Abstract
Two gene clusters are tightly linked in a narrow region of chromosome 22q11.23: the macrophage migration inhibitory factor (MIF) gene family and the glutathione S-transferase theta class. Within 120 kb in this region, two 30-kb deletions reach high frequencies in human populations. This gives rise to four haplotypic arrangements, which modulate the number of genes in both families. The variable patterns of linkage disequilibrium (LD) between these copy number variants (CNVs) in diverse human populations remain poorly understood. We analyzed 2469 individuals belonging to 27 human populations with different ethnic origins. Then we correlated the genetic variability of 22q11.23 CNVs with environmental variables. We confirmed an increasing strength of LD from Africa to Asia and to Europe. Further, we highlighted strongly significant correlations between the frequency of one of the haplotypes and pigmentation-related variables: skin color (R2=0.675, P<0.001), distance from the equator (R2=0.454, P<0.001), UVA radiation (R2=0.439, P<0.001), and UVB radiation (R2=0.313, P=0.002). The fact that all MIF-related genes are retained on this haplotype and the evidences gleaned from experimental systems seem to agree with the role of MIF-related genes in melanogenesis. As such, we propose a model that explains the geographic and ethnic distribution of 22q11.23 CNVs among human populations, assuming that MIF-related gene dosage could be associated with adaptation to low UV radiation.
Introduction
Recent investigations have demonstrated that copy number variants (CNVs) are recurrent elements in the human genome and are strongly associated with human phenotypic variation.1, 2 CNVs can be either common or rare, and they may account for disease susceptibility and population diversity.3, 4 The frequencies of common CNVs vary across human populations, and in some cases these variants can influence adaptation processes and exposure to natural selection.5, 6
In 2009, Zhao et al7 analyzed two high-frequency CNVs, which were located on chromosome 22q11.23. This genomic region is a complex locus; the genes for glutathione S-transferase (GST) theta class and the macrophage migration inhibitory factor (MIF) gene family are located within it.8 The entire region likely originated from an ancestral duplication event.7 The GST theta class includes three genes (GSTT1, GSTT2, and GSTT2B) that encode for the GST theta enzymes. Molecular studies on the GST theta class have indicated that these genes may affect an individual's predisposition to diseases related to oxidative stress.9, 10 The MIF gene family is less characterized than the GST theta class. However, several studies have analyzed the role MIF has in various molecular pathways, such as the innate and the adaptive immune response, the regulation of redox balance, and melanogenesis.11 The gene cluster also includes DDT (D-dopachrome tautomerase), which encodes for an enzyme initially involved in melanin biosynthesis.12 Recently, some authors have concluded that DDT is also a functional homolog of MIF and have proposed to designate it MIF-2.13, 14 Furthermore, another gene similar to MIF and DDT, DDTL (D-dopachrome tautomerase-like), is also located in this region. Unfortunately, information on its product is currently limited. Regarding the evolution of MIF gene family, different studies have analyzed this topic in different species, indicating its complex phylogenetic history.15, 16, 17
The two high-frequency CNVs in chromosome 22q11.23 encompass two 30-kb regions separated by 55 kb. The first deletion polymorphism (hereafter indicated as CNV_22q11.23_A; chr22:g.24273998_24311657del in GRCh37/hg19) removes the entire GSTT2B and part of DDTL gene, whereas the second CNV (hereafter indicated as CNV_22q11.23_B; chr22: g.24366215_24395305del) removes GSTT1 and the provisional locus LOC391322 (Supplementary Figure S1). According to the NCBI Gene database, LOC391322 is a protein-coding gene, described as another DDTL (information available at http://www.ncbi.nlm.nih.gov/gene/391322). The analyses by Zhao et al7 have indicated that alleles at each of these CNVs reach high frequencies among human populations. Most interestingly, the two CNVs maintain a strong linkage disequilibrium (LD) across populations of European and Asian descent.7 The presence of a high LD between deletion polymorphisms is rare,7 and the pattern observed in chromosome 22q11.23 suggests that an adaptation mechanism may be linked to the function of the GST theta class or the MIF gene family or both.7
In order to understand the mechanisms that contribute to the current pattern of genetic diversity observed in 22q11.23 CNVs, we considered 2469 individuals belonging to 27 human populations. Then we correlated the observed genetic variability with variables that summarize physical environmental conditions. Our analysis led us to conclude that the pattern of haplotype arrangements observed in chromosome 22q11.23 and their frequencies in world populations are in agreement with a model based on pigmentation-related mechanisms.
Materials and methods
Samples
A total of 874 unrelated adult individuals of both sexes from different human populations were typed: Amhara (n=103) and Oromo (n=98) from Ethiopia; Bamileke (n=30) from Cameroon; Cayapas (n=182), Colorados (n=83), and African Ecuadorians (n=185) from Ecuador; Italians from Italy (n=105); and three ethnically diverse samples from Russia, ie, Perm Russians (n=36), Khanty&Mansi (n=28), and Chukchee (n=24). Information about the classification of these human groups and DNA extraction procedures is available through previous studies.18, 19, 20, 21 We exported additional data from the databases of HapMap and 1000 Genomes Projects.22, 23 Specifically, we extracted the HapMap CNV data from a bulk download (available at ftp://ftp.ncbi.nlm.nih.gov/hapmap/cnv_data/). For 1000 Genomes Project, we extracted the information about the 22q11.23 CNVs from a VCF (Variant Call Format) of the chromosomal region chr22:g.24000000_24500000. Thereafter, we considered the 11 HapMap populations: ASW (African Ancestry in Southwest US), CEU (Utah residents with northern and western European ancestry from the CEPH collection), CHB (Han Chinese in Beijing), CHD (Chinese in Metropolitan Denver, Colorado), GIH (Gujarati Indians in Houston, Texas), JPT (Japanese in Toyko, Japan), LWK (Luhya in Webuye, Kenya), MEX (Mexican Ancestry in Los Angeles, CA), MKK (Maasai in Kinyawa, Kenya), TSI (Tuscan in Italy), and YRI (Yoruban in Ibadan, Nigeria). From the 1000 Genomes Project, we considered the following populations: CHS (Han Chinese South), CLM (Colombian in Medellin, Colombia), GBR (British from England and Scotland), IBS (Iberian populations in Spain), FIN (Finnish from Finland), and PUR (Puerto Rican in Puerto Rico). The combination of genotyped and database samples allowed us to reach 2469 individuals, belonging to 27 human populations. In order to perform an ethnicity-based analysis, we stratified the populations according to their ethno-geographic origin: African origin (Amhara, African Ecuadorians, Bamileke, Oromo, ASW, LWK, MKK, YRI); American origin (Cayapa, Colorado, CLM, MEX, PUR); Central Asian origin (Khanty&Mansi, GIH); East Asian origin (Chukchee, CHB, CHD, CHS, JPT); and European origin (Italians, CEU, FIN, GBR, IBS, PER).
Genotyping
Genotyping of 22q11.23 CNVs was achieved by the TaqMan Copy Number Assay from Applied Biosystems Inc. (Foster City, CA, USA). For CNV_22q11.23_A (chr22:g.24273998_24311657del), a TaqMan Custom Assay was used (Applied Biosystems). The design of the TaqMan Custom Assay was performed using GeneAssist (Applied Biosystems) and considering GSTT2B locus as target region. For CNV_22q11.23_B (chr22:g.24366215_24395305del), the predesigned Copy Number Assay ID is Hs00705316 (Applied Biosystems). Details about experimental conditions were described previously.24 Two blank controls in each 96-well plate were used for the assay quality control. The genotypes call was performed using the CopyCaller software version 1.0 (Applied Biosystems). CNV genotyping is well known to be problematic, particularly so when data are combined from studies using DNA samples of different sources (eg, cell lines versus whole blood) and typed using different CNV detection assays. To ensure reliability of the results, approximately 10% of the samples were randomly selected and analyzed independently using PCR-based methods: in all cases, the outcome was concordant. Moreover, we also analyzed four HapMap individuals (NA10839, NA12753, NA12145, and NA12003) with TaqMan Copy Number Assays (Supplementary Table S1), in order to confirm the publicly available CNV genotypes. In all cases, our outcomes confirmed the HapMap data. Further details about the reliability of CNV genotypes in HapMap and 1000 Genomes Project are available in their reference articles.25, 26
Estimation of geographical distances
To estimate the geographical distance of the populations, we considered the geographical coordinates of their present (eg, Tuscany for TSI) or original (eg, Liberia for African-Ecuadorians) location, as appropriate (Supplementary Table S2). To evaluate the distance from the equator, we considered the latitude of the geographical coordinates of each population. Regarding the distance from East Africa (Afar region in Ethiopia; coordinates: 11.921103, 40.968018), we considered known routes of human migrations across land masses and used the ArcGIS software from Environmental Systems Research Institute, Inc. (Redlands, CA, USA).
Estimation of skin color classes
To quantify the skin color for each population, we took into account the data available in Biasutti.27 A map of the world obtained from the same data set is reported by Barsh.28 Considering the distribution of the skin color across the globe, we assigned color skin classes from 1 (darkest skin) to 8 (lightest skin) to the analyzed populations.
Estimation of UVA and UVB radiation classes
To quantify the UVA and UVB radiation class for each considered population, we used the data available in Jablonski and Chaplin.29 Considering the distribution of the UVA and UVB radiation across the globe, we assigned UV radiation classes from 1 (highest radiation) to 10 (lowest radiation) to the analyzed populations.
Statistical analyses
Haploview version 4.2 was used to estimate allele and haplotype frequencies, to calculate coefficients of pairwise LD (D', R2), and to verify the Hardy-Weinberg equilibrium.30 Pairwise FST values were calculated using Arlequin 3.5.1.2.31 Multi-dimensional scaling analysis, using FST values and correlation analysis were performed using the SPSS 15.0 (SPSS, Chicago, IL, USA). BLAST was used to align and calculate the amino-acid similarity of the proteins encoded by MIF, DDT, DDTL, and LOC391322. Fisher's exact test was used to verify the significance of the co-expression of MIF-related genes. To analyze the allele differences (DFs) of loss-of function variants, we used the method proposed by Hofer et al.32 For each allele i, we computed the average allele frequency pij within each geographic origin group j, as well as the difference in the average frequency computed over all other populations via the equation IDFI=Ipij−p-ijI, where p-ij is the average frequency of allele i in all populations not belonging to the geographic region j. We used the data of 1000 Genomes Project to calculate the allele frequencies of all variants located in MIF-related genes in the four ethnic groups (ie, Africa, America, East Asia, and Europe).
Results
Supplementary Figure S1 displays the genomic arrangement of the CNVs analyzed here and their haplotype combinations: the A*1_B*1 haplotype, in which four MIF-related genes (MIF, DDT, DDTL, and LOC391322) are present; the A*1_B*0 haplotype, in which DDTL is disrupted and three MIF-related genes remain; the A*0_B*1 haplotype, in which LOC391322 is deleted and three MIF-related genes remain; and the A*0_B*0 haplotype, in which LOC391322 is deleted, DDTL is disrupted and only two MIF-related genes are preserved.
Genotype distributions (Table 1) were consistent with Hardy-Weinberg expectations for both loci and populations. As to the inter-ethnic differences, the allele *0 of CNV_22q11.23_A ranged from 0.383 (Amhara) to 0.714 (Cayapas), whereas the allele *0 of CNV_22q11.23_B ranged from 0.271 (Chukchee) to 0.670 (Amhara).
Table 1. Genotype and allele frequency distributions of CNVs in chromosome 22q11.23 among 10 human populations originally examined in this study.
| CNV_22q11.23_A | CNV_22q11.23_B | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Population | N | *1/*1 | *1/*0 | *0/*0 | *0 | *1/*1 | *1/*0 | *0/*0 | *0 |
| Amhara | 103 | 42 | 43 | 18 | 0.383 | 7 | 54 | 42 | 0.670 |
| Oromo | 98 | 38 | 38 | 22 | 0.418 | 18 | 46 | 34 | 0.582 |
| Italians | 105 | 20 | 37 | 48 | 0.633 | 22 | 49 | 34 | 0.557 |
| Bamileke | 30 | 8 | 10 | 12 | 0.567 | 11 | 14 | 5 | 0.400 |
| Cayapas | 182 | 18 | 68 | 96 | 0.714 | 69 | 94 | 19 | 0.362 |
| Colorados | 83 | 19 | 27 | 37 | 0.608 | 25 | 46 | 12 | 0.422 |
| Afroecuadorians | 185 | 38 | 95 | 52 | 0.538 | 57 | 102 | 26 | 0.416 |
| Perm Russians | 36 | 4 | 27 | 4 | 0.500 | 1 | 18 | 17 | 0.722 |
| Khanty&Mansi | 28 | 4 | 17 | 4 | 0.500 | 3 | 12 | 10 | 0.64 |
| Chukchee | 24 | 1 | 17 | 6 | 0.604 | 2 | 9 | 13 | 0.729 |
Abbreviations: N, number of analyzed individuals; *1, presence allele; *0, deletion allele.
Table 2 reports the estimated haplotype frequencies and LD parameters for the same populations along with 17 populations for which individual genotype data were available in public databases. The lowest LD coefficients were observed in populations of African descent (D'=0.015–0.247; R2=0–0.072), whereas populations of European descent showed the highest degree of LD (D'=0.624-1; R2=0.210–0.743). East Asian populations had a lower LD (D'=0.433–0.573; R2=0.085–0.139) than European populations. Among American and Central Asian populations, a marked heterogeneity related to geography was observed. In America, Central populations (CLM, MEX, PUR) showed relatively high LD coefficients between 22q11.23 CNVs (D'=0.293–0.622; R2=0.068–0.114), whereas no LD (D'=0.049–0.051; R2=0.001–0.001) was present in Southern populations (Cayapas, Colorados). In Central Asia, Khanty&Mansi, two North-Western Siberian indigenous populations with high affinities, showed no LD between 22q11.23 CNVs, whereas GIH, a Southern population, showed high LD coefficients (D'=0.777; R2=0.330). In the vast majority of cases, the LD between 22q11.23 CNVs was due to a lower than expected frequency of the A*1_B*1 haplotype (Table 2, column 4).
Table 2. Haplotype frequency distribution and LD indexes for CNVs in chromosome 22q11.23 among the analyzed human populations and populations of HapMap and 1000 Genomes Projects.
| Haplotypes | ||||||||
|---|---|---|---|---|---|---|---|---|
| Population | Source | Origin | A*1_B*1 | A*0_B*1 | A*1_B*0 | A*0_B*0 | D' | R2 |
| Afroecuadorians | Present study | Africa | 0.221 (0.270) | 0.344 | 0.241 | 0.194 | 0.172 | 0.027 |
| Amhara | Present study | Africa | 0.158 (0.204) | 0.172 | 0.459 | 0.211 | 0.224 | 0.040 |
| ASW | HapMap | Africa | 0.238 (0.304) | 0.322 | 0.305 | 0.135 | 0.329 | 0.072 |
| Bamileke | Present study | Africa | 0.257 (0.260) | 0.343 | 0.177 | 0.223 | 0.015 | 0 |
| LWK | HapMap | Africa | 0.270 (0.265) | 0.202 | 0.292 | 0.236 | 0.024 | 0 |
| MKK | HapMap | Africa | 0.179 (0.238) | 0.254 | 0.372 | 0.195 | 0.247 | 0.058 |
| Oromo | Present study | Africa | 0.202 (0.243) | 0.216 | 0.38 | 0.202 | 0.168 | 0.028 |
| YRI | HapMap | Africa | 0.246 (0.243) | 0.194 | 0.306 | 0.254 | 0.015 | 0 |
| Cayapas | Present study | America | 0.184 (0.182) | 0.456 | 0.096 | 0.264 | 0.049 | 0.001 |
| CLM | 1000 Genomes | America | 0.152 (0.216) | 0.239 | 0.398 | 0.211 | 0.293 | 0.068 |
| Colorados | Present study | America | 0.238 (0.226) | 0.34 | 0.159 | 0.263 | 0.051 | 0.001 |
| MEX | HapMap | America | 0.130 (0.273) | 0.415 | 0.369 | 0.086 | 0.622 | 0.327 |
| PUR | 1000 Genomes | America | 0.096 (0.178) | 0.34 | 0.313 | 0.251 | 0.461 | 0.114 |
| GIH | HapMap | Central Asia | 0.036 (0.163) | 0.515 | 0.267 | 0.182 | 0.777 | 0.330 |
| Khanty&Mansi | Present study | Central Asia | 0.179 (0.180) | 0.179 | 0.321 | 0.321 | 0 | 0 |
| CHB | HapMap | East Asia | 0.073 (0.172) | 0.266 | 0.432 | 0.229 | 0.573 | 0.171 |
| CHD | HapMap | East Asia | 0.081 (0.168) | 0.26 | 0.414 | 0.245 | 0.52 | 0.138 |
| CHS | 1000 Genomes | East Asia | 0.070 (0.030) | 0.215 | 0.035 | 0.68 | 0.538 | 0.085 |
| Chukchee | Present study | East Asia | 0.178 (0.107) | 0.093 | 0.218 | 0.511 | 0.433 | 0.106 |
| JPT | HapMap | East Asia | 0.074 (0.166) | 0.269 | 0.403 | 0.254 | 0.555 | 0.139 |
| CEU | HapMap | Europe | 0.03 (0.205) | 0.652 | 0.289 | 0.029 | 0.869 | 0.743 |
| FIN | 1000 Genomes | Europe | 0.046 (0.135) | 0.615 | 0.158 | 0.181 | 0.659 | 0.218 |
| GBR | 1000 Genomes | Europe | 0.054 (0.156) | 0.502 | 0.227 | 0.217 | 0.656 | 0.210 |
| IBS | 1000 Genomes | Europe | 0 (0.116) | 0.464 | 0.25 | 0.286 | 1 | 0.289 |
| Italians | Present study | Europe | 0.044 (0.162) | 0.399 | 0.36 | 0.197 | 0.752 | 0.306 |
| Perm Russians | Present study | Europe | 0.023 (0.139) | 0.254 | 0.463 | 0.26 | 0.826 | 0.248 |
| TSI | HapMap | Europe | 0.089 (0.237) | 0.496 | 0.322 | 0.093 | 0.624 | 0.389 |
For A*1_B*1 haplotype, the frequencies expected under null LD are also reported in parentheses. For the definition of the acronyms, see Material and Methods.
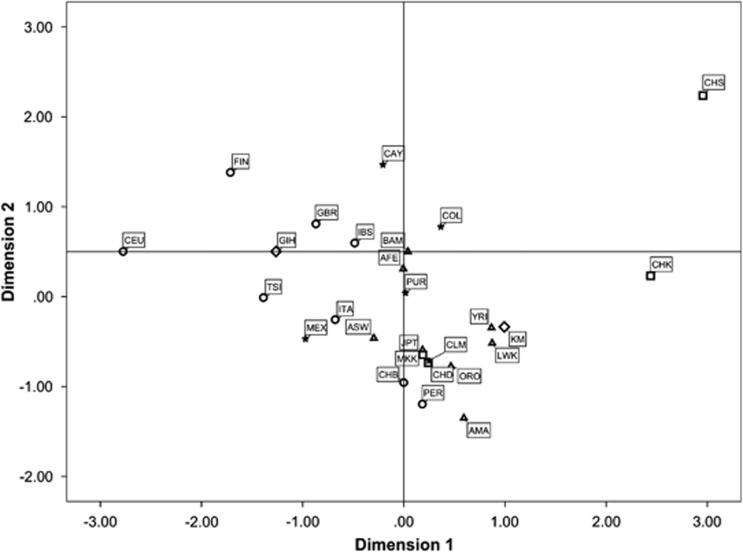
Multidimensional scaling analysis based on FST values confirmed that shared ethnicity is not a fundamental factor for explaining the similarities in 22q11.23 CNV haplotype frequencies (Figure 1). Indeed, only a loose European cluster (circles) seems to be present, whereas the remaining populations do not cluster according to their ethnic/continental provenance. The proximity of Europeans in the plot can be attributed to the sharing of low frequencies of A*1_B*1 haplotype (range 0–0.089) and A*0_B*0 (range 0.029–0.286), thus generating among the highest D' values.
Figure 1.
Multidimensional scaling based on FST distances. African populations are represented by triangles, Americans by stars, Central Asians by diamonds, East Asians by squares, and Europeans by circles.
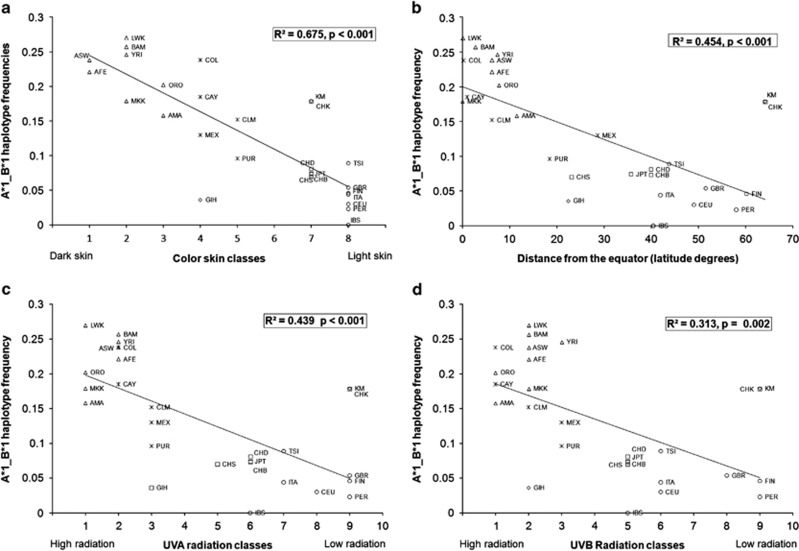
In order to search for possible correlates of the pattern of diversity observed for 22q11.23 CNVs, we regressed the haplotype frequencies against environmental variables obtained for each population based on its present or original place of residence, as appropriate (Supplementary Table S2). Highly significant correlations were observed between the frequency of the haplotype carrying both *1 alleles (A*1_B*1) and pigmentation-related variables (Figure 2).
Figure 2.
Correlation analyses between the frequency of haplotype A*1_B*1 and pigmentation-related variables. African populations are represented by triangles, Americans by stars, Central Asians by diamonds, East Asians by squares, and Europeans by circles. (a) Correlation between the frequency of A*1_B*1 and skin color classes as proposed by Biasutti.27 (b) Correlation between the frequency of A*1_B*1 and distance from the equator. (c, d) Correlations between the frequency of A*1_B*1 and UVA and UVB radiation classes, respectively, as defined by Jablonsky and Chaplin.29 AFE: African-Ecuadorians; BAM: BAmileke; ORO: Oromo; AMA: Amhara; COL: Colorados; CAY: Cayapas; KM: Kanthy&Mansi; CHK: Chukchee; ITA: Italians; PER: Perm Russians. For the definition of the others acronyms, see Material and Methods.
Among the variables, the highest correlation was found with skin color (R2=0.675, P<0.001), when using the skin color distribution by Biasutti.27 Darker skins were correlated with higher A*1_B*1 haplotype frequencies. A significant correlation was also observed with the distance from the equator: less distance was associated with high A*1_B*1 haplotype frequencies (R2=0.454, P<0.001). We also analyzed the correlation of A*1_B*1 haplotype frequencies with UV radiation, using data on the UVA and UVB radiation classes.29 High UVA and UVB radiations were correlated with high frequencies of the A*1_B*1 haplotype (UVA: R2=0.439, P<0.001; UVB: R2=0.313, P=0.002).
In Supplementary Table S3, we report the pairwise correlations between pigmentation-related variables. Although all correlations were strong, interestingly the correlations with skin color were not as strong as the others. Thus, the fact that this variable is the most strongly correlated with the frequencies of haplotype A*1_B*1 reinforces the idea that it is coupled with this phenotypic trait.
In order to evaluate the above correlations in the context of other non-linked loci (SNPs) whose association with human pigmentation is also supported by functional studies,33 we correlated the allele frequencies recorded in the 17 populations of the HapMap and 1000 Genomes Projects at these loci, utilizing the same pigmentation-related variables (Table 3). In this subset of populations, none of these SNPs showed correlations as high as that of the A*1_B*1 haplotype. This is partly because some of the assayed SNPs follow a pattern of variation related to pigmentation, but this is restricted to certain continents.34, 35 Frequencies of the haplotype A*1_B*1 above average are shared by the heavily pigmented UV-exposed populations of both Africa and South America, whereas frequencies below average are found among lightly pigmented populations living at middle latitudes in both Asia and Europe. Also, the correlation coefficients for A*1_B*1 were even higher in the subset of 17 populations than they were in the full set due to the presence, in the latter, of some outliers (Figure 2). This observation denotes the presence of quite expected confounding factors.
Table 3. Correlations between pigmentation-related variables and genetic variants previously associated with human pigmentation in the 17 populations of the HapMap and 1000 Genomes Projects.
| Gene/location | Variants | Skin color | Distance from the equator | UVA | UVB |
|---|---|---|---|---|---|
| 22q11.23 | A*1_B*1 | 0.749 (<0.001) | 0.652 (<0.001) | 0.585 (<0.001) | 0.406 (0.006) |
| TYR | rs1042602 | 0.251 (0.057) | 0.163 (0.136) | 0.165 (0.133) | 0.081 (0.304) |
| rs1126809 | 0.326 (0.033) | 0.362 (0.023) | 0.342 (0.028) | 0.252 (0.068) | |
| TYRP1 | rs1408799 | 0.111 (0.191) | 0.165 (0.105) | 0.172 (0.098) | 0.166 (0.104) |
| rs2733832 | 0.241 (0.045) | 0.334 (0.015) | 0.304 (0.022) | 0.262 (0.036) | |
| OCA2 | rs1800401 | 0.376 (0.020) | 0.195 (0.114) | 0.220 (0.091) | 0.153 (0.167) |
| rs1800407 | 0.258 (0.044) | 0.170 (0.112) | 0.173 (0.109) | 0.103 (0.225) | |
| rs1800414 | 0.102 (0.227) | 0.026 (0.551) | 0.035 (0.491) | 0.024 (0.569) | |
| SLC45A2 | rs26722 | 0.015 (0.637) | 0.001 (0.926) | 0.007 (0.758) | 0.024 (0.550) |
| rs16891982 | 0.348 (0.026) | 0.392 (0.017) | 0.406 (0.014) | 0.318 (0.036) | |
| SLC24A5 | rs1426654 | 0.186 (0.083) | 0.222 (0.056) | 0.205 (0.068) | 0.139 (0.141) |
| KITLG | rs642742 | 0.695 (<0.001) | 0.517 (0.001) | 0.476 (0.002) | 0.282 (0.028) |
The results for the 22q11.23 CNVs on the same population subset are reported on top. For each entry, the R2 value and the corresponding P value (in parentheses) are reported. For the definitions of pigmentation-related variables (ie, skin color, distance from the equator, UVA, and UVB), see Material and Methods.
Additionally, we noticed that the considered variables follow a trend that coincides the distribution of land masses. Specifically, land masses were located along the same major axes of the dispersal routes out of Africa and the associated serial founder effects.36, 37 We wanted to test the correlation between A*1_B*1 haplotype frequency and the geographical distance from East Africa (Afar region in Ethiopia). This did not indicate any regular trend of frequency change (Supplementary Figure S2), as expected under the founder model, supporting that the pigmentation-related variables were better predictors than distance of A*1_B*1 frequencies.
Finally, we verified whether there are loss-of-function variants with large allele differences (DFs) among human populations that may confound our model (Supplementary Table S4). No loss-of-function variants with large DF are present in MIF-related genes.
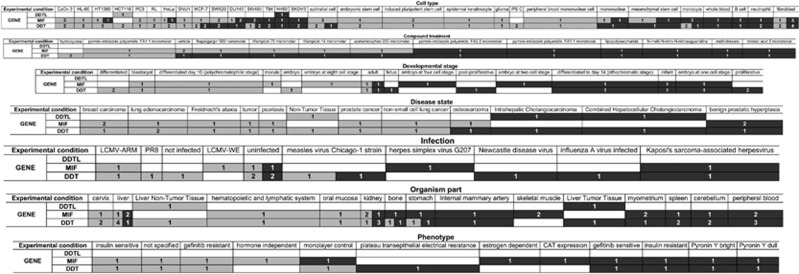
In order to achieve insight into the functional homology of the provisional locus LOC391322, deleted by CNV_22q11.23_B (Supplementary Figure S1), and the other MIF-related genes, we analyzed the similarity of the amino-acid sequences. The protein encoded by LOC391322 showed a 95% amino-acid similarity with the products of both DDT and DDTL and a lower similarity with MIF (39%) (Figure 3). Furthermore, we performed a comparative analysis of expression of MIF-related genes using the Gene Expression Atlas database (available at http://www.ebi.ac.uk/gxa/). The heatmap is reported in Figure 4. Most of the data relates to MIF and DDT expression, with a little relating to DDTL, and none to LOC391322. When counting the number of experimental conditions in which the three (or two) genes showed a similar shift in expression, we observed significant signs of co-regulation (P<0.001). Overall the MIF-related genes were co-expressed in 77% of the experimental conditions (Cell type: 64% Compound treatment: 58% Developmental stage: 100% Disease state: 92% Infection: 100% Organism part: 86% Phenotype: 88%).
Figure 3.
Alignment of the proteins encoded by the four members of the MIF gene family residing in 22q11.23. The alignment was obtained with BLAST using the following sequences: MIF: P14174 – human MIF; DDT: P30046 – human DOPD; DDTL: A6NHG4 – human DDTL; and LOC391322: 2013031826IQRD3TSY. The amino-acid sequence of LOC391322 (2013031826IQRD3TSY) was downloaded from the UCSC database. Asterisks represent sequence similarity, colons indicate conservation between groups of strongly similar properties, and periods highlighted conservation between groups of weakly similar properties.
Figure 4.
Heatmap of expression analysis of MIF-related genes obtained from the Gene Expression Atlas database. In each cell, the number of studies is reported. Overexpression is represented by light grey, and the underexpression is represented by dark grey. Cells split with light and dark grey represent experimental conditions with conflicting data.
Discussion
In 2009, Zhao et al7 indicated that European populations have a high LD between 22q11.23 CNVs. Our data confirmed that European populations do have a high LD between 22q11.23 CNVs, whereas African populations do not display an LD between 22q11.23 CNVs, and Asian populations have an intermediate LD. These data are compatible with the effect of the younger history for the Asian and European as compared with the African population, further enhanced by a bottleneck that affected mainly the Europeans.38 However, they are also compatible with a concurrent selection acting against one of the four CNV-based haplotypes (the A*1_B*1 haplotype) out of the African continent. In fact, in most cases the LD between 22q11.23 CNVs results in a lower than expected A*1_B*1 haplotype frequency.
Through analysis of the genetic diversity of human populations, we observed that ethnicity does not represent a determinant of the haplotype frequency distribution. Conversely, we found that the frequencies of haplotype without deletions (A*1_B*1) are significantly correlated with pigmentation-related variables. Skin color variation among the analyzed populations explains 67.5% of the variance of the A*1_B*1 haplotype frequencies,while distance from the equator explains only 45.4%. Besides, UVA and UVB radiations explain 43.9 and 31.9% of the variance, respectively. These outcomes strongly support the hypothesis that the 22q11.23 chromosomal region might be involved in a selection mechanism related to pigmentation. As a first test to rule out that the correlations observed are not solely due to population structure or demography, we considered A*1_B*1 frequencies as a function of the distance from eastern Africa with a not significant dependence.
Experimental evidence suggests that the MIF gene family is involved in a molecular pathway related to melanogenesis. Shimizu et al39 observed that MIF production in cultured keratinocytes was increased by UVB exposure. Sonesson et al40 demonstrated that DDT is also expressed in the skin and its activity increased approximately twofold after UVB exposure. In 2004, Watanabe et al41 indicated that MIF is involved in the expression of matrix metalloproteinases (MMPs), which are responsible for dermal photoaging in the human skin and that MIF-deficient mice achieved a greater reduction of the UVA-related expression of MMP-13 than wild-type mice. Furthermore, several clinical studies have supported the role of MIF in the skin, specifically in relation to skin inflammation, immune response, disease, tumorigenesis, and cutaneous wound healing.42 Finally, a recent study by Enomoto et al43 indicated that UVB induces MIF-mediated melanogenesis in keratinocytes. This literature suggests that MIF and DDT have a role in the skin's response to UV, confirming that DDT is a functional homolog of MIF.13, 14 Our data on the amino-acid sequences of the MIF gene family indicated that both DDTL and LOC391322 proteins have a 95% amino-acid similarity with DDT, supporting a strong homology with DDT and MIF functions. Furthermore, our gene expression analysis indicated that similar patterns are still present among MIF-related genes in different experimental conditions.
In accordance with these functional data, chromosome 22q11.23 has four MIF-related genes whose products may be related to melanogenesis and its adaptive value, possibly in multiple ways. Our hypothesis, which is based on the inducibility of MIF-mediated melanogenesis, posits that the output of four MIF-related genes is excessive in populations exposed and adapted to low UV radiation (ie, Europeans and Asians). Our correlation analysis indicated that a haplotype with four MIF-related genes (A*1_B*1) has the highest frequency in strongly pigmented populations, and its frequencies decrease in populations with lighter skin. This suggests that haplotype A*1_B*1 may be disadvantageous to populations exposed and adapted to low UV radiation. Thus this haplotype is likely lost over generations.
In our model, in the presence of multiple pathways contributing to skin pigmentation, exposure to high UV radiations in Africa did not directionally select any of the four haplotypes. The inducibility of the MIF system could lead to a balanced functionality of the four haplotypes under high UV radiation, justifying the coexistence of all four haplotypes at polymorphic frequencies in African populations and the absence of LD. In this scenario, haplotype frequencies evolved along two different paths, as populations became exposed and adapted to low UV radiation in other continents. In Europeans, the reduction of the A*1_B*1 haplotype was associated with an increase of the A*0_B*1, generating a strong LD. In East Asians, the reduction of the A*1_B*1 haplotype was associated with an increase of the A*1_B*0 haplotype, also generating an LD between 22q11.23 CNVs (although it was weaker than the LD observed in Europeans).
As to America, the haplotype with four MIF-related genes (ie, A*1_B*1) was likely present among its colonizers. Indeed, in the Northern Central Asian and Beringian populations analyzed here, this haplotype reaches frequencies higher than expected based on their geographic location. This makes plausible two non-mutually exclusive scenarios for the evolution of the A*1_B*1 frequency in the Americas, where the first colonizers of the tropical belt encountered high UV radiation and benefitted from any mechanism capable of efficient melanogensis. The first scenario is one of randomly drifting frequencies, mostly determined by the enhanced and repeated founder effects associated with the fast North-to-South peopling of the continent.44 The allowable range of the A*1_B*1 frequencies again included high values, now compatible with the reactivation of the mechanisms related to adaptation to high UV radiation. The second scenario assumes that A*1_B*1 haplotype played a more obvious role in melanogenesis. Specifically, it suggests that it may have become adaptive and slightly advantageous during the colonization of Southern America, thus increasing in frequency once again.
To be best of our knowledge, the data presented on 22q11.23 and our proposed model to explain the observed pattern of genetic diversity at this locus represent the first instance of a pigmentation-related human adaptation based on CNVs. Indeed, no studies have investigated the role CNVs play in human pigmentation, meaning that potentially important adaptation mechanisms have remained unexplored. Conversely, several genome-wide association studies have identified SNPs in genes related to pigmentation (ie, TYR, TYRP1, OCA2, SLC45A2, SLC24A5, and KITLG), which show strong correlations with skin color.33 As shown in Table 3, the strength of the association between the previously identified SNPs and skin color in 17 of the 27 populations here examined is not as high as that reported for A*1_B*1 in the present study, supporting the hypothesis that the 22q11.23 chromosomal region harbors a yet unidentified pigmentation-related locus in populations across the globe. However, the pattern of selection proposed here for the CNV-based haplotype A*1_B*1 is different from that valid for SNPs: the A*1_B*1haplotype may be subjected to a purifying selection in worldwide populations on a background of other three nearly neutral haplotypes (A*1_B*0, A*0_B*1, and A*0_B*0), whereas the previously identified pigmentation-related SNPs are subjected to a directional positive selection specific for certain populations.
Finally, the best correlation of A*1_B*1 haplotype distribution was with skin color rather than UVA or UVB radiation. Indeed, melanogenesis is a complex process in which multiple factors interact. UV radiation is the most relevant, but other factors, such as diet, also have important roles.29, 45 The correlations here discussed do not allow us to distinguish whether the 22q11.23 MIF-related gene products directly contribute to the skin color phenotype or interact with it once this adaptation is already established. Nevertheless, we identify and separate populations that share high frequencies of the A*1_B*1 haplotype and, presumably, MIF-related gene output onto otherwise remarkably differentiated genomic backgrounds.
In conclusion, our study offers new insight into human adaptation, the function of the MIF gene family, and the 22q11.23 CNVs. Our data strongly support a dosage effect for the overall output of MIF-related genes. Accordingly, the study of 22q11.23 CNVs may be useful in understanding the role of the MIF family in human health.
Acknowledgments
We are grateful to the research groups of HapMap and 1000 Genomes Projects. The subjects of the investigation were adequately informed about the aims of the study and gave their approval, which is also gratefully acknowledged. Human studies have been performed in accordance with the ethical standards as laid down by law. This study was supported by PRIN 2009–2011 (grant numbers 200975T9EW and 2009P2CNKK_003 to MF and AN respectively) from MIUR (Italian Ministry of Instruction, University, and Research).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Girirajan S, Campbell CD, Eichler EE. Human copy number variation and complex genetic disease. Annu Rev Genet. 2011;45:203–226. doi: 10.1146/annurev-genet-102209-163544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietilainen OP, Rehnstrom K, Jakkula E, et al. Phenotype mining in CNV carriers from a population cohort. Hum Mol Genet. 2011;20:2686–2695. doi: 10.1093/hmg/ddr162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Altshuler DM. Copy-number variation and association studies of human disease. Nat Genet. 2007;39:S37–S42. doi: 10.1038/ng2080. [DOI] [PubMed] [Google Scholar]
- Campbell CD, Sampas N, Tsalenko A, et al. Population-genetic properties of differentiated human copy-number polymorphisms. Am J Hum Genet. 2011;88:317–332. doi: 10.1016/j.ajhg.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskow RC, Gokcumen O, Lee C. Exploring the role of copy number variants in human adaptation. Trends Genet. 2012;28:245–257. doi: 10.1016/j.tig.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Marotta M, Eichler EE, Eng C, Tanaka H. Linkage disequilibrium between two high-frequency deletion polymorphisms: implications for association studies involving the glutathione-S transferase (GST) genes. PLoS Genet. 2009;5:e1000472. doi: 10.1371/journal.pgen.1000472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coggan M, Whitbread L, Whittington A, Board P. Structure and organization of the human theta-class glutathione S-transferase and D-dopachrome tautomerase gene complex. Biochem J. 1998;334 (Pt 3:617–623. doi: 10.1042/bj3340617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polimanti R, Piacentini S, Lazzarin N, Re MA, Manfellotto D, Fuciarelli M. Glutathione S-transferase variants as risk factor for essential hypertension in Italian patients. Mol Cell Biochem. 2011;357:227–233. doi: 10.1007/s11010-011-0893-3. [DOI] [PubMed] [Google Scholar]
- Thorn CF, Ji Y, Weinshilboum RM, Altman RB, Klein TE. PharmGKB summary: very important pharmacogene information for GSTT1. Pharmacogenet Genomics. 2012;22:646–651. doi: 10.1097/FPC.0b013e3283527c02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savaskan NE, Fingerle-Rowson G, Buchfelder M, Eyupoglu IY. Brain miffed by macrophage migration inhibitory factor. Int J Cell Biol. 2012;2012:139573. doi: 10.1155/2012/139573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin DJ. Biochemistry of human skin—our brain on the outside. Chem Soc Rev. 2005;35:52–67. doi: 10.1039/b505793k. [DOI] [PubMed] [Google Scholar]
- Merk M, Zierow S, Leng L, et al. The D-dopachrome tautomerase (DDT) gene product is a cytokine and functional homolog of macrophage migration inhibitory factor (MIF) Proc Natl Acad Sci USA. 2011;108:E577–E585. doi: 10.1073/pnas.1102941108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merk M, Mitchell RA, Endres S, Bucala R. D-dopachrome tautomerase (D-DT or MIF-2): doubling the MIF cytokine family. Cytokine. 2012;59:10–17. doi: 10.1016/j.cyto.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozza M, Kolakowski LF, Jr, Jenkins NA, et al. Structural characterization and chromosomal location of the mouse macrophage migration inhibitory factor gene and pseudogenes. Genomics. 1995;27:412–419. doi: 10.1006/geno.1995.1071. [DOI] [PubMed] [Google Scholar]
- Du J, Xie X, Chen H, et al. Macrophage migration inhibitory factor (MIF) in chinese amphioxus as a molecular marker of immune evolution during the transition of invertebrate/vertebrate. Dev Comp Immunol. 2004;28:961–971. doi: 10.1016/j.dci.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Parisi MG, Toubiana M, Mangano V, Parrinello N, Cammarata M, Roch P. MIF from mussel: coding sequence, phylogeny, polymorphism, 3D model and regulation of expression. Dev Comp Immunol. 2012;36:688–696. doi: 10.1016/j.dci.2011.10.014. [DOI] [PubMed] [Google Scholar]
- De Stefano GF, Martinez-Labarga C, Casalotti R, et al. Analysis of three RFLPs of the COL1A2 (Type I Collagen) in the Amhara and the Oromo of Ethiopia. Ann Hum Biol. 2002;29:432–441. doi: 10.1080/03014460110101440. [DOI] [PubMed] [Google Scholar]
- Luca F, Bubba G, Basile M, et al. Multiple advantageous amino acid variants in the NAT2 gene in human populations. PLoS One. 2008;3:e3136. doi: 10.1371/journal.pone.0003136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polimanti R, Piacentini S, Porreca F, Fuciarelli M. Glutathione S-transferase omega class (GSTO) polymorphisms in a sample from Rome (Central Italy) Ann Hum Biol. 2010;37:585–592. doi: 10.3109/03014460903508520. [DOI] [PubMed] [Google Scholar]
- Polimanti R, Piacentini S, De Angelis F, De Stefano GF, Fuciarelli M. Human GST loci as markers of evolutionary forces: GSTO1*E155del and GSTO1*E208K polymorphisms may be under natural selection induced by environmental arsenic. Dis Markers. 2011;31:231–239. doi: 10.3233/DMA-2011-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium. Altshuler DM, Gibbs RA, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium. Abecasis GR, Altshuler D, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piacentini S, Polimanti R, De Angelis F, Iorio A, Fuciarelli M. Phenotype versus genotype methods for copy number variant analysis of glutathione S-transferases M1. Ann Hum Genet. 2013;77:409–415. doi: 10.1111/ahg.12025. [DOI] [PubMed] [Google Scholar]
- McCarroll SA, Kuruvilla FG, Korn JM, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- Mills RE, Walter K, Stewart C, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasutti R. Le Razze e i Popoli Della Terra. 4. Edizione riveduta e aggiornata. Torino: Unione Tipografico-Editrice Torinese; 1967. [Google Scholar]
- Barsh GS. What controls variation in human skin color. PLoS Biol. 2003;1:E27. doi: 10.1371/journal.pbio.0000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski NG, Chaplin G. Colloquium paper: human skin pigmentation as an adaptation to UV radiation. Proc Natl Acad Sci USA. 2010;107 (Suppl 2:8962–8968. doi: 10.1073/pnas.0914628107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Hofer T, Ray N, Wegmann D, Excoffier L. Large allele frequency differences between human continental groups are more likely to have occurred by drift during range expansions than by selection. Ann Hum Genet. 2009;73:95–108. doi: 10.1111/j.1469-1809.2008.00489.x. [DOI] [PubMed] [Google Scholar]
- Sturm RA. Molecular genetics of human pigmentation diversity. Hum Mol Genet. 2009;18:R9–17. doi: 10.1093/hmg/ddp003. [DOI] [PubMed] [Google Scholar]
- Candille SI, Absher DM, Beleza S, et al. Genome-wide association studies of quantitatively measured skin, hair, and eye pigmentation in four european populations. PLoS One. 2012;7:e48294. doi: 10.1371/journal.pone.0048294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton HL, Kittles RA, Parra E, et al. Genetic evidence for the convergent evolution of light skin in Europeans and East Asians. Mol Biol Evol. 2007;24:710–722. doi: 10.1093/molbev/msl203. [DOI] [PubMed] [Google Scholar]
- Ramachandran S, Deshpande O, Roseman CC, Rosenberg NA, Feldman MW, Cavalli-Sforza LL. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci USA. 2005;102:15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriver MD, Mei R, Parra EJ, et al. Large-scale SNP analysis reveals clustered and continuous patterns of human genetic variation. Hum Genomics. 2005;2:81–89. doi: 10.1186/1479-7364-2-2-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller KE, Indap AR, Schmidt S, et al. Proportionally more deleterious genetic variation in European than in African populations. Nature. 2008;451:994–997. doi: 10.1038/nature06611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Abe R, Ohkawara A, Nishihira J. Ultraviolet B radiation upregulates the production of macrophage migration inhibitory factor (MIF) in human epidermal keratinocytes. J Invest Dermatol. 1999;112:210–215. doi: 10.1046/j.1523-1747.1999.00486.x. [DOI] [PubMed] [Google Scholar]
- Sonesson B, Rosengren E, Hansson AS, Hansson C. UVB-induced inflammation gives increased d-dopachrome tautomerase activity in blister fluid which correlates with macrophage migration inhibitory factor. Exp Dermatol. 2003;12:278–282. doi: 10.1034/j.1600-0625.2003.120307.x. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Shimizu T, Nishihira J, et al. Ultraviolet A-induced production of matrix metalloproteinase-1 is mediated by macrophage migration inhibitory factor (MIF) in human dermal fibroblasts. J Biol Chem. 2004;279:1676–1683. doi: 10.1074/jbc.M303650200. [DOI] [PubMed] [Google Scholar]
- Shimizu T. Role of macrophage migration inhibitory factor (MIF) in the skin. J Dermatol Sci. 2005;37:65–73. doi: 10.1016/j.jdermsci.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Enomoto A, Yoshihisa Y, Yamakoshi T, et al. UV-B radiation induces macrophage migration inhibitory factor-mediated melanogenesis through activation of protease-activated receptor-2 and stem cell factor in keratinocytes. Am J Pathol. 2011;178:679–687. doi: 10.1016/j.ajpath.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke DH, Raff JA. The human genetic history of the Americas: the final frontier. Curr Biol. 2010;20:R202–R207. doi: 10.1016/j.cub.2009.11.051. [DOI] [PubMed] [Google Scholar]
- Jablonski NG.Skin colorationin Muehlenbein MP (ed): Human Evolutionary Biology Cambridge, UK: Cambridge University Press; 2010 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.