Abstract
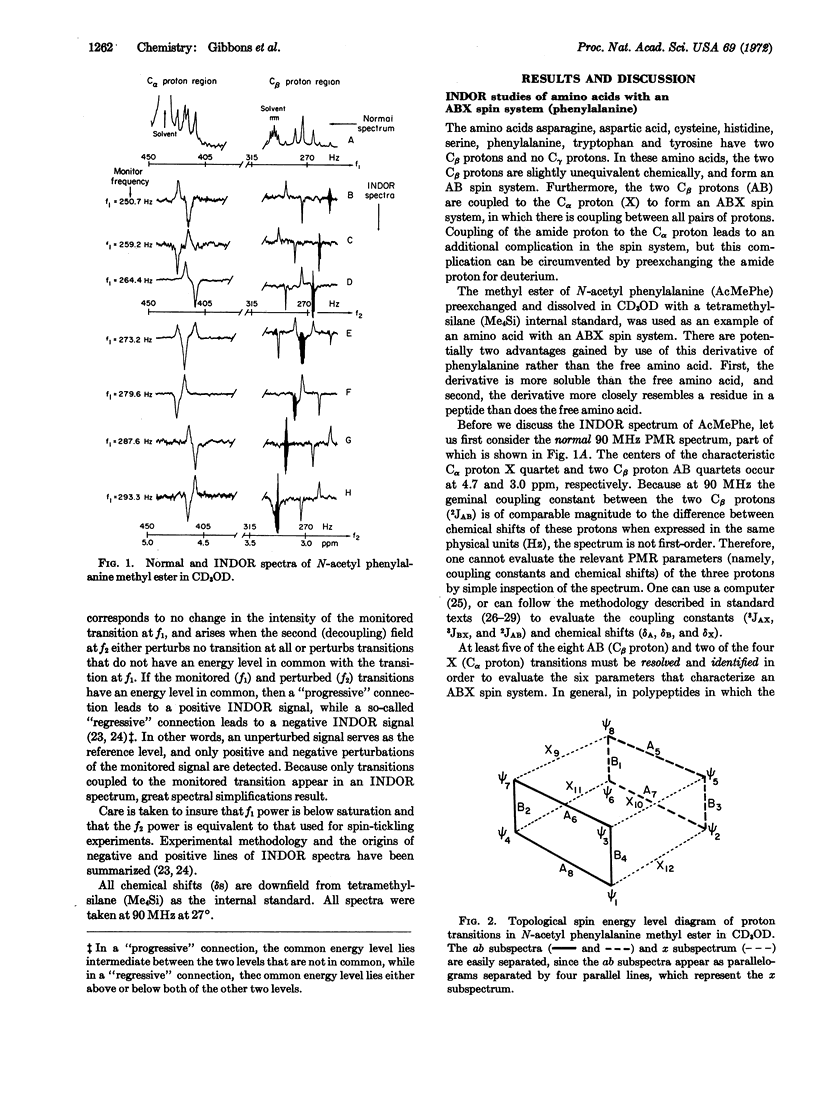
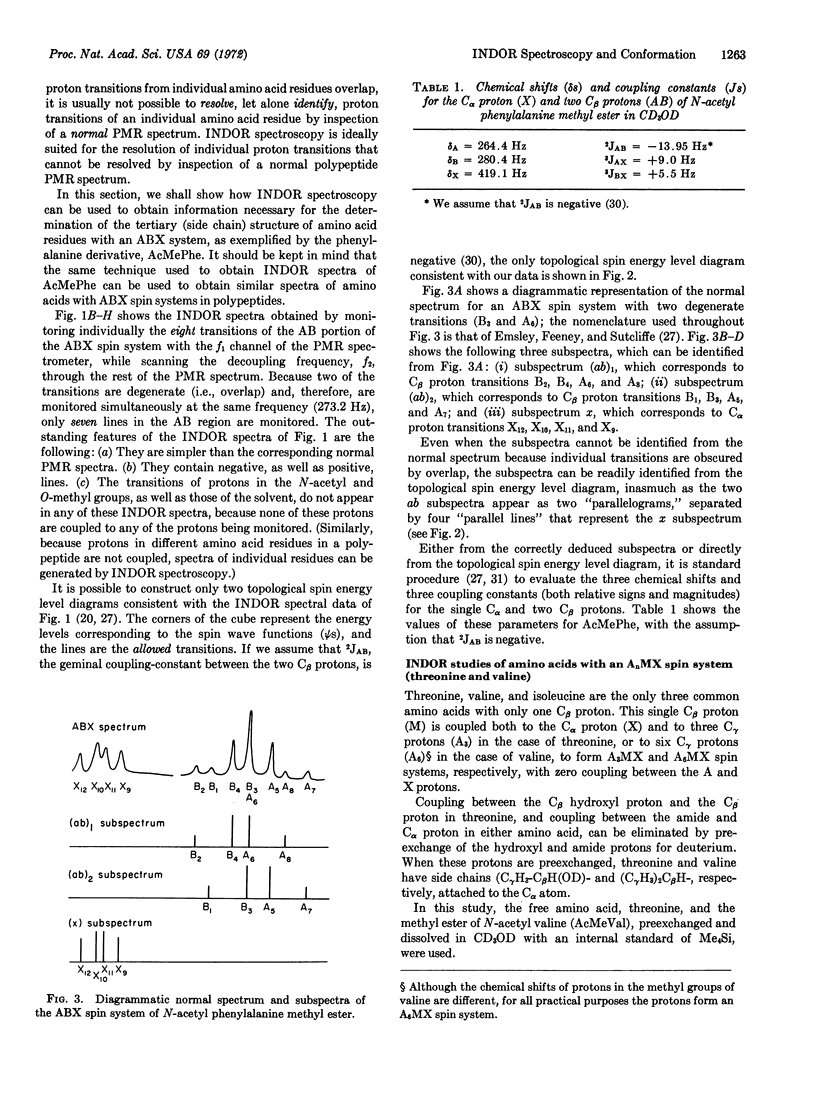
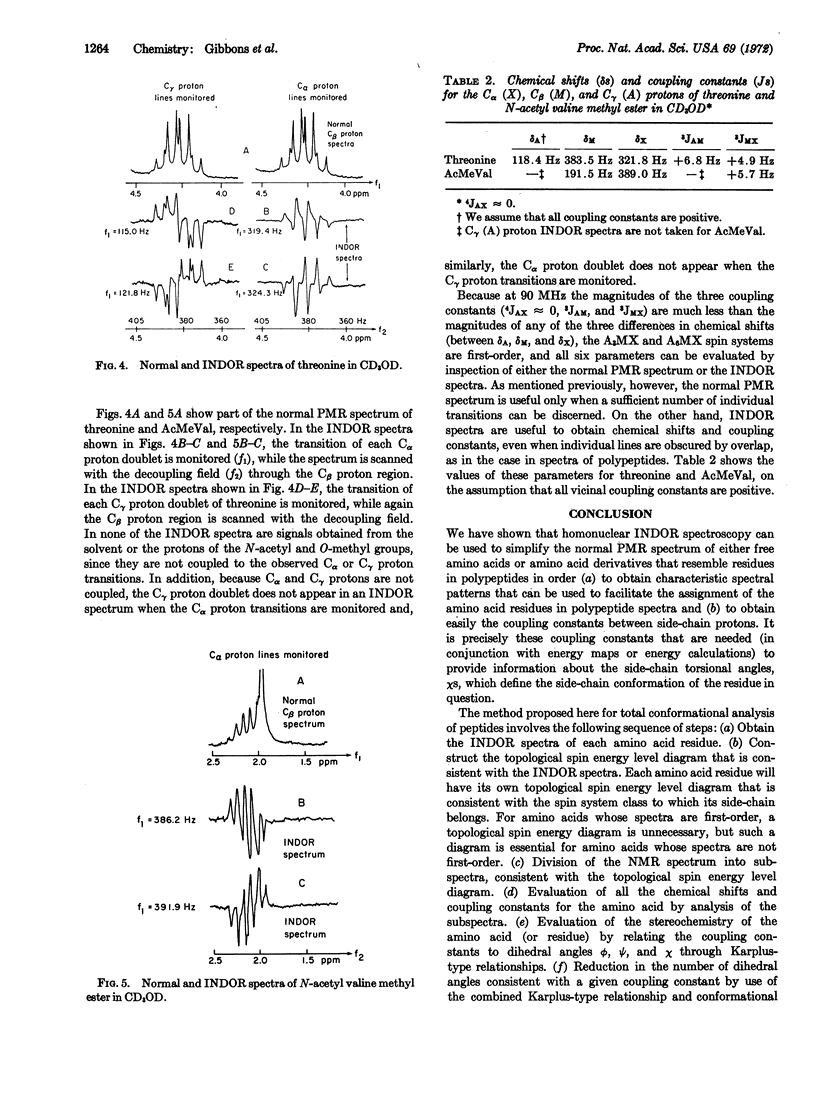
INDOR (Internuclear Double Resonance) spectroscopy is shown to be superior to conventional (spectra obtained not by sweeping, but by maintaining constant the decoupling frequency) nuclear single- or double-resonance techniques for conformational studies of amino acids and amino acid residues in the following ways: (a) INDOR spectra of amino acids are inherently simpler than conventional proton magnetic resonance spectra of amino acids, and INDOR spectra of individual amino acid residues are slightly, if at all, complicated by overlap with either solvent peaks or the transitions of nuclei in other residues. (b) For each amino acid, the side-chain and Cα proton belong to a particular class of spin system characterized by unique INDOR spectra, the pattern of which aids in the proper assignment of spectral lines. (c) For an amino acid with a first-order spin system, INDOR spectra directly reveal hidden chemical shifts and coupling constants. For an amino acid with a spin system other than first-order, INDOR spectra indirectly reveal values for chemical shifts and coupling constants as follows: INDOR spectra permit construction of a topological spin energy level diagram which, in turn, allows division of the PMR spectrum of the spin system into subspectra that easily yield values for chemical shifts and coupling constants.
Although we only report INDOR spectra of free amino acids or amino acid derivatives that resemble amino acid residues in polypeptides, we, in effect, demonstrate a novel method to obtain total polypeptide conformation based on INDOR spectroscopy, inasmuch as the total conformation is the sum of the individual residue conformations.
Keywords: free amino acids, amino acid residues, NMR, PMR, INDOR spectroscopy, molecular conformation
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bystrov V. F., Portnova S. L., Balashova T. A., Tsetlin V. I., Ivanov V. T., Kostetzky P. V., Ovchinnikov Y. A. Conformational studies of peptide systems. NMR and IR spectra of N-acetyl-alanyl-phenylalanine and N-acetyl-phenylalanyl-alanine methyl esters. Tetrahedron Lett. 1969 Dec;(60):5283–5286. doi: 10.1016/s0040-4039(01)88944-9. [DOI] [PubMed] [Google Scholar]
- Gibbons W. A., Némethy G., Stern A., Craig L. C. An approach to conformational analysis of peptides and proteins in solution based on a combination of nuclear magnetic resonance spectroscopy and conformational energy calculations. Proc Natl Acad Sci U S A. 1970 Sep;67(1):239–246. doi: 10.1073/pnas.67.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes D. H., Kowalsky A., Pressman B. C. Application of nuclear magnetic resonance to the conformational changes in valinomycin during complexation. J Biol Chem. 1969 Jan 25;244(2):502–505. [PubMed] [Google Scholar]
- Horsley W. J., Sternlicht H. Carbon-13 magnetic resonance studies of amino acids and peptides. J Am Chem Soc. 1968 Jul 3;90(14):3738–3748. doi: 10.1021/ja01016a025. [DOI] [PubMed] [Google Scholar]
- Hruby V. J., Brewster A. I., Glasel J. A. NMR studies on the conformation of derivatives of the side chain of oxytocin: examples of cis-trans isomerism. Proc Natl Acad Sci U S A. 1971 Feb;68(2):450–453. doi: 10.1073/pnas.68.2.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L. F., Schwartz I. L., Walter R. Oxytocin and neurohypophyseal peptides: spectral assignment and conformational analysis by 220 MHz nuclear magnetic resonance. Proc Natl Acad Sci U S A. 1969 Dec;64(4):1269–1275. doi: 10.1073/pnas.64.4.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopple K. D., Marr D. H. Conformation of cyclic peptides. The folding of cyclic dipeptides containing an aromatic side chain. J Am Chem Soc. 1967 Nov 22;89(24):6193–6200. doi: 10.1021/ja01000a035. [DOI] [PubMed] [Google Scholar]
- Kopple K. D., Ohnishi M. Conformations of cyclic peptides. II. Side-chain conformation and ring shape in cyclic dipeptides. J Am Chem Soc. 1969 Feb 12;91(4):962–975. doi: 10.1021/ja01032a029. [DOI] [PubMed] [Google Scholar]
- Llinás M., Klein M. P., Neilands J. B. Solution conformation of ferrichrome, a microbial iron transport cyclohexapeptide, as deduced by high resolution proton magnetic resonance. J Mol Biol. 1970 Sep 28;52(3):399–414. doi: 10.1016/0022-2836(70)90409-2. [DOI] [PubMed] [Google Scholar]
- Ohnishi M., Urry D. W. Temperature dependence of amide proton chemical shifts: the secondary structures of gramicidin S and valinomycin. Biochem Biophys Res Commun. 1969 Jul 23;36(2):194–202. doi: 10.1016/0006-291x(69)90314-3. [DOI] [PubMed] [Google Scholar]
- Stern A., Gibbons W. A., Craig L. C. A conformational analysis of gramicidin S-A by nuclear magnetic resonance. Proc Natl Acad Sci U S A. 1968 Oct;61(2):734–741. doi: 10.1073/pnas.61.2.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonelli A. E., Patel D. J., Goodman M., Naider F., Faulstich H., Wieland T. Experimental and calculated conformational characteristics of the cyclic decapeptide antamanide. Biochemistry. 1971 Aug 17;10(17):3211–3217. doi: 10.1021/bi00793a008. [DOI] [PubMed] [Google Scholar]
- Urry D. W., Ohnishi M., Walter R. Secondary structure of the cyclic moiety of the peptide hormone oxytocin and its deamino analog. Proc Natl Acad Sci U S A. 1970 May;66(1):111–116. doi: 10.1073/pnas.66.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urry D. W., Walter R. Proposed conformation of oxytocin in solution. Proc Natl Acad Sci U S A. 1971 May;68(5):956–958. doi: 10.1073/pnas.68.5.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victor T. A., Hruska F. E., Hikichi K., Danyluk S. S., Bell C. L. Nuclear magnetic resonance study of the structure and interactions of actinomycin D: temperature and solvent effects on the N--H and NH2 groups. Nature. 1969 Jul 19;223(5203):302–303. doi: 10.1038/223302a0. [DOI] [PubMed] [Google Scholar]



