Abstract
MicroRNAs (miRNAs) are small, non-coding RNAs that can act as oncogenes or tumor suppressor genes in human cancer. Increasing evidences indicate that deregulation of miRNAs contributes to the hepatocarcinogenesis. In this study, we demonstrated that the levels of miR-543 were dramatically increased in clinical hepatocellular carcinoma (HCC) tissues and cell lines. Moreover, forced expression of miR-543 promoted the proliferative and invasive potential of HepG2. We also identified PAQR3 as a direct target gene for miR-543 using a fluorescent reporter assay and western blot. The levels of PAQR3 were dramatically decreased in clinical hepatocellular carcinoma (HCC) tissues and cell lines. The mRNA levels of PAQR3 were inversely correlated with the miR-543 expression level.Thus, our finding provides a new insight into the mechanism of hepatocarcinogenesis, indicating a therapeutic potential of miR-543 in the treatment of HCC.
Keywords: miR-543, PAQR3, hepatocellular carcinoma, oncogene
Introduction
Cancer accounts for the highest mortality in developed countries and the second highest in developing countries, making it a worldwide health problem [1,2]. Hepatocellular carcinoma (HCC) is the fifth most prevalent malignancy and the third leading cause of death from cancer worldwide [3,4]. Nearly 600,000 people die of HCC each year [5]. Despite the clinical implementation of numerous therapeutic strategies, the 5-year survival rate of HCC patients is only approximately 5% [6]. The bad outcome is mainly due to late presentation at advanced stages, frequent tumour metastasis and tumour recurrence after surgical intervention [7]. Metastasis is the major cause of tumour recurrence and effective therapies for HCC metastasis are not available [8]. Thus, an understanding of the causes and mechanisms behind the progression and metastasis of HCC is crucial to more effective therapeutic approaches.
MicroRNAs (miRNAs) are endogenous small, evolutionarily conserved, non-coding RNAs which act as negative regulators for mRNA expression [9]. As a result, miRNAs have been shown to be profoundly involved in pathogenesis of many human diseases such as cancer and cardiovascular diseases [10,11]. MiRNAs play an important role in cell proliferation, apoptosis, gene regulation and the maintenance of cell differentiation [12-14]. Researchers have aslo found that deregulation expression of some miRNAs may be involved in tumor initiation, progression, and invasion [15-17]. Indeed, some miRNAs havebeen shown to act as oncogenes or tumour suppressor genes [18-20]. However, there still remains a lot to understand as regards to the involvement of miRNAs in hepatocarcinogenesis and progression of HCC.
In the present study, we demonstrated that the levels of miR-543 were dramatically increased in clinical hepatocellular carcinoma (HCC) tissues and cell lines. Moreover, we found that forced expression of miR-543 promoted the proliferative and invasive potential of HCC cell line HepG2. We also identified PAQR3 as a direct target gene for miR-543.
Material and methods
Ethics statement
All patients agreed to participate in the study and gave written informed consent. This study was approved by the ethical board of the institute of The Fourth Hospital of Harbin Medical University and complied with Declaration of Helsinki.
Samples and cases
HCC and their morphologically normal tissues (located > 3 cm away from the tumor) were obtained between 2009 and 2012 from 60 HCC patients undergoing surgery at The Fourth Hospital of Harbin Medical University. Tissue samples were cut into two parts, one was fixed with 10% formalin for histopathological diagnosis, and the other was immediately snap-frozen in liquid nitrogen, and stored in liquid nitrogen until RNA extraction. None of the patients received radiotherapy or chemotherapy before surgery. The characteristics of patients are described in Table 1.
Table 1.
Clinicopathologic characteristics of patients with HCC
| Parameter | Total samples | miR-543 expression | P | |
|---|---|---|---|---|
|
| ||||
| High (n%) | Low (n%) | |||
| Age (years) | 0.76 | |||
| ≥ 50 | 35 | 29 | 6 | |
| < 50 | 25 | 22 | 3 | |
| Gender | 0.61 | |||
| Male | 48 | 41 | 7 | |
| Female | 12 | 10 | 2 | |
| AFP (ug/L) | 0.76 | |||
| ≥ 400 | 28 | 22 | 6 | |
| < 400 | 32 | 29 | 3 | |
| Metastasis | 0.01 | |||
| Present | 13 | 13 | 0 | |
| Absent | 47 | 38 | 9 | |
| With cirrhosis | 0.85 | |||
| Present | 31 | 26 | 5 | |
| Absent | 29 | 25 | 4 | |
Cell lines and cell culture
The following human cell lines were used in this study: HepG2, Hep3B, Bel7402, SMMC-7721 and HL-7702. All of these cell lines were purchased from American Type Culture Collection (ATCC, Mannasas, VA, USA). These cells were culture and maintained at RPMI 1640 medium (PAA) supplemented with 10% FBS (GIBCO, NY, USA). Cells were maintained at 37°C in a humidified chamber with 95% air and 5% CO2.
Cell transfection
Oligonucleotides were synthesized by GenePharma (Shanghai, China) and transfected into the cells to a final oligonucleotide concentration of 20 nmol/L. Transfection was performed with Lipofectamine 2000 Reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. Briefly, cells were trypsinized, counted and seeded in plates on the day before transfection to ensure suitable cell confluency on the day of transfection.
Quantitative RT-PCR
Total RNA was extracted from the cells or tumor tissues using Trizol (Invitrogen) according to the manufacturer’s protocol. cDNA synthesis was carried out from 1 μg of total RNA in 12 μl of final volume containing 2 M stem-loop primer, 10 mM dNTP Mix (Invitrogen, USA). MicroRNAs were quantitated by real-time PCR using TaqMan MicroRNA assay (Invitrogen, USA) according to the protocol provided by the company. Small nuclear RNA U6 was used for normalisation. For the analysis of CADM1 expression, GAPDH was used for normalization. All of the reactions were run in triplicate. The ∆∆Ct method was used for relative quantification of gene expression to determinemiR-543 and PAQR3 mRNA expression levels. The primers are listed in Table 2.
Table 2.
Primer sequences
| Name | Sequence (5’-3’) |
|---|---|
| miRNA reverse transcription prime | |
| miRNA-543 | CAGACAGGAAACCGGTGCTAGTTTTGCCCTATTCCT |
| U6 snRNA | AAAATATGGAACGCTTCACGAATTTG |
| Real-time PCR primer sequence | |
| miRNA-543 | CCAGCTACACTGGGCAGCAGCAATTCATGTTT |
| CTCAACTGGTGTCGTGGA | |
| U6 snRNA | CTCGCTTCGGCAGCACATATACT |
| ACGCTTCACGAATTTGCGTGTC | |
| GAPDH | AATGGGCAGCCGTTAGGAAA |
| TGAAGGGGTCATTGATGGCA | |
| PAQR3 | TTTGGTCCCCTTCAACC |
| GTGCAGGGTCCGAGGT |
Cell proliferation assay
Cells were incubated in 10% CCK-8 (Dojindo; Kumamoto, Japan) diluted in normal cultured medium at 37°C until visual color conversion occurred. Proliferation rates were determined at 0, 24, 48 and 72 hours after transfection.
Cell migration and invasion assays
Cell migration and invasion ability were determined by the ability of cells passing through Matrigel-coated membrane matrix (BD Biosciences). Cells were harvested and resuspended in serum-free medium after transfection. About 3 × 104 (migration assay) or 1 × 105 (invasion assay) prepared cells were added into the chamber and incubated for 24 h at 37°C. Cells that had migrated or invaded through the membrane were fixed with 20% methanol and stained with 0.1% crystal violet (Sigma), imaged and counted. The migration assay is the same with invasion assay excepting no matrigel was used.
Luciferase assays
About 2 × 105 HEK293T cells per well were seeded in 24-well plates for 24 h before transfection. About 100 ng of wild-type or mutant PAQR3 3’-UTR psiCHECK-2 plasmid (Promega) was transiently cotransfected with miR-543 mimic or NC into 293T cells using 1.44 μl Lipofectamine reagent (Invitrogen). Cell lysates were harvested 24 h after transfection and then firefly and Renilla luciferase activities were measured by the Dual-Luciferase Reporter Assay System (Promega) on aBerthold AutoLumat LB9507 rack luminometer. Renilla luciferase activities were normalized to firefly luciferase activities to control for transfection efficiency.
Western blotting analysis
For the protein expression analyses, standard western blotting was carried out. Whole cell protein lysates were electrophoresed on 10% sodium dodecylsulfate-polyacrylamide gels and transferred onto polyvinylidene difluoride membranes (Millipore). The membranes were incubated with primary antibodies overnight at 4°C and then with the appropriate horseradish peroxidase-conjugated secondary antibody. The following antibodies wereused: anti-PAQR3 antibody (Abcam, England); anti-GAPDH antibody (Proteintech, Chicago, USA).
Statistical analysis
Each experiment was repeated at least three times. Statistical analysis was performed using SPSS 15.0. Data are presented as the mean ± standard deviation. Statistical analyses were done by analysis of variance (ANOVA) or Student’s t test and statistical significance level was set at α = 0.05 (two-side).
Result
MiR-543 is highly expressed in HCC
Real-time quantitative PCR demonstrated that miR-543 was significantly upregulated in the HCC tissues in comparison to the adjacent tissues (Figure 1A and 1B, p < 0.01). We also observed that the higher level of miR-543 were associated with pM stage (p ≤ 0.01, metastasis vs. no metastasis) in HCC patients (Figure 1C). We also determined the expression levels of miR-543 in normal hepatocytes cell lines (HL-7792) and HCC cells lines (HepG2, Hep3B, Bel7402, SMMC-7721). MiR-543 was significantly upregulated in HCC cells lines (HepG2, Hep3B, Bel7402, SMMC-7721) when compared with normal hepatocytes cell lines (HL-7792) (Figure 1D).
Figure 1.
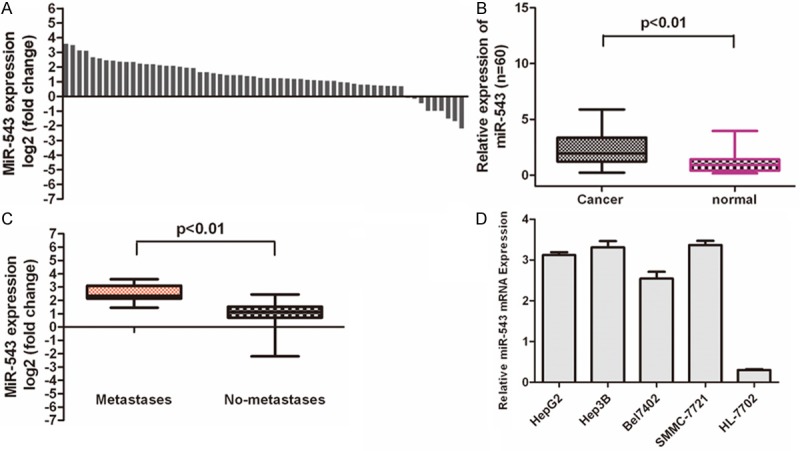
MiR-543 is over-expressed in hepatocellular carcinoma tissues. A. qRT-PCR analysis of miR-543 expression in 60 pairs HCC tissues and their corresponding adjacent nontumorous livers. The expression of miR-543 was normalized to U6 snRNA. B. Relative miR-543 expression levels in HCC tissues and adjacent normal regions; C. Statistical analysis of the association between miRNA level, pM stage (No metastasis and Metastasis, respectively). D. qRT-PCR analysis of miR-543 expression in HCC cells (HepG2, Hep3B, Bel7402, SMMC-7721) and normal hepatocytes (HL-7792 cells).
MiR-543 promotes HCC cell growth and invasion
To determine the function of miR-543 in the progression of HCC, we sought to determine whether miR-543 affect the proliferation of HCC cells. HepG2 cells were transfected with control oligo or miR-543 mimics and inhibitors, which exhibited high transfection efficiency (Figure 2A and 2B). The proliferation rates of HepG2 cells with forced expression of miR-543 were significantly increased compared with those of control miRNA transfected or no treated cells (Figure 2C). Conversely, miR-543 inhibitor significantly decreased the cell proliferation of HepG2 (Figure 2D). Matrigel invasion assays demonstrated that overexpression of miR-543 significantly induced the invasion capacity of the HepG2 cells; conversely, miR-543 inhibitor significantly decreased the cell invasion of the HepG2 cells (Figure 2E).
Figure 2.
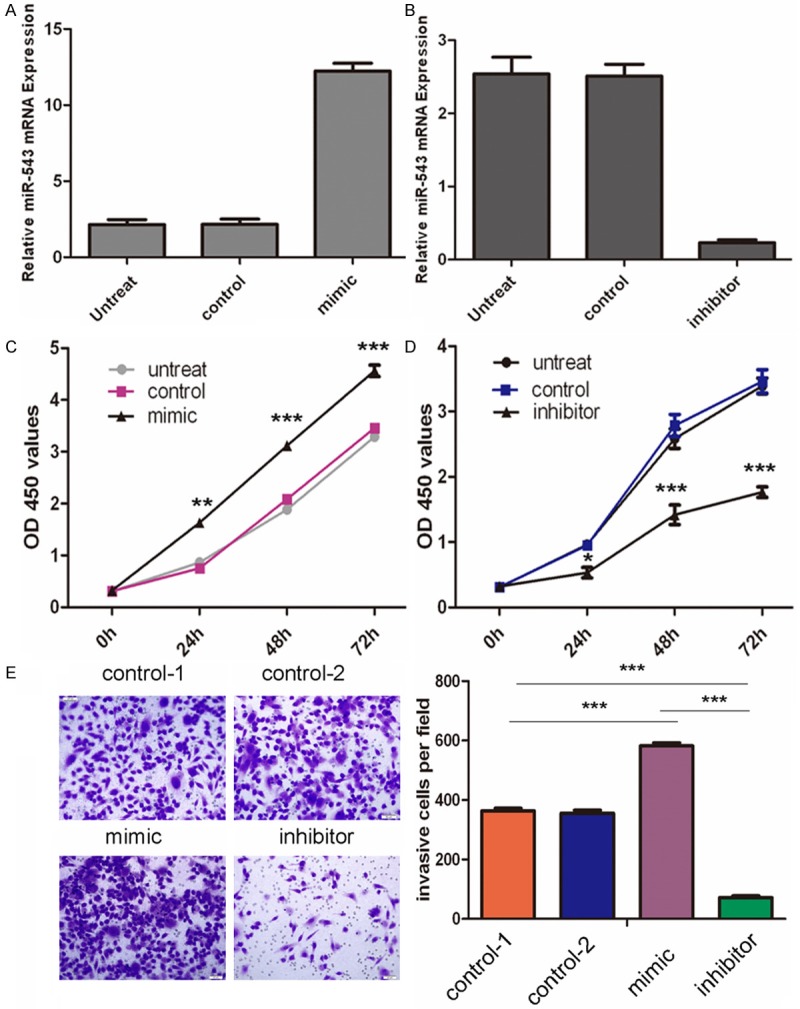
MiR-543 promotes hepatocellular carcinoma cell growth and invasion. A. qRT-PCR analysis of miR-543 expression after the transfection of miR-543 mimic, or control or no transfection. The expression of miR-543 was normalized to U6 snRNA. B. qRT-PCR analysis of miR-543 expression after the transfection of miR-543 inhibitor, or control or no transfection. The expression of miR-543 was normalized to U6 snRNA. C. The CCK8 assay analysis was used to evaluate the proliferation of HepG2 cells after transfection with the miR-543 mimic, or scramble or no transfection. D. The CCK8 assay analysis was used to evaluate the proliferation of HepG2 cells after transfection with the miR-543 inhibitor, or scramble or no transfection. E. Transwell analysis of the HepG2 cells after treatment with miR-543 mimics, inhibitors or control or no transfection; the invasive cells per field is shown below, *p < 0.05, **p < 0.01, and ***p < 0.001.
PAQR3 was direct target of miR-543
To identify effectors of miR-543, we used two algorithms that predict the mRNA targets of a miRNA-PicTar and TargetScan. PAQR was one of the multiple putative target genes that were predicted by both of the algorithms. Using TargetScan, we located potential binding sites for miR-543 at the 3’UTR of PAQR3 mRNAs (Figure 3A). Overexpression of miR-543 significantly decreased the luciferase activity of wild-type PAQR3 3’UTR but not binding site mutants PAQR3 3’UTR (Figure 3B). The mRNA level of PAQR3 was significantly decreased in the HepG2 cells with forced expression of miR-543 using real-time quantitative PCR. In addition, forced expression of miR-543 significantly reduced PAQR3 protein levels in the HepG2 cells, using western blot analyses (Figure 3D).
Figure 3.
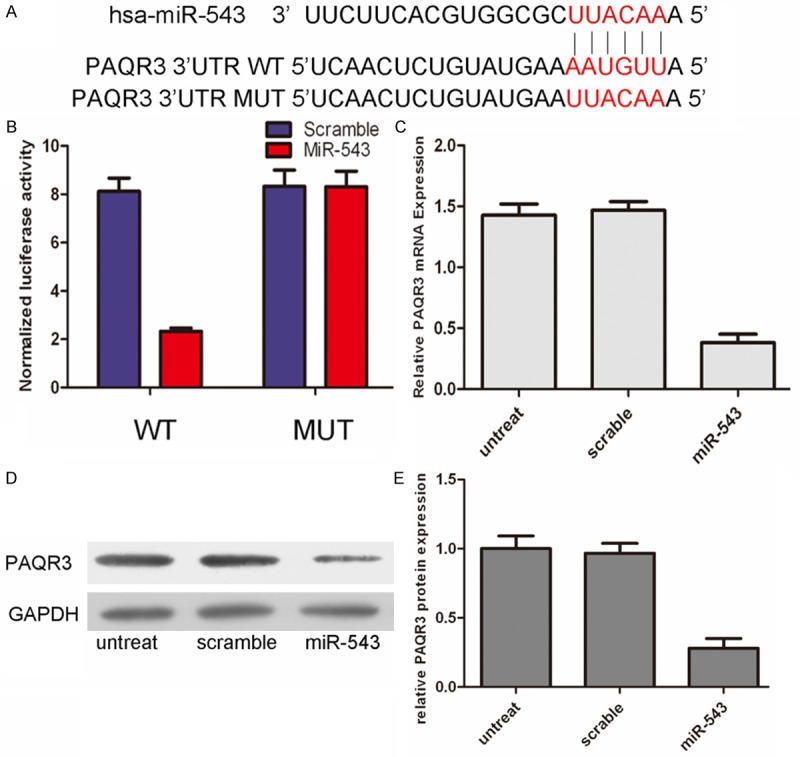
PAQR3 was direct target of miR‑543. A. The sequences of miR-543 binding sites within the human PAQR3 3’UTRs and schematic reporter constructs, in this panel, PAQR3-WT represent the reporter constructs containing the entire 3’UTR sequences of PAQR3. PAQR3-MUT represent the reporter constructs containing mutated nucleotides. B. The analysis of the relative luciferase activities of PAQR3-WT, PAQR3-MUT in 293T cells. The error bars are derived from triplicate experiments. C. qRT-PCR analysis of PAQR3 mRNA expression in HepG2 cells after treatment with miRNA mimics or scramble or no transfection. The expression of PAQR3 was normalized to GAPDH. D, E. Western blot analysis of PAQR3 expression in HepG2 cells transfected with miR-543 mimics or scramble or no transfection. GAPDH was also detected as a loading control.
MiR-543 expression inversely correlated with PAQR3 mRNA in HCC samples
Real-time quantitative PCR demonstrated that PAQR3 was significantly downregulated in the HCC tissues in comparison to in the adjacent tissues (Figure 4A and 4B, p < 0.01). We also observed that the lower level of PAQR3 were also associated with pM stage (p ≤ 0.01, metastasis vs. no metastasis) in HCC patients (Figure 4C). A markedly inversed correlation between PAQR3 mRNA expression and miR-543 levels was also detected in patient samples (Figure 4D). PAQR3 was significantly downregulated in HCC cells lines (HepG2, Hep3B, Bel7402, and SMMC-7721) when compared with normal hepatocytes cell lines (HL-7792) (Figure 4D). Western blot analysis demonstrated that the protein of PAQR3 was also downregulated in HCC cells lines when compared with normal hepatocytes cell lines (Figure 4E). We also found that PAQR3 levels were significantly decreased in HCC tissue compared with normal tissues in three samples (Figure 4E).
Figure 4.
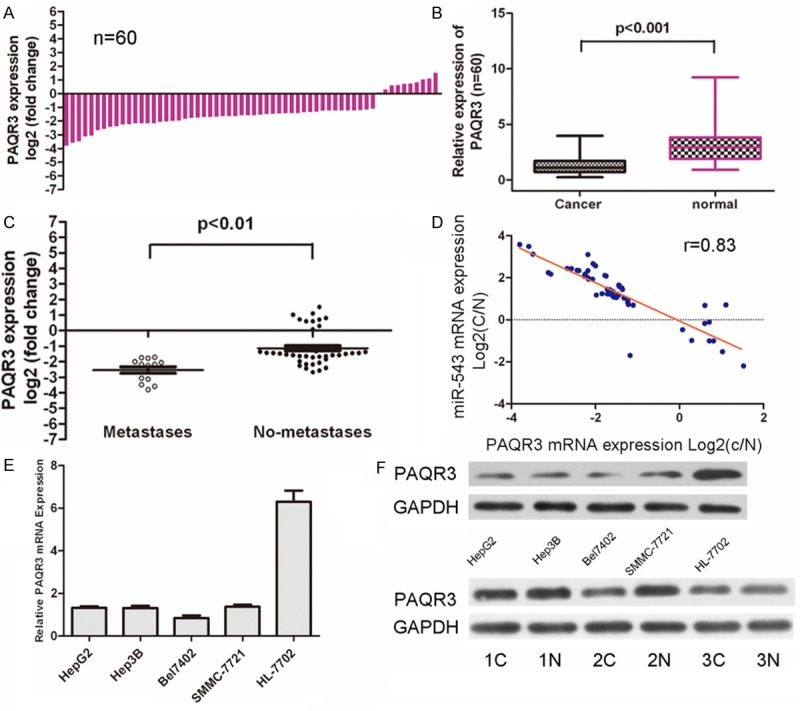
MiR‑543 expression inversely correlated with PAQR3 mRNA in HCC samples. A. qRT-PCR analysis of PAQR3 expression in 60 pairs HCC tissues and their corresponding adjacent nontumorous livers. The expression of PAQR3 was normalized to GAPDH mRNA. B. Relative PAQR3 expression levels in HCC tissues and adjacent normal regions; C. Statistical analysis of the association between PAQR3 level, pM stage (No metastasis and Metastasis, respectively). D. The mRNA expression of PAQR3 was inversed correlation the expression of miR-543 levels in patient samples. E. qRT-PCR analysis of PAQR3 expression in HCC cells (HepG2, Hep3B, Bel7402, SMMC-7721) and normal hepatopcytes (HL-7792 cells). F. Western blot analysis of PAQR3 expression in the HCC cells (HepG2, Hep3B, Bel7402, SMMC-7721) and normal hepatopcytes (HL-7792 cells); and 3 pairs NSCLC tissues and their corresponding adjacent normal lung tissues.
Discussion
MicroRNA expression correlates with various cancers [21-24]. Many microRNAs contribute to tumor initiation and progression, functioning as tumour suppressors or oncogenes [25,26]. The current work showed that miR-543 expression is significantly increased in human HCC compared with matching adjacent nontumoral tissue. PAQR3 is negatively regulated by miR-543 at the posttranscriptional level, via a specific target site within the 3’UTR. Moreover, we showed that miR-543 promoted HCC cell proliferation, invasion and migration in vitro. Furthermore, the expression level of PAQR3 was significantly decreased in HCC samples in comparison with adjacent normal tissues and the expression level of PAQR3 was inversely associated with the expression of miR-543. The identification of miR-543 as an important regulator of HCC cell migration and invasion emphasizes an essential role of this miRNA in mediating hepatic oncogenesis.
Previous studies have showed that miR-543 could inhibit cell proliferation, migration and invasion, which might due to its ability to regulate genes involved in diverse aspects of proliferation and metastasis [27,28]. Moreover, recent findings have showed that miR-543 inhibited epithelial to mesenchymal transition (EMT) through reducing TWIST1 signaling in the colorectal cancer [27]. Furthermore, miR-543 expression was proved to be decreased in endometrial cancer and serves as a tumor suppressor by targeting FAK and TWIST1 expression [27]. However, the expression and the biological function of miR-543 in HCC are still unclear. Our results have showed that the expression of miR-543 in HCC tissues was significantly higher than that in adjacent tissues. Moreover, overexpression of miR-543 significantly promoted cell proliferation, migration and invasion and down-regulation of miR-543 significantly inhibited cell proliferation, migration and invasion. In summary, our results suggested that miR-543 contributed to the progression and metastasis of HCC, acting as potential oncogene.
In this study, PAQR3 was identified as a direct target of miR-543 in HepG2 cells, and this conclusion is supported by the following reasons: complementary sequence of miR-543 is identified in the 3’UTR of PAQR3 mRNA; overexpression of miR-543 led to a significant reduction in PAQR3 at both mRNA and protein level; miR-543 overexpression suppressed PAQR3 3’UTR luciferase report activity and this effect was abolished by mutation of the miR-543 seed binding site. These results indicated that miR-543 may function as a tumor suppressor in HCC partly mediated by repressing PAQR3 expression.
PAQR3 (Progestin and AdipoQ Receptors) is a member of the progestin and adipoQ receptor (PAQR) family and is a seven-transmembrane protein specifically located at the Golgi apparatus in mammalian cells [29,30]. Previous studies have showed that PAQR3 function as cell surface receptors for adiponectin, an adipocyte-secreted cytokine that regulates glucose and lipid metabolism [31]. PAQR3 expression level is closely associated with the progression and metastasis of cancers [32]. The expression level of PAQR3 was significantly decreased in colorectal cancer and gastric cancer [32-34]. In line with these, our results have showed that the expression of PAQR3 is decreased in human HCC. Moreover, the expression of PAQR3 was negatively correlated with miR-543 levels in HCC. Upregulation of these miR-543 in HCC may inhibit the expression of PAQR3, leading to enhanced metastasis of the cancer.
In conclusion, the current study provides novel evidence that miR-543 promotes HCC cell proliferationand migration partly through repression of PAQR3. Our findings on miR-543 suggest that this miRNA-543 might be a potential target for the diagnosis and treatment of HCC in future.
Acknowledgements
This manuscript was supported by the Post-doctoral Fund of Heilongjiang Province (No. LBH-Z12195) and the Technological innovation Special Foundation of Harbin science and Technology Bureau (No. 2014RFQGJ188).
Disclosure of conflict of interest
None.
References
- 1.Caffarelli E, Filetici P. Epigenetic regulation in cancer development. Front Biosci (Landmark Ed) 2011;16:2682–2694. doi: 10.2741/3880. [DOI] [PubMed] [Google Scholar]
- 2.Rosell R, Cuello M, Cecere F, Santarpia M, Reguart N, Felip E, Taron M. Usefulness of predictive tests for cancer treatment. Bull Cancer. 2006;93:E101–108. [PubMed] [Google Scholar]
- 3.Wang PR, Xu M, Toffanin S, Li Y, Llovet JM, Russell DW. Induction of hepatocellular carcinoma by in vivo gene targeting. Proc Natl Acad Sci U S A. 2012;109:11264–11269. doi: 10.1073/pnas.1117032109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zucman-Rossi J. Molecular classification of hepatocellular carcinoma. Dig Liver Dis. 2010;42(Suppl 3):S235–241. doi: 10.1016/S1590-8658(10)60511-7. [DOI] [PubMed] [Google Scholar]
- 5.Banaudha KK, Verma M. The role of microRNAs in the management of liver cancer. Methods Mol Biol. 2012;863:241–251. doi: 10.1007/978-1-61779-612-8_14. [DOI] [PubMed] [Google Scholar]
- 6.Fornari F, Milazzo M, Chieco P, Negrini M, Marasco E, Capranico G, Mantovani V, Marinello J, Sabbioni S, Callegari E, Cescon M, Ravaioli M, Croce CM, Bolondi L, Gramantieri L. In hepatocellular carcinoma miR-519d is up-regulated by p53 and DNA hypomethylation and targets CDKN1A/p21, PTEN, AKT3 and TIMP2. J Pathol. 2012;227:275–285. doi: 10.1002/path.3995. [DOI] [PubMed] [Google Scholar]
- 7.Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31:766–776. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Zhang JB, Qin Y, Wang W, Wei L, Teng Y, Guo L, Zhang B, Lin Z, Liu J, Ren ZG, Ye QH, Xie Y. PROX1 promotes hepatocellular carcinoma metastasis by way of up-regulating hypoxia-inducible factor 1alpha expression and protein stability. Hepatology. 2013;58:692–705. doi: 10.1002/hep.26398. [DOI] [PubMed] [Google Scholar]
- 9.Yu X, Li Z, Shen J, Wu WK, Liang J, Weng X, Qiu G. MicroRNA-10b Promotes Nucleus Pulposus Cell Proliferation through RhoC-Akt Pathway by Targeting HOXD10 in Intervetebral Disc Degeneration. PLoS One. 2013;8:e83080. doi: 10.1371/journal.pone.0083080. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.He W, Li Y, Chen X, Lu L, Tang B, Wang Z, Pan Y, Cai S, He Y, Ke Z. miR-494 acts as an anti-oncogene in gastric carcinoma by targeting c-myc. J Gastroenterol Hepatol. 2014;29:1427–34. doi: 10.1111/jgh.12558. [DOI] [PubMed] [Google Scholar]
- 11.Cheng Y, Liu X, Yang J, Lin Y, Xu DZ, Lu Q, Deitch EA, Huo Y, Delphin ES, Zhang C. MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circ Res. 2009;105:158–166. doi: 10.1161/CIRCRESAHA.109.197517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gong M, Ma J, Guillemette R, Zhou M, Yang Y, Hock JM, Yu X. MiR-335 Inhibits Small Cell Lung Cancer Bone Metastases via IGF-IR and RANKL Pathways. Mol Cancer Res. 2014;12:101–110. doi: 10.1158/1541-7786.MCR-13-0136. [DOI] [PubMed] [Google Scholar]
- 13.Jung CJ, Iyengar S, Blahnik KR, Ajuha TP, Jiang JX, Farnham PJ, Zern M. Epigenetic modulation of miR-122 facilitates human embryonic stem cell self-renewal and hepatocellular carcinoma proliferation. PLoS One. 2011;6:e27740. doi: 10.1371/journal.pone.0027740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin J, Cai L, Liu ZM, Zhou XS. miRNA-218 Inhibits Osteosarcoma Cell Migration and Invasion by Down-regulating of TIAM1, MMP2 and MMP9. Asian Pac J Cancer Prev. 2013;14:3681–3684. doi: 10.7314/apjcp.2013.14.6.3681. [DOI] [PubMed] [Google Scholar]
- 15.Jin Y, Peng D, Shen Y, Xu M, Liang Y, Xiao B, Lu J. MicroRNA-376c inhibits cell proliferation and invasion in osteosarcoma by targeting to transforming growth factor-alpha. DNA Cell Biol. 2013;32:302–309. doi: 10.1089/dna.2013.1977. [DOI] [PubMed] [Google Scholar]
- 16.Bou Kheir T, Futoma-Kazmierczak E, Jacobsen A, Krogh A, Bardram L, Hother C, Gronbaek K, Federspiel B, Lund AH, Friis-Hansen L. miR-449 inhibits cell proliferation and is down-regulated in gastric cancer. Mol Cancer. 2011;10:29. doi: 10.1186/1476-4598-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan SH, Wu CW, Li AF, Chi CW, Lin WC. miR-21 microRNA expression in human gastric carcinomas and its clinical association. Anticancer Res. 2008;28:907–911. [PubMed] [Google Scholar]
- 18.Cho WJ, Shin JM, Kim JS, Lee MR, Hong KS, Lee JH, Koo KH, Park JW, Kim KS. miR-372 regulates cell cycle and apoptosis of ags human gastric cancer cell line through direct regulation of LATS2. Mol Cells. 2009;28:521–527. doi: 10.1007/s10059-009-0158-0. [DOI] [PubMed] [Google Scholar]
- 19.Furuta M, Kozaki K, Tanimoto K, Tanaka S, Arii S, Shimamura T, Niida A, Miyano S, Inazawa J. The tumor-suppressive miR-497-195 cluster targets multiple cell-cycle regulators in hepatocellular carcinoma. PLoS One. 2013;8:e60155. doi: 10.1371/journal.pone.0060155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lang Q, Ling C. MiR-124 suppresses cell proliferation in hepatocellular carcinoma by targeting PIK3CA. Biochem Biophys Res Commun. 2012;426:247–252. doi: 10.1016/j.bbrc.2012.08.075. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Guo Z, Wang J, Mao Y, Gao Q. Serum miR-18a: A Potential Marker for Hepatitis B Virus-Related Hepatocellular Carcinoma Screening. Dig Dis Sci. 2012;57:2910–2916. doi: 10.1007/s10620-012-2317-y. [DOI] [PubMed] [Google Scholar]
- 22.Li C, Li JF, Cai Q, Qiu QQ, Yan M, Liu BY, Zhu ZG. MiRNA-199a-3p in Plasma as a Potential Diagnostic Biomarker for Gastric Cancer. Ann Surg Oncol. 2013;3:S397–405. doi: 10.1245/s10434-012-2600-3. [DOI] [PubMed] [Google Scholar]
- 23.He C, Xiong J, Xu X, Lu W, Liu L, Xiao D, Wang D. Functional elucidation of MiR-34 in osteosarcoma cells and primary tumor samples. Biochem Biophys Res Commun. 2009;388:35–40. doi: 10.1016/j.bbrc.2009.07.101. [DOI] [PubMed] [Google Scholar]
- 24.Gu H, Yang T, Fu S, Chen X, Guo L, Ni Y. MicroRNA-490-3p inhibits proliferation of A549 lung cancer cells by targeting CCND1. Biochem Biophys Res Commun. 2014;444:104–108. doi: 10.1016/j.bbrc.2014.01.020. [DOI] [PubMed] [Google Scholar]
- 25.Du B, Wang Z, Zhang X, Feng S, Wang G, He J, Zhang B. MicroRNA-545 Suppresses Cell Proliferation by Targeting Cyclin D1 and CDK4 in Lung Cancer Cells. PLoS One. 2014;9:e88022. doi: 10.1371/journal.pone.0088022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fei X, Qi M, Wu B, Song Y, Wang Y, Li T. MicroRNA-195-5p suppresses glucose uptake and proliferation of human bladder cancer T24 cells by regulating GLUT3 expression. FEBS Lett. 2012;586:392–397. doi: 10.1016/j.febslet.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Bing L, Hong C, Li-Xin S, Wei G. MicroRNA-543 suppresses endometrial cancer oncogenicity via targeting FAK and TWIST1 expression. Arch Gynecol Obstet. 2014;3:533–41. doi: 10.1007/s00404-014-3219-3. [DOI] [PubMed] [Google Scholar]
- 28.Haga CL, Phinney DG. MicroRNAs in the imprinted DLK1-DIO3 region repress the epithelial-to-mesenchymal transition by targeting the TWIST1 protein signaling network. J Biol Chem. 2012;287:42695–42707. doi: 10.1074/jbc.M112.387761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garitaonandia I, Smith JL, Kupchak BR, Lyons TJ. Adiponectin identified as an agonist for PAQR3/RKTG using a yeast-based assay system. J Recept Signal Transduct Res. 2009;29:67–73. doi: 10.1080/10799890902729456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Wang L, Zhu L, Pan Y, Xiao F, Liu W, Wang Z, Guo F, Liu Y, Thomas WG, Chen Y. PAQR3 modulates insulin signaling by shunting phosphoinositide 3-kinase p110alpha to the Golgi apparatus. Diabetes. 2013;62:444–456. doi: 10.2337/db12-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, Wang X, Li Z, Xia T, Zhu L, Liu B, Zhang Y, Xiao F, Pan Y, Liu Y, Guo F, Chen Y. PAQR3 has modulatory roles in obesity, energy metabolism, and leptin signaling. Endocrinology. 2013;154:4525–4535. doi: 10.1210/en.2013-1633. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Li X, Fan F, Jiao S, Wang L, Zhu L, Pan Y, Wu G, Ling ZQ, Fang J, Chen Y. PAQR3 plays a suppressive role in the tumorigenesis of colorectal cancers. Carcinogenesis. 2012;33:2228–2235. doi: 10.1093/carcin/bgs245. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Jiang X, Qin X, Ye D, Yi Z, Liu M, Bai O, Liu W, Xie X, Wang Z, Fang J, Chen Y. RKTG inhibits angiogenesis by suppressing MAPK-mediated autocrine VEGF signaling and is downregulated in clear-cell renal cell carcinoma. Oncogene. 2010;29:5404–5415. doi: 10.1038/onc.2010.270. [DOI] [PubMed] [Google Scholar]
- 34.Ling ZQ, Guo W, Lu XX, Zhu X, Hong LL, Wang Z, Chen Y. A Golgi-specific protein PAQR3 is closely associated with the progression, metastasis and prognosis of human gastric cancers. Ann Oncol. 2014;25:1363–1372. doi: 10.1093/annonc/mdu168. [DOI] [PubMed] [Google Scholar]


