Abstract
Since target therapy with mTOR inhibitors plays an important role in the current management of clear cell renal cell carcinoma (RCC), there is an increasing demand for predictive biomarkers, which may help to select patients that are most likely to benefit from personalized treatment. When dealing with formalin-fixed paraffin-embedded (FFPE) cancer tissue specimens, several techniques may be used to identify potential molecular markers, yielding different outcome in terms of accuracy. We sought to investigate and compare the capability of three main techniques to detect molecules performing an active function in mTOR pathway in RCC. Immunohistochemistry (IHC), Western blot (WB) and immunofluorescence (IF) analyses were performed on FFPE RCC tissue specimens from 16 patients by using the following mTOR pathway-related: mTOR (Ser235/236), phospho-mTOR (p-mTOR/Ser2448), phospho-p70S6k (p-p70S6k/Thr389), both monoclonal and polyclonal, phospho-S6Rb (p-S6Rb) and phospho-4EBP1 (p-4EBP1/Thr37/46). No single molecule was simultaneously revealed by all three techniques. Only p-p70S6k was detected by two methods (IHC and IF) using a monoclonal antibody. The other molecules were detected exclusively by one technique, as follows: p-mTOR and polyclonal p-p70S6K by IHC, p70S6K, p-S6Rb and p-4EBP1 by WB, and, finally, mTOR by IF. We found significant differences in detecting mTOR pathway-related active biomarkers by using three common techniques such as IHC, WB and IF on RCC samples. Such results have important implications in terms of predictive biomarker testing, and need to be related to clinical end-points such as responsiveness to targeted drugs by prospective studies.
Keywords: mTOR, clear cell renal cell carcinoma, immunohistochemical, Western blot, immunofluorescence analysis, p-mTOR, p-p70S6k, methods
Introduction
mTOR -mammalian target of rapamycin- inhibitors play an important role in the targeted treatment of clear cell renal cell carcinoma (RCC) [1,2], however biomarkers able to predict responsiveness to such drugs, thus providing pre-treatment patient stratification, are lacking. The use of different techniques and scoring systems affect the accuracy of results when assessing active mTOR pathway-related molecules on formalin-fixed paraffin-embedded (FFPE) tissue samples, the most common being immunohistochemistry (IHC), Western blotting (WB) and immunofluorescence (IF). Unlike the targeted Her-2/neu gene for breast or gastric cancer, ALK gene for pulmonary adenocarcinoma, 1p/19 chromosomes for oligodendroglioma or EGFR gene testing, no consensus recommendations are available to proper assessment of molecular alterations in mTOR pathway as predictive biomarkers.
Overall, cancer therapy has begun to shift from a standardized global treatment regimen, which included the administration of conventional chemotherapy, including actin cytostatic molecules, to a more personalized approach, targeting specific proteins in individual tumors. In order to limit the toxic effects, maximize efficacy and avoid cost-intensive treatments with little benefit for patients, it is highly desirable to develop technologies, which reliably detect and validate cancer-specific targets. Efforts to target the therapy patient level include the identification of informative biomarkers, which could be used to improve early diagnosis, lead the choice on which patients could be treated and to identify the best personalized therapy with a predictive biomarkers assay [3-6]. In patients affected by metastatic clear cell RCC the use of mTOR inhibitors are of routine practice in addition to other drugs, such as sorafenib or sutent. Different potential mechanisms may also lead to resistance to mTOR inhibitors and actually no consensus has been reached to which of the several mTOR pathway-related molecules, which methods and which tissues are suitable to predict responsiveness to targeted mTOR therapies [7-10].
Thus, detecting such biological parameters with clinical relevance to predict the activity of mTOR inhibitors still represent a major challenge and it may also help the ongoing development of new generations of mTOR inhibitors [11].
Aim of this study is to investigate and compare the capability of several techniques in identifying mTOR pathway-related molecules carrying active function in clear cell RCC.
Materials and methods
Patients
Sixteen patients affected by metastatic (stage IV) clear cell RCC were retrieved from Verona Uroncological Database. Tissue samples included 4 pulmonary, 4 pancreatic, 2 hepatic, 2 skin, 2 adrenal, 1 epidural and 1 cerebellar metastases.
The expression of mTOR (rabbit monoclonal, dilution 1:1,000; EPITOMICS, Burlingame, CA), phosphorylated mTOR (p-mTOR, rabbit monoclonal, dilution 1:1,000; EPITOMICS, Burlinga-me, CA), p70S6k, phosphorylated p70S6k (p-p70S6k), both monoclonal and polyclonal (dilution 1:1,000; EPITOMICS, Burlingame, CA), ph-osphorylated S6Rb (p-S6Rb, rabbit monoclonal, dilution 1:1,000; EPITOMICS, Burlingame, CA) and phosphorylated 4EBP1 (p-4EBP1, rabbit monoclonal, dilution 1:1,000; EPITOMICS, Burlingame, CA) was examined by immunohistochemistry (IHC ), Western blot (WB) and immunofluorescence (IF).
Western blot analysis
Frozen tissue samples
Section obtained from fresh frozen tissue samples were collected into an Eppendorf tube. Cell Lysis Buffer (Cell Signaling, Beverly, MA) was added prior to heating at 100°C for 5 min. Samples were then cooled for 5 min on ice, centrifuged at 140000 x g for 15 minutes and supernatants were subsequently transferred to a new collection tube and stored at -20°C.
Formalin-fixed and paraffin-embedded tissue samples
After deparaffinization and rehydration of tissue sections, proteins were extracted using Qproteome FFPE Tissue Kit (Qiagen, Milan, Italy). Briefly, the area of interest was excised with a needle and transferred to a collection tube containing 100 μl of extraction buffer. The sample was vortexed and incubated in 100°C water bath for 20 minutes and then in 80°C thermomixer for 2 h with shake at 750 rpm. After heat-treatment, the sample was cooled to 4°C for 5 minutes and centrifuged (14000 g, 15 minutes, 4°C). The supernatant was transferred to a new collection tube and stored at -20°C). Using the Bio-Rad protein assay kit according to manufacturer’s instructions performed protein quantification. 25 μg of extracted lysates was resolved in 10% polyacrylamide SDS-PAGE gel in a BioRad Mini Protean tetra cell system at 150 V for 1 h.
Electrophoresed proteins were transferred into a nitrocellulose membrane at 250 mA for 90 minutes. The membranes were blocked in TBST (100 mM Tris pH 7.5, 0.9% NaCl, 0.1% Tween 20) plus 5% non-fat dry milk for 1 h at RT with constant shaking and probed overnight at 4°C with abovementioned antibodies. The sections were incubated O.N. at 4°C with the indicated antibodies, washed three time with TBST and incubated with the specific secondary anti-mouse or anti-rabbit peroxidase-conjugated anti IgG antibody. After three washes with TBST the immunoblots were visualized with ECL plus (Amersham/GE Healthcare Europe GmgH, Munchen, Germany).
Expression levels of each marker were quantified with ImageJ densitometric analysis.
Immunohistochemical analysis
Paraffin-embedded tissue block were cut into 2-3 μm sections and mounted on adhesion microscope glass slides.
For immunohistochemical staining, sections were stained in an autostainer Leica Bond System with the abovementioned antibodies. Briefly, slides were deparaffinized twice in xylene for 5 minutes and rehydrated through graded ethanol solutions to distilled water. Antigen retrieval was performed by heating sections in citrate buffer, ethylenediaminetetraacetic acid buffer, or enzymatically with proteinase K. Inactivation of endogenous peroxidase activity was obtained by incubating sections in 3% H2O2 for 15 minutes. Localization of bound antibodies was performed with a peroxidase-labeled streptavidin-biotin system (DAKO, LSAB2 Kit) with 3, 3-diaminobenzidine as a chromogen. Appropriate positive controls for each antibody were run concurrently and showed adequate immunostaining.
A case was interpreted positive if at least 10% of neoplastic cells showed immunoexpression. The staining score for each sample counting the intensity of neoplastic positive cells, was grade as 1=lightly perceptible antibody signal, 2=average perceptible antibody signal and 3 clearly perceptible antibody signal.
Immunofluorescence analysis
FFPE tissue blocks were cut into 2-3 μm sections and mounted on adhesion microscope glass slides. After, the sections were dewaxed and rehydrated. Antigen retrieval was performed in prewarmed citrate buffer (pH 6 temp. 95°C) for 30 minutes. The sections were cooled to room temperature and then incubated with a protein block serum free solution for 15 minutes at RT to block non-specific binding.
For immunofluorescence staining, sections were incubated with primary anti-human antibody. Slides were then incubated with the corresponding Alexa 488-conjugated antibody (1:1000 INVITROGEN Molecular Probe, Milan, Italy). Reduction of the autofluorescence background was performed by incubation with Sudan Black B 0.1% (Sigma-Aldrich). Nuclei were stained with Prolong Gold antifade reagent with DAPI (INVITROGEN Molecular Probe, Milan, Italy). Slides were analysed by a Olympus BX61 microscope.
Results
No one molecules of the mTOR pathway was simultaneously revealed by all three techniques. p-p70S6k monoclonal was detected by two (IHC and IF) methods (Table 1 and Figure 1). The other molecules were detected by only one technique as follows: IHC showed the sensitivity to detecting the expression of p-mTOR (Ser2448) and p-p70S6K (Thr389) molecules, Western blot analysis was the sensitive method to detect the expression of p70S6K molecules (Thr389), p-S6Rb and p-4EBP1 ones (Thr37/46) and immunofluorescence was sensitive in detecting the expression of mTOR (Ser235/236).
Table 1.
mTOR pathway: sensibility in detecting biomarkers by three tissue-based techniques
| 1 Molecules | Immunohistochemistry (IHC) | Western blot (WB) | Immunofluorescence (IF) |
|---|---|---|---|
| mTOR | - | - | ++ |
| p-mTOR | ++ | - | // |
| p70S6k | - | ++ | // |
| p-p70S6k monoclonal | ++ | - | + |
| p-p70S6k polyclonal | - | - | // |
| p-S6RP | - | ++ | // |
| p-4EBP1 | - | ++ | // |
//: not recommend.
Figure 1.

Testing mTOR pathway active biomarkers in clear cell renal cell carcinoma: preferable methods to be used to stratify patients for more likely response to mTOR inhibitors.
Western blot findings
Western blot analysis did not show any positivity for the p-mTOR and p-p70S6k monoclonal, and also for the mTOR and p-p70S6k polyclonal biomarker testing. Differently, Western blot analysis showed strong positivity for p70S6k (band visible at 70kDa), p-S6 Rb (band visible at 32kDa) and p-4EBP1 molecules (band visible at 13-15kDa) (Figure 2).
Figure 2.

Western blot positive findings in metastatic clear cell RCC for the p70S6k, p-S6 Rb and p-4EBP1 molecules. A. Evidence for the band for p70S6k: E1-clear cell RCC, G1-clear cell RCC, B1-clear cell RCC, B2-pulmonary metastasis, F1-skin metastasis, F2-skin metastasis, A4-contralateral metastasis, A5-epidural metastasis, A3-cerebral metastasis, H2-clear cell renal cell carcinoma, H3- pulmonary metastasis; B. Evidence for the band for p-S6Rb: H1 normal kidney, H2 clear cell RCC, H3 pulmonary metastasis, B1 clear cell RCC, B2 pulmonary metastasis, F1 skin metastasis, F2 skin metastasis, A4 contralateral metastasis, A5 epidural metastasis, A3 cerebellar metastasis; C. Evidence for the band for p-4EBP1: G1 clear cell RCC, F1 skin metastasis, F2 skin metastasis, A2 adrenal metastasis, A3 cerebellar metastasis, A4 contralateral metastasis, A5 epidural metastasis, B1 clear cell RCC, B2 pulmonary metastasis.
Immunophenotypical findings
The immunohistochemical testing revealed in all cases the positive immunoexpression of p- mTOR and p-p70S6k monoclonal molecules; respectively 12 cases scored 2+ and 4 scored 1+ for the p-mTOR test and 14 cases scored 2+ and 4 scored 1+ for the p-p70S6k test (Figure 3). The IHC analysis did not show any positivity in all cases for mTOR, p70S6k, p-p70S6k polyclonal, p-S6 Rb and p-4EBP1 molecules.
Figure 3.
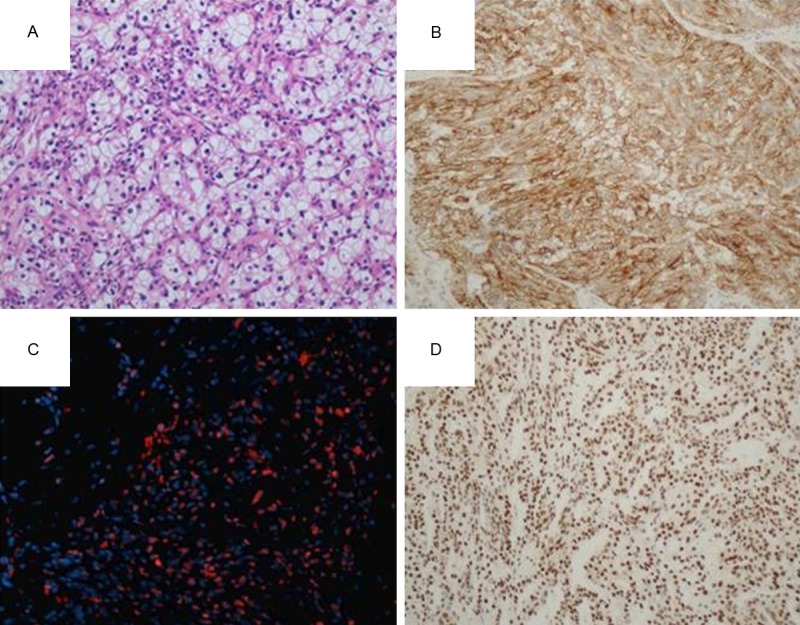
Immunohistochemical and immunofluorescence findings in metastatic clear cell RCC. A. Clear cell RCC (hematoxylin and eosin); B. 2+ positive p-mTOR immunoexpression in a pulmonary metastases from clear cell RCC; C. Pancreatic metastasis from clear cell RCC with positive expression after incubation with primary anti-human p-p70S6k monoclonal antibody (immunofluorescence, orange); D. 2+ positive p-p70S6k monoclonal immunoexpression in a skin metastases from clear cell RCC.
Immunofluorescence findings
The immunofluorescence analysis revealed the positive expression for mTOR molecule and for p-p70S6k monoclonal molecules. Notably, the mTOR was not evidenced by any other two techniques such as the immunohistochemical and Western blot analysis. The remaining were not performed because they are not recommended by datasheet.
Discussion
In our study we concluded that: 1) there is a significant difference in detecting mTOR pathway’s active biomarkers by using three common techniques such as IHC, WB and IF analysis; 2) the methods to detect active molecules of the mTOR pathway are important when justifying responsiveness or resistance to targeted drugs; 3) clinical trials need an agreement for standard methods to use for tissue testing in order to improve accuracy when correlation is performed between scoring the mTOR pathway activation and clinical end-points such as efficacy to targeted drugs.
We observed significant differences in detecting the mTOR pathway protein expression by using and comparing three major common tissue-based techniques such as the IHC, WB and IF analysis. Of note, the methods to detect active molecules may justify at least in part the correlation in between molecular testing used and responsiveness or resistance to targeted drugs.
mTOR has presented itself as a valid target for the treatment of cancer in clear cell RCC [12,13] and activation of this pathway has been suggested to correlate with aggressive behavior and unfortunate prognosis in clear cell RCC [14]. mTOR also regulates the translation of mRNA for p70S6 kinase (p70S6k) in cancer cells and overexpression of p70S6k is observed in ~60% of patients with clear cell RCC and seems to be predictive of response and treatment outcomes [15,16]. Phosphorylation of downstream targets p70S6k and eukaryotic translation initiation factor 4E (eIF4E) binding protein (4E-BP1) is also inhibited by this pathway [17]. For the reasons aforementioned, mTOR inhibitors play an important role in the targeted treatment of clear cell RCC, however no pre-selection of patients is available to predictive high efficacy to targeted inhibitors in a metastatic setting. The mTOR pathway is also of particular relevance to clear cell RCC also for anti-angiogenetic pathway, as it has been shown that HIF protein expression is dependent by mTOR in certain cellular contexts. Inappropriate accumulation of HIF-1α and HIF-2α as a result of biallelic alterations in the von Hippel-Lindau (VHL) gene observed in the majority of clear cell RCC is believed to be a critical step in RCC tumorigenesis as a result of increased expression of HIF-regulated gene products including VEGF, PDGF and TGF-α.
In our study we tested metastatic clear cell RCC with several molecules (mTOR, p-mTOR, p70-S6K, p-p70S6K, 4EBP1) because different potential mechanisms may also lead to resistance to mTOR inhibitors. The rationale to choose these molecules is due to the fact that rampamycin gains function by binding to FKBP12 and the resultant FKBP12-RAP complex inhibits the mTOR kinase activity, which, in turn, blocks the activation of two critical downstream signaling elements. By inhibiting the phosphorylation of mTOR, the activation of the 40S ribosomal protein S6 kinase (p70s6k) is blocked, leading to reduced translation of 5’-terminal oligopyrimidine (5’TOP) mRNAs that encode for essential components of the protein synthesis machinery [18]. The rapamycin mTOR inhibition also blocks phosphorylation of the eukaryotic initiation factor 4E binding protein-1 (4EBP1), which is also known as PHAS-1 (phosphorylated heat- and acid-stable protein 1). In its dephosphorylated state, 4EBP1 binds tightly to eIF-4E, thereby inhibiting the translation of mRNAs with regulatory elements in their 5’-untranslated regions (5’UTR) that encode for critical regulatory proteins such as growth factors, oncoproteins, and other cell cycle regulators [19].
Evidence of activation of predictive biomarkers by molecular testing on FFPE cancerous tissue blocks does also depend to different technique used. The visualization of active mTOR pathway has not been tested and compared on the same cohort of patients by using simultaneously the most common techniques such as IHC, WB and IF analysis. This cause to confusion about what is an effective technique for the analysis of important mTOR pathway proteins, thus we investigated and compared the capability of three major techniques in evidencing active function of molecules related to mTOR pathway on 16 patients’ samples affected by cell clear RCC in a metastatic setting. We used IHC, WB and IF on several molecules related to mTOR pathways such as mTOR, p-mTOR, p70S6k, p-p70S6k polyclonal and monoclonal, p-S6Rb and p-4EBP1. Importantly, we found that no one molecules was simultaneously revealed by all three techniques: only p-p70S6k monoclonal was detected by two methods (IHC and IF), while the other proteins were detected by only one technique: p-mTOR (Ser2448) and p-p70S6K (Thr389) proteins were detected to IHC method, WB analysis was the sensitive method to detect the expression of p70-S6K protein (Thr389), p-S6Rb and p-4EBP1 ones (Thr37/46) and the IF was sensitive in detecting the expression of mTOR.
These techniques are useful in order to determination and quantification of specific proteins directly on a histological preparation or a lysate. Each of these techniques takes advantage of the high specificity in the formation immune complex Antigen-Antibody and its detection by use of antibodies marked, with the development of a different signal depending on the method used. Because generally characterized antibodies are added antiglobulin secondarily to the test, the primary antibodies, specific antibody for the protein in question, can theoretically be used in each of the three techniques mentioned above, in according with the instructions for use of the company manufacturer.
Overall, we observed stratification in the magnitude of mTOR pathway molecular activation, showing a range of expression and visualization. A consensus in appropriate methods to be used to test protein expression and recommendations has to be obtained at least for comparing huge amount of data published or to initialize prospective trials with new generation mTOR inhibitors. The importance in such a consensus regard to the key node of mTOR pathways and the inhibitors that are going to be improved in the oncological clinical practice. mTOR is acknowledged as a master switch of cellular metabolism, modulating cell growth and proliferation. mTOR is located at the interface of two different signals such as growth/survival factors and nutritional/stress response. Since energy-, oxygen- and nutrient-deprivation is common in malignant tumors; cancer cells insensitive to these stresses may display selective growth and survival advantage. In fact, in malignant tumors, mTOR is considered as a crucial effector in the regulation of cell survival and proliferation, as well as in tumor angiogenesis [20].
Li et al. studied paraffin-embedded tumor tissue specimens derived from 18 metastatic RCC patients before everolimus treatment, who participated the phase 1b trial of everolimus in VEGF receptor (VEGFR)-tyrosine kinase inhibitor (TKI)-refractory. Chinese patients with metastatic RCC were examined for the expression levels of phosphorylated AKT, mTOR, eukaryotic initiation factor 4E (eIF4E) binding protein-1 (4EBP1) and 40S ribosomal protein S6 (S6RP) by immunohistochemistry. They concluded that the expression levels of p-mTOR and p-S6RP may be potential predictive biomarkers for efficacy of everolimus in patients with metastatic RCC and propose that combining examinations of phosphorylated mTOR, S6RP and/or 4EBP1 may be a potential strategy to select metastatic RCC patients sensitive to mTOR inhibitor treatment [21]. Conversely, Darwish et al. studied hundreds of clear cell RCC patients and showed that cumulative number of altered biomarkers in mTOR pathway is an independent predictor of outcome in patients with clear cell RCC [22]. Moreover, Cho et al. studied tissue specimens obtained from 20 patients affected by advanced clear cell RCC and suggested that phospho-S6 and pAkt expression are promising predictive biomarkers for response to temsirolimus [23].
The mTOR inhibitors are rapamycin and its analogs such as temsirolimus, everolimus and ridaforolimus. They are structural derivatives of the macrocyclic lactone rapamycin (also known as sirolimus) and originally shown to have fungicidal, immunosuppressive and antiproliferative properties. Sirolimus was first approved as an immunosuppressant for patients with solid organ transplants, followed by usage in sirolimus-eluting stents for the prevention of coronary artery restenosis [24]. Recent phase I and II trials have also shown sirolimus to reduce the size of angiomyolipomas in patients with tuberous sclerosis complex and lymphangioleiomyomatosis [25,26]. Notably, temsirolimus, everolimus and ridaforolimus inhibit mTOR by binding to the cytosolic protein FKBP12. All three agents have been evaluated in clinical cancer trials [13,24]. Temsirolimus has been investigated as a treatment for advanced cancer, including metastatic RCC, locally advanced or metastatic breast cancer and mantle cell lymphoma [27,28]. Everolimus has been assessed as a treatment for patients with advanced cancer, including pancreatic neuroendocrine tumors, metastatic breast cancer and metastatic RCC [13,29]. To analyze the effect of those inhibitors will be necessary to evaluate the activation of the main molecular signaling mTOR pathway, in order to identify the possible biomarkers with future predictive value of therapeutic response.
In conclusion, testing mTOR pathway active biomarkers in clear cell RCC may lead to different results based on different techniques used. Clinical trials need an agreement for standard methods on tissue in order to correlate mTOR biomarkers and clinical end-points. The efficacy of new generation of mTOR inhibitors will depend, at least in part, by the methods used. Combining tests of multiple biomarkers will improve the predictiveness of such markers to targeted therapy with mTOR inhibitors. Therefore, we propose a rationale approach to stratify patients affected by clear cell RCC for more likely response to mTOR inhibitors.
Acknowledgements
Internal fundings Matteo Brunelli 2013.
Disclosure of conflict of interest
None to declare.
References
- 1.Hudes GR. Targeting mTOR in renal cell carcinoma. Cancer. 2009;115:2313–2320. doi: 10.1002/cncr.24239. [DOI] [PubMed] [Google Scholar]
- 2.Konings IR, Verweij J, Wiemer EA, Sleijfer S. The applicability of mTOR inhibition in solid tumors. Curr Cancer Drug Targets. 2009;9:439–450. doi: 10.2174/156800909788166556. [DOI] [PubMed] [Google Scholar]
- 3.Brennan DJ, O’Connor DP, Rexhepaj E, Ponten F, Gallagher WM. Antibody-based proteomics: fast-tracking molecular diagnostics in oncology. Nat Rev Cancer. 2010;10:605–617. doi: 10.1038/nrc2902. [DOI] [PubMed] [Google Scholar]
- 4.Gonzalez-Angulo AM, Hennessy BT, Mills GB. Future of personalized medicine in oncology: a systems biology approach. J. Clin. Oncol. 2010;28:2777–2783. doi: 10.1200/JCO.2009.27.0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sawyers CL. The cancer biomarker problem. Nature. 2008;452:548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- 6.Van’t Veer LJ, Bernards R. Enabling personalized cancer medicine through analysis of gene-expression patterns. Nature. 2008;452:564–570. doi: 10.1038/nature06915. [DOI] [PubMed] [Google Scholar]
- 7.Juengel E, Makarević J, Reiter M, Mani J, Tsaur I, Bartsch G, Haferkamp A, Blaheta RA. Resistance to the mTOR Inhibitor Temsirolimus Alters Adhesion and Migration Behavior of Renal Cell Carcinoma Cells through an Integrin alpha5-and Integrin beta3-Dependent Mechanism. Neoplasia. 2014;16:291–300. doi: 10.1016/j.neo.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calvo E, Ravaud A, Bellmunt J. What is the optimal therapy for patients with metastatic renal cell carcinoma who progress on an initial VEGFr-TKI? Cancer Treat Rev. 2013;39:366–374. doi: 10.1016/j.ctrv.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 9.Eisengart LJ, MacVicar GR, Yang XJ. Predictors of response to targeted therapy in renal cell carcinoma. Arch Pathol Lab Med. 2012;136:490–495. doi: 10.5858/arpa.2010-0308-RA. [DOI] [PubMed] [Google Scholar]
- 10.Carew JS, Kelly KR, Nawrocki ST. Mechanisms of mTOR inhibitor resistance in cancer therapy. Target Oncol. 2011;6:17–27. doi: 10.1007/s11523-011-0167-8. [DOI] [PubMed] [Google Scholar]
- 11.Albert S, Serova M, Dreyer C, Sablin MP, Faivre S, Raymond E. New inhibitors of the mammalian target of rapamycin signaling pathway for cancer. Expert Opin Investig Drugs. 2010;19:919–930. doi: 10.1517/13543784.2010.499121. [DOI] [PubMed] [Google Scholar]
- 12.Benjamin D, Colombi M, Moroni C, Hall MN. Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat Rev Drug Discov. 2011;10:868–880. doi: 10.1038/nrd3531. [DOI] [PubMed] [Google Scholar]
- 13.Husseinzadeh HD, Garcia JA. Therapeutic rationale for mTOR inhibition in advanced renal cell carcinoma. Curr Clin Pharmacol. 2011;6:214–221. doi: 10.2174/157488411797189433. [DOI] [PubMed] [Google Scholar]
- 14.Pantuck AJ, Seligson DB, Klatte T, Yu H, Leppert JT, Moore L, O’Toole T, Gibbons J, Belldegrun AS, Figlin RA. Prognostic relevance of the mTOR pathway in renal cell carcinoma: implications for molecular patient selection for targeted therapy. Cancer. 2007;109:2257–2267. doi: 10.1002/cncr.22677. [DOI] [PubMed] [Google Scholar]
- 15.Robb VA, Karbowniczek M, Klein-Szanto AJ, Henske EP. Activation of the mTOR signaling pathway in renal clear cell carcinoma. J Urol. 2007;177:346–352. doi: 10.1016/j.juro.2006.08.076. [DOI] [PubMed] [Google Scholar]
- 16.Cho D, Signoretti S, Regan M, Mier JW, Atkins MB. The role of mammalian target of rapamycin inhibitors in the treatment of advanced renal cancer. Clin Cancer Res. 2007;13:758s–763s. doi: 10.1158/1078-0432.CCR-06-1986. [DOI] [PubMed] [Google Scholar]
- 17.Beuvink I, Boulay A, Fumagalli S, Zilbermann F, Ruetz S, O’Reilly T, Natt F, Hall J, Lane HA, Thomas G. The mTOR inhibitor RAD001 sensitizes tumor cells to DNA-damaged induced apoptosis through inhibition of p21 translation. Cell. 2005;120:747–759. doi: 10.1016/j.cell.2004.12.040. [DOI] [PubMed] [Google Scholar]
- 18.Jefferies HB, Fumagalli S, Dennis PB, Reinhard C, Pearson RB, Thomas G. Rapamycin suppresses 5’TOP mRNA translation through inhibition of p70s6k. EMBO J. 1997;16:3693–3704. doi: 10.1093/emboj/16.12.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 20.Faivre S, Kroemer G, Raymond E. Current development of mTOR inhibitors as anticancer agents. Nat Rev Drug Discov. 2006;5:671–688. doi: 10.1038/nrd2062. [DOI] [PubMed] [Google Scholar]
- 21.Li S, Kong Y, Si L, Chi Z, Cui C, Sheng X, Guo J. Phosphorylation of mTOR and S6RP predicts the efficacy of everolimus in patients with metastatic renal cell carcinoma. BMC Cancer. 2014;14:376. doi: 10.1186/1471-2407-14-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darwish OM, Kapur P, Youssef RF, Bagrodia A, Belsante M, Alhalabi F, Sagalowsky AI, Lotan Y, Margulis V. Cumulative number of altered biomarkers in mammalian target of rapamycin pathway is an independent predictor of outcome in patients with clear cell renal cell carcinoma. Urology. 2013;81:581–586. doi: 10.1016/j.urology.2012.11.030. [DOI] [PubMed] [Google Scholar]
- 23.Cho D, Signoretti S, Dabora S, Regan M, Seeley A, Mariotti M, Youmans A, Polivy A, Mandato L, McDermott D, Stanbridge E, Atkins M. Potential histologic and molecular predictors of response to temsirolimus in patients with advanced renal cell carcinoma. Clin Genitourin Cancer. 2007;5:379–385. doi: 10.3816/CGC.2007.n.020. [DOI] [PubMed] [Google Scholar]
- 24.Dancey JE. Therapeutic targets: MTOR and related pathways. Cancer Biol Ther. 2006;5:1065–1073. doi: 10.4161/cbt.5.9.3175. [DOI] [PubMed] [Google Scholar]
- 25.Bissler JJ, McCormack FX, Young LR, Elwing JM, Chuck G, Leonard JM, Schmithorst VJ, Laor T, Brody AS, Bean J, Salisbury S, Franz DN. Sirolimus for angiomyolipoma in tuberous sclerosis complex or lymphangioleiomyomatosis. N Engl J Med. 2008;358:140–151. doi: 10.1056/NEJMoa063564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davies DM, de Vries PJ, Johnson SR, McCartney DL, Cox JA, Serra AL, Watson PC, Howe CJ, Doyle T, Pointon K, Cross JJ, Tattersfield AE, Kingswood JC, Sampson JR. Sirolimus therapy for angiomyolipoma in tuberous sclerosis and sporadic lymphangioleiomyomatosis: a phase 2 trial. Clin Cancer Res. 2011;17:4071–4081. doi: 10.1158/1078-0432.CCR-11-0445. [DOI] [PubMed] [Google Scholar]
- 27.Atkins MB, Hidalgo M, Stadler WM, Logan TF, Dutcher JP, Hudes GR, Park Y, Liou SH, Marshall B, Boni JP, Dukart G, Sherman ML. Randomized phase II study of multiple dose levels of CCI-779, a novel mammalian target of rapamycin kinase inhibitor, in patients with advanced refractory renal cell carcinoma. J. Clin. Oncol. 2004;22:909–918. doi: 10.1200/JCO.2004.08.185. [DOI] [PubMed] [Google Scholar]
- 28.Hess G, Herbrecht R, Romaguera J, Verhoef G, Crump M, Gisselbrecht C, Laurell A, Offner F, Strahs A, Berkenblit A, Hanushevsky O, Clancy J, Hewes B, Moore L, Coiffier B. Phase III study to evaluate temsirolimus compared with investigator's choice therapy for the treatment of relapsed or refractory mantle cell lymphoma. J. Clin. Oncol. 2009;27:3822–3829. doi: 10.1200/JCO.2008.20.7977. [DOI] [PubMed] [Google Scholar]
- 29.Yao JC, Shah MH, Ito T, Bohas CL, Wolin EM, Van Cutsem E, Hobday TJ, Okusaka T, Capdevila J, de Vries EG, Tomassetti P, Pavel ME, Hoosen S, Haas T, Lincy J, Lebwohl D, Öberg K RAD001 in Advanced Neuroendocrine Tumors, Third Trial (RADIANT-3) Study Group. Everolimus for advanced pancreatic neuroendocrine tumors. N Engl J Med. 2011;364:514–523. doi: 10.1056/NEJMoa1009290. [DOI] [PMC free article] [PubMed] [Google Scholar]


