Graphical abstract
Keywords: Anthelmintic resistance, Anthelmintic drugs, Molecular markers, Anthelmintic targets, Receptors
Highlights
-
•
We report on the Consortium for Anthelmintic Resistance and Susceptibility 2013 meeting.
-
•
Recent advances in the identification of markers for anthelmintic resistance are described.
-
•
The use of markers for benzimidazole resistance in field studies with veterinary and human nematodes.
-
•
The application of the newest high-throughput sequencing technologies to the study of anthelmintic resistance.
Abstract
Anthelmintic resistance has a great impact on livestock production systems worldwide, is an emerging concern in companion animal medicine, and represents a threat to our ongoing ability to control human soil-transmitted helminths. The Consortium for Anthelmintic Resistance and Susceptibility (CARS) provides a forum for scientists to meet and discuss the latest developments in the search for molecular markers of anthelmintic resistance. Such markers are important for detecting drug resistant worm populations, and indicating the likely impact of the resistance on drug efficacy. The molecular basis of resistance is also important for understanding how anthelmintics work, and how drug resistant populations arise. Changes to target receptors, drug efflux and other biological processes can be involved. This paper reports on the CARS group meeting held in August 2013 in Perth, Australia. The latest knowledge on the development of molecular markers for resistance to each of the principal classes of anthelmintics is reviewed. The molecular basis of resistance is best understood for the benzimidazole group of compounds, and we examine recent work to translate this knowledge into useful diagnostics for field use. We examine recent candidate-gene and whole-genome approaches to understanding anthelmintic resistance and identify markers. We also look at drug transporters in terms of providing both useful markers for resistance, as well as opportunities to overcome resistance through the targeting of the transporters themselves with inhibitors. Finally, we describe the tools available for the application of the newest high-throughput sequencing technologies to the study of anthelmintic resistance.
1. Introduction
The Consortium for Anthelmintic Resistance and Susceptibility (CARS) met for the fifth time on the 25th of August, 2013 in Perth, Australia. The group meets every two years to discuss the latest research aiming to elucidate mechanisms of resistance to anthelmintic drugs. The CARS group aims to promote research on anthelmintic resistance, with a view towards the development of molecular markers for anthelmintic resistance diagnosis, and to assist in the development of new drugs. The meeting was attended by approximately 90 delegates drawn from universities, government primary industry departments, national research institutes (medical and veterinary), animal health companies, and veterinary service providers.
Anthelmintic resistance has great impact on sheep and cattle production systems worldwide (Kaplan, 2004; Sutherland and Leathwick, 2011), while the impact on equine health is also increasing (von Samson-Himmelstjerna, 2012). In addition, there is some concern that resistance may arise in soil-transmitted helminth parasites infecting humans (Vercruysse et al., 2011), and there is evidence of ivermectin resistance emerging already in Onchocerca volvulus (Osei-Atweneboana et al., 2011) and praziquantel resistance has been reported in Schistosoma haematobium (reviewed in Wang et al., 2012). Resistance also has an impact on parasite control in companion animals; for example, resistance to nicotinic agonist anthelmintics has been reported in canine hookworms (Kopp et al., 2007), and there is some evidence that resistance to macrocyclic lactones is emerging in canine heartworms (Geary et al., 2011). Clearly, there is a significant need for better management of drug use in helminth control. If means existed to reliably identify the presence of parasite populations resistant to specific drugs, improvements in drug-use decisions could be made to avoid ineffective treatment and, hence, slow the selection for resistance. Sensitive molecular diagnostics are an attractive option for providing the basis for such drug-use decisions.
Research on resistance markers is not only useful for developing diagnostic tools, but can also help increase our understanding of drug effects: for example, the interaction of drugs with their molecular targets. Also, an understanding of drug resistance mechanisms, can assist in revealing the nature of the interactions of anthelmintics with parasite defensive systems, including drug efflux pumps (e.g. P-glycoprotein, P-gp) and detoxification enzymes (e.g. cytochrome P450). The development of resistance markers is, therefore, seen as an enabling science which will help counter the impact of anthelmintic resistance in multiple ways; providing new drug targets, new synergistic or combination drug preparations, and a better understanding of how parasites evolve to cope with any potential xenobiotic, including the next generation of anthelmintic drugs.
The last review paper produced by the CARS group was in 2011, covering the 2009 meeting (Beech et al., 2011), and hence the present review represents a timely ‘state of play’ document describing the most recent research into anthelmintic resistance mechanisms and molecular markers. The CARS2013 meeting covered the latest research on anthelmintic resistance against each of the main drug classes, focusing on the development of markers for resistance to each class. For drugs where resistance has not yet been reported (the cyclooctadepsipeptides, e.g. emodepside) or has only recently been reported (monepantel- Scott et al., 2013), the talks described research on the drug’s mode of action, revealing that changes at the receptor site(s) are likely resistance mechanisms. Drug transporters (e.g. P-glycoproteins, P-gps) were examined from two perspectives, firstly, in terms of their potential use as markers for resistance, and secondly, analysis of the potential to use P-gp inhibitors as synergists to overcome transporter-mediated anthelmintic resistance. The meeting focused both on candidate gene-based (Fig. 1) and worm genetics and genomics-based approaches to elucidate resistance mechanisms. Finally, the meeting was presented with an update on the genomic resources available to researchers studying molecular aspects of nematode and trematode biology.
Fig. 1.
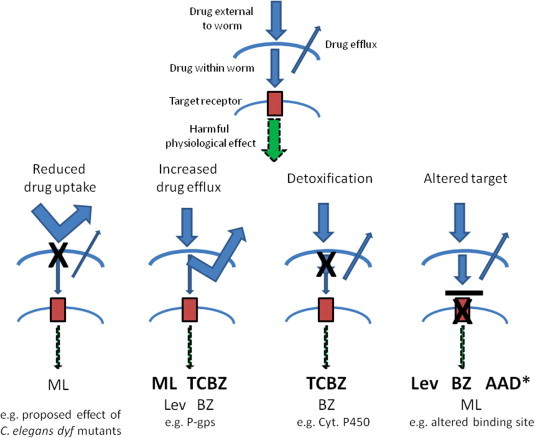
Schematic representation of principal known anthelmintic resistance pathways, and their relevance to each of the current anthelmintic drug classes. The ability of the drug to enter the worm and interact with its target receptor in order to trigger a harmful physiological effect (shown at top for a drug- susceptible worm) is diminished through four principal mechanisms. These mechanisms apply to varying degrees to the major anthelmintic drug classes, as indicted by the relative font of the drug class names at the base of the figure; ML = macrocyclic lactones, TCBZ = triclabendazole, Lev = levamisole (as a representative of the nicotinic agonist drug class), BZ = benzimidazoles, AAD = amino-acetonitrile derivatives; ∗denotes that resistance to the AADs is only characterised in laboratory-selected isolates.
2. Macrocyclic lactones (MLs): the target site
The biological targets for MLs are glutamate-gated chloride ion channel receptors (GluClRs) expressed in the neurons and muscle cells of nematodes (Cully et al., 1994). ML drugs irreversibly activate these channels, thereby inhibiting neuronal activity and muscle contractility, and thus inducing flaccid paralysis and death. MLs also activate other ligand-gated ion channel receptors, namely the γ-aminobutyric acid (GABA) and glycine (Gly) receptors, however, this activation requires much higher drug concentrations than required for the GluClRs (Adelsberger et al., 2000), and hence the GluClRs are considered to be the principal target for this class of anthelmintics.
Early reports on the mechanism of ivermectin resistance in parasitic nematodes highlighted the presence of mutations in GluClRs (Blackhall et al., 1998; Njue et al., 2004). Blackhall et al. (1998) reported an increased frequency for an allele of a GluCl α-subunit gene in ivermectin- and moxidectin-resistant Haemonchus contortus isolates, suggesting that a mutation in this gene was associated with ML resistance. Importantly, these resistant isolates had been generated by repeated selections with either ivermectin or moxidectin at sub-therapeutic levels rather than being field-derived. Njue et al. (2004) detected a number of mutations in the GluClα3 subunit gene of an isolate of Cooperia oncophora recovered originally from the field in Somerset, UK (Coles et al., 1998). They found that one of these mutations, L256F, accounted for differences in the response to ivermectin shown by homomeric channels composed of GluClα3 subunit genes from the resistant and susceptible isolates expressed in Xenopus oocytes. McCavera et al. (2009) confirmed the importance of the L256F mutation in interactions of ivermectin with GluClRs, by showing that substitutions of various aromatic amino acid residues for L256 in H. contortus GluClα3B genes transfected into COS-7 cell membranes caused a reduction in the binding of ivermectin to membrane preparations. They also stated that they were looking to see whether any mutation at position 256 was present in field isolates of H. contortus, and although the extent of this effort was not described, they reported ‘we have had no success’.
As described above, ivermectin is also known to interact with GABA receptors, and there is some evidence that GABA receptors may contribute to the nematocidal activity of MLs (McCavera et al., 2007). Changes in GABA receptors have been implicated in ML resistance in parasitic nematodes (Feng et al., 2002; Blackhall et al., 2003). However, these resistant isolates were derived from repeated selections at sub-therapeutic drug doses rather than being derived from the type of drug selection pressure occurring in the field.
The last couple of years have seen great advances in the understanding of how ivermectin interacts with Cys-loop domains within receptor proteins (Lynagh and Lynch 2010, 2012a; Hibbs and Gouaux, 2011). Lynagh and Lynch (2010) showed that the presence or absence of a glycine residue in the third transmembrane domain of these receptors (denoted M3-Gly) could predict their ivermectin sensitivity. For instance, the M3-Gly residue is present in GluClRs of various ivermectin-sensitive helminths such as H. contortus, C. oncophora, and Dirofilaria immitis, but is replaced by larger residues in ivermectin –insensitive trematodes such as Schistosoma mansoni, S. japonica and Clonorchis sinensis (Lynagh and Lynch, 2012a). Importantly, in terms of its possible reflection of a ML resistance mechanism, mutation of this glycine residue in the H. contortus GluClα3B protein expressed on HEK293 cells resulted in the loss of ivermectin sensitivity (Lynagh and Lynch, 2010). However, while explaining ivermectin sensitivity differences between different Cys-loop receptors within species, and between different species possessing different receptor types, the question remained as to whether changes in the M3-Gly residue were involved in the reduced sensitivity to ivermectin seen in field isolates of many helminth species. Importantly, Lynagh and Lynch (2010) noted that GluClα3 from resistant C. oncophora (Njue et al., 2004) did have the M3 glycine residue, suggesting that changes at M3-Gly are not necessary for reduced ivermectin sensitivity arising as a result of drug selection pressure in the field.
Indeed, recent studies have failed to find a link between the M3-Gly residue, or mutations equivalent to L256F in C. oncophora, and ivermectin resistance in various field-derived isolates of several parasitic nematode species. Further, these recent reports have failed to find any polymorphisms in GluClRs that can be specifically linked to the observed ML resistances. These reports have examined: (i) the avr-14B gene in field-derived C. oncophora (including two isolates showing 0% ivermectin efficacy) and laboratory-selected O. ostertagi (El-Abdellati et al., 2011); (ii) a number of ligand gated chloride ion channels, including both GluCl and GABA channels (avr-14B, glc-5, lgc-37 and glc-6), in field-derived resistant H. contortus (Williamson et al., 2011), and (iii) the avr-14B gene in field-derived Teladorsagia circumcincta (Martínez-Valladares et al., 2012). More recently, no polymorphisms linked to resistance were found in the glc-6 gene of two of the C. oncophora isolates examined earlier by El-Abdellati et al. (2011) (Geldhof, unpublished data). Hence, to date, there is no evidence that polymorphisms of GluClRs or GABA receptors can explain the observed resistance to ML drugs in most field isolates of a number of parasitic nematodes.
Some attention has been paid in the last couple of years to the possible role of changes in expression levels of the ivermectin target in conferring resistance. El-Abdellati et al. (2011) showed that transcription levels of avr-14B were decreased in resistant isolates of both C. oncophora and O. ostertagi, however, the decreases were ‘relatively modest’. Similarly, Williamson et al. (2011) found that transcription of two GluCls (glc-3 and glc-5) was slightly reduced in a resistant H. contortus isolate, however, again, the changes were modest. In contrast, Martínez-Valladares et al. (2012) reported that avr-14B transcript levels were slightly higher in resistant isolates of T. circumcincta compared to susceptible isolates. These modest changes in transcription levels of GluClR subunits in both upward and downward directions in resistant isolates may suggest that the differences are related to differences in the genetic background of the resistant and susceptible isolates examined in each study, rather than specific responses to drug selection pressure in the resistant isolates.
In conclusion, despite some early indications that resistance to MLs may be due to specific polymorphisms in the drug target receptors, there are not yet any mutations identified that can explain the resistance phenotypes observed in most field isolates of the studied parasitic nematode species. Further, recent studies on receptor transcript levels have been inconclusive. Although the various studies on the role of the ivermectin target in resistance have not yet been exhaustive, together they suggest that target site changes are not involved in most cases of field resistance to the ML drug class. Attention therefore turns to other potential resistance mechanisms such as P-gps (Lespine et al., 2012) or drug uptake pathways (for example, as suggested for C. elegans dyf mutants, Dent et al., 2000). The role of P-gps in ML resistance is examined later in this review.
3. Macrocyclic lactones: genetic marker association mapping
The work of Carl Johnson at Nemapharm in the 1980s established that in C. elegans a few major genes could confer resistance to high concentrations of ivermectin (>50 ng/mL in media) (Rand and Johnson, 1995). Further analysis revealed that many other loci could be mutated to impart resistance to ivermectin at lower levels (<50 ng/mL). Later, this work was confirmed by others and published (Dent et al., 2000). The observation of a multitude of potential genes conferring resistance to ivermectin opened the possibility of a more quantitative, additive genetic mechanism. In the parasite H. contortus, some early work with the Australian Chiswick avermectin resistant strain (CAVR) suggested that a single dominant allele might be responsible for resistance, so the research focus for identifying ivermectin resistance markers in parasites concentrated on the possibility of single genes with large effects (Le Jambre et al., 2000).
In more recent years, the continued absence of useful molecular markers for ivermectin and other ML resistances in field populations of parasites (as described in the previous section) has led to a re-examination of the likely genetic mechanisms of resistance. A study of F2 populations of H. contortus crosses between a field resistant isolate (Wallangra2003) and a laboratory cultured susceptible parent (McMaster 1931), indicated that the level of IVM resistance displayed by the F2 (and derived from Wallangra2003) could not be explained using a single gene model (Hunt et al., 2010). This has recently been supported by two serial backcross experiments between the susceptible strain MHco3 (ISE) and two ivermectin resistant strains MHco10 (ISE) and MHco4 (WRS) in which the proportion of the phenotypically resistant individuals present after the fourth backcross was consistent with a multi-genic basis of resistance (Redman et al., 2012). A recent study also suggests ivermectin resistance in C. elegans is multi-genic when selected in field populations (Ghosh et al., 2012). In this work, naturally occurring abamectin resistant populations of C. elegans were isolated from the field and genetic mapping identified a four amino acid deletion in the ligand-binding domain of GLC-1, the alpha subunit of a glutamate gated chloride channel, was an important genetic determinant of resistance. However, Quantitative Trait loci (QTL) analysis showed this locus contributed to 26% of the variance of the resistance phenotype, suggesting other loci were also involved.
In parallel to drug resistance research, advances in DNA sequencing technology, in marker association analysis techniques, and the advent of genome projects for a range of parasite species, have provided new opportunities. Using yeast as a model for drug resistance, Ehrenreich et al. (2012) established some new approaches for discovering resistance genes using genetic markers derived from known single nucleotide polymorphisms (SNP) analysed using high throughput SNP analysis platforms, or a range of polymorphisms assayed directly from high throughput genome sequencing, so called extreme QTL (X-QTL) methods. To some degree these approaches are being adopted in other species (for example Plasmodium reviewed by Anderson et al., 2011). Some of the characteristics of resistance alleles identified in the yeast work have parallels in parasite drug resistance phenomena. For example, the notion of multi-drug resistance via detoxification or efflux pump mechanisms has been widely posited as an alternative to drug-specific mechanisms. The yeast X-QTL study of Ehrenreich et al. (2012) revealed more than 800 chemical resistance loci and 20% of these were associated with resistance to four or more compounds, whilst 40% were specific to a single compound. Therefore, it should not be surprising when we find that both specific and cross-resistance mechanisms are at play in anthelmintic resistance. Similarly, mutations effecting drug resistance have been found which are identical across a wide range of populations (see BZ resistance section below), whilst other examples of drug resistance mechanisms differing between populations are also emerging. The yeast X-QTL studies have revealed the same phenomenon, with 32% of identified resistance loci arising from only a single parent strain from the four studied, whilst 7% of the loci clearly originated from multiple parent strains.
Mapping anthelmintic resistance using marker association has been a consistent topic at CARS meetings and has its origin with some seminal work on microsatellite markers in H. contortus, Teladorsagia circumcincta and Trichostrongylus colubriformis (Hoekstra et al., 1997; Otsen et al., 2000; Grillo et al., 2006; Redman et al., 2008) and the use of a H. contortus/placei species-hybrid system (Le Jambre et al., 1999). Though SNP markers and small indels may well replace the use of microsatellite markers in the near future, these markers are still useful and have been used to establish the methodology for mapping anthelmintic resistance in H. contortus (Hunt et al., 2010; Redman et al., 2012). The most recent of these studies (Redman et al., 2012) used two different ivermectin resistant populations, MHco10(CAVRS) (a derivative of CAVR) and MHco4(WRS), and crossed these with an inbred susceptible strain MHco3(ISE), which has also been used for genome sequencing in the species (Laing et al., 2013). Each of the parental strains was highly genetically divergent from the others, allowing parental genotypes to be discriminated in backcross progeny. Multiple backcrosses to MHco3(ISE) were conducted with ivermectin selection for resistance at each generation, and the parental strains and the fourth backcross generation were analysed using a panel of microsatellite markers. In addition, a study of survivors of ivermectin treatment from the fourth backcross was undertaken. Analysis of the ivermectin surviving backcross progeny, using a panel of nine discriminatory microsatellite markers, showed their allelic frequencies to be very similar to those of the MHco3(ISE) susceptible parental strain, and dissimilar to those in the resistant parental strains, demonstrating ivermectin resistance loci had been introgressed into the MHco3(ISE) genetic background. Wright’s FST analysis and the presence of alleles specific to the resistant parental strain at high frequency in fourth generation progeny of both backcrosses suggested one of the microsatellite markers, Hcms8a20, was located in an introgressed region, and hence, was genetically linked to an ivermectin resistance-conferring locus. This is an exciting finding that should be pursued, but further work is needed to identify the causal mutation and it is likely that this will be just one of several important loci. Genome-wide sequencing analysis of both backcrossed lines using Illumina deep sequencing is currently underway to map and delineate the introgressed regions across the genome.
The speaker for this section of the CARS2013 meeting (PWH) revealed some unpublished observations from their own work to demonstrate what might be done using high throughput sequencing in place of microsatellite or SNP markers. Although these results await peer review, it can be revealed that a cross using Wallangra2003 and McMaster1931 was the basis of the work (as in Hunt et al., 2010), and so there was an expectation of polygenic ivermectin resistance. SNP and short indel markers were readily revealed by analysis of a reduced representation genomic DNA sequence, and association with ivermectin resistance was explored using three differing statistical approaches. None of these analyses supported a mono-genic model of ivermectin resistance in Wallangra2003. The methodology employed did not rely on extensive backcrossing, but rather the creation of linkage disequilibrium between McMaster1931 and Wallangra2003 genomic segments in a single large scale cross, comparing the F2 generation with congeneric survivors of ivermectin treatment.
Future comparisons between the methods employed and the results of the Mhco3(ISE)/MHco10(CAVR) and MHco3(ISE)/MHco4(WRS) backcross studies as compared to the Wallangra2003/McMaster1931 study would seem most desirable. The occurrence of a successful CARS meeting coinciding with the release of two assemblies of the H. contortus genome (Laing et al. 2103; Schwarz et al., 2013) creates a major opportunity for future work. The assemblies have been produced using an inbred version of the MHco3 (ISE) strain resulting from a single pair mating – MHco3(ISE).N1 (Laing et al., 2013) and separately using McMaster 1931 (Schwarz et al., 2013). This assembled sequence information will hopefully enable the pace of method development and therefore drug resistance gene discovery to accelerate.
In conclusion, recent work on genome tools and ivermectin resistance in H. contortus has opened possibilities for faster and less expensive discovery of drug resistance genes in the future. It is hoped that these advances not only assist those interested in this species but also those studying other nematode and trematode parasites of humans and livestock.
4. Benzimidazole resistance: taking SNP-based tests to the field
It is widely accepted that the major molecular target site for the BZ anthelmintics within nematodes is β-tubulin, which plays a vital role in a number of sub-cellular processes. Furthermore, the major genetic determinant of BZ resistance (BZ-R) in most, if not all, trichostrongylid nematode species is the possession of single nucleotide polymorphisms (or SNPs), in the parasite’s isotype-1 β-tubulin gene. Pivotal amongst these is a tyrosine for phenylalanine substitution at codon 200 (the so-called F200Y SNP), encoded by a change from TTC to TAC. Its role in BZ-R was elegantly demonstrated by Kwa et al. (1994) in the mid-1990s through transfection of BZ-R C. elegans with a BZ-susceptible (BZ-S) isotype-1 β-tubulin transcript from H. contortus, conferring a BZ-S phenotype on the transgenic C. elegans. Subsequent in vitro mutagenesis of these transgenic worms resulted in the acquisition of the F200Y mutation, thus restoring their BZ-R phenotype. Since then, further BZ-R-associated SNPs have been discovered at codons 167 (F167Y) and 198 (E198A) in isotype-1 β-tubulin in a number of nematode species (Silvestre and Cabaret, 2002; Ghisi et al., 2007, respectively), but F200Y would still appear to be the most important with respect to BZ-R phenotype.
Thus, resistance to the BZs is, by some distance, the best understood anthelmintic resistance at the molecular level. So, armed with this precise genetic information defining the BZ-R genotype and phenotype of key nematode pathogens, and analytical tools for resistance detection, how far have we advanced towards improved diagnosis and management of BZ-R in the field? This was one of the questions posed at CARS2013. Approximately 90 publications on the general topic of ‘benzimidazole resistance’ have appeared since CARS2011. Encouragingly, a number of these publications have attempted to evaluate DNA-based testing of BZ-R under field/farm conditions. In the first of these studies, Barrère et al. (2013a) evaluated the efficiency of a genetic test for detection of BZ-R in H. contortus on sheep farms in Quebec, Canada. Eleven farms were recruited in total, and faecal samples were collected from 10 animals per group, before and after fenbendazole treatment. H. contortus was identified by fluorescent peanut agglutinin staining of eggs and found to be present on 8 of the 11 farms. Pyrosequencing assays targeting the aforementioned F200Y, E198A and F167Y SNPs were used to genotype individual H. contortus eggs. BZ-R was found on each of the 8 farms where H. contortus was present and this was backed up by faecal egg count reduction tests (FECRTs) carried out on the same farms (FEC reductions ranged from 26–54%). The average percentage of BZ-R parasites on the 8 farms was 77.7%, with the TTC to TAC at codon 200 responsible for most of the observed BZ-R genotypes (85.4% codon 200 versus 6% codon 167). The authors concluded that the genetic test brought substantial savings over FECRT in terms of cost, labour and time and could detect BZ-R in the resting parasite population before treatment, which FECRT could not. In a similar study in Ontario, Barrère et al. (2013b) tested H. contortus eggs isolated from 16 farms using the same panel of pyrosequencing assays, targeting the F200Y, E198A and F167Y SNPs. BZ-R was detected on all 16 farms, above an arbitrary threshold of 10% BZ-R alleles, the average BZ-R allele frequency was 68.5%. A further field-based study on BZ-R in H. contortus was carried out in Sao Paulo State, Brazil, by Niciura et al. (2012), using a different molecular method, ARMS-PCR. The frequency of the F200Y SNP was determined in L3 derived from 33 sheep flocks in the region. Resistant allele frequencies ranged from 9% to 74%, with resistance genotype frequencies of 0–66.7%. Resistant genotype frequencies >40% were associated with multiple risk factors, based on farm questionnaire data. These included being new sheep enterprises, the absence of farm records, the use of Dorper and Suffolk breeds, the frequent introduction of new animals into the flock, the use of whole flock treatments and failure to use treatment indicators, such as FAMACHA anaemia charts. These studies raise interesting practical considerations for the deployment of molecular genetic tests in the field, in that we do not currently have agreed guidelines in relation to resistant allele frequency, and what management interventions should follow, if any. These studies also raise technological issues with the tests themselves in that the H. contortus eggs tested had to be identified, isolated and assayed individually, adding greatly to labour, time and cost of testing. If such tests are to see widespread uptake by testing laboratories and/or the industry, they must be able to deal with multi-species, pooled samples. This is a significant technological challenge and a major bottleneck at this time, however pooled samples have been used to estimate β-tubulin allele frequencies (von Samson-Himmelstjerna et al., 2009) and the extraction of DNA from field-derived, multispecies samples is improving (McNally et al., 2013).
Assembly and annotation of the draft H. contortus genome (Laing et al., 2013) recently confirmed that the complete β-tubulin gene family in this species comprises four members (termed isotypes), with isotype -3 and -4 genes (Hco-tbb-iso-3 and Hco-tbb-iso-4) being identified as ‘new’ β-tubulin genes to add to the previously identified isotype-1 and -2 genes (Saunders et al., 2013). Although, the potential role of these two new H. contortus β-tubulin genes in BZ-R has not been directly studied, Saunders et al. (2013) argued that they are unlikely to be major determinants of resistance since they are orthologous to the C. elegans mec-7 and tbb-4 genes, respectively, which appear not to have a role in BZ-R in that organism. They are likely to have specialized functions, as they are both expressed at extremely low levels compared to isotypes 1 and 2 (∼1000-fold lower) with Hco-tbb-iso-3 expression being confined to just seven cells, thought to be touch receptor neurons (Saunders et al., 2013).
Recent population genetic studies, using field samples collected from sheep farms across the UK, have shed light on the possible origins and spread of BZ-R alleles (Redman et al., submitted). Seven farms with both H. contortus and T. circumcincta present were chosen for population genetic analysis to allow direct comparison of the two species under the same anthelmintic treatment regime without confounding environmental and management factors. High levels of F200Y were seen on most farms for both species. F167Y was almost as common as F200Y for H. contortus but was extremely rare in T. circumcincta. A previously unreported mutation E198L was identified in T. circumcincta on several farms. Interestingly, haplotypes comprising 167F:200Y and 167Y:200F were prevalent, but not 167Y:200Y, suggesting that this combination does not provide any additional advantage under BZ selection, and may not occur as previously found for H. contortus (Barrère et al., 2013b). Analysis of the distribution and phylogenetic relationships of resistance haplotypes demonstrated multiple independent origins of BZ-R in both species, even on these seven farms. The data suggested that recurrent new mutations are an important source of resistance mutations rather than pre-existing mutations alone, which has been the dogma until now.
Another recent population genetics study that may have implications for the spread of BZ-R alleles involves the identification of H. contortus/H. placei F1 hybrid worms in field populations from Pakistan (Chaudhry et al., unpublished). The F200Y BZ-R haplotype was found to be present in an F1 hybrid worm, opening up the potential for introgression of BZ-R alleles from H. contortus into H. placei. Finally, the first case of BZ-R has recently been reported in Nematodirus battus in the UK (Mitchell et al., 2011). This parasite is responsible for severe outbreaks of disease in lambs in spring and has always been considered to be naturally susceptible to BZ anthelmintics. Preliminary DNA sequence analysis has shown that BZ-R individuals, harvested from donor lambs post BZ treatment, carry the same F200Y mutation as most other BZ-R nematode parasites. There is no evidence yet of the presence of F167Y or E198A (Morrison et al., unpublished).
Since 2011, two publications have reported on the molecular analysis of BZ-resistance in human soil transmitted helminths. In an extension of their earlier work (Diawara et al., 2009), during which they had developed pyrosequencing assays for the analysis of F200Y for Ascaris lumbricoides and Trichuris trichiura, the authors established assays for F167Y and E198A for these parasites and assays for all three SNPs for the hookworm, Necator americanus (Diawara et al., 2013a). A mean 200Y allele frequency of 36% was observed in 25 pooled hookworm egg samples (each sample with 10 eggs) originating from a region in Haiti where BZ treatment was previously performed. Taking advantage of these molecular assays, Diawara et al. (2013b) conducted β-tubulin codon 167, 198 and 200 SNP allele frequency analysis in A. lumbricoides, T. trichiura and hookworm eggs isolated pre- and post-BZ treatment during worm control campaigns in Haiti, Kenya and Panama. All three above-mentioned sites were found to be polymorphic in T. trichiura eggs isolated pre- and post-treatment. Significant increases in heterozygous and homozygous resistant eggs were mainly found at codon 200. Changes in the 198 SNP were less pronounced and only documented in eggs isolated in Haiti. Noteworthy, for the 167 SNP, nearly 80% of the eggs from Panama showed the homozygous resistant genotype. With respect to A. lumbricoides eggs for all three study sites, polymorphism was only seen at position 167 for which the predominant genotype found pre- and post-treatment was the homozygous resistant genotype. With over 95%, the highest pre- and post-treatment codon 167Y frequencies were seen in ascarid eggs isolated in Panama, while in Haiti and Kenya, these ranged between 40 and over 70%, respectively. These findings are particularly noteworthy since, at the same time, the drug efficacy evaluated as ascarid egg reduction rate (ERR) was found to be high, with approximately 99%, 97% and 89% for samples from Haiti, Kenya and Panama, respectively. To the best of our knowledge, this is the first report where drug susceptibility rather than resistance was correlated with a high frequency of β-tubulin 167Y. However, the genotyping of A. lumbricoides needs to be interpreted with some caution as there are a large number of β-tubulin genes in the A. suum genome (at least nine, Gilleard unpublished) and hence more work is required to identify which of these may be involved in any observed field resistance to BZ drugs in Ascaris spp. Finally, in hookworms from all three countries, the frequency of eggs with the homozygous susceptible genotype 200F was at least 97%. All eggs analysed were 198E and 167F, which was consistent with good drug efficacy according to the faecal egg count data.
BZ resistance in parasitic nematodes of large ruminants is less prevalent than in parasites from small ruminants or horses. Accordingly, early detection of resistance is important since prevention of widespread BZ resistance is still an option. In this context, it seems an advantage to have molecular pyrosequencing-based assays for the analysis of the β-tubulin codon 167, 198 and 200 alleles now developed for some of the most prevalent and important gastrointestinal nematodes of cattle, for example C. oncophora, Ostertagia ostertagi and H. placei (Demeler et al., 2013a; Chaudhry et al., in press). The testing of field isolates originating from multiple countries revealed polymorphism at all three SNP sites in C. oncophora, O. ostertagi (Demeler et al., 2013a). The molecular data were in good agreement with the respective phenotype of all isolates as examined either by egg hatch assays or faecal egg count reduction tests. Interestingly, the codon 167Y allele frequencies were at least 70% in three out of five resistant isolates, whilst codon 200Y exceeded 70% in one isolate, and the fifth isolate displayed low resistance allele frequencies of 17% and 22% for 167Y and 200Y, respectively. This study also found that the isotype-1 and -2 β-tubulins of these two species of parasitic nematodes clustered with the C. elegans ben-1. Importantly, in terms of its relationship to drug resistance studies in human nematodes, this study also showed that the single β-tubulin gene in T. trichiura clustered with C. elegans tbb-4 and mec-7 β-tubulins, rather than with the C. elegans ben-1, suggesting that this T. trichiura gene may represent the first non-ben-1-like β-tubulin to be involved in BZ resistance. Recently, the P200Y polymorphism was detected at low frequency in six out of nine populations of H. placei isolated from cattle in southern and mid-west USA, indicating the risk of resistance emerging in this parasite should BZs be intensively used for parasite control in US cattle (Chaudhry et al., in press).
To conclude, thanks to various ongoing genome and transcriptome projects, our knowledge of the diversity of the tubulin gene family has been increased for some important parasitic nematodes (e.g. H. contortus, A. suum) and it seems appropriate to conclude that the level of complexity is higher than previously anticipated. We certainly need further insight into the identity of currently unknown tubulin genes and isotypes but, more importantly, into their biological role and their role, if any, in BZ-R. The recent development of tools for quantitative analysis of BZ-R-associated β-tubulin SNPs and their successful application in the field in parasites of human and veterinary importance, should encourage further progress towards routine molecular monitoring of the BZ-resistance status in the major parasite species, wherever this drug class is being used. Today, it is evident that the relevance for the BZ-resistance phenotype of the three β-tubulin SNPs, i.e. at positions 167, 198 and 200, respectively, differs between different parasite species and even between isolates of the same species. Accordingly, it is vital that we examine the allele frequencies of all three SNPs in parallel. However, more experimental data is still needed to better understand the functional significance of each of the three SNPs, that is, the correlation between the individual SNP allele frequencies (and potential combinations) and the phenotypic consequences. Two recent studies have highlighted this issue: Kotze et al. (2012) examined a population of H. contortus consisting of various genotypes at the 198 and 200 SNP positions and found that the most resistant individuals (as assessed in larval development assays) were homozygous resistant at the 198 position alone; in the second study, Barrère et al. (2012) examined a population of H. contortus consisting of a mixture of genotypes at the 167 and 200 SNP positions and found that the percentage of individuals homozygous resistant at the 200 SNP was significantly higher in a sub-population able to survive a high dose of albendazole in vivo compared to the population as a whole (in the absence of any drug treatment). On another note, it may also prove wise to remain open to the possibility of the combined presence of target-associated (i.e. β-tubulin), and non-specific (e.g. P-gps, drug metabolism) resistance mechanisms.
5. Pharmacology of levamisole, derquantel and abamectin: recent observations
Levamisole and pyrantel target the nicotinic acetylcholine receptors (nAChRs) of nematodes. nAChRs are composed of 5 subunits that together form a transmembrane ion-channel. The channel receptor is opened by a ligand, which is normally acetylcholine, to allow entry of Na+ and Ca++, producing a physiological response. The nAChRs that have been studied most in nematodes are found on the somatic muscle cells, but nAChRs are also present on pharyngeal muscle (McKay et al., 2004) and on the nerve cells (Segerberg and Stretton, 1993). Opening of the nAChRs on the somatic muscle gives rise to muscle depolarization and contraction. A large (30+) number of different nAChRs subunits are present in nematode parasites (Laing et al., 2013), and these may combine in different pentameric structures, to give rise to an even larger number of possible pentameric nAChRs. We do not yet know which receptor subunit combinations are permissible, and which are not. The various subunit combinations give rise to receptors that are pharmacologically different, and sensitive to different cholinergic anthelmintics (Williamson et al., 2009; Boulin et al., 2011; Buxton et al., 2014).
The levamisole-activated nAChR receptor is believed to be composed of four different receptor subunits (UNC-29, UNC-38, UNC-63 and ACR-8), with one repeated to make up the pentamer, in H. contortus (Boulin et al., 2011) and O. dentatum (Buxton et al., 2014) (Fig. 2A). The expression of O. dentatum subunits in Xenopus oocytes gives rise to receptors that are activated by levamisole (Fig. 2B). One of the interesting observations is that different combinations of receptor subunits, give rise to receptors that have different pharmacological properties. For example, expression of the subunits UNC-63 and UNC-29 gives rise to a receptor that is sensitive to the drugs pyrantel and tribendimidine, but a receptor which is not very sensitive to levamisole or acetylcholine. If the receptor subunit ACR-8 is co-expressed with UNC-63 and UNC-29, acetylcholine becomes more potent. If the receptor subunits UNC-63, UNC-29 and UNC-38 are expressed, a receptor more sensitive to pyrantel than levamisole is observed. If all four receptor subunits are co-expressed, then a receptor that is most sensitive to levamisole is produced. We conclude from these observations that different pharmacological receptor subtypes can be produced by different combinations of subunits.
Fig. 2.
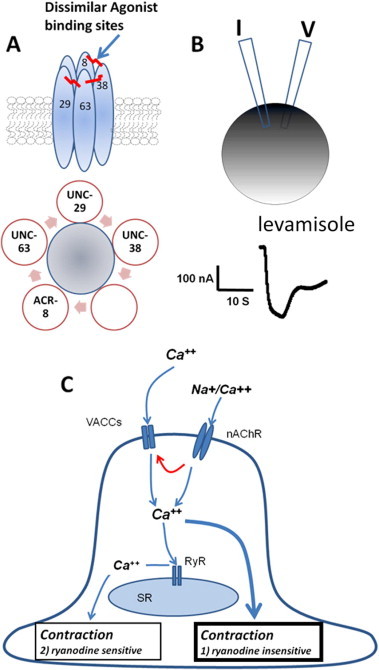
Mechanistic and structural features of muscle nematode somatic muscle ion channels. (A) Diagram of the putative pentameric subunit composition of the levamisole receptor in Oesophagostomum dentatum composed of one or more subunits of UNC-63, UNC-29, UNC-38, and ACR-8. (B) Two-micropipettes used for a two electrode voltage-clamp oocyte recordings of expressed nAChRs from O. dentatum. (C) Diagram of a proposed mechanism of calcium entry and muscle contraction in Ascaris suum muscle, with entry through the sarcolemma via calcium permeable nicotinic acetylcholine receptors (nAChRs activated by levamisole) and voltage-activated calcium channels (VACCs) which produce the biggest component of contraction (ryanodine-insensitive), and another component of contraction (ryanodine-sensitive) mediated by a calcium-induced calcium-release via the ryanodine receptors in the sarcoplasmic reticulum.
5.1. Levamisole receptor contraction coupling: ryanodine receptors are also involved
The coupling process between opening of levamisole-activated nAChRs and muscle contraction is not direct, and is subject to physiological control. The amplitude of contraction produced by levamisole is dependent on extracellular calcium, and is inhibited partially, but not completely, by ryanodine and dantrolene, indicating a role for ryanodine receptors (Robertson et al., 2010a; Puttachary et al., 2010). These ryanodine receptors have the potential to be exploited as therapeutic targets, as they have in insects (Isaacs et al., 2012). Nematode muscle contraction may also be produced by high concentrations of caffeine, which is known to cause calcium release from the endoplasmic reticulum (Puttachary et al., 2010). The potency and effect of levamisole is also altered by FMRFamide peptides: the neuropeptide AF2 increases the amplitude of contraction produced by activation of nAChRs (Trailovic et al., 2005). These observations show that the effects of levamisole, although mediated via nicotinic receptors, are under more complex regulation (Fig. 2C). This coupling between contraction and levamisole receptor opening may be subject to modification, and may contribute to the development of resistance.
5.2. Derquantel: potency on different nAChR subtypes, and effects alone or in combination with abamectin on Ascaris muscle nAChRs and pharyngeal GluCls.
Derquantel is a selective antagonist of nematode nicotinic acetylcholine receptors (Qian et al., 2006). The compound has a greater antagonistic effect on the pyrantel-sensitive receptor than on the levamisole-sensitive receptor (Buxton et al., 2014). The implication of this observation is that derquantel may remain active as an anthelmintic in the presence of some types of levamisole resistance.
Derquantel and abamectin are used in combination in the oral drench product StartectR. We were interested to determine the effects of derquantel and abamectin alone and in combination on different tissues of the nematode parasite Ascaris suum (Puttachary et al., 2013). Acetylcholine caused dose-dependent isometric contractions in an Ascaris muscle flap preparation (Fig. 3A). The application of 1 μM derquantel caused inhibition; this inhibition increased in the presence of 0.3 μM abamectin. The antagonism showed some reversal on washing. The effect of abamectin was to increase the antagonism of derquantel in a dose-dependent manner (Fig. 3B). Puttachary et al. (2013) also used a two micro-electrode current-clamp technique to observe the effects of derquantel on acetylcholine responses in Ascaris muscle. Derquantel at 0.1 μM significantly reduced the responses to 3 μM acetylcholine applied to the Ascaris muscle preparations. The inhibitory effects of derquantel on the acetylcholine responses were reversible; washing returned the acetylcholine currents to control values. The IC50 of derquantel was 20 nM demonstrating that derquantel is a potent antagonist of acetylcholine receptors. Both derquantel and abamectin produced antagonism of the acetylcholine responses. Again, like the effects on muscle contraction described above, abamectin potentiated the effects of derquantel on acetylcholine depolarisations (Puttachary et al., 2013). These authors also tested the effects of abamectin and derquantel on Ascaris pharynx GluCl receptors. They found that abamectin produced hyperpolarization, and an increase in membrane conductance which was dose-dependent. On the other hand, derquantel had no effect on pharynx membrane potential or conductance.
Fig. 3.
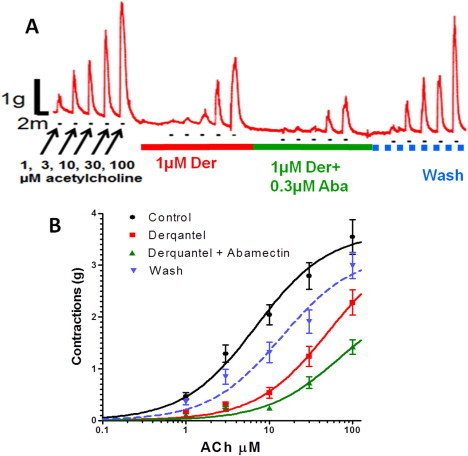
Effects of derquantel and abamectin on Ascaris suum muscle strips. (A) Isometric contraction of Ascaris suum muscle strips produced by application of increasing concentrations of acetylcholine, and antagonism by 1 μM derquantel (red bar), 1 μM derquantel+0.3 μM abamectin (green bar), and wash (blue bar). Note that derquantel decreases the responses to acetylcholine and that the addition of abamectin increases the inhibition. (B) The concentration-depolarizing-response plot of acetylcholine showing mean ± S.E. (n = 11). Control (black); in the presence of 1 μM derquantel (red); 1 μM derquantel+0.3 μM abamectin (green) and wash (blue). Note that abamectin increases the inhibition produced by derquantel (Figure modified from Puttachary et al., 2013). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
5.3. Levamisole resistance
The mechanisms of laboratory-produced levamisole resistance have been studied in the model nematode C. elegans. There are a large number of genes that are involved in the observed resistance. These genes code for a levamisole-sensitive muscle nAChR (Fleming et al., 1997), the contraction signaling pathway, (eg. Kagawa et al., 1997), the processing and assembly of subunit ancillary proteins (Gottschalk et al., 2005), and the support and maintenance of the levamisole-receptor (Hobert, 2013). However, the mechanism of clinical levamisole resistance in parasitic nematodes is less well understood, and may include some different mechanisms and genes. Kopp et al. (2009) reported reduced expression of several nAChR subunit genes associated with a decreased sensitivity to pyrantel in an isolate of A. caninum. Neveu et al. (2010) reported the existence of abbreviated isoforms of unc-63, referred to as unc-63b, in resistant isolates of H. contortus, T. circumcincta and T. colubriformis. The Hco-unc-63b gene encodes a truncated isoform that was subsequently shown to have a dominant negative effect on the levamisole nAChR expression in Xenopus oocytes (Boulin et al., 2011). In addition, a truncated acr-8 mRNA splice variants of the acr-8 gene, referred to as Hco-acr-8b, was found to be expressed in several H. contortus levamisole-resistant isolates, highlighting its potential as a marker for levamisole resistance (Fauvin et al., 2010; Williamson et al., 2011). Recently, the genetic basis giving rise to the expression of this Hco-acr-8b truncated transcript was reported to be likely associated with an insertion/deletion of 63 bp located just downstream from the splice acceptor site for the alternative third exon of the Hco-acr-8 gene (Barrère et al., 2014). Neveu et al. (2010) reported the presence of 4 paralogues of the gene unc-29: Hco-unc-29.1, Hco-unc-29.2, Hco-unc-29.3 and Hco-unc-29.4. These observations suggest that the levamisole receptor of some parasitic nematodes may not be the same as those found in C. elegans. The subunits that arise from these 4 paralogues may not function in the same way, with some rendering the receptor pentamer less sensitive to levamisole. Sarai et al. (2013) examined the expression of levamisole receptor subunit genes of H. contortus and found evidence of reduced expression levels of Hco-unc-63a and all four paralogues of Hco-unc-29 in resistant isolates. Sarai et al. (2014) subsequently examined subpopulations within a heterogeneous isolate of H. contortus and found that expression of several P-gp genes was increased in larvae showing a low level of resistance to levamisole, while expression of some receptor subunit genes, as well as ancillary protein genes, were decreased in the most resistant larvae in the population. This suggested the presence of multiple mechanisms of resistance within the same population (drug efflux and altered target site), with the various mechanisms conferring different levels of resistance. Recently, Romine et al. (2014) compared levamisole-sensitive O. dentatum adult males with levamisole-resistant O. dentatum adult males and found that expression of Ode-unc-63 was reduced, and that expression of acr-21 and acr-25 increased, perhaps as a compensatory mechanism. There were also 4 SNPs that were associated with the resistant isolate. Taken together, this body of literature suggests that resistance to cholinergic anthelmintics in parasitic nematodes is polygenic rather than a simple single-gene mechanism, and involves changes in expression of nAChR subunits, truncated receptor subunits, and mutations in receptor subunits, as well as a possible contribution from P-gps. This situation complicates our ability to develop effective molecular-based diagnostic tools for the detection of resistance to this drug class.
6. Amino-acetonitrile derivatives: mode of action
Monepantel was discovered through an intensive drug screening program by Novartis Animal Health (Hosking et al., 2009). The first anthelmintic lead of the amino-acetonitrile derivatives (AADs) in 2000 was the starting point for the synthesis and subsequent in vitro and in vivo evaluation of more than 700 analogues of AADs, arriving finally at monepantel (Kaminsky et al., 2008b). This compound is remarkably efficient against numerous multidrug-resistant nematodes (Kaminsky et al., 2008a; Sager et al., 2012), pointing to a different mode of action than other anthelmintic drugs. Monepantel was introduced to the market in 2009 in New Zealand and is today commercially available as Zolvix® in most sheep farming countries of the world.
The mode of action of monepantel was primarily investigated by identifying the drug’s target in the genetic model organism C. elegans. In vivo studies demonstrated that monepantel interfered with the worm’s movement, growth and viability (Kaminsky et al., 2008a). This manifests as a hyper-contraction of the body wall muscles leading to paralysis, spasmodic contractions of the anterior portion of the pharynx, and ultimately death (Rufener et al., 2013). In genetic screens for resistance to monepantel, 27 independent mutations were identified in acr-23, a gene coding for a putative nicotinic acetylcholine receptor (nAChR) subunit. Therefore, acr-23 was considered likely to be a major candidate contributor to the AAD response in C. elegans (Kaminsky et al., 2008a). The ACR-23 protein belongs to the nematode-specific DEG-3 subfamily of nAChR subunits (Rufener et al., 2010). The nAChR subunits targeted by the currently available nicotinic agonist anthelmintics (e.g. levamisole) are different from ACR-23, explaining the absence of cross-resistance between monepantel and the other anthlemintics (Kaminsky et al., 2008a).
To investigate the function of ACR-23, a green fluorescent protein (GFP) fused in-frame with acr-23 was used to localize ACR-23 in vivo expression in C. elegans (Rufener et al., 2013). Integrated lines expressed GFP::ACR-23 in various tissues, including body wall muscles, and head and tail neurons (Fig. 4A, B); no expression was observed in eggs or first stage larvae. This expression pattern (muscles and neurons) is in agreement with the paralysis phenotype observed with nematodes exposed to monepantel. More insights into the expression and possible physiological role of acr-23 came from a recent publication reporting that the acr-23 gene was strongly expressed in the six mechanosensory neurons in C. elegans (Peden et al., 2013). These neurons innervate the locomotory command neurons to stimulate touch-induced movement as well as spontaneous locomotion (Chalfie et al., 1985).
Fig. 4.
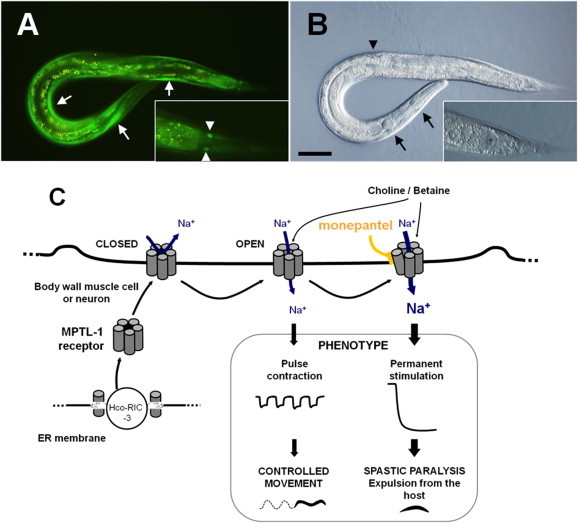
Expression pattern of monepantel receptor in C. elegans, and model for the interaction of the drug and its receptor in H. contortus. (A) Expression of ACR-23. Transgenic L4 larva containing an integrated array expressing the acr-23 open reading frame fused to the green fluorescent protein gene. Transgene expression was mainly visible in the body wall muscle bundles (white arrows), and in two unidentified cells, which are neither the PLM neurons nor body wall muscle cells nuclei (white arrowheads in the inset, which shows a magnification of the tail). Gut granules emit yellow autofluorescence. (B) Image taken by differential interference contrast microscopy. Black arrows and arrowhead indicate the pharyngeal bulbs and the position of the developing vulva, respectively. The inset shows a detail of the tail, ventral view. The rectal opening (asterisk) is immediately anterior to the two GFP labelled cells in A. Bar, 50 μm. (C) Hypothetical model for the interaction of monepantel with its target receptor in H. contortus, Hco-MPTL-1. In the resting situation, the MPTL-1 receptor is closed and no ion is flowing through the channel. The neurons or muscle cells are silent respectively not contracted. When the receptor-agonist (e.g. choline or betaine) is released from a presynaptic or potentially an epidermal cell, it binds to the MPTL-1 receptors present at the postsynaptic nerve cell or at the body wall muscle cell. An inflow of Na+ ions enters the cell through the pore formed by the opened receptor, creating a depolarization of the cell membrane. This leads to the stimulation of the nerve cell or to the pulse contraction of the muscle cell and finally a controlled movement. The interaction of monepantel with MPTL-1 results in a permanent stimulation or contraction creating a spastic paralysis of the nematode and its expulsion from the host. The ancillary protein RIC-3, which is resident in the endoplasmic reticulum (ER), may play a role for the assembly of the receptor containing MPTL-1. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In order to functionally characterize the ACR-23 protein, which we hypothesized to function as a membrane receptor, Xenopus laevis oocytes were injected with acr-23 cRNA and inward current was recorded using a Two Electrodes Voltage Clamp (TEVC) after addition of an agonist (Rufener et al., 2013). Choline proved to be a more potent agonist than acetylcholine or nicotine. Currents measured with choline were characterized by a fast channel opening followed by a slow desensitization. Oocytes rapidly recovered to the initial resting membrane potential once the agonist was washed away. A more recent publication describes betaine, a ubiquitous non-canonical amino acid, as the natural agonist of the ACR-23 receptor in C. elegans (Peden et al., 2013).
The potential of monepantel to modulate the ACR-23 receptor was tested both in the absence and presence of an agonist. In the absence of any agonist, monepantel showed a strong agonistic effect on the channel at concentrations higher than 0.3 mM, enhancing the observed maximum current peak measured as well as the kinetics of the channel, producing characteristic “V” shape like currents (Rufener et al., 2013). At lower concentrations, monepantel was found to act as an allosteric modulator of choline and betaine gating (Rufener et al., 2013; Peden et al., 2013). For instance, at 300 pM, a concentration at which monepantel does not activate the ion channel by itself, the desensitization of the ACR-23 receptor is blocked when coupled with 1 mM betaine.
The first investigation to understand the mode of action of monepantel in H. contortus was performed using freshly harvested adult nematodes exposed in vitro to various concentrations of the drug. Treated worms were almost completely paralyzed but still able to move head and tail sections. This was in agreement with the localization of the ACR-23 receptors in C. elegans and a strong indicator that monepantel was targeting or interfering with neuromuscular signal transmission in C. elegans and in parasitic nematodes. In order to elucidate the molecular mechanism of monepantel, a forward genetic approach was followed (Rufener et al., 2009a). The primary step was to select in the laboratory a H. contortus population able to survive a full-dose treatment of monepantel in sheep. For this, a new “in vitro selection”–“in vivo propagation” protocol was developed (Rufener et al., 2009a,b), which allowed the successful selection of two independent AAD-mutant lines, Hc-CRA AADM and Hc-Howick AADM (Kaminsky et al., 2008a; Rufener et al., 2009b). With the discovery of acr-23 in C. elegans, it was possible to identify a potential homologue in H. contortus, named mptl-1. A panel of loss-of-function mutations were identified in the Hco-mptl-1 gene in AAD-mutant lines, providing further evidence that this subunit is the most likely target for AAD action against H. contortus (Rufener et al., 2009a).
The identification of monepantel-resistant T. circumcincta and T. colubriformis recovered from a goat farm in New Zealand provides a new means to investigate potential target sites in these species (Scott et al., 2013). Given our knowledge of MPTL-1 in H. contortus, and the presence of mutations in the Hco-mptl-1 gene associated with resistance in a laboratory isolate of this species, it will be of interest to determine whether changes in the homologous genes in these two other species are associated with the observed resistance.
Our current understanding of the molecular mode of action of monepantel is illustrated in Fig. 4C. Briefly, infected sheep are treated orally with monepantel; the drug may affect the nematodes either by direct contact or after absorption via the blood. Monepantel reaches the acetylcholine receptors containing the MPTL-1 subunits present in body wall muscle cells or neurons. Upon binding of the drug, the monepantel-sensitive receptors remain open allowing an unrestricted inflow of cations that produces a constant depolarization of the membrane of muscle or neuronal cells. Consequently, spastic paralysis of the nematodes leads to their expulsion from the host, and ultimately to their death.
7. Cyclooctadepsipeptides: mode of action
The cyclooctadepsipeptides are a relatively new class of drugs, and have up to now only been licensed as a combination of the semi-synthetic emodepside with either praziquantel (Profender®) or toltrazuril (Procox®), both for use in cats and dogs. The parental component PF1022A is a fermentation product of the fungus Rosellinia spp. PF1022, forming a sterilia mycelia on the leaves of the plant Camelia japonica (Krücken et al., 2012). The parent compound, PF1022A, which is much cheaper than emodepside, has recently been compared with emodepside for treatment of Trichuris muris, as a model for the human whipworm (T. trichiura), which is the dose-limiting gastrointestinal nematode in humans and particularly problematic due to its long period of prepatency. Although emodepside was much more effective than PF1022A after intraperitoneal and subcutaneous application, only a small difference was detected after oral application (Kulke et al., 2014). It was also shown to be able to eliminate all stages of T. muris including histotropic L1. Using an optimised formulation will probably allow reduced dosages (and costs) and suggests that PF1022A could be a prospect for the treatment of the soil transmitted helminths which infect humans.
As this drug class has been on the market for only a limited number of years, and has not yet been marketed for those host species where anthelmintic resistance has been observed to arise most rapidly for other drug classes (i.e. small ruminants and horses), it is not surprising that there are currently no reports of resistance towards emodepside. Nevertheless, the body of research dealing with the mode of action of cyclooctadepsipeptides is seen as very relevant to the study of anthelmintic resistance markers as the receptors involved in the drugs’ action provide a likely site for molecular changes which may confer resistance if it does arise in the field. Over the last couple of years there have been considerable efforts to summarise the current knowledge on the mode of action of this drug group, and to bring the results of different studies together to build new hypotheses (Holden-Dye et al., 2012; Krücken et al., 2012; Martin et al., 2012b). SLO-1 is widely accepted as the major target of emodepside, particularly since Crisford et al. (2011) showed that ectopic expression of SLO-1 in the pharynx muscle of slo-1 deficient C. elegans rendered pharyngeal pumping susceptible to emodepside. LAT-1 is also considered to be a secondary target. However, older data presented for the first time in the review of Holden-Dye et al. (2012) indicate that C. elegans mutants deficient in signalling downstream of LAT-1 (but also of many other G-protein coupled receptors such as G proteins and their interaction partners) are more resistant to emodepside than strains deficient in the receptor gene lat-1 itself. This suggests that not only signalling through LAT-1 but maybe also through other unidentified G protein coupled receptors (Buxton et al., 2011) might be able to modify the effects of emodepside. One possibility is that these signalling pathways alter SLO-1 by posttranslational modifications such as phosphorylation.
The involvement of GABAA receptors as immediate targets of emodepside was discussed more controversially at the CARS2013 meeting. Although very early work using A. suum preparations suggested binding of emodepside in competition to the GABAA receptor agonist methyl-bicuculline (Chen et al., 1996), the fact that the latter drug does not act as a GABA agonist on Ascaris muscle preparations (Holden-Dye et al., 1988) is an important argument against binding to ionotropic GABA receptors. In contrast, slightly increased emodepside susceptibility of C. elegans deficient in the GABAA receptor unc-49, which could be reversed by transgenic expression of unc-49b cDNA of Toxocara canis, suggests that ionotropic GABA signalling somehow also modulates emodepside effects on nematodes (Miltsch et al., 2012).
8. Triclabendazole: mapping resistance loci in genetically recombinant Fasciola hepatica
The BZ compound, triclabendazole (TCBZ), is the drug of choice for controlling Fasciola hepatica infection in livestock and humans due to its ability to target the parasite at the earliest stages of infection as it migrates through the liver. Since TCBZ resistance was first reported nearly 20 years ago in Victoria, Australia, in 1995 (Overend and Bowen, 1995) cases of resistance in both sheep and cattle have been reported in multiple countries, most recently in naturally infected Australian beef and dairy cattle herds (Brockwell et al., 2013). The first report of the failure of TCBZ in humans has also recently emerged (Winkelhagen et al., 2012). Despite significant advances in our understanding of BZ resistance, and identification of BZ resistance markers for several nematode species, the specific nature of the mode of action of TCBZ against F. hepatica, and mechanism of TCBZ resistance, remain unknown.
The majority of TCBZ resistance studies have relied on comparative analyses of phenotypically TCBZ-resistant (TCBZ-R) and TCBZ-susceptible (TCBZ-S) laboratory and/or field isolates following exposure to TCBZ either in vitro or in vivo. These studies have primarily focused on changes in biological parameters (reviewed by Fairweather, 2011) or on defining biochemical pathways involved in drug uptake, efflux and metabolism (e.g. Robinson et al., 2004; Alvarez et al., 2005). Most recently, detecting apoptotic events in the reproductive organs of F. hepatica was proposed as an aid in the diagnosis of TCBZ resistance in field outbreaks of fasciolosis (Hanna et al., 2013). However, very few studies have aimed to identify genetic markers for TCBZ resistance. Given that TCBZ is a BZ, these analyses have echoed candidate marker gene studies (primarily tubulins) that proved successful for BZ resistance in nematodes (see BZ section above). However, there remains little evidence for a role for tubulins in TCBZ resistance (Ryan et al., 2008; Fuchs et al., 2013). Similarly to anthelmintic resistance studies in other helminths, drug efflux mechanisms mediated by P-gps have been highlighted as potential contributors to TCBZ resistance (Reed et al., 1998; Meaney et al., 2103). Wilkinson et al. (2012) reported single nucleotide polymorphisms (SNPs) in the nucleotide binding domain of P-gp genes between a small number of TCBZ-S and TCBZ-R adult F. hepatica from the field. However, further data presented here at the CARS2013 meeting, using an increased sample size of adult F. hepatica, found no evidence to support the involvement of the P-gp T687G SNP in TCBZ resistance (Skuce et al., unpublished).
Given the lack of genetic markers for TCBZ resistance in F. hepatica, the speaker for this section of CARS2013 (JH) described a novel project to map loci involved in conferring TCBZ resistance using a genome-wide analysis of genetically recombinant F. hepatica that was the subject of a recent review paper (Hodgkinson et al., 2013). Key to the success of the project was the ability to exploit the parasite life-cycle and produce genetic crosses of TCBZ-R and TCBZ-S isolates. Given the diversity of F. hepatica populations in the field (Elliott et al., 2014; Hodgkinson, unpublished) the speaker reported the generation of clonal isolates of F. hepatica derived by infecting sheep with metacercariae shed from snails experimentally infected with single miracidia. The production of three TCBZ-R clonal isolates, using parasites recovered from naturally infected UK sheep, was described in a series of, as yet unpublished, experimental studies. Similarly, a TCBZ-S clonal isolate was generated using a laboratory maintained isolate known as the Shrewsbury isolate (Ridgeway Research, UK). For subsequent mapping of resistance markers, genetic crosses of TCBZ-S and TCBZ-R clonal isolates were reported as underway, assisted by the use of neutral microsatellite markers to dissect the complex reproductive biology of F. hepatica. In the absence of a genome for F. hepatica within the international parasitology research community, the speaker reported that they had generated a ∼1.3 Gb draft genome from a single adult parasite of the Shrewsbury clonal isolate. Future production of F2 progeny was reported to be in progress along with genome-wide SNP profiling of F2 populations exposed to TCBZ in vitro and in vivo. Pooled genotyping of phenotyped F2 progeny to localise regions of the genome associated with TCBZ resistance was proposed.
In conclusion, recent work on genomic and genetic tools for F. hepatica and production of clonal TCBZ-S and TCBZ –R parasite isolates has forged the way for non-biased discovery of drug resistance markers and genes conferring resistance in trematodes of humans and livestock in the future. Importantly, the clonal isolates described here represent the first such resource for this important parasite species and are the current focus of complementary studies involving specific TCBZ-R candidate genes. Although the focus of the project was mapping anthelmintic markers, a number of valuable genomic, genetic and parasite resources were generated that are already proving invaluable to the wider community of F. hepatica researchers. This highlights the fact that studies to detect drug resistance markers are relevant in their broadest context and can be the driving force for advancing knowledge of many aspects of the biological systems under study.
9. Drug transporters: potential markers for resistance, and therapeutic targets
9.1. Role of MDR transporters in multidrug resistance in mammals, and anthelmintic resistance in nematodes
Multidrug resistance (MDR) ABC transporters belong to an evolutionarily well-conserved family of ATP-binding-cassette membrane proteins. They comprise the so-called P-glycoprotein (P-gp or MDR1 of the ABCB family), the Multidrug Resistance-associated Proteins (MRPs of the ABCC family) and Breast Cancer Resistance Protein (BCRP) from the ‘half-transporter’ subfamily (ABCG2). Their main function is the active transport of a number of structurally unrelated endogenous and exogenous compounds including a large range of drugs (Gottesman and Pastan, 1993). They provide complementary and overlapping activities as multispecific drug efflux pumps, and are involved in multidrug resistance in cancer cells. Interestingly, homologues of these transporters are found in many pathogens such as protozoa, fungi, bacteria and also in insects, and have been involved in drug resistance through an increase of MDR transporter gene expression.
Ivermectin and other MLs are transported by mammalian P-gp which is responsible for their elimination by the intestinal and biliary tract (Ballent et al., 2006; Kiki-Mvouaka et al., 2010) and for maintaining low drug concentration inside the brain, guaranteeing the safety of these drugs (Mealey et al., 2001). In addition, the high affinity of MLs for P-gp (Lespine et al., 2007) and to a lesser extent for MRP (Lespine et al., 2006) and BCRP (Jani et al., 2011), suggests that combined efflux-pumping activities of these transporters may have an impact on pharmacokinetics, efficacy and safety of these drugs.
Nematodes have a number of genes homologous to ABCB, ABCCs and half-transporters, and there is increasing evidence that the products of at least some of these genes, are involved in drug transport. The crystal structure of C. elegans P-gp1 has provided valuable information on the three dimensional structure of the transporter, and several putative substrate and drug binding sites, similar to those found in human P-gp, have been identified (Jin et al., 2012), which strongly suggests that this protein is able to transport chemicals. Loss of function of individual P-gps in C. elegans results in significant increases in ivermectin susceptibility, revealing that all the P-gps in this species interact with this ML, with P-gp1, 2, 8, 9, 11 and 12 being more important than the others (Janssen et al., 2013b). Thus, P-gps of nematodes clearly contribute to lower drug efficacy, probably by diminishing exposure of the target to the drug, and this may be the basis for favouring the development of multidrug resistance not only against MLs but also other anthelmintics.
Modulation of ABC transporter genes has been reported in free-living and parasitic nematodes exposed to ivermectin (Ardelli and Prichard, 2013; Lespine et al., 2012). In C. elegans, several P-gp genes are overexpressed after short-time exposure to ivermectin or moxidectin (Ardelli and Prichard, 2013). Similarly, step-wise exposure to increasing doses of ivermectin results in multidrug resistance and overexpression of several ABC transporters (James and Davey, 2009). Furthermore, silencing of ABC transporter gene expression by RNA interference induced a change in phenotypic response to ivermectin exposure (Yan et al., 2012). In addition, constitutive overexpression of a number of P-gp genes has been reported in multi-drug resistant H. contortus isolates (Williamson et al., 2011; Sarai et al., 2014). A constitutive increased expression of Tci-pgp9, with increased gene sequence polymorphism, was reported in multidrug-resistant T. circumcincta (Dicker et al., 2011). Similarly, pgp-11 showed increased expression in ivermectin-exposed resistant populations of Parascaris equorum (Janssen et al., 2013a) and C. oncophora (De Graef et al., 2013; Demeler et al., 2013b). Bygarski et al. (2014) have recently used ML-resistant and P-gp deletion strains of C. elegans to show that resistance to moxidectin in this species is mediated at least in part by P-gps. Moreover, decreased ivermectin susceptibility has also been associated with genetic variation of ABC transporter homologues in P. equorum (Janssen et al., 2013a), Onchocerca volvulus (Ardelli and Prichard, 2007; Osei-Atweneboana et al., 2007, 2011) and Dirofilaria immitis (Bourguinat et al., 2011). All these studies are consistent with a role for some P-gps in ivermectin susceptibility in nematodes, and together they raise the possibility that P-gp activity/gene expression/SNPs could be a component of a set of diagnostic tools for the detection and quantification of anthelmintic resistance, particularly for the MLs. However, the likely polygenic nature of resistance to MLs will mean that tests based on drug transporters will most-likely be only a part of such a diagnostic kit.
9.2. Therapeutic potential: targeting drug transporters in order to increase the efficacy of anthelmintics
Although ML resistance is polygenic, the final concentration of an anthelmintic in the parasite is a key determinant for efficacy (Lloberas et al., 2013), making the drug efflux transporters of central interest. The challenge is to minimize the drug transporter pumping action in order to increase drug sensitivity (Nobili et al., 2006). Inhibition or modulation of P-gp activity therefore represents a possible strategy for worm control. A large number of compounds have been shown to inhibit MDR transporters. Co-administration of MLs and these inhibitors is presumed to lead to an increase of ML concentration in the plasma and tissues of the host and hopefully in the parasite, thus offering the expectation of increased anthelmintic efficacy (Lespine et al., 2008). Such a strategy has been investigated in vivo in animals infected with ivermectin-resistant nematodes. When combined with loperamide, ML efficacy was improved in cattle and sheep (Lifschitz et al., 2010a,b). In addition, pluronic P85 enhanced the anthelmintic activity of ivermectin against ivermectin-resistant H. contortus isolates in sheep (Bartley et al., 2012). These results strongly support the hypothesis that MDR transporter inhibitors enhance efficacy of MLs by blocking host P-gp, and possibly by exerting direct effects on the nematode MDR transporters. Accordingly, the effectiveness of inhibitors of MDR transporters has been demonstrated in vitro and in vivo using ML-resistant C. elegans (James and Davey, 2009), H. contortus, T. circumcincta (Bartley et al., 2009) and C. onchophora (Demeler et al., 2013b). In these studies, the combination of inhibitors of P-gp, such as verapamil, partially or fully restored drug sensitivity, offering hope for future sustainable anthelmintic-based therapy.
However, in looking to develop such a therapeutic approach to worm control it is important to separate the potential effects on the target worm from the effects on the host; that is, to separate the desirable aim of inhibiting the nematode’s drug transporters (hence increasing the amount of drug reaching its target within the worm) from the undesirable effect of inhibiting the host animal’s drug transporters, with the negative consequences this may have in terms of neurotoxicity and maintenance of homeostasis. Hence, the current dogma is that for a reversing agent to be successful, nematode specificity is crucial in order to preserve the activity of related transporters in mammals. The speaker for this session at CARS2013 (AL) described unpublished work from their laboratory aiming to address this need. While ML drugs have themselves been shown to be potent inhibitors of P-gps, and hence have been proposed as reversing agents for multidrug resistance (Lespine et al., 2008), the commercial ML compounds are not suitable for this purpose due to neurotoxic effects in mammals (Mealey et al., 2001). Indeed, when co-administrated with a P-gp substrate drug, they may accumulate in the brain at high concentrations and be neurotoxic due to their effects in opening mammalian GABA receptors in the central nervous system (Menez et al., 2012). However, many non-commercial ML derivatives exist, and it was reasoned that among these may exist some compounds with low affinity for nematode glutamate and GABA receptors (hence of no use as commercial anthelmintics), and hence likely to also exhibit low affinity for mammalian GABA receptors, alongside useful P-gp-inhibiting activity. One such candidate has been identified as ivermectin aglycone (Lespine et al, unpublished data). This compound has little anthelmintic activity against parasitic nematodes (Shoop et al., 1995), suggesting that it has low activity on glutamate-gated chloride channels, and perhaps on GABA-receptors. The speaker reported that ivermectin aglycone inhibits P-gp transport activity in cells overexpressing human P-gp with a similar efficiency as ivermectin. When co-incubated with vinblastin, ivermectin aglycone restored drug sensitivity in vinblastin-resistant cells overexpressing P-gp with a similar degree of reversal in potency as ivermectin. Ivermectin aglycone by itself had little anthelmintic effect on ivermectin-resistant C. elegans, but used in combination with ivermectin it restored sensitivity to the latter. Moreover, unlike ivermectin, ivermectin aglycone had low affinity for rat GABA receptors and had no observable neurotoxicity in P-gp-deficient mice. Its ability to reverse P-gp-mediated drug resistance in mammalian cells and in nematodes and its very low neurotoxicity make ivermectin aglycone an interesting P-gp reversing agent for use in vivo. Combining ivermectin aglycone with MLs to increase drug efficacy and to reverse ML resistance opens a new perspective for sustaining anthelmintic control when ML resistance has arisen.
The speaker also noted that because many ABC transporters are present in nematodes, it is difficult at this stage to anticipate which transporters may be involved in resistance to specific ML drugs. Targeting upstream transcriptional regulators of ABC transporters might control the expression of several transporters at the same time, and hence improve anthelmintic drug efficacy in susceptible and resistant nematodes. The success of such approaches relies on improving knowledge of the proteins involved in ML transport within nematodes, and the mechanism of their regulation.
10. Advances in high-throughput sequencing and analyses
Recent years have seen major advances in the development and application of high-throughput sequencing (HTS) technologies and their potential application in exploring the development of anthelmintic resistance (Fig. 5). Draft genome sequences now exist for a number of animal and plant parasitic nematode species and more genome projects are being completed at an accelerating pace. Beginning with the sequencing of the Brugia malayi genome in 2007 (Ghedin et al., 2007), draft genome sequences have been published for Meloidogyne hapla (Opperman et al., 2008), M. incognito (Abad et al., 2008), Trichinella spiralis (Mitreva et al., 2011), A. suum (Jex et al., 2011), D. immitis (Godel et al., 2012), H. contortus McMaster (Schwarz et al., 2013) and H. contortus MHco3(ISE) (Laing et al., 2013), Loa loa (Desjardins et al., 2013) and Trichuris suis (Jex, unpublished). A large number of additional genome studies are currently under-way (see 959 Nematode Genome Project (www.nematodes.org) and 50HGI Project (www.sanger.ac.uk)), including a number of projects on ruminant strongyle species of major importance for veterinary health and for which anthelmintic resistance is prevalent (e.g., C. oncophora, O. dentatum and T. circumcincta: see www.nematode.net).
Fig. 5.
Utility of high-throughput sequencing (HTS) to explore the development of anthelmintic resistance. Four separate HTS applications shown as coloured boxes in columns A–D, with single horizontal black box representing generalized features of library construction, sequencing and post-sequencing quality control applicable to most HTS applications (∗fragmentation not required for small RNA sequencing). (A) Blue boxes present the detection of genetic mutations through de novo assembly and subsequent comparative alignment, or in comparison with a reference genome. Analyses relevant to genomic sequencing include the identification of single nucleotide polymorphisms (SNP), as well as, structural (SV) and gene copy number variation (CNV). (B) Green boxes outline the use of RNA-seq to define differentially expressed genes (DEG) through de novo transcriptomic assembly, or a referenced transcriptome assembly through alignment to an existing genome. (C) Orange boxes demonstrate the identification of small non-coding RNAs by their mapping to a reference genome and exploration of their potential role in regulating differentially expressed genes. These analyses include the identification of micro-RNAs (miRNAs) by prediction of their hair-pin precursor, and their annotation using data in miRBase (www.miRBase.org). Additional small RNAs can be explored by identifying ncRNA precursor transcripts (represented by large, contiguous clusters of small-RNA reads mapping to a reference genome) and their annotation through comparisons [e.g., by hidden Markov modeling (HMM)] with the Rfam database (rfam.sanger.ac.uk); α small-RNAs are also identified by their length, sequence and relative mapping position (e.g., with respect to coding genes, gene regulatory and/or transposable elements). (D) Purple boxes describe the use of ChIP-seq and bisulfite sequencing (BSS) to explore epigenetic changes, including histone modification and DNA methylation respectively, that might impact on gene regulation. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
These genomic data-sets, and their accompanying transcriptomic data, provide major insights into the biology of parasitic nematodes because they are the basis for characterizing SNPs (e.g., DNA-seq; (Metzker, 2010)), changes in transcription and alternative splicing patterns (i.e., RNA-seq: (Ozsolak and Milos, 2011)) and genetic regulation through epigenetic modifications [e.g., CHIP-seq (Furey, 2012) or bisulphite sequencing (Krueger et al., 2012)] and/or the activity of non-coding RNAs (Jacquier, 2009; Wang et al., 2011). Resources for accessing and analysing these data-sets are developing (Box 1). WormBase (www.wormbase.org) has expanded its data repository to include parasitic nematodes, with the genomes, annotated gene models and inferred function, and related files available for public access via ftp download (ftp://ftp.sanger.ac.uk/pub2/wormbase/live_release). Nematode.net provides a wide variety of resources for users to explore genomic and functional information for parasitic nematodes (Martin et al., 2012a), including specific data-mining and comparative analysis tools such as NemaBLAST, NemaBrowse, NemaPATH (which allows exploration of many of the functional pathways encoded by parasitic nematodes) and NemaSNP (which allows the user to explore SNPs detected in short read sequences aligned to published nematode transcriptomic assemblies). Tools for accessing transcriptomic data for nematodes, particularly those lacking a draft genome, are needed, and have lagged behind genomic resources. NEMBASE is one such example of an improved resource for these data (Elsworth et al., 2011). HelmDB (Mangiola et al., 2013) was recently constructed to further improve the accessibility and annotation of transcriptomic information for parasitic worms, specifically including data-sets for species lacking a published genomic sequence. Often, de novo assembled transcriptomes will contain a large number of incomplete transcripts that cannot be functionally annotated. HelmDB uses gene clustering, data integration and annotation transfer (Defoin-Platel et al., 2011) to provide a functional inference for these sequences in helminth transcriptomes. This approach involves clustering gene/protein sequences from 3 or more organisms based on their multiple pair-wise alignment e.g. (Di Tommaso et al., 2011) and can be optimized by adjusting the inclusion parameters for each cluster, including alignment length and/or percent identity, or through the use of more sophisticated orthology prediction algorithms (Kuzniar et al., 2010). Once clustered, functional information can be transferred from annotated to unannotated sequences, often with high confidence. In HelmDB, the annotation transfer approach allowed the improved functional characterization of 8–20% of the transcripts available for each of 11 species incorporated into the database.
Box 1. Key resources and applications for high-throughput sequencing and bioinformatic analysis.
| Tool | Description | Source |
| Guidelines | ||
| MINSEQE | RNA-seq guidelines | http://www.fged.org/projects/minseqe/ |
| Encode Genome Annotation Assessment Project (EGASP) | Genome annotation guidelines | Guigo et al. (2006) |
| RNA-seq Genome Annotation Assessment Project (RGASP) | RNA-seq analyses guidance | Engstrom et al. (2013) and Steijger et al. (2013) |
| Advice/Protocols | ||
| SEQAnswers | HTS and bioinformatics | www.seqanswers.com |
| Galaxy Project | Bioinformatic advice/lectures | www.galaxyproject.org |
| Current Protocols in Bioinformatics | Command-based protocols | http://cda.currentprotocols.com/ |
| Nature Protocols | Command-based protocols | www.nature.com/nprot/ |
| Applications | ||
| Read quality | FastQC/Trimmomatic | http://www.bioinformatics.babraham.ac.uk/projects/fastqc// Bolger et al. (2014) |
| De novo genome assembly | Velvet/SOAPdenovo/SGA Assembler | Zerbino (2010)/Li et al. (2010)/Simpson and Durbin (2012) |
| Read alignment | BWA/Bowtie2 | Li and Durbin (2010)/Langmead and Salzberg (2012) |
| De novo transcriptome assembly | Trinity/TransABYSS | Grabherr et al. (2011)/Robertson et al. (2010b) |
| Referenced transcriptome assembly | Tophat-Cufflinks | Trapnell et al. (2012) |
| Differential transcriptional analysis | Tophat-Cufflinks | Trapnell et al. (2012) |
| SNP detection | SAMtools/GATK | Li et al. (2009)/McKenna et al. (2010) |
| Comparative alignments | MUMMER/BLAT | Delcher et al. (2003)/Kent (2002) |
| microRNA identification | miRDeep2 | Friedlander et al. (2012) |
| Pipelines | ||
| Geneious | Graphic-based bioinformatics | Kearse et al. (2012) |
| Galaxy project | Web-based bioinformatics | www.galaxyproject.org |
Although each of these resources improves accessibility to existing data and can help users conduct comparative analyses of published and/or their own data, none provide specific help for researchers interested in conducting HTS projects of their own. Fortunately, a rapid expansion in the number of tools, community resources, protocols, data handling and storage and experimental design standards is occurring. The Functional Genomics Data Society have recently developed the MINSEQE guidelines (http://www.fged.org/projects/minseqe/) for sample information, and experimental design relating to HTS; for example, advice on the minimum number of biological or technical replicates needed for RNA-seq experiments. A wide-variety of freely available software and scripts exist for analysis of HTS applications. SGA Assembler (Simpson and Durbin, 2012), for example, is able to conduct de novo assembly of complete human genomes using less than 50 Gb of RAM, compared with the ∼1000 Gb of RAM required for de novo assembly of large genomes using standard assemblers, such as Velvet (Zerbino and Birney, 2008) or SOAPdenovo (Li et al., 2010). Numerous resources now exist to aid researchers in accessing these programs and applying them to helminth research. Examples range from large, community-based blog sites, such as SEQanswers.com, to dedicated protocol-based publications, such as Current Protocols in Bioinformatics. The latter provides step-by-step command-based instructions for everything from extracting information from public databases, to assembling and annotating genomes and transcriptomes, exploring CHIP-seq data, conducting comparative genomic investigations of SNPs and structural variants, modelling regulatory elements, identifying small non-coding RNAs, and processing proteomic, metabolomic and other small molecular data-sets. Although not specifically dedicated to bioinformatics, Nature Protocols also represents an excellent resource for some of the major bioinformatic applications currently available; key examples including excellent protocols for refining/finishing draft genomes using the Post-Assembly Genome Improvement Tool (Swain et al., 2012) and the reconstruction and differential analysis of transcriptomic data using Tuxedo suite (i.e., Tophat/Cufflinks: (Trapnell et al., 2012)).
Just as guidelines are being established for experimental design (e.g., MINSEQE), projects are under-way to assess the specificity and sensitivity of these many software applications and establish ‘gold-standard’ methods for data analysis. A key example is the RNA-seq Genome Annotation Assessment Project (RGASP), which benchmarks leading RNA-seq analysis tools for their ability to accurately reconstruct the transcriptome, identify splice isoforms and quantify transcriptional abundance (see Engstrom et al., 2013; Steijger et al., 2013). The increasing availability of bioinformatic applications, reduced demands for computing power, and development of guidelines and protocols for bioinformatic analyses are vastly improving the accessibility of this field to the global scientific community. Nonetheless, knowledge of Unix-commands and some access to custom Perl/Python scripting is needed, particularly to take the output from one application and reformat it for another. This remains a significant obstacle for many research groups. The development of bioinformatic pipelines with a Graphical User Interface (GUI) is of significant interest to many wishing to enter this field. An example is the proprietary software Geneious (Kearse et al., 2012), which allows users to perform many standard bioinformatic analyses using menu-based commands on a personal computer. These include pipelines to conduct de novo assemblies, align reads to a reference genome/transcriptome, and conduct/visualize a variety of comparative analyses. The Galaxy Project (http://galaxyproject.org/) provides arguably the best of GUI-based resources currently available, allowing users free access to a remote server for their analyses and a web-based interface. Major strengths of the Galaxy Project is that it utilizes a growing variety of leading open source bioinformatic applications, including SAMtools (Li et al., 2009), GATK (McKenna et al., 2010) and TopHat/Cufflinks (Trapnell et al., 2010), and is supported by a large community of bioinformaticians seeking to expand the available applications accessible to users. Users can build custom pipelines linking these applications to create semi-automated workflows for their analyses and, through the Galaxy Project community, have access to a wealth of presentations, lectures and blog-based articles on conducting bioinformatics research. Collectively, these resources improve accessibility to bioinformatic platforms for non-bioinformaticians.
A key message from these platforms is that although experimental design remains critical and data analysis/handling is not trivial, many resources now exist to provide entry into this field for researchers wishing to utilize the power of HTS to explore helminths and the development of anthelmintic resistance.
11. Conclusion
Our knowledge of the mechanisms of anthelmintic resistance has increased greatly over the last few years (Table 1). However, in most cases this has led to a realisation of greater than expected complexity, especially for resistances that have arisen in the field. Consequently, simple molecular-based tests for resistance to most of the drug groups will not be possible for some years. Encouragingly, more complex molecular-based tests such as high throughput SNP tools, are becoming more affordable and their use more routine. Drug resistance may be a complex, polygenic or quantitative trait in many instances, but the exception to this is BZ resistance where most studies point to a small number of beta-tubulin SNPs. Hence, some steps have recently been taken to translate SNP-based tests for BZ resistance into the field for veterinary applications. However, this process has revealed that in some instances even BZ resistance can be more complex. BZ efficacy cannot in all cases be explained by beta-tubulin allele frequencies, suggesting that other mechanisms, perhaps P-gps or drug metabolism, may also contribute to the observed field resistances. For MLs, the situation is far more uncertain, with the early promise of SNP-based tests based on GluClR genes not being realised, and some attention turning to the role of P-gps and other potential mechanisms. For the nicotinic agonist group, evidence is emerging of complex polygenic resistances in which various changes in gene structure or expression levels are associated with a reduction in drug sensitive nAChRs in resistant worms via many different specific mechanisms. Not only does this suggest polygenic resistance within single field-selected populations, but also the existence of completely distinct resistance-conferring molecular changes in separate populations within a species. Hence, while most advanced for BZ drugs, the potential for the widespread use of molecular-based tests for detection and quantification of anthelmintic resistance remains a research goal rather than a clinical reality.
Table 1.
Recent (2011-present) publications on the molecular basis of anthelmintic resistances and drug/receptor interactions.
| Drug group | Aspect covered | References |
|---|---|---|
| Macrocyclic lactones | Interactions with GluClRs and other Cys-loop receptors | Hibbs and Gouaux (2011), Lynagh et al. (2011), Lynagh and Lynch (2012a) and Lynagh and Lynch (2012b) |
| The lack of GluClR SNPs or gene transcription patterns able to explain observed resistances | El-Abdellati et al. (2011), Williamson et al. (2011) and Martínez-Valladares et al. (2012) | |
| Amino acid deletion in a GluClR conferring resistance in C. elegans | Ghosh et al. (2012) | |
| Introgression of resistance genes into a susceptible isolate of H. contortus; an important advance for resistance gene mapping | Redman et al. (2012) | |
| P-gp SNPs or gene transcription patterns associated with drug resistance | Williamson et al. (2011), Bourguinat et al. (2011), Dicker et al. (2011), Lespine et al. (2012), Yan et al. (2012), Janssen et al. (2013a), Janssen et al. (2013b), Ardelli and Prichard (2013), De Graef et al. (2013), Demeler et al. (2013b), Sarai et al. (2014) and Bygarski et al. (2014) | |
| Benzimidazoles | Relative role of different SNPs in conferring resistance | Kotze et al. (2012) and Barrère et al. (2012) |
| Pyrosequencing assays for SNPs in cattle worms | Demeler et al. (2013a) and Chaudhry et al. (in press) | |
| Evaluation of SNP-based tests in the field for livestock | Barrère et al. (2013a,b) and Niciura et al. (2012) | |
| Molecular analysis in human soil-transmitted helminths | Diawara et al. (2013a,b) | |
| Description of all beta-tubulins in H. contortus | Saunders et al. (2013) | |
| Nicotinic agonists | Altered nAChR subunit gene transcription patterns associated with resistance | Williamson et al. (2011), Boulin et al. (2011), Sarai et al. (2013, 2014) and Romine et al. (2014) |
| Putative DNA marker for levamisole resistance | Barrère et al. (2014) | |
| Derquantel/abamectin | Description of derquantel-sensitive nAChRs | Buxton et al. (2014) |
| Interaction of the two drugs at nAChRs on the somatic muscle; suggestion of synergism | Puttachary et al. (2013) | |
| Amino-acetonitrile derivatives | Role of C. elegans ACR-23 receptor in drug action | Rufener et al. (2013) |
| Cyclooctadepsi-peptides | Evidence for role of SLO-1 in drug action | Crisford et al. (2011), Buxton et al. (2011) |
| Role for GABA signalling in drug action | Miltsch et al. (2012) | |
| Reviews on mode of action, focusing on SLO-1 | Krücken et al. (2012), Holden-Dye et al. (2012) and Martin et al. (2012b) | |
| Triclabendazole | P-gp inhibitor and SNP studies linking drug efflux and resistance | Wilkinson et al. (2012) and Meaney et al. (2103) |
| Lack of association between beta-tubulin and resistance | Fuchs et al. (2013) | |
| Genome-wide approach to mapping resistance loci | Hodgkinson et al. (2013) |
With the publication of helminth genomes and the rapid development of tools for analysis of high throughput sequencing data, the prospect of utilizing whole-genome approaches to discover markers for anthelmintic resistance becomes more real. The hope that such approaches will provide a suite of markers for anthelmintic resistance detection seems realistic. Such tools will hopefully have a broader application than the specific candidate gene-based SNPs or gene expression changes described to date. However, despite the development of molecular tools with application across many fields of biology, there are specific features of helminth biology that will make the use of some of these tools for resistance marker discovery more problematic. Some of these challenges are described in Box 2; much work is needed before the complete analysis of many helminth genomes will be a reality. Some aspects of worm biology, such as genetic crossing methods and the costs associated with generating inbred parental lines, also present difficulties.
Box 2. Key challenges for the mapping of genetic markers for anthelmintic resistance in helminth parasites.
| 1. | Current status of genome sequences |
| The parasite genome assemblies available need further improvement. Assembly has been complicated because the genomes are generally highly polymorphic, containing a high number of repetitive DNA sequences, and some are very large; Statistical methods and genome assembly methods which can better deal with repetitive DNA sequence are needed; Genetic methods in parasites are difficult and so genetic recombination maps are not available to help genome assembly for most species; In addition, genome projects have not progressed for some species (most notably the various Trichostrongylus species); |
|
| 2. | Genetic methodology |
| Improvements are needed in our ability to perform genetic crosses in some important species; In some cases the development of alternative animal host models or in vitro life cycle culture may be required; Some consideration of the value of mixing populations rather than deliberate crosses should be made; |
|
| 3. | Inbred parental lines |
| In most instances the existence of a highly inbred susceptible parent strain would assist discovery, removing noise from the statistical analysis; The MHco3(ISE).N1 is available for H. contortus, but this cannot be used worldwide, and no such resource exists for other species of interest; The production of clonal isolates of F. hepatica has been a significant boost for resistance studies with this species; |
|
| 4. | Polygenic resistance |
| Evidence that resistance to most anthelmintic classes is most likely polygenic, and the potential for multiple origins of resistance to occur in parasite populations, indicates that a suite of markers may be required for use across different field isolates within single helminth species. | |
| 5. | Functional genomics methods |
| Functional studies using RNAi are problematic in strongylid species as the technique has only limited usefulness (Geldhof et al. 2007; Zawadzki et al., 2012); F. hepatica is however much more amenable to the use of RNAi (McGonigle et al., 2008; Rinaldi et al., 2008). |
Nevertheless, as described in this paper, there have been significant recent advances in the application of genomic and genetic tools to the study of anthelmintic resistance in H. contortus and F. hepatica. It is anticipated that these advances will establish a pathway for anthelmintic resistance marker discovery for these species, and with more genomes available, marker discovery in other veterinary and human helminth parasites should progress within the next few years.
Acknowledgements
Funds to support the CARS2013 meeting were provided by Zoetis, and Virbac Animal Health. Alessandro Puoti is thanked for providing Figs. 4A and B. Support to RJM was via NIH grant R01 AI047194 National Institute of Allergy and Infectious Diseases. The funding agency had no role in the design, execution or publication of these studies. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases.
References
- Abad P., Gouzy J., Aury J.M., Castagnone-Sereno P., Danchin E.G., Deleury E., Perfus-Barbeoch L., Anthouard V., Artiguenave F., Blok V.C., Caillaud M.C., Coutinho P.M., Dasilva C., De Luca F., Deau F., Esquibet M., Flutre T., Goldstone J.V., Hamamouch N., Hewezi T., Jaillon O., Jubin C., Leonetti P., Magliano M., Maier T.R., Markov G.V., McVeigh P., Pesole G., Poulain J., Robinson-Rechavi M., Sallet E., Segurens B., Steinbach D., Tytgat T., Ugarte E., van Ghelder C., Veronico P., Baum T.J., Blaxter M., Bleve-Zacheo T., Davis E.L., Ewbank J.J., Favery B., Grenier E., Henrissat B., Jones J.T., Laudet V., Maule A.G., Quesneville H., Rosso M.N., Schiex T., Smant G., Weissenbach J., Wincker P. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat. Biotechnol. 2008;26:909–915. doi: 10.1038/nbt.1482. [DOI] [PubMed] [Google Scholar]
- Adelsberger H., Lepier A., Dudel J. Activation of rat recombinant alpha(1)beta(2)gamma(2S) GABA(A) receptor by the insecticide ivermectin. Eur. J. Pharmacol. 2000;394:163–170. doi: 10.1016/s0014-2999(00)00164-3. [DOI] [PubMed] [Google Scholar]
- Alvarez L.L., Solana H.D., Mottier M.L., Virkel G.L., Fairweather I., Lanusse C.E. Altered drug influx/efflux and enhanced metabolic activity in triclabendazole-resistant liver flukes. Parasitology. 2005;131:501–510. doi: 10.1017/S0031182005007997. [DOI] [PubMed] [Google Scholar]
- Anderson T., Nkhoma S., Ecker A., Fidock D. How can we identify parasite genes that underlie antimalarial drug resistance? Pharmacogenomics. 2011;12:59–85. doi: 10.2217/pgs.10.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardelli B.F., Prichard R.K. Reduced genetic variation of an Onchocerca volvulus ABC transporter gene following treatment with ivermectin. Trans. R. Soc. Trop. Med. Hyg. 2007;101:1223–1232. doi: 10.1016/j.trstmh.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Ardelli B.F., Prichard R.K. Inhibition of P-glycoprotein enhances sensitivity of Caenorhabditis elegans to ivermectin. Vet. Parasitol. 2013;191:264–275. doi: 10.1016/j.vetpar.2012.09.021. [DOI] [PubMed] [Google Scholar]
- Ballent M., Lifschitz A., Virkel G., Sallovitz J., Lanusse C. Modulation of the P-glycoprotein-mediated intestinal secretion of ivermectin: in vitro and in vivo assessments. Drug Metab. Dispos. 2006;34:457–463. doi: 10.1124/dmd.105.007757. [DOI] [PubMed] [Google Scholar]
- Barrère V., Alvarez L., Suarez G., Ceballos L., Moreno L., Lanusse C., Prichard R.K. Relationship between increased albendazole systemic exposure and changes in single nucleotide polymorphisms on the β-tubulin isotype 1 encoding gene in Haemonchus contortus. Vet. Parasitol. 2012;186:344–349. doi: 10.1016/j.vetpar.2011.11.068. [DOI] [PubMed] [Google Scholar]
- Barrère V., Keller K., von Samson-Himmelstjerna G., Prichard R.K. Efficiency of a genetic test to detect benzimidazole resistant Haemonchus contortus nematodes in sheep farms in Quebec. Canada Parasitol. Int. 2013;62:464–470. doi: 10.1016/j.parint.2013.06.001. [DOI] [PubMed] [Google Scholar]
- Barrère V., Falzon L.C., Shakya K.P., Menzies P.I., Peregrine A.S., Prichard R.K. Assessment of benzimidazole resistance in Haemonchus contortus in sheep flocks in Ontario, Canada: comparison of detection methods for drug resistance. Vet. Parasitol. 2013;198:159–165. doi: 10.1016/j.vetpar.2013.07.040. [DOI] [PubMed] [Google Scholar]
- Barrère V., Beech R.N., Charvet C.L., Prichard R.K. Novel assay for the detection and monitoring of levamisole resistance in Haemonchus contortus. Int. J. Parasitol. 2014;44:235–241. doi: 10.1016/j.ijpara.2013.12.004. [DOI] [PubMed] [Google Scholar]
- Bartley D.J., McAllister H., Bartley Y., Dupuy J., Menez C., Alvinerie M., Jackson F., Lespine A. P-glycoprotein interfering agents potentiate ivermectin susceptibility in ivermectin sensitive and resistant isolates of Teladorsagia circumcincta and Haemonchus contortus. Parasitology. 2009;136:1081–1088. doi: 10.1017/S0031182009990345. [DOI] [PubMed] [Google Scholar]
- Bartley D.J., Morrison A.A., Dupuy J., Bartley Y., Sutra J.F., Menez C., Alvinerie M., Jackson F., Devin L., Lespine A. Influence of Pluronic 85 and ketoconazole on disposition and efficacy of ivermectin in sheep infected with a multiple resistant Haemonchus contortus isolate. Vet. Parasitol. 2012;187:464–472. doi: 10.1016/j.vetpar.2012.02.011. [DOI] [PubMed] [Google Scholar]
- Beech R.N., Skuce P., Bartley D.J., Martin R.J., Prichard R.K., Gilleard J.S. Anthelmintic resistance: markers for resistance, or susceptibility? Parasitology. 2011;138:160–174. doi: 10.1017/S0031182010001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackhall W.J., Pouliot J.F., Prichard R.K., Beech R.N. Haemonchus contortus: selection at a glutamate-gated chloride channel gene in ivermectin- and moxidectin-selected strains. Exp. Parasitol. 1998;90:42–48. doi: 10.1006/expr.1998.4316. [DOI] [PubMed] [Google Scholar]
- Blackhall W.J., Prichard R.K., Beech R.N. Selection at a gamma-aminobutyric acid receptor gene in Haemonchus contortus resistant to avermectins/milbemycins. Mol. Biochem. Parasitol. 2003;131:137–145. doi: 10.1016/s0166-6851(03)00201-9. [DOI] [PubMed] [Google Scholar]
- Bolger A.M., Lohse A., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulin T., Fauvin A., Charvet C., Cortet J., Cabaret J., Bessereau J.-L., Neveu C. Functional reconstitution of Haemonchus contortus acetylcholine receptors in Xenopus oocytes provides mechanistic insights into levamisole resistance. Br. J. Pharm. 2011;164:1421–1432. doi: 10.1111/j.1476-5381.2011.01420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguinat C., Keller K., Bhan A., Peregrine A., Geary T., Prichard R. Macrocyclic lactone resistance in Dirofilaria immitis. Vet. Parasitol. 2011;181:388–392. doi: 10.1016/j.vetpar.2011.04.012. [DOI] [PubMed] [Google Scholar]
- Brockwell Y.M., Elliott T.P., Anderson G.R., Stanton R., Spithill T.W., Sangster N.C. Confirmation of Fasciola hepatica resistant to triclabendazole in naturally infected Australian beef and dairy cattle. Int. J. Parasitol. Drugs Drug Res. 2013;4:48–54. doi: 10.1016/j.ijpddr.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxton S.K., Neveu C., Charvet C.L., Robertson A.P., Martin R.J. On the mode of action of emodepside: slow effects on membrane potential and voltage-activated currents in Ascaris suum. Br. J. Pharmacol. 2011;164:453–470. doi: 10.1111/j.1476-5381.2011.01428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxton S.K., Charvet C.L., Neveu C., Cabaret J., Cortet J., Peineau N., Abongwa M., Courtot E., Robertson A.P., Martin R.J. Investigation of acetylcholine receptor diversity in a nematode parasite leads to characterization of tribendimidine- and derquantel-sensitive nAChRs. PLoS Pathog. 2014;10:e1003870. doi: 10.1371/journal.ppat.1003870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bygarski E.E., Prichard R.K., Ardelli B.F. Resistance to the macrocyclic lactone moxidectin is mediated in part by membrane transporter P-glycoproteins: Implications for control of drug resistant parasitic nematodes. Int. J. Parasitol.: Drugs Drug Resis. 2014;4:143–151. doi: 10.1016/j.ijpddr.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M., Sulston J.E., White J.G., Southgate E., Thomson J.N., Brenner S. The neural circuit for touch sensitivity in Caenorhabditis elegans. J. Neurosci. 1985;5:956–964. doi: 10.1523/JNEUROSCI.05-04-00956.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry, U., Miller, M., Yazwinski, T., Kaplan, R., Gilleard, J., in press. The presence of benzimidazole resistance mutations in Haemonchus placei of US. Cattle. Vet. Parasitol. [DOI] [PubMed]
- Chen W., Terada M., Cheng J.T. Characterization of subtypes of gamma-aminobutyric acid receptors in an Ascaris muscle preparation by binding assay and binding of PF1022A, a new anthelmintic, on the receptors. Parasitol. Res. 1996;82:97–101. doi: 10.1007/s004360050077. [DOI] [PubMed] [Google Scholar]
- Coles G.C., Stafford K.A., MacKay P.H. Ivermectin-resistant Cooperia species from calves on a farm in Somerset. Vet. Rec. 1998;142:255–256. [PubMed] [Google Scholar]
- Crisford A., Murray C., O’Connor V., Edwards R.J., Krüger N., Welz C., von Samson-Himmelstjerna G., Harder A., Walker R.J., Holden-Dye L. Selective toxicity of the anthelmintic emodepside revealed by heterologous expression of human KCNMA1 in Caenorhabditis elegans. Mol. Pharmacol. 2011;79:1031–1043. doi: 10.1124/mol.111.071043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cully D.F., Vassilatis D.K., Liu K.K., Paress P.S., Van der Ploeg L.H., Schaeffer J.M., Arena J.P. Cloning of an avermectin-sensitive glutamate-gated chloride channel from Caenorhabditis elegans. Nature. 1994;371:707–711. doi: 10.1038/371707a0. [DOI] [PubMed] [Google Scholar]
- Defoin-Platel M., Hassani-Pak K., Rawlings C. Gaining confidence in cross-species annotation transfer: from simple molecular function to complex phenotypic traits. Asp. Appl. Biol. 2011;107:79–87. [PMC free article] [PubMed] [Google Scholar]
- De Graef J., Demeler J., Skuce P., Mitreva M., Von Samson-Himmelstjerna G., Vercruysse J., Claerebout E., Geldhof P. Gene expression analysis of ABC transporters in a resistant Cooperia oncophora isolate following in vivo and in vitro exposure to macrocyclic lactones. Parasitology. 2013;140:499–508. doi: 10.1017/S0031182012001849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher, A.L., Salzberg, S.L., Phillippy, A.M., 2003. Using MUMmer to identify similar regions in large sequence sets. Curr. Protoc. Bioinf. Chapter 10, Unit 10 13. [DOI] [PubMed]
- Demeler J., Krüger N., Krücken J., von der Heyden V.C., Ramünke S., Küttler U., Miltsch S., López Cepeda M., Knox M., Vercruysse J., Geldhof P., Harder A., von Samson-Himmelstjerna G. Phylogenetic characterization of β-tubulins and development of pyrosequencing assays for benzimidazole resistance in cattle nematodes. PLoS One. 2013;8:e70212. doi: 10.1371/journal.pone.0070212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeler J., Krücken J., AlGusbi S., Ramunke S., De Graef J., Kerboeuf D., Geldhof P., Pomroy W.E., von Samson-Himmelstjerna G. Potential contribution of P-glycoproteins to macrocyclic lactone resistance in the cattle parasitic nematode Cooperia oncophora. Mol. Biochem. Parasitol. 2013;188:10–19. doi: 10.1016/j.molbiopara.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Dent J.A., Smith M.M., Vassilatis D.K., Avery L. The genetics of ivermectin resistance in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2000;97:2674–2679. doi: 10.1073/pnas.97.6.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins C.A., Cerqueira G.C., Goldberg J.M., Dunning Hotopp J.C., Haas B.J., Zucker J., Ribeiro J.M., Saif S., Levin J.Z., Fan L., Zeng Q., Russ C., Wortman J.R., Fink D.L., Birren B.W., Nutman T.B. Genomics of Loa loa, a Wolbachia-free filarial parasite of humans. Nat. Genet. 2013;45:495–500. doi: 10.1038/ng.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diawara A., Drake L.J., Suswillo R.R., Kihara J., Bundy D.A., Scott M.E., Halpenny C., Stothard J.R., Prichard R.K. Assays to detect beta-tubulin codon 200 polymorphism in Trichuris trichiura and Ascaris lumbricoides. PLoS Negl. Trop. Dis. 2009;3:e397. doi: 10.1371/journal.pntd.0000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diawara A., Halpenny C.M., Churcher T.S., Mwandawiro C., Kihara J., Kaplan R.M., Streit T.G., Idaghdour Y., Scott M.E., Basáñez M.G., Prichard R.K. Association between response to albendazole treatment and β-tubulin genotype frequencies in soil-transmitted helminths. PLoS Negl. Trop. Dis. 2013;7:e2247. doi: 10.1371/journal.pntd.0002247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diawara A., Schwenkenbecher J.M., Kaplan R.M., Prichard R.K. Molecular and biological diagnostic tests for monitoring benzimidazole resistance in human soil-transmitted helminths. Am. J. Trop. Med. Hyg. 2013;88:1052–1061. doi: 10.4269/ajtmh.12-0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicker A.J., Nisbet A.J., Skuce P.J. Gene expression changes in a P-glycoprotein (Tci-pgp-9) putatively associated with ivermectin resistance in Teladorsagia circumcincta. Int. J. Parasitol. 2011;41:935–942. doi: 10.1016/j.ijpara.2011.03.015. [DOI] [PubMed] [Google Scholar]
- Di Tommaso P., Moretti S., Xenarios I., Orobitg M., Montanyola A., Chang J.M., Taly J.F., Notredame C. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011;39:W13–17. doi: 10.1093/nar/gkr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich I.M., Bloom J., Torabi N., Wang X., Jia Y., Kruglyak L. Genetic architecture of highly complex chemical resistance traits across four yeast strains. PLoS Genet. 2012;8:e1002570. doi: 10.1371/journal.pgen.1002570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Abdellati A., De Graef J., Van Zeveren A., Donnan A., Skuce P., Walsh T., Wolstenholme A., Tait A., Vercruysse J., Claerebout E., Geldhof P. Altered avr-14B gene transcription patterns in ivermectin-resistant isolates of the cattle parasites, Cooperia oncophora and Ostertagia ostertagi. Int. J. Parasitol. 2011;41:951–957. doi: 10.1016/j.ijpara.2011.04.003. [DOI] [PubMed] [Google Scholar]
- Elliott T., Muller A., Brockwell Y., Murphy N., Grillo V., Toet H.M., Anderson G., Sangster N., Spithill T.W. Evidence for high genetic diversity of NAD1 and COX1 mitochondrial haplotypes among triclabendazole resistant and susceptible populations and field isolates of Fasciola hepatica (liver fluke) in Australia. Vet. Parasitol. 2014;200:90–96. doi: 10.1016/j.vetpar.2013.11.019. [DOI] [PubMed] [Google Scholar]
- Elsworth B., Wasmuth J., Blaxter M. NEMBASE4: the nematode transcriptome resource. Int. J. Parasitol. 2011;41:881–894. doi: 10.1016/j.ijpara.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Engstrom P.G., Steijger T., Sipos B., Grant G.R., Kahles A., Alioto T., Behr J., Bertone P., Bohnert R., Campagna D., Davis C.A., Dobin A., Gingeras T.R., Goldman N., Guigo R., Harrow J., Hubbard T.J., Jean G., Kosarev P., Li S., Liu J., Mason C.E., Molodtsov V., Ning Z., Ponstingl H., Prins J.F., Ratsch G., Ribeca P., Seledtsov I., Solovyev V., Valle G., Vitulo N., Wang K., Wu T.D., Zeller G. Systematic evaluation of spliced alignment programs for RNA-seq data. Nat. Methods. 2013;10:1185–1191. doi: 10.1038/nmeth.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairweather I. Liver fluke isolates: a question of provenance. Vet. Parasitol. 2011;176:1–8. doi: 10.1016/j.vetpar.2010.12.011. [DOI] [PubMed] [Google Scholar]
- Fauvin A., Charvet C., Issouf M., Cortet J., Cabaret J., Neveu C. CDNA-AFLP analysis in levamisole-resistant Haemonchus contortus reveals alternative splicing in a nicotinic acetylcholine receptor subunit. Mol. Biochem. Parasitol. 2010;170:105–107. doi: 10.1016/j.molbiopara.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Feng X.P., Hayashi J., Beech R.N., Prichard R.K. Study of the nematode putative GABA type-A receptor subunits: evidence for modulation by ivermectin. J. Neurochem. 2002;83:870–878. doi: 10.1046/j.1471-4159.2002.01199.x. [DOI] [PubMed] [Google Scholar]
- Fleming J.T., Squire M.D., Barnes T.M., Tornoe C., Matsuda K., Ahnn J., Fire A., Sulston J.E., Barnard E.A., Sattelle D.B., Lewis J.A. Caenorhabditis elegans levamisole resistance genes lev-1, unc-29, and unc-38 encode functional nicotinic acetylcholine receptor subunits. J. Neurosci. 1997;17:5843–5857. doi: 10.1523/JNEUROSCI.17-15-05843.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander M.R., Mackowiak S.D., Li N., Chen W., Rajewsky N. MiRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs M.A., Ryan L.A., Chambers E.L., Moore C.M., Fairweather I., Trudgett A., Timson D.J., Brennan G.P., Hoey E.M. Differential expression of liver fluke β-tubulin isotypes at selected life cycle stages. Int. J. Parasitol. 2013;43:1133–1139. doi: 10.1016/j.ijpara.2013.08.007. [DOI] [PubMed] [Google Scholar]
- Furey T.S. ChIP-seq and beyond: new and improved methodologies to detect and characterize protein-DNA interactions. Nat. Rev. Genet. 2012;13:840–852. doi: 10.1038/nrg3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geary T.G., Bourguinat C., Prichard R.K. Evidence for macrocyclic lactone anthelmintic resistance in Dirofilaria immitis. Top. Companion Anim. Med. 2011;26:186–192. doi: 10.1053/j.tcam.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Geldhof P., Visser A., Clark D., Saunders G., Britton C., Gilleard J., Berriman M., Knox D. RNA interference in parasitic helminths: current situation, potential pitfalls and future prospects. Parasitolology. 2007;134:609–619. doi: 10.1017/S0031182006002071. [DOI] [PubMed] [Google Scholar]
- Ghedin E., Wang S., Spiro D., Caler E., Zhao Q., Crabtree J., Allen J.E., Delcher A.L., Guiliano D.B., Miranda-Saavedra D., Angiuoli S.V., Creasy T., Amedeo P., Haas B., El-Sayed N.M., Wortman J.R., Feldblyum T., Tallon L., Schatz M., Shumway M., Koo H., Salzberg S.L., Schobel S., Pertea M., Pop M., White O., Barton G.J., Carlow C.K., Crawford M.J., Daub J., Dimmic M.W., Estes C.F., Foster J.M., Ganatra M., Gregory W.F., Johnson N.M., Jin J., Komuniecki R., Korf I., Kumar S., Laney S., Li B.W., Li W., Lindblom T.H., Lustigman S., Ma D., Maina C.V., Martin D.M., McCarter J.P., McReynolds L., Mitreva M., Nutman T.B., Parkinson J., Peregrin-Alvarez J.M., Poole C., Ren Q., Saunders L., Sluder A.E., Smith K., Stanke M., Unnasch T.R., Ware J., Wei A.D., Weil G., Williams D.J., Zhang Y., Williams S.A., Fraser-Liggett C., Slatko B., Blaxter M.L., Scott A.L. Draft genome of the filarial nematode parasite Brugia malayi. Science. 2007;317:1756–1760. doi: 10.1126/science.1145406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisi M., Kaminsky R., Maser P. Phenotyping and genotyping of Haemonchus contortus isolates reveals a new putative candidate mutation for benzimidazole resistance in nematodes. Vet. Parasitol. 2007;144:313–320. doi: 10.1016/j.vetpar.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Ghosh R., Andersen E.C., Shapiro J.A., Gerke J.P., Kruglyak L. Natural variation in a chloride channel subunit confers avermectin resistance in C. elegans. Science. 2012;335:574–578. doi: 10.1126/science.1214318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godel C., Kumar S., Koutsovoulos G., Ludin P., Nilsson D., Comandatore F., Wrobel N., Thompson M., Schmid C.D., Goto S., Bringaud F., Wolstenholme A., Bandi C., Epe C., Kaminsky R., Blaxter M., Maser P. The genome of the heartworm, Dirofilaria immitis, reveals drug and vaccine targets. FASEB J. 2012;26:4650–4661. doi: 10.1096/fj.12-205096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman M.M., Pastan I. Biochemistry of multidrug resistance mediated by the multidrug transporter. Annu. Rev. Biochem. 1993;62:385–427. doi: 10.1146/annurev.bi.62.070193.002125. [DOI] [PubMed] [Google Scholar]
- Gottschalk A., Almedom R.B., Schedletzky T., Anderson S.D., Yates J.R., III, Schafer W.R. Identification and characterization of novel nicotinic receptor-associated proteins in Caenorhabditis elegans. EMBO J. 2005;24:2566–2578. doi: 10.1038/sj.emboj.7600741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillo V., Jackson F., Gilleard J.S. Characterisation of Teladorsagia circumcincta microsatellites and their development as population genetic markers. Mol. Biochem. Parasitol. 2006;148:181–189. doi: 10.1016/j.molbiopara.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Guigo R., Flicek P., Abril J.F., Reymond A., Lagarde J., Denoeud F., Antonarakis S., Ashburner M., Bajic V.B., Birney E., Castelo R., Eyras E., Ucla C., Gingeras T.R., Harrow J., Hubbard T., Lewis S.E., Reese M.G. EGASP: the human ENCODE Genome Annotation Assessment Project. Genome Biol. 2006;7(Suppl 1):21–31. doi: 10.1186/gb-2006-7-s1-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna R.E., Forster F.I., Brennan G.P., Fairweather I. Fasciola hepatica: histological demonstration of apoptosis in the reproductive organs of flukes of triclabendazole-sensitive and triclabendazole-resistant isolates, and in field-derived flukes from triclabendazole-treated hosts, using in situ hybridisation to visualise endonuclease-generated DNA strand breaks. Vet. Parasitol. 2013;191:240–251. doi: 10.1016/j.vetpar.2012.09.014. [DOI] [PubMed] [Google Scholar]
- Hibbs R.E., Gouaux E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature. 2011;474:54–60. doi: 10.1038/nature10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert, O. 2013. The neuronal genome of Caenorhabditis elegans. WormBook, ed. The C. elegans Research Community, WormBook. pp. 1-106. doi: 10.1895/wormbook.1.161.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Hodgkinson J., Cwiklinski K., Beesley N.J., Paterson S., Williams D.J. Identification of putative markers of triclabendazole resistance by a genome-wide analysis of genetically recombinant Fasciola hepatica. Parasitology. 2013;140:1523–1533. doi: 10.1017/S0031182013000528. [DOI] [PubMed] [Google Scholar]
- Hoekstra R., Criado-Fornelio A., Fakkeldij J., Bergman J., Roos M.H. Microsatellites of the parasitic nematode Haemonchus contortus: polymorphism and linkage with a direct repeat. Mol. Biochem. Parasitol. 1997;89:97–107. doi: 10.1016/s0166-6851(97)00108-4. [DOI] [PubMed] [Google Scholar]
- Holden-Dye L., Hewitt G.M.K.T.W., Krogsgard-Larsen P., Walker R.J. Studies involving avermectin and the 4-aminobutyric acid (GABA) receptor of Ascaris suum muscle. Pest Manag. Sci. 1988;24:231–245. [Google Scholar]
- Holden-Dye L., Crisford A., Welz C., von Samson-Himmelstjerna G., Walker R.J., O’Connor V. Worms take to the slo lane: a perspective on the mode of action of emodepside. Invert. Neurosci. 2012;12:29–36. doi: 10.1007/s10158-012-0133-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosking B.C., Dobson D.P., Stein P.A., Kaminsky R., Bapst B., Mosimann D., Mason P.C., Seewald W., Strehlau G., Sager H. Dose confirmation studies for monepantel, an amino-acetonitrile derivative, against fourth stage gastro-intestinal nematode larvae infecting sheep. Vet. Parasitol. 2009;160:251–257. doi: 10.1016/j.vetpar.2008.11.028. [DOI] [PubMed] [Google Scholar]
- Hunt P.W., Kotze A.C., Knox M.R., Anderson L.J., McNally J., Le Jambre L.F. The use of DNA markers to map anthelmintic resistance loci in an intraspecific cross of Haemonchus contortus. Parasitology. 2010;137:705–717. doi: 10.1017/S0031182009991521. [DOI] [PubMed] [Google Scholar]
- Isaacs A., Qi S., Sarpong R., Casida J. Insect ryanodine receptor: distinct but coupled insecticide binding sites for [N-(CH3)-H-3]chlorantraniliprole, flubendiamide, and [H-3]ryanodine. Chem. Res. Toxicol. 2012;25:1571–1573. doi: 10.1021/tx300326m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat. Rev. Genet. 2009;10:833–844. doi: 10.1038/nrg2683. [DOI] [PubMed] [Google Scholar]
- James C.E., Davey M.W. Increased expression of ABC transport proteins is associated with ivermectin resistance in the model nematode Caenorhabditis elegans. Int. J. Parasitol. 2009;39:213–220. doi: 10.1016/j.ijpara.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Jani M., Makai I., Kis E., Szabo P., Nagy T., Krajcsi P., Lespine A. Ivermectin interacts with human ABCG2. J. Pharm. Sci. 2011;100:94–97. doi: 10.1002/jps.22262. [DOI] [PubMed] [Google Scholar]
- Janssen I.J., Krücken J., Demeler J., Basiaga M., Kornas S., von Samson-Himmelstjerna G. Genetic variants and increased expression of Parascaris equorum P-glycoprotein-11 in populations with decreased ivermectin susceptibility. PLoS One. 2013;8:e61635. doi: 10.1371/journal.pone.0061635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen I.J., Krücken J., Demeler J., von Samson-Himmelstjerna G. Caenorhabditis elegans: modest increase of susceptibility to ivermectin in individual P-glycoprotein loss-of-function strains. Exp. Parasitol. 2013;134:171–177. doi: 10.1016/j.exppara.2013.03.005. [DOI] [PubMed] [Google Scholar]
- Jex A.R., Liu S., Li B., Young N.D., Hall R.S., Li Y., Yang L., Zeng N., Xu X., Xiong Z., Chen F., Wu X., Zhang G., Fang X., Kang Y., Anderson G.A., Harris T.W., Campbell B.E., Vlaminck J., Wang T., Cantacessi C., Schwarz E.M., Ranganathan S., Geldhof P., Nejsum P., Sternberg P.W., Yang H., Wang J., Gasser R.B. Ascaris suum draft genome. Nature. 2011;479:529–533. doi: 10.1038/nature10553. [DOI] [PubMed] [Google Scholar]
- Jin M.S., Oldham M.L., Zhang Q., Chen J. Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans. Nature. 2012;490:566–569. doi: 10.1038/nature11448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa H., Takuwa K., Sakube Y. Mutations and expressions of the tropomyosin gene and the troponin C gene of Caenorhabditis elegans. Cell Struct. Funct. 1997;22:213–218. doi: 10.1247/csf.22.213. [DOI] [PubMed] [Google Scholar]
- Kaminsky R., Ducray P., Jung M., Clover R., Rufener L., Bouvier J., Weber S.S., Wenger A., Wieland-Berghausen S., Goebel T., Gauvry N., Pautrat F., Skripsky T., Froelich O., Komoin-Oka C., Westlund B., Sluder A., Mäser P. A new class of anthelmintics effective against drug-resistant nematodes. Nature. 2008;452:176–180. doi: 10.1038/nature06722. [DOI] [PubMed] [Google Scholar]
- Kaminsky R., Gauvry N., Schorderet Weber S., Skripsky T., Bouvier J., Wenger A., Schroeder F., Desaules Y., Hotz R., Goebel T., Hosking B.C., Pautrat F., Wieland-Berghausen S., Ducray P. Identification of the amino-acetonitrile derivative monepantel (AAD 1566) as a new anthelmintic drug development candidate. Parasitol. Res. 2008;103:931–939. doi: 10.1007/s00436-008-1080-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan R.M. Drug resistance in nematodes of veterinary importance: a status report. Trends Parasitol. 2004;20:477–481. doi: 10.1016/j.pt.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Meintjes P., Drummond A. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiki-Mvouaka S., Menez C., Borin C., Lyazrhi F., Foucaud-Vignault M., Dupuy J., Collet X., Alvinerie M., Lespine A. Role of P-glycoprotein in the disposition of macrocyclic lactones: A comparison between ivermectin, eprinomectin, and moxidectin in mice. Drug Metab. Dispos. 2010;38:573–580. doi: 10.1124/dmd.109.030700. [DOI] [PubMed] [Google Scholar]
- Kopp S.R., Kotze A.C., McCarthy J.S., Coleman G.T. High-level pyrantel resistance in the hookworm Ancylostoma caninum. Vet. Parasitol. 2007;143:299–304. doi: 10.1016/j.vetpar.2006.08.036. [DOI] [PubMed] [Google Scholar]
- Kopp S.R., Coleman G.T., Traub R.J., McCarthy J.S., Kotze A.C. Acetylcholine receptor subunit genes from Ancylostoma caninum: altered transcription patterns associated with pyrantel resistance. Int. J. Parasitol. 2009;39:435–441. doi: 10.1016/j.ijpara.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Kotze A.C., Cowling K., Bagnall N.H., Hines B.M., Ruffell A.P., Hunt P.W., Coleman G.T. Relative level of thiabendazole resistance associated with the E198A and F200Y SNPs in larvae of a multi-drug resistant isolate of Haemonchus contortus. Int. J. Parasitol. Drugs Drug. Resist. 2012;2:92–97. doi: 10.1016/j.ijpddr.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krücken J., Harder A., Jeschke P., Holden-Dye L., O’Connor V., Welz C., von Samson-Himmelstjerna G. Anthelmintic cyclooctadepsipeptides: complex in structure and mode of action. Trends Parasitol. 2012;28:385–394. doi: 10.1016/j.pt.2012.06.005. [DOI] [PubMed] [Google Scholar]
- Krueger F., Kreck B., Franke A., Andrews S.R. DNA methylome analysis using short bisulfite sequencing data. Nat. Methods. 2012;9:145–151. doi: 10.1038/nmeth.1828. [DOI] [PubMed] [Google Scholar]
- Kulke D., Krücken J., Harder A., von Samson-Himmelstjerna G. Efficacy of cyclooctadepsipeptides and aminophenylamidines against larval, immature and mature adult stages of a parasitologically characterized trichurosis model in mice. PLoS Negl. Trop. Dis. 2014;8:e2698. doi: 10.1371/journal.pntd.0002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzniar A., Roeland C.H.J.v.H., Pongor S., Leunissen J.A.M. The quest for orthologs: finding the corresponding gene across genomes. Trends Genet. 2010;24:539–551. doi: 10.1016/j.tig.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Kwa M.S.G., Veenstra J.G., Roos M.H. Benzimidazole resistance in Haemonchus contortus is correlated with a conserved mutation at amino acid 200 in [beta]-tubulin isotype-1. Mol. Biochem. Parasitol. 1994;63:299–303. doi: 10.1016/0166-6851(94)90066-3. [DOI] [PubMed] [Google Scholar]
- Laing R., Kikuchi T., Martinelli A., Tsai I.J., Beech R.N., Redman E., Holroyd N., Bartley D.J., Beasley H., Britton C., Curran D., Devaney E., Gilabert A., Hunt M., Jackson F., Johnston S.L., Kryukov I., Li K., Morrison A.A., Reid A.J., Sargison N., Saunders G.I., Wasmuth J.D., Wolstenholme A., Berriman M., Gilleard J.S., Cotton J.A. The genome and transcriptome of Haemonchus contortus, a key model parasite for drug and vaccine discovery. Genome Biol. 2013;14:R88. doi: 10.1186/gb-2013-14-8-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Jambre L.F., Lenane I.J., Wardrop A.J. A hybridisation technique to identify anthelmintic resistance genes in Haemonchus. Int. J. Parasitol. 1999;29:1979–1985. doi: 10.1016/s0020-7519(99)00157-5. [DOI] [PubMed] [Google Scholar]
- Le Jambre L.F., Gill J.H., Lenane I.J., Baker P. Inheritance of avermectin resistance in Haemonchus contortus. Int. J. Parasitol. 2000;30:105–111. doi: 10.1016/s0020-7519(99)00172-1. [DOI] [PubMed] [Google Scholar]
- Lespine A., Dupuy J., Orlowski S., Nagy T., Glavinas H., Krajcsi P., Alvinerie M. Interaction of ivermectin with multidrug resistance proteins (MRP1, 2 and 3) Chem. Biol. Interact. 2006;159:169–179. doi: 10.1016/j.cbi.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Lespine A., Martin S., Dupuy J., Roulet A., Pineau T., Orlowski S., Alvinerie M. Interaction of macrocyclic lactones with P-glycoprotein: structure-affinity relationship. Eur. J. Pharm. Sci. 2007;30:84–94. doi: 10.1016/j.ejps.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Lespine A., Alvinerie M., Vercruysse J., Prichard R.K., Geldhof P. ABC transporter modulation: a strategy to enhance the activity of macrocyclic lactone anthelmintics. Trends Parasitol. 2008;24:293–298. doi: 10.1016/j.pt.2008.03.011. [DOI] [PubMed] [Google Scholar]
- Lespine A., Menez C., Bourguinat C., Prichard R.K. P-glycoproteins and other multidrug resistance transporters in the pharmacology of anthelmintics: prospects for reversing transport-dependent anthelmintic resistance. Int. J. Parasitol. Drugs Drug. Resist. 2012;2:58–75. doi: 10.1016/j.ijpddr.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Hu Y., Bolund L., Wang J. State of the art de novo assembly of human genomes from massively parallel sequencing data. Hum. Genom. 2010;4:271–277. doi: 10.1186/1479-7364-4-4-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschitz A., Entrocasso C., Alvarez L., Lloberas M., Ballent M., Manazza G., Virkel G., Borda B., Lanusse C. Interference with P-glycoprotein improves ivermectin activity against adult resistant nematodes in sheep. Vet. Parasitol. 2010;172:291–298. doi: 10.1016/j.vetpar.2010.04.039. [DOI] [PubMed] [Google Scholar]
- Lifschitz A., Suarez V.H., Sallovitz J., Cristel S.L., Imperiale F., Ahoussou S., Schiavi C., Lanusse C. Cattle nematodes resistant to macrocyclic lactones: comparative effects of P-glycoprotein modulation on the efficacy and disposition kinetics of ivermectin and moxidectin. Exp. Parasitol. 2010;125:172–178. doi: 10.1016/j.exppara.2010.01.009. [DOI] [PubMed] [Google Scholar]
- Lloberas M., Alvarez L., Entrocasso C., Virkel G., Ballent M., Mate L., Lanusse C., Lifschitz A. Comparative tissue pharmacokinetics and efficacy of moxidectin, abamectin and ivermectin in lambs infected with resistant nematodes: impact of drug treatments on parasite P-glycoprotein expression. Int. J. Parasitol. Drugs Drug. Resist. 2013;3:20–27. doi: 10.1016/j.ijpddr.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynagh T., Lynch J.W. A glycine residue essential for high ivermectin sensitivity in Cys-loop ion channel receptors. Int. J. Parasitol. 2010;40:1477–1481. doi: 10.1016/j.ijpara.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Lynagh T., Webb T.I., Dixon C.L., Cromer B.A., Lynch J.W. Molecular determinants of ivermectin sensitivity at the glycine receptor chloride channel. J. Biol. Chem. 2011;286:43913–43924. doi: 10.1074/jbc.M111.262634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynagh T., Lynch J.W. Ivermectin binding sites in human and invertebrate Cys-loop receptors. Trends Pharmacol. Sci. 2012;33:432–441. doi: 10.1016/j.tips.2012.05.002. [DOI] [PubMed] [Google Scholar]
- Lynagh T., Lynch J.W. Molecular mechanisms of Cys-loop ion channel receptor modulation by ivermectin. Front. Mol. Neurosci. 2012;5:60. doi: 10.3389/fnmol.2012.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiola S., Young N.D., Korhonen P., Mondal A., Scheerlinck J.P., Sternberg P.W., Cantacessi C., Hall R.S., Jex A.R., Gasser R.B. Getting the most out of parasitic helminth transcriptomes using HelmDB: implications for biology and biotechnology. Biotechnol. Adv. 2013;31:1109–1119. doi: 10.1016/j.biotechadv.2012.12.004. [DOI] [PubMed] [Google Scholar]
- Martin J., Abubucker S., Heizer E., Taylor C.M., Mitreva M. Nematode.net update 2011: addition of data sets and tools featuring next-generation sequencing data. Nucleic Acids Res. 2012;40:D720–728. doi: 10.1093/nar/gkr1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R.J., Buxton S.K., Neveu C., Charvet C.L., Robertson A.P. Emodepside and SL0-1 potassium channels: a review. Exp. Parasitol. 2012;132:40–46. doi: 10.1016/j.exppara.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Valladares M., Geldhof P., Jonsson N., Rojo-Vázquez F.A., Skuce P. Teladorsagia circumcincta: molecular characterisation of the avr-14B subunit and its relatively minor role in ivermectin resistance Int. J. Parasitol. Drugs Drug Res. 2012;2:154–161. doi: 10.1016/j.ijpddr.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCavera S., Walsh T.K., Wolstenholme A.J. Nematode ligand-gated chloride channels: an appraisal of their involvement in macrocyclic lactone resistance and prospects for developing molecular markers. Parasitology. 2007;134:1111–1121. doi: 10.1017/S0031182007000042. [DOI] [PubMed] [Google Scholar]
- McCavera S., Rogers A.T., Yates D.M., Woods D.J., Wolstenholme A.J. An ivermectin-sensitive glutamate-gated chloride channel from the parasitic nematode Haemonchus contortus. Mol. Pharmacol. 2009;75:1347–1355. doi: 10.1124/mol.108.053363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGonigle L., Mousley A., Marks N.J., Brennan G.P., Dalton J.P., Spithill T.W., Day T.A., Maule A.G. The silencing of cysteine proteases in Fasciola hepatica newly excysted juveniles using RNA interference reduces gut penetration. Int. J. Parasitol. 2008;38:149–155. doi: 10.1016/j.ijpara.2007.10.007. [DOI] [PubMed] [Google Scholar]
- McKay J.P., Raizen D.M., Gottschalk A., Schafer W.R., Avery L. Eat-2 and eat-18 are required for nicotinic neurotransmission in the Caenorhabditis elegans pharynx. Genetics. 2004;166:161–169. doi: 10.1534/genetics.166.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally J., Callan D., Andronicos N.M., Bott N., Hunt P.W. DNA-based methodology for the quantification of gastrointestinal nematode eggs in sheep faeces. Vet. Parasitol. 2013;198:325–335. doi: 10.1016/j.vetpar.2013.09.014. [DOI] [PubMed] [Google Scholar]
- Mealey K.L., Bentjen S.A., Gay J.M., Cantor G.H. Ivermectin sensitivity in collies is associated with a deletion mutation of the mdr1 gene. Pharmacogenetics. 2001;11:727–733. doi: 10.1097/00008571-200111000-00012. [DOI] [PubMed] [Google Scholar]
- Meaney, M., Savage, J., Brennan, G.P., Hoey, E., Trudgett, A., Fairweather, I., 2103. Increased susceptibility of a triclabendazole (TCBZ)-resistant isolate of Fasciola hepatica to TCBZ following co-incubation in vitro with the P-glycoprotein inhibitor, R(+)-verapamil. Parasitology 140, 1287–303. [DOI] [PubMed]
- Menez C., Sutra J.F., Prichard R., Lespine A. Relative neurotoxicity of ivermectin and moxidectin in Mdr1ab (−/−) mice and effects on mammalian GABA(A) channel activity. PLoS Negl. Trop. Dis. 2012;6:e1883. doi: 10.1371/journal.pntd.0001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzker M.L. Sequencing technologies - the next generation. Nat. Rev. Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- Miltsch S.M., Krücken J., Demeler J., Janssen I.J., Krüger N., Harder A., von Samson-Himmelstjerna G. Decreased emodepside sensitivity in unc-49 gamma-aminobutyric acid (GABA)-receptor-deficient Caenorhabditis elegans. Int. J. Parasitol. 2012;42:761–770. doi: 10.1016/j.ijpara.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Mitchell S., Mearns R., Richards I., Donnan A.A., Bartley D.J. Benzimidazole resistance in Nematodirus battus. Vet. Rec. 2011;168:623–624. doi: 10.1136/vr.d3584. [DOI] [PubMed] [Google Scholar]
- Mitreva M., Jasmer D.P., Zarlenga D.S., Wang Z., Abubucker S., Martin J., Taylor C.M., Yin Y., Fulton L., Minx P., Yang S.P., Warren W.C., Fulton R.S., Bhonagiri V., Zhang X., Hallsworth-Pepin K., Clifton S.W., McCarter J.P., Appleton J., Mardis E.R., Wilson R.K. The draft genome of the parasitic nematode Trichinella spiralis. Nat. Genet. 2011;43:228–235. doi: 10.1038/ng.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveu C., Charvet C., Fauvin A., Cortet J., Beech R., Cabaret J. Genetic diversity of levamisole receptor subunits in parasitic nematode species and abbreviated transcripts associated with resistance. Pharmacogenet. Genom. 2010;20:414–425. doi: 10.1097/FPC.0b013e328338ac8c. [DOI] [PubMed] [Google Scholar]
- Niciura S.C., Veríssimo C.J., Gromboni J.G., Rocha M.I., de Mello S.S., Barbosa C.M., Chiebao D.P., Cardoso D., Silva G.S., Otsuk I.P., Pereira J.R., Ambrosio L.A., Nardon R.F., Ueno T.E., Molento M.B. F200Y polymorphism in the β-tubulin gene in field isolates of Haemonchus contortus and risk factors of sheep flock management practices related to anthelmintic resistance. Vet. Parasitol. 2012;190:608–612. doi: 10.1016/j.vetpar.2012.07.016. [DOI] [PubMed] [Google Scholar]
- Njue A.I., Hayashi J., Kinne L., Feng X.P., Prichard R.K. Mutations in the extracellular domains of glutamate-gated chloride channel alpha3 and beta subunits from ivermectin-resistant Cooperia oncophora affect agonist sensitivity. J. Neurochem. 2004;89:1137–1147. doi: 10.1111/j.1471-4159.2004.02379.x. [DOI] [PubMed] [Google Scholar]
- Nobili S., Landini I., Giglioni B., Mini E. Pharmacological strategies for overcoming multidrug resistance. Curr. Drug Targets. 2006;7:861–879. doi: 10.2174/138945006777709593. [DOI] [PubMed] [Google Scholar]
- Opperman C.H., Bird D.M., Williamson V.M., Rokhsar D.S., Burke M., Cohn J., Cromer J., Diener S., Gajan J., Graham S., Houfek T.D., Liu Q., Mitros T., Schaff J., Schaffer R., Scholl E., Sosinski B.R., Thomas V.P., Windham E. Sequence and genetic map of Meloidogyne hapla: a compact nematode genome for plant parasitism. Proc. Natl. Acad. Sci. USA. 2008;105:14802–14807. doi: 10.1073/pnas.0805946105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osei-Atweneboana M.Y., Eng J.K., Boakye D.A., Gyapong J.O., Prichard R.K. Prevalence and intensity of Onchocerca volvulus infection and efficacy of ivermectin in endemic communities in Ghana: a two-phase epidemiological study. Lancet. 2007;369:2021–2029. doi: 10.1016/S0140-6736(07)60942-8. [DOI] [PubMed] [Google Scholar]
- Osei-Atweneboana MY, Awadzi K, Attah SK, Boakye DA, Gyapong JO, Prichard RK. 2011. Phenotypic evidence of emerging ivermectin resistance in Onchocerca volvulus. PLoS Negl. Trop, Dis. 5, e998. [DOI] [PMC free article] [PubMed]
- Otsen M., Plas M.E., Lenstra J.A., Roos M.H., Hoekstra R. Microsatellite diversity of isolates of the parasitic nematode Haemonchus contortus. Mol. Biochem. Parasitol. 2000;110:69–77. doi: 10.1016/s0166-6851(00)00257-7. [DOI] [PubMed] [Google Scholar]
- Overend D.J., Bowen F.L. Resistance of Fasciola hepatica to triclabendazole. Aust. Vet. J. 1995;72:275–276. doi: 10.1111/j.1751-0813.1995.tb03546.x. [DOI] [PubMed] [Google Scholar]
- Ozsolak F., Milos P.M. RNA sequencing: advances, challenges and opportunities. Nat. Rev. Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden A.S., Mac P., Fei Y.J., Castro C., Jiang G., Murfitt K.J., Miska E.A., Griffin J.L., Ganapathy V., Jorgensen E.M. Betaine acts on a ligand-gated ion channel in the nervous system of the nematode C. elegans. Nat. Neurosci. 2013;16:1794–1801. doi: 10.1038/nn.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttachary S., Robertson A.P., Clark C.L., Martin R.J. Levamisole and ryanodine receptors. II: an electrophysiological study in Ascaris suum. Mol. Biochem. Parasitol. 2010;171:8–16. doi: 10.1016/j.molbiopara.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttachary S., Trailovic S.M., Robertson A.P., Thompson D.P., Woods D.J., Martin R.J. Derquantel and abamectin: effects and interactions on isolated tissues of Ascaris suum. Mol. Biochem. Parasitol. 2013;188:79–86. doi: 10.1016/j.molbiopara.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H., Martin R.J., Robertson A.P. Pharmacology of N-, L- and B-subtypes of nematode nAChR resolved at the single-channel level in Ascaris suum. FASEB J. 2006;20:2606–2608. doi: 10.1096/fj.06-6264fje. [DOI] [PubMed] [Google Scholar]
- Rand, J.B., Johnson, C.D. 1995. Genetic Pharmacology. In: Epstein H.F., Shakes, D.C. (Eds.), C. elegans: Modern Biological Analysis of an Organism, Academic Press, New York 187–201.
- Redman E., Packard E., Grillo V., Smith J., Jackson F., Gilleard J.S. Microsatellite analysis reveals marked genetic differentiation between Haemonchus contortus laboratory isolates and provides a rapid system of genetic fingerprinting. Int. J. Parasitol. 2008;38:111–122. doi: 10.1016/j.ijpara.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Redman E., Sargison N., Whitelaw F., Jackson F., Morrison A., Bartley D.J., Gilleard J.S. Introgression of ivermectin resistance genes into a susceptible Haemonchus contortus strain by multiple backcrossing. PLoS Pathog. 2012;8:e1002534. doi: 10.1371/journal.ppat.1002534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed M.B., Spithill T.W., Strugnell R.A., Panaccio M. Fasciola hepatica: stage-specific expression of novel gene sequences as identified by differential display. Exp. Parasitol. 1998;89:169–179. doi: 10.1006/expr.1998.4287. [DOI] [PubMed] [Google Scholar]
- Rinaldi G., Morales M.E., Cancela M., Castillo E., Brindley P.J., Tort J.F. Development of functional genomic tools in trematodes: RNA interference and luciferase reporter gene activity in Fasciola hepatica. PLoS Negl. Trop. Dis. 2008;9:e260. doi: 10.1371/journal.pntd.0000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson A.P., Clark C.L., Martin R.J. Levamisole and ryanodine receptors. I: a contraction study in Ascaris suum. Mol. Biochem. Parasitol. 2010;171:1–7. doi: 10.1016/j.molbiopara.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G., Schein J., Chiu R., Corbett R., Field M., Jackman S.D., Mungall K., Lee S., Okada H.M., Qian J.Q. De novo assembly and analysis of RNA-seq data. Nat. Methods. 2010;7:909–912. doi: 10.1038/nmeth.1517. [DOI] [PubMed] [Google Scholar]
- Robinson M.W., Lawson J., Trudgett A., Hoey E.M., Fairweather I. The comparative metabolism of triclabendazole sulphoxide by triclabendazole-susceptible and triclabendazole-resistant Fasciola hepatica. Parasitol. Res. 2004;92:205–210. doi: 10.1007/s00436-003-1003-6. [DOI] [PubMed] [Google Scholar]
- Romine N.M., Martin R.J., Beetham J.K. Transcriptomic evaluation of the nicotinic acetylcholine receptor pathway in levamisole-resistant and -sensitive Oesophagostomum dentatum. Mol. Biochem. Parasitol. 2014;193:66–70. doi: 10.1016/j.molbiopara.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufener L., Maser P., Roditi I., Kaminsky R. Haemonchus contortus acetylcholine receptors of the DEG-3 subfamily and their role in sensitivity to monepantel. PLoS Pathog. 2009;5:e1000380. doi: 10.1371/journal.ppat.1000380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufener L., Kaminsky R., Maser P. In vitro selection of Haemonchus contortus for benzimidazole resistance reveals a mutation at amino acid 198 of beta-tubulin. Mol. Biochem. Parasitol. 2009;168:120–122. doi: 10.1016/j.molbiopara.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Rufener L., Keiser J., Kaminsky R., Maser P., Nilsson D. Phylogenomics of ligand-gated ion channels predicts monepantel effect. PLoS Pathog. 2010;6:e1001091. doi: 10.1371/journal.ppat.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufener L., Bedoni N., Baur R., Rey S., Glauser D.A., Bouvier J., Beech R., Sigel E., Puoti A. Acr-23 encodes a monepantel-sensitive channel in Caenorhabditis elegans. PLoS Pathog. 2013;9:e1003524. doi: 10.1371/journal.ppat.1003524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan L.A., Hoey E., Trudgett A., Fairweather I., Fuchs M., Robinson M.W., Chambers E., Timson D., Ryan E., Fetwell T., Ivens A., Bentley G., Johnston D. Fasciola hepatica expresses multiple alpha- and beta-tubulin isotypes. Mol. Biochem. Parasitol. 2008;159:73–78. doi: 10.1016/j.molbiopara.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager H., Bapst B., Strehlau G.A., Kaminsky R. Efficacy of monepantel, derquantel and abamectin against adult stages of a multi-resistant Haemonchus contortus isolate. Parasitol. Res. 2012;111:2205–2207. doi: 10.1007/s00436-012-2949-z. [DOI] [PubMed] [Google Scholar]
- Sarai R., Kopp S., Coleman G., Kotze A.C. Acetylcholine receptor subunit and P-glycoprotein transcription patterns in levamisole-susceptible and -resistant Haemonchus contortus. Int. J. Parasitol. Drugs Drug Resist. 2013;3:51–58. doi: 10.1016/j.ijpddr.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarai R., Kopp S., Coleman G., Kotze A.C. Drug-efflux and target-site gene expression patterns in Haemonchus contortus larvae able to survive increasing concentrations of levamisole in vitro. Int. J. Parasitol. Drugs Drug Resist. 2014;4:77–84. doi: 10.1016/j.ijpddr.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders G.I., Wasmuth J.D., Beech R., Laing R., Hunt M., Naghra H., Cotton J.A., Berriman M., Britton C., Gilleard J.S. Characterization and comparative analysis of the complete Haemonchus contortus β-tubulin gene family and implications for benzimidazole resistance in strongylid nematodes. Int. J. Parasitol. 2013;43:465–475. doi: 10.1016/j.ijpara.2012.12.011. [DOI] [PubMed] [Google Scholar]
- Schwarz E.M., Korhonen P.K., Campbell B.E., Young N.D., Jex A.R., Jabbar A., Hall R.S., Mondal A., Howe A.C., Pell J., Hofmann A., Boag P.R., Zhu X.Q., Gregory T.R., Loukas A., Williams B.A., Antoshechkin I., Brown C.T., Sternberg P.W., Gasser R.B. The genome and developmental transcriptome of the strongylid nematode Haemonchus contortus. Genome Biol. 2013;14:R89. doi: 10.1186/gb-2013-14-8-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott I., Pomroy W.E., Kenyon P.R., Smith G., Adlington B., Moss A. Lack of efficacy of monepantel against Teladorsagia circumcincta and Trichostrongylus colubriformis. Vet. Parasitol. 2013;198:166–171. doi: 10.1016/j.vetpar.2013.07.037. [DOI] [PubMed] [Google Scholar]
- Segerberg M.A., Stretton A.O. Actions of cholinergic drugs in the nematode Ascaris suum. Complex pharmacology of muscle and motorneurons. J. Gen. Physiol. 1993;101:271–296. doi: 10.1085/jgp.101.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoop W.L., Mrozik H., Fisher M.H. Structure and activity of avermectins and milbemycins in animal health. Vet. Parasitol. 1995;59:139–156. doi: 10.1016/0304-4017(94)00743-v. [DOI] [PubMed] [Google Scholar]
- Silvestre A., Cabaret J. Mutation in position 167 of isotype 1 beta-tubulin gene of Trichostrongylid nematodes: role in benzimidazole resistance? Mol. Biochem. Parasitol. 2002;120:297–300. doi: 10.1016/s0166-6851(01)00455-8. [DOI] [PubMed] [Google Scholar]
- Simpson J.T., Durbin R. Efficient de novo assembly of large genomes using compressed data structures. Genome Res. 2012;22:549–556. doi: 10.1101/gr.126953.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steijger T., Abril J.F., Engstrom P.G., Kokocinski F., Akerman M., Alioto T., Ambrosini G., Antonarakis S.E., Behr J., Bertone P., Bohnert R., Bucher P., Cloonan N., Derrien T., Djebali S., Du J., Dudoit S., Gerstein M., Gingeras T.R., Gonzalez D., Grimmond S.M., Guigo R., Habegger L., Harrow J., Hubbard T.J., Iseli C., Jean G., Kahles A., Lagarde J., Leng J., Lefebvre G., Lewis S., Mortazavi A., Niermann P., Ratsch G., Reymond A., Ribeca P., Richard H., Rougemont J., Rozowsky J., Sammeth M., Sboner A., Schulz M.H., Searle S.M., Solorzano N.D., Solovyev V., Stanke M., Stevenson B.J., Stockinger H., Valsesia A., Weese D., White S., Wold B.J., Wu J., Wu T.D., Zeller G., Zerbino D., Zhang M.Q. Assessment of transcript reconstruction methods for RNA-seq. Nat. Methods. 2013;10:1177–1184. doi: 10.1038/nmeth.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland I.A., Leathwick D.M. Anthelmintic resistance in nematode parasites of cattle: a global issue? Trends Parasitol. 2011;27:176–181. doi: 10.1016/j.pt.2010.11.008. [DOI] [PubMed] [Google Scholar]
- Swain M.T., Tsai I.J., Assefa S.A., Newbold C., Berriman M., Otto T.D. A post-assembly genome-improvement toolkit (PAGIT) to obtain annotated genomes from contigs. Nat. Protoc. 2012;7:1260–1284. doi: 10.1038/nprot.2012.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trailovic S.M., Clark C.L., Robertson A.P., Martin R.J. Brief application of AF2 produces long lasting potentiation of nAChR responses in Ascaris suum. Mol. Biochem. Parasitol. 2005;139:51–64. doi: 10.1016/j.molbiopara.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercruysse J., Albonico M., Behnke J.M., Kotze A.C., Prichard R.K., McCarthy J.S., Montresor A., Levecke B. Is anthelmintic resistance a concern for the control of human soil-transmitted helminths? Int. J. Parasitol. Drugs Drug Res. 2011;1:14–27. doi: 10.1016/j.ijpddr.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Samson-Himmelstjerna G. Anthelmintic resistance in equine parasites - detection, potential clinical relevance and implications for control. Vet. Parasitol. 2012;185:2–8. doi: 10.1016/j.vetpar.2011.10.010. [DOI] [PubMed] [Google Scholar]
- von Samson-Himmelstjerna G., Walsh T.K., Donnan A.A., Carriere S., Jackson F., Skuce P.J., Rohn K., Wolstenholme A.J. Molecular detection of benzimidazole resistance in Haemonchus contortus using real-time PCR and pyrosequencing. Parasitology. 2009;136:349–358. doi: 10.1017/S003118200800543X. [DOI] [PubMed] [Google Scholar]
- Wang J., Czech B., Crunk A., Wallace A., Mitreva M., Hannon G.J., Davis R.E. Deep small RNA sequencing from the nematode Ascaris reveals conservation, functional diversification, and novel developmental profiles. Genome Res. 2011;21:1462–1477. doi: 10.1101/gr.121426.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Wang L., Liang Y.S. Susceptibility or resistance of praziquantel in human schistosomiasis: a review. Parasitol. Res. 2012;111:1871–1877. doi: 10.1007/s00436-012-3151-z. [DOI] [PubMed] [Google Scholar]
- Wilkinson R., Law C.J., Hoey E.M., Fairweather I., Brennan G.P., Trudgett A. An amino acid substitution in Fasciola hepatica P-glycoprotein from triclabendazole-resistant and triclabendazole-susceptible populations. Mol. Biochem. Parasitol. 2012;186:69–72. doi: 10.1016/j.molbiopara.2012.08.008. [DOI] [PubMed] [Google Scholar]
- Williamson S.M., Robertson A.P., Brown L., Williams T., Woods D.J., Martin R.J., Sattelle D.B., Wolstenholme A.J. The nicotinic acetylcholine receptors of the parasitic nematode Ascaris suum: formation of two distinct drug targets by varying the relative expression levels of two subunits. PLoS Pathog. 2009;5:e1000517. doi: 10.1371/journal.ppat.1000517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson S.M., Storey B., Howell S., Harper K.M., Kaplan R.M., Wolstenholme A.J. Candidate anthelmintic resistance-associated gene expression and sequence polymorphisms in a triple-resistant field isolate of Haemonchus contortus. Mol. Biochem. Parasitol. 2011;180:99–105. doi: 10.1016/j.molbiopara.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Winkelhagen A.J., Mank T., de Vries P.J., Soetekouw R. Apparent triclabendazole-resistant human Fasciola hepatica infection, the Netherlands. Emerg. Infect. Dis. 2012;18:1028–1109. doi: 10.3201/eid1806.120302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Urdaneta-Marquez L., Keller K., James C.E., Davey M.W., Prichard R.K. The role of several ABC transporter genes in ivermectin resistance in Caenorhabditis elegans. Vet. Parasitol. 2012;190:519–529. doi: 10.1016/j.vetpar.2012.06.038. [DOI] [PubMed] [Google Scholar]
- Zawadzki J.L., Kotze A.C., Fritz J.A., Johnson N.M., Hemsworth J.E., Hines B.M., Behm C.A. Silencing of essential genes by RNA interference in Haemonchus contortus. Parasitology. 2012;139:613–629. doi: 10.1017/S0031182012000121. [DOI] [PubMed] [Google Scholar]
- Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using be Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino, D.R., 2010. Using the Velvet de novo assembler for short-read sequencing technologies. Curr. Protoc. Bioinformatics. Chapter 11, Unit 11 15. [DOI] [PMC free article] [PubMed]


