chang WS, Wu MT, Yu CC, Wu T, Lee YH, Huang JK, Wu WJ, Huang CH. Androgen receptor gene polymorphism and prostate cancer in Taiwan. J Formos Med Assoc. 2003;102:680–686. [PubMed] [Google Scholar]
- 11.Irvine RA, Yu MC, Ross RK, Coetzee GA. The CAG and GGC microsatellites of the androgen receptor gene are in linkage disequilibrium in men with prostate cancer. Cancer Res. 1995;55:1937–1940. [PubMed] [Google Scholar]
- 12.Lange EM, Sarma AV, Ray A, Wang Y, Ho LA, Anderson SA, Cunningham JM, Cooney KA. The androgen receptor CAG and GGN repeat polymorphisms and prostate cancer susceptibility in African-American men: results from the Flint Men’s Health Study. J Hum Genet. 2008;53:220–226. doi: 10.1007/s10038-007-0240-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Brien TG, Guo Y, Visvanathan K, Sciulli J, McLaine M, Helzlsouer KJ, Watkins-Bruner D. Differences in ornithine decarboxylase and androgen receptor allele frequencies among ethnic groups. Mol Carcinog. 2004;41:120–123. doi: 10.1002/mc.20047. [DOI] [PubMed] [Google Scholar]
- 14.Platz EA, Rimm EB, Willett WC, Kantoff PW, Giovannucci E. Racial variation in prostate cancer incidence and in hormonal system markers among male health professionals. J Natl Cancer Inst. 2000;92:2009–2017. doi: 10.1093/jnci/92.24.2009. [DOI] [PubMed] [Google Scholar]
- 15.Price DK, Chau CH, Till C, Goodman PJ, Baum CE, Ockers SB, English BC, Minasian L, Parnes HL, Hsing AW, Reichardt JK, Hoque A, Tangen CM, Kristal AR, Thompson IM, Figg WD. Androgen receptor CAG repeat length and association with prostate cancer risk: results from the prostate cancer prevention trial. J Urol. 2010;184:2297–2302. doi: 10.1016/j.juro.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sartor O, Zheng Q, Eastham JA. Androgen receptor gene CAG repeat length varies in a race-specific fashion in men without prostate cancer. Urology. 1999;53:378–380. doi: 10.1016/s0090-4295(98)00481-6. [DOI] [PubMed] [Google Scholar]
- 17.Hsing AW, Gao YT, Wu G, Wang X, Deng J, Chen YL, Sesterhenn IA, Mostofi FK, Benichou J, Chang C. Polymorphic CAG and GGN repeat lengths in the androgen receptor gene and prostate cancer risk: a population-based case-control study in China. Cancer Res. 2000;60:5111–5116. [PubMed] [Google Scholar]
- 18.Lindstrom S, Ma J, Altshuler D, Giovannucci E, Riboli E, Albanes D, Allen NE, Berndt SI, Boeing H, Bueno-de-Mesquita HB, Chanock SJ, Dunning AM, Feigelson HS, Gaziano JM, Haiman CA, Hayes RB, Henderson BE, Hunter DJ, Kaaks R, Kolonel LN, Le Marchand L, Martinez C, Overvad K, Siddiq A, Stampfer M, Stattin P, Stram DO, Thun MJ, Trichopoulos D, Tumino R, Virtamo J, Weinstein SJ, Yeager M, Kraft P, Freedman ML. A large study of androgen receptor germline variants and their relation to sex hormone levels and prostate cancer risk. Results from the National Cancer Institute Breast and Prostate Cancer Cohort Consortium. J Clin Endocrinol Metab. 2010;95:E121–127. doi: 10.1210/jc.2009-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esteban E, Rodon N, Via M, Gonzalez-Perez E, Santamaria J, Dugoujon JM, Chennawi FE, Melhaoui M, Cherkaoui M, Vona G, Harich N, Moral P. Androgen receptor CAG and GGC polymorphisms in Mediterraneans: repeat dynamics and population relationships. J Hum Genet. 2006;51:129–136. doi: 10.1007/s10038-005-0336-7. [DOI] [PubMed] [Google Scholar]
- 20.Gu M, Dong X, Zhang X, Niu W. The CAG repeat polymorphism of androgen receptor gene and prostate cancer: a meta-analysis. Mol Biol Rep. 2012;39:2615–2624. doi: 10.1007/s11033-011-1014-9. [DOI] [PubMed] [Google Scholar]
- 21.Zeegers MP, Kiemeney LA, Nieder AM, Ostrer H. How strong is the association between CAG and GGN repeat length polymorphisms in the androgen receptor gene and prostate cancer risk? Cancer Epidemiol Biomarkers Prev. 2004;13:1765–1771. [PubMed] [Google Scholar]
- 22.Akinloye O, Gromoll J, Simoni M. Variation in CAG and GGN repeat lengths and CAG/GGN haplotype in androgen receptor gene polymorphism and prostate carcinoma in Nigerians. Br J Biomed Sci. 2011;68:138–142. doi: 10.1080/09674845.2011.11730341. [DOI] [PubMed] [Google Scholar]
- 23.Kuasne H, Rodrigues IS, Fuganti PE, Losi-Guembarovski R, Ito K, Kishima MO, Rodrigues MA, Rogatto SR, Santos RM, Colus IM. Polymorphisms in the AR and PSA genes as markers of susceptibility and aggressiveness in prostate cancer. Cancer Invest. 2010;28:917–924. doi: 10.3109/07357907.2010.483509. [DOI] [PubMed] [Google Scholar]
- 24.Mittal RD, Mishra DK, Thangaraj K, Singh R, Mandhani A. Is there an inter-relationship between prostate specific antigen, kallikrein-2 and androgen receptor gene polymorphisms with risk of prostate cancer in north Indian population? Steroids. 2007;72:335–341. doi: 10.1016/j.steroids.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 25.Andersson P, Varenhorst E, Soderkvist P. Androgen receptor and vitamin D receptor gene polymorphisms and prostate cancer risk. Eur J Cancer. 2006;42:2833–2837. doi: 10.1016/j.ejca.2006.06.030. [DOI] [PubMed] [Google Scholar]
- 26.Li C, Gronberg H, Matsuyama H, Weber G, Nordenskjold M, Naito K, Bergh A, Bergerheim U, Damber JE, Larsson C, Ekman P. Difference between Swedish and Japanese men in the association between AR CAG repeats and prostate cancer suggesting a susceptibility-modifying locus overlapping the androgen receptor gene. Int J Mol Med. 2003;11:529–533. [PubMed] [Google Scholar]
- 27.Mononen N, Ikonen T, Autio V, Rokman A, Matikainen MP, Tammela TL, Kallioniemi OP, Koivisto PA, Schleutker J. Androgen receptor CAG polymorphism and prostate cancer risk. Hum Genet. 2002;111:166–171. doi: 10.1007/s00439-002-0776-5. [DOI] [PubMed] [Google Scholar]
- 28.Stanford JL, Just JJ, Gibbs M, Wicklund KG, Neal CL, Blumenstein BA, Ostrander EA. Polymorphic repeats in the androgen receptor gene: molecular markers of prostate cancer risk. Cancer Res. 1997;57:1194–1198. [PubMed] [Google Scholar]
- 29.Giovannucci E, Stampfer MJ, Krithivas K, Brown M, Dahl D, Brufsky A, Talcott J, Hennekens CH, Kantoff PW. The CAG repeat within the androgen receptor gene and its relationship to prostate cancer. Proc Natl Acad Sci U S A. 1997;94:3320–3323. doi: 10.1073/pnas.94.7.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buchanan G, Irvine RA, Coetzee GA, Tilley WD. Contribution of the androgen receptor to prostate cancer predisposition and progression. Cancer Metastasis Rev. 2001;20:207–223. doi: 10.1023/a:1015531326689. [DOI] [PubMed] [Google Scholar]
- 31.Nelson KA, Witte JS. Androgen receptor CAG repeats and prostate cancer. Am J Epidemiol. 2002;155:883–890. doi: 10.1093/aje/155.10.883. [DOI] [PubMed] [Google Scholar]
- 32.Salinas CA, Austin MA, Ostrander EO, Stanford JL. Polymorphisms in the androgen receptor and the prostate-specific antigen genes and prostate cancer risk. Prostate. 2005;65:58–65. doi: 10.1002/pros.20230. [DOI] [PubMed] [Google Scholar]
- 33.Freedman ML, Pearce CL, Penney KL, Hirschhorn JN, Kolonel LN, Henderson BE, Altshuler D. Systematic evaluation of genetic variation at the androgen receptor locus and risk of prostate cancer in a multiethnic cohort study. Am J Hum Genet. 2005;76:82–90. doi: 10.1086/427224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santos ML, Sarkis AS, Nishimoto IN, Nagai MA. Androgen receptor CAG repeat polymorphism in prostate cancer from a Brazilian population. Cancer Detect Prev. 2003;27:321–326. doi: 10.1016/s0361-090x(03)00106-5. [DOI] [PubMed] [Google Scholar]
- 35.Gsur A, Preyer M, Haidinger G, Zidek T, Madersbacher S, Schatzl G, Marberger M, Vutuc C, Micksche M. Polymorphic CAG repeats in the androgen receptor gene, prostate-specific antigen polymorphism and prostate cancer risk. Carcinogenesis. 2002;23:1647–1651. doi: 10.1093/carcin/23.10.1647. [DOI] [PubMed] [Google Scholar]
- 36.Chen C, Lamharzi N, Weiss NS, Etzioni R, Dightman DA, Barnett M, DiTommaso D, Goodman G. Androgen receptor polymorphisms and the incidence of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2002;11:1033–1040. [PubMed] [Google Scholar]
- 37.Latil AG, Azzouzi R, Cancel GS, Guillaume EC, Cochan-Priollet B, Berthon PL, Cussenot O. Prostate carcinoma risk and allelic variants of genes involved in androgen biosynthesis and metabolism pathways. Cancer. 2001;92:1130–1137. doi: 10.1002/1097-0142(20010901)92:5<1130::aid-cncr1430>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 38.Beilin J, Harewood L, Frydenberg M, Mameghan H, Martyres RF, Farish SJ, Yue C, Deam DR, Byron KA, Zajac JD. A case-control study of the androgen receptor gene CAG repeat polymorphism in Australian prostate carcinoma subjects. Cancer. 2001;92:941–949. doi: 10.1002/1097-0142(20010815)92:4<941::aid-cncr1404>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 39.Edwards SM, Badzioch MD, Minter R, Hamoudi R, Collins N, Ardern-Jones A, Dowe A, Osborne S, Kelly J, Shearer R, Easton DF, Saunders GF, Dearnaley DP, Eeles RA. Androgen receptor polymorphisms: association with prostate cancer risk, relapse and overall survival. Int J Cancer. 1999;84:458–465. doi: 10.1002/(sici)1097-0215(19991022)84:5<458::aid-ijc2>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 40.Yoo S, Pettersson A, Jordahl KM, Lis RT, Lindstrom S, Meisner A, Nuttall EJ, Stack EC, Stampfer MJ, Kraft P, Brown M, Loda M, Giovannucci EL, Kantoff PW, Mucci LA. Androgen receptor CAG repeat polymorphism and risk of TMPRSS2:ERG positive prostate cancer. Cancer Epidemiol Biomarkers Prev. 2014;23:2027–31. doi: 10.1158/1055-9965.EPI-14-0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Das K, Cheah PY, Lim PL, Zain YB, Stephanie FC, Zhao Y, Cheng C, Lau W. Shorter CAG repeats in androgen receptor and non-GG genotypes in prostate-specific antigen loci are associated with decreased risk of benign prostatic hyperplasia and prostate cancer. Cancer Lett. 2008;268:340–347. doi: 10.1016/j.canlet.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 42.Mao X, Yu Y, Boyd LK, Ren G, Lin D, Chaplin T, Kudahetti SC, Stankiewicz E, Xue L, Beltran L, Gupta M, Oliver RT, Lemoine NR, Berney DM, Young BD, Lu YJ. Distinct genomic alterations in prostate cancers in Chinese and Western populations suggest alternative pathways of prostate carcinogenesis. Cancer Res. 2010;70:5207–5212. doi: 10.1158/0008-5472.CAN-09-4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bastus NC, Boyd LK, Mao X, Stankiewicz E, Kudahetti SC, Oliver RT, Berney DM, Lu YJ. Androgen-induced TMPRSS2:ERG fusion in nonmalignant prostate epithelial cells. Cancer Res. 2010;70:9544–9548. doi: 10.1158/0008-5472.CAN-10-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haffner MC, Aryee MJ, Toubaji A, Esopi DM, Albadine R, Gurel B, Isaacs WB, Bova GS, Liu W, Xu J, Meeker AK, Netto G, De Marzo AM, Nelson WG, Yegnasubramanian S. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat Genet. 2010;42:668–675. doi: 10.1038/ng.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, Zhang J, Rose DW, Fu XD, Glass CK, Rosenfeld MG. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mani RS, Tomlins SA, Callahan K, Ghosh A, Nyati MK, Varambally S, Palanisamy N, Chinnaiyan AM. Induced chromosomal proximity and gene fusions in prostate cancer. Science. 2009;326:1230. doi: 10.1126/science.1178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mao X, Shaw G, James SY, Purkis P, Kudahetti SC, Tsigani T, Kia S, Young BD, Oliver RT, Berney D, Prowse DM, Lu YJ. Detection of TMPRSS2:ERG fusion gene in circulating prostate cancer cells. Asian J Androl. 2008;10:467–473. doi: 10.1111/j.1745-7262.2008.00401.x. [DOI] [PubMed] [Google Scholar]
- 48.Boyd LK, Mao X, Xue L, Lin D, Chaplin T, Kudahetti SC, Stankiewicz E, Yu Y, Beltran L, Shaw G, Hines J, Oliver RT, Berney DM, Young BD, Lu YJ. High-resolution genome-wide copy-number analysis suggests a monoclonal origin of multifocal prostate cancer. Genes Chromosomes Cancer. 2012;51:579–589. doi: 10.1002/gcc.21944. [DOI] [PubMed] [Google Scholar]
- 49.Ren G, Liu X, Mao X, Zhang Y, Stankiewicz E, Hylands L, Song R, Berney DM, Clark J, Cooper C, Lu YJ. Identification of frequent BRAF copy number gain and alterations of RAF genes in Chinese prostate cancer. Genes Chromosomes Cancer. 2012;51:1014–1023. doi: 10.1002/gcc.21984. [DOI] [PubMed] [Google Scholar]
- 50.Boyd LK, Mao X, Lu YJ. The complexity of prostate cancer: genomic alterations and heterogeneity. Nat Rev Urol. 2012;9:652–664. doi: 10.1038/nrurol.2012.185. [DOI] [PubMed] [Google Scholar]


