Abstract
Objective: The present study was conducted to elucidate the prognostic prediction value of the methylation of the RASSF10 promoter in gastric cancer (GC). Methods: A total of 300 patients with GC revealed the methylation degrees of the DNA of the RASSF10 promoter. Methylation-specific PCR (MSP) analysis was performed to qualitatively detect the methylated degrees of the DNA of the RASSF10 promoter of 300 patients with GC. Associations between molecular, clinicopathological and survival data were analyzed. Results: The protein and mRNA expressions of RASSF10 in GC tissues were lower than those in normal gastric mucosal tissues. In the MSP analysis cohort, patients with methylated RASSF10 promoter exhibited significantly shorter median OS than those with unmethylated RASSF10 promoter (P < 0.001). Multivariate survival analysis results showed that methylated RASSF10 promoter was an independent predictor of the survival of patients with GC. Conclusions: The methylation of the RASSF10 promoter could be applied for the clinical prediction of the prognosis of GC.
Keywords: Stomach, neoplasm, ras association domain protein, survival, methylation
Introduction
Development of the malignant diseases is via a multistage process, including gene DNA methylation of CpG islands. Many researchers demonstrated that genetic alterations were the important tumorigenic events, which may be involved in gastric tumorigenesis [1-6]. Promoter methylation is one of the major mechanisms to inactivate tumour-related genes, thereby causing gastric carcinogenesis in theory [7].
The RAS-association domain family (RASSF) proteins can interact with a wide variety of downstream effectors to regulate several signalling pathways important for normal cellular growth and apoptosis [8]. RASSF10 was characterized by the inclusion of an N-terminus, which was described from a predicted sequence with homology to RASSF9/P-CIP1 [9]. RASSF10 gene was identified as a two-exon gene giving rise to a pair transcript encoding a 615 amino acid protein or a single exon gene encoding a shorter 507 amino acid protein [10]. Recently, the expression status of RASSF10 was reported in a wide variety of tissues, including thyroid, pancreas, placenta, heart, lung, and kidney [11]. In addition, researchers also reported that the expression of RASSF10 was found in human bone marrow RNA and amplified the RASSF10 gene from a human liver cDNA library [10,12].
Although there is little published data hinting at RASSF10 function, RASSF10 methylation was demonstrated to be correlated with loss of gene expression in many human tumors. Underhill-Day N et al [12] had assessed the role of RASSF10 in childhood acute lymphoblastic leukemia (ALL) by analysis of the methylation status of CpG islands associated with RASSF promoter regions, and then they found high frequency of methylation (88%) of RASSF10 in childhood T-ALL as well as in leukemia cell lines (100%) [12]. Further, they also found that the lower methylation frequency (16%) of RASSF promoter in childhood B-ALL derived from peripheral blood lymphocytes and bone marrow [12]. Additionally, frequent methylation of the RASSF10 promoter had been detected in both thyroid carcinomas and gliomas [11,13].
In this study, we initially detected the expression differences of RASSF10 in between gastric cancer (GC) tissues and normal gastric mucosal tissues, and then assessed the methylation degrees of RASSF10 promoter in the large scale GC tissues (n = 300) by methylation-specific PCR (MSP) analyses to determine the precise prognostic prediction values of RASSF10 promoter methylation in patients with GC.
Patients and methods
Data source
After the institutional review board of Tianjin Medical University Cancer Hospital (China) approved our study, data from the cancer registry of the Tianjin Cancer Institute were obtained. Data obtained from the registry were listed as follows: age; gender, tumour location, tumour size, depth of tumour invasion (T stage, according to the Sixth Edition of UICC TNM Classification for GC), number of metastatic lymph nodes (N stage, according to the Sixth Edition of UICC TNM Classification for GC), extent of lymph node metastasis, Lauren classification, and follow-up vital status. Oral and written informed consents were also obtained from patients who were included in this study.
Patients and study samples
To analyze RASSF10 promoter methylation, we collected 300 fresh GC tissues from patients with GC and who underwent curative gastrectomy between April 2003 and December 2007 at the Department of Gastroenterology, Tianjin Medical University Cancer Hospital. A cohort of 25 normal gastric mucosal epithelial tissues from normal people was also obtained between 2004 and 2007 at the Department of Endoscopic Examination and Treatment, Tianjin Medical University Cancer Hospital. The tumour and normal gastric mucosal epithelial tissue samples were histologically verified. The patients were not subjected to radiation, chemical or biological treatment before potentially curative gastrectomy was performed. Adjuvant chemotherapy or radiotherapy was not routinely administered to the patients. The clinicopathological characteristics of the two cohorts are summarized in Table 1. Consent regarding the use of tissue samples and records was obtained from each patient. The Institutional Research Ethics Committee of Tianjin Medical University Cancer Hospital approved the study protocol and provided permission to use patient clinical data.
Table 1.
Patient information
| Cohort | RASSF10 promoter methylation | RASSF10 promoter unmethylation |
|---|---|---|
| Gender | ||
| Male | 58 | 143 |
| Female | 33 | 66 |
| Age at surgery | ||
| ≤ 60 | 61 | 119 |
| > 60 | 30 | 90 |
| Tumor size | ||
| < 4.0 | 11 | 28 |
| ≥ 4.0 | 80 | 181 |
| Tumor location | ||
| Upper third | 22 | 49 |
| Middle third | 29 | 52 |
| Lower third | 37 | 93 |
| More than 2/3 stomach | 3 | 15 |
| Depth of tumor invasion (T stage) | ||
| T1 | 0 | 3 |
| T2 | 9 | 19 |
| T3 | 56 | 130 |
| T4 | 26 | 57 |
| Number of metastatic lymph nodes (N stage) | ||
| N0 | 18 | 61 |
| N1 | 33 | 73 |
| N2 | 20 | 44 |
| N3 | 20 | 31 |
| Location of lymph node metastasis | ||
| No | 18 | 61 |
| Perigastric | 30 | 65 |
| Extragastric | 43 | 83 |
| Lauren classification | ||
| Intestinal | 20 | 62 |
| Diffuse | 68 | 143 |
| Mixed | 3 | 4 |
Surgical treatment
Curative resection was defined as the complete absence of grossly visible tumour tissues and metastatic lymph nodes remaining after resection was performed with pathologically negative resection margins. Primary tumors were resected en bloc with limited or extended lymphadenectomy (D1 or D2-3 according to the Japanese Gastric Cancer Association). Surgical specimens were evaluated according to the Sixth UICC TNM Classification for GC.
DNA and RNA extraction
Genomic DNA was extracted from the 300 GC tissues and the 25 normal gastric mucosal tissues by using a QIAamp DNA mini kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Genomic DNA was modified using sodium bisulphite in EZ DNA Methylation-GoldTM kit (Zymo Research, Hornby, Canada). RNA was extracted from 25 of the 300 GC tissues and 25 normal gastric mucosal tissues by using Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions.
Western blotting and semi-quantitative reverse transcription PCR (RT-PCR)
A total of the 25 of 300 GC tissues and 25 normal gastric mucosal tissues were each added to 1 mL of 100 mmol/L Tris/HCl (pH 7.5), 100 mmol/L NaCl, 0.5% sodium deoxycholate, 1 mmol/L ethylenediaminetetraacetic acid, 1% Nonidet P-40, 0.1% sodium dodecyl sulphate and protease inhibitor. After blocking was performed, 50 µg of the sample was incubated for 60 min with goat anti-RASSF10 (Santa Cruz, sc-243928, 1:1000 dilution) at room temperature. A gel imager system (Asia Xingtai Mechanical and Electrical Equipment Company, Beijing, China) was used to analyze images and determine gray values.
The mRNA expression of RASSF10 was detected by subjecting the 25 of 300 GC tissues and the 25 normal gastric mucosal tissues to RT-PCR. Total RNA was reverse-transcribed to cDNA in a 20 µl solution by using a reverse transcription kit (Invitrogen, Carlsbad, CA). The primers designed and utilized for RASSF10 were listed as follows: forward sequence 5’-AGGGTGAGGGTAGAAGAAGTAGG-3’ and reverse sequence 5’-GTTCTAGCTGCCATCAGTCCTT-3’. The GAPDH gene was used as an endogenous control of semi-quantitative DNA-PCR. Primers designed and utilized for GAPDH were listed as follows: forward sequence 5’-GAAGGTGAAGGTCGGAGTC-3’ and reverse sequence 5’-GAAGATG GTGATGGGATTTC-3’. The following PCR cycling conditions were applied: 35 cycles of denaturation at 95°C for 3 min; annealing at 94°C for 30 s; and extension at 56°C for 30 s; and a final extension at 72°C for 8 min. PCR products were electrophoresed on 2% agarose gel with ethidium bromide and visualized using a gel imager system (Asia Xingtai Mechanical and Electrical Equipment Company, Beijing, China).
Methylation-specific PCR (MSP)
A total of 300 GC tissues and 25 normal gastric mucosal tissues were subjected to qualitative methylation analysis of the RASSF10 promoter by methylation-specific PCR (MSP). The following RASSF10 primers were used to detect the methylated (M) or unmethylated (U) alleles of the RASSF10 promoter: for methylated alleles, RASSF10-MF 5’-TAGAGCGTAGTCGTAATCGC-3’ and RASSF10-MR 5’-CCGAAATCTACTAAAACGACG-3’; for unmethylated alleles, RASSF10-UF 5’-TGGGTAGAGTGTAGTTGTAATTGT-3’ and RASSF10-UR 5’-CAACCAAAA TCTACTAAAACAACA-3’. A total of 25 cycles of MSP were performed using Ampli Taq-Gold (methylation-specific primers, annealing temperature 600°C; unmethylation-specific primers, annealing temperature 580°C). MSP primers were initially evaluated to verify whether or not any unbisulphited DNA is amplified, and the specificity of MSP was further confirmed by directly sequencing some PCR products. PCR was resolved using 2% agarose gel. Image J software was used to calculate the relative values of the methylation of the RASSF10 promoter in the 300 GC tissues. The hypermethylation of the RASSF10 promoter was defined as the calculated methylated value of GC tissue but not less than the positive control value.
Follow up
After curative surgery, all of the patients were followed up every three months or six months for two years at the outpatient department; these patients were also followed up every year from the third year to the fifth year. Thereafter, these patients were followed up annually until their death. The median follow up of the entire cohort was 41 months, ranging from 1 month to 104 months. The follow up of the patients included in this study was completed in December 2012. Ultrasonography, CT scans, chest X-ray and endoscopy were performed in every visit.
Statistical analysis
Median overall survival (OS) was determined using Kaplan-Meier method, and log-rank test was performed to determine significance. Potentially important factors in univariate analyses (P < 0.05) were included in multivariate analyses. OS was subjected to multivariate analysis by using Cox proportional hazard model with forward step procedures. Hazard ratios (HR) and 95% confidence interval were calculated. Bayesian information criterion (BIC) in a Cox proportional hazard regression model was calculated in terms of different categories to quantify their discriminatory ability. A small BIC value corresponds to an efficient model to predict outcomes. Significance was set at P < 0.05. Statistical analyses were performed using SPSS 18.0.
Results
Patient demographics
The clinicopathological characteristics of the patients and the methylation status of the RASSF10 promoter are listed in Table 1. The median OS of all patients was 21 months, ranging from 2 months to 95 months. Only 61 (13.32%) patients with GC are alive even after follow up was completed.
Protein and mRNA expressions of RASSF10 in GC tissues and normal gastric mucosal tissues
The mRNA expression of RASSF10 was detected in the 25 of 300 GC tissues and the 25 normal gastric mucosal tissues by RT-PCR (Figure 1A). The relative mRNA expression of RASSF10 in the GC tissues was significantly lower than that in the normal gastric mucosal tissues (0.574 ± 0.082 vs. 1.317 ± 0.202, P = 0.015). The protein expression of RASSF10 was also simultaneously detected in the 25 of 300 GC tissues and the 25 normal gastric mucosal tissues by western blot (Figure 1B). The relative protein expression value of RASSF10 in the GC tissues was significantly lower than that in the normal gastric mucosal tissues (0.336 ± 0.137 vs. 1.547 ± 0.179, P = 0.008).
Figure 1.

A. RASSF10 mRNA expression (RT-PCR) in GC tissues and that in normal gastric mucosal tissues; B. Western Blot analysis for RASSF10 protein expression in GC tissues and that in normal gastric mucosal tissues. (Representation: T, GC tissues; N: normal gastric mucosal tissues).
Methylation of the RASSF10 promoter in GC tissues
We conducted MSP analysis and detected the methylation of the RASSF10 promoter in 300 GC tissues; by contrast, the RASSF10 promoter was not methylated in the 25 normal gastric mucosal tissues (Figure 2). Among the 300 GC tissues, 91 (30.33%) presented methylated RASSF10 promoter, and 209 (69.67%) presented with unmethylated RASSF10 promoter.
Figure 2.
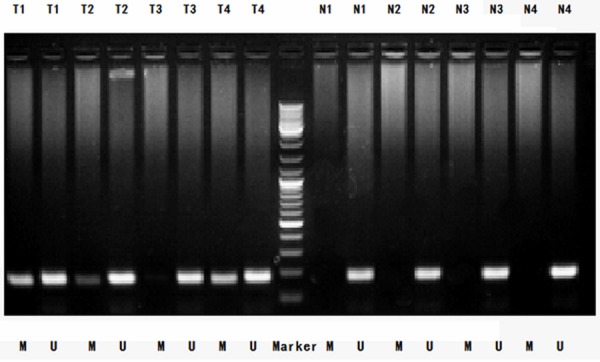
MSP detection of RASSF10 promoter methylation in different GC tissues and normal gastric mucosal tissues. (Representation: T, GC tissues; N, normal gastric mucosal tissues; M, methylation; U, unmethylation).
Survival analysis
In univariate survival analysis, three clinicopathological characteristics were significantly associated with the survival of 300 patients with GC. In MSP analysis, methylated RASSF10 promoter was observed in the same patients. The following characteristics were observed (Table 2): T stage (P < 0.001); N stage (P < 0.001); and location of lymph node metastasis (P < 0.001). In addition, the methylation of the RASSF10 promoter was significantly associated with the OS of patients, as indicated in Kaplan-Meier curves (P < 0.001; Table 2; Figure 3). These factors were included in a multivariate Cox proportional hazard model to adjust the effects of covariates. The methylation of the RASSF10 promoter (HR = 1.687, P < 0.001), N stage (HR = 1.531, P < 0.001), and T stage (HR = 1.433, P = 0.001) were identified as the independent predictors of the OS of patients with GC, as revealed by the multivariate survival analysis. In addition, the methylation of the RASSF10 promoter yielded a smaller BIC value than N stage and T stage, representing the optimal prognostic predictor of GC (Table 2).
Table 2.
Survival analysis of 300 GC patients with MSP detection of RASSF10 promoter methylation
| Variables | Median OS (mo) | X 2 value | Univariate value | HR value | Multivariate P value | BIC value |
|---|---|---|---|---|---|---|
| Gender | ||||||
| Male | 23 | 3.699 | 0.054 | |||
| Female | 18 | |||||
| Age at surgery (years) | ||||||
| ≤ 60 | 19 | 0.139 | 0.709 | |||
| > 60 | 23 | |||||
| Tumor location | ||||||
| Upper third | 22 | 2.232 | 0.526 | |||
| Middle third | 16 | |||||
| Lower third | 24 | |||||
| ≥ 2/3 stomach | 10 | |||||
| Tumor size (cm) | ||||||
| < 4.0 | 26 | 1.032 | 0.310 | |||
| ≥ 4.0 | 21 | |||||
| Lauren classification | ||||||
| Intestinal | 24 | 3.456 | 0.178 | |||
| Diffuse | 20 | |||||
| Mixed | 24 | |||||
| Depth of tumor invasion (T stage) | ||||||
| T1 | 47 | 21.042 | < 0.001 | 1.433 (1.161-1.769) | 0.001 | 77.789 |
| T2 | 24 | |||||
| T3 | 24 | |||||
| T4 | 14 | |||||
| Number of metastatic lymph nodes (N stage) | ||||||
| N0 | 35 | 60.358 | < 0.001 | 1.531 (1.351-1.736) | < 0.001 | 83.090 |
| N1 | 22 | |||||
| N2 | 17 | |||||
| N3 | 9 | |||||
| Location of lymph node metastasis | ||||||
| No | 35 | 29.047 | < 0.001 | |||
| Perigastric | 20 | |||||
| Extragastric | 15 | |||||
| Methylated status of RASSF10 promoter | ||||||
| Methylation | 14 | 17.340 | < 0.001 | 1.687 (1.297-2.196) | < 0.001 | 74.909 |
| Unmethylation | 25 | |||||
Figure 3.

Kaplan-Meier survival curves comparing months of survival in gastric cancer patients are shown for methylated status of RASSF10 promoter (MSP).
Discussion
DNA methylation is a major mechanism by which tumor suppressor genes are inactivated in many kinds of cancer cells, including GC [14]. Tumor suppressor genes involved in cell proliferation are aberrantly inactivated by promoter methylation, which is frequently involved in the processes of canceration [15]. RASSF tumor suppressor genes are elucidated to include 10 members, including an RAS-association domain at C-terminus (RASSF1-6) or N-terminus (RASSF7-10). RASSF10 appears to be the most frequently methylated of the N-terminal RASSFs. To date, authors have elucidated a potential tumor-suppressor role for RASSF10 through frequent epigenetic inactivation of the RASSF10 gene in the three types of cancers [10,11,13]. RASSF10 methylation correlated with loss of gene expression in leukemia and thyroid cancer cell lines, which could be reversed after treatment with 5-aza-2’deoxycytidine [10,11]. Schagdarsurengin et al [11] analyzed the methylation status of RASSF10 promoter to found methylation in 66% (21/32) of primary thyroid carcinomas. Of these, the highest methylation frequency (100%) was observed in medullary thyroid carcinoma and the lowest in undifferentiated thyroid cancer (40%) [11]. None of the normal thyroid controls showed RASSF10 methylation [11]. Hill et al 13 also reported that RASSF10 promoter methylation was detected in 80% grade III and 60% grade IV primary glioblastomas. To our knowledge, few authors investigated the expression and function of RASSF10 gene in GC [16,17]. Wei et al [16] revealed that RASSF10 was silenced in six out of eight gastric cancer cell lines and RASSF10 inhibited tumor growth by blocking activation of the Wnt/β-catenin signaling pathway. Li et al [17] demonstrated that RASSF10 is an epigenetically silenced gene involved in tumor invasion and metastasis in gastric cancer, suggesting that the methylation status of RASSF10 may be a useful indicator to predict the malignant degree of gastric cancer. Similarly, we also demonstrated that RASSF10 expression in GC tissues significantly lower than that in normal gastric mucosal tissues in this study. In addition, we found that 91 of 300 (30.33%) GC tissues presented methylated RASSF10 promoter, and none of normal gastric mucosal tissues presented with methylated RASSF10 promoter. Therefore, we thought these findings have indicated that the tumour-specific methylation of RASSF10 can be performed as an epigenetic biomarker to diagnose cancer.
Currently, promoter methylation has been proposed as a potential diagnostic or prognostic biomarker in various cancers [18]. Theoretically, an optimal prognostic predictor should be detected in large scale patients suffering from all stages of cancer.
To date, the impact of RASSF10 promoter methylation on evaluation the prognosis of GC is not elucidated clearly. With the univariate survival analysis, we found that the survival rate of patients with methylated RASSF10 promoter was significantly different from that of patients with unmethylated RASSF10 promoter; the patients with methylated RASSF10 promoter exhibited shorter OS than the other patients (P < 0.001). Subsequently, the Cox proportional hazard model analysis showed that RASSF10 promoter methylation was an independent indicator of GC patients’ prognosis, indicating the importance of RASSF10 promoter methylation. Additionally, our finding of the methylated status of RASSF10 promoter exhibited a smaller BIC value than those of N stage and T stage. This result indicated that the RASSF10 promoter methylation was the most effective potential prognostic predictor of GC.
Acknowledgements
This study was supported in part by grants from the National Basic Research Program of China (973 Program) (NO. 2010CB529301), the Anticancer Major Projects of Tianjin Municipal Science and Technology Commission (NO. 12ZCDZSY16400), the Key Project of Tianjin Municipal Science and Technology Commission (NO. 13ZCZCSY20300) and the Science Found Program of Tianjin Medical University (NO. 2012KYM01).
Disclosure of conflict of interest
None.
References
- 1.Zang ZJ, Cutcutache I, Poon SL, Zhang SL, McPherson JR, Tao J, Rajasegaran V, Heng HL, Deng N, Gan A, Lim KH, Ong CK, Huang D, Chin SY, Tan IB, Ng CC, Yu W, Wu Y, Lee M, Wu J, Poh D, Wan WK, Rha SY, So J, Salto-Tellez M, Yeoh KG, Wong WK, Zhu YJ, Futreal PA, Pang B, Ruan Y, Hillmer AM, Bertrand D, Nagarajan N, Rozen S, Teh BT, Tan P. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44:570–574. doi: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- 2.Wang K, Kan J, Yuen ST, Shi ST, Chu KM, Law S, Chan TL, Kan Z, Chan AS, Tsui WY, Lee SP, Ho SL, Chan AK, Cheng GH, Roberts PC, Rejto PA, Gibson NW, Pocalyko DJ, Mao M, Xu J, Leung SY. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. 2011;43:1219–1223. doi: 10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- 3.Jones S, Li M, Parsons DW, Zhang X, Wesseling J, Kristel P, Schmidt MK, Markowitz S, Yan H, Bigner D, Hruban RH, Eshleman JR, Iacobuzio-Donahue CA, Goggins M, Maitra A, Malek SN, Powell S, Vogelstein B, Kinzler KW, Velculescu VE, Papadopoulos N. Somatic mutations in the chromatin remodeling gene ARID1A occur in several tumor types. Hum Mutat. 2012;33:100–103. doi: 10.1002/humu.21633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corso G, Velho S, Paredes J, Pedrazzani C, Martins D, Milanezi F, Pascale V, Vindigni C, Pinheiro H, Leite M, Marrelli D, Sousa S, Carneiro F, Oliveira C, Roviello F, Seruca R. Oncogenic mutations in gastric cancer with microsatellite instability. Eur J Cancer. 2011;47:443–451. doi: 10.1016/j.ejca.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 5.Shi J, Yao D, Liu W, Wang N, Lv H, Zhang G, Ji M, Xu L, He N, Shi B, Hou P. Highly frequent PIK3CA amplification is associated with poor prognosis in gastric cancer. BMC Cancer. 2012;12:50. doi: 10.1186/1471-2407-12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi J, Yao D, Liu W, Wang N, Lv H, He N, Shi B, Hou P, Ji M. Frequent gene amplification predicts poor prognosis in gastric cancer. Int J Mol Sci. 2012;13:4714–4726. doi: 10.3390/ijms13044714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qu Y, Dang S, Hou P. Gene methylation in gastric cancer. Clin Chim Acta. 2013;424:53–65. doi: 10.1016/j.cca.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 9.Sherwood V, Manbodh R, Sheppard C, Chalmers AD. RASSF7 is a member of a new family of RAS association domain-containing proteins and is required for completing mitosis. Mol Biol Cell. 2008;19:1772–1782. doi: 10.1091/mbc.E07-07-0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Underhill-Day N, Hill V, Latif F. N-terminal RASSF family: RASSF7-RASSF10. Epigenetics. 2011;6:284–292. doi: 10.4161/epi.6.3.14108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schagdarsurengin U, Richter AM, Wohler C, Dammann RH. Frequent epigenetic inactivation of RASSF10 in thyroid cancer. Epigenetics. 2009;4:571–576. doi: 10.4161/epi.4.8.10056. [DOI] [PubMed] [Google Scholar]
- 12.Hesson LB, Dunwell TL, Cooper WN, Catchpoole D, Brini AT, Chiaramonte R, Griffiths M, Chalmers AD, Maher ER, Latif F. The novel RASSF6 and RASSF10 candidate tumour suppressor genes are frequently epigenetically inactivated in childhood leukaemias. Mol Cancer. 2009;8:42. doi: 10.1186/1476-4598-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill VK, Underhill-Day N, Krex D, Robel K, Sangan CB, Summersgill HR, Morris M, Gentle D, Chalmers AD, Maher ER, Latif F. Epigenetic inactivation of the RASSF10 candidate tumour suppressor gene is a frequent and early event in gliomagenesis. Oncogene. 2011;30:978–989. doi: 10.1038/onc.2010.471. [DOI] [PubMed] [Google Scholar]
- 14.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 15.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 16.Wei Z, Chen X, Chen J, Wang W, Xu X, Cai Q. RASSF10 is epigenetically silenced and functions as a tumor suppressor in gastric cancer. Biochem Biophys Res Commun. 2013;432:632–637. doi: 10.1016/j.bbrc.2013.02.033. [DOI] [PubMed] [Google Scholar]
- 17.Li Z, Chang X, Dai D, Deng P, Sun Q. RASSF10 is an epigenetically silenced tumor suppressor in gastric cancer. Oncol Rep. 2014;31:1661–1668. doi: 10.3892/or.2014.3039. [DOI] [PubMed] [Google Scholar]
- 18.Meng W, Huebner A, Shabsigh A, Chakravarti A, Lautenschlaeger T. Combined RASSF1A and RASSF2A Promoter Methylation Analysis as Diagnostic Biomarker for Bladder Cancer. Mol Biol Int. 2012;2012:701814. doi: 10.1155/2012/701814. [DOI] [PMC free article] [PubMed] [Google Scholar]


