SUMMARY
Loss of Pax3, a developmentally regulated transcription factor expressed in pre-migratory neural crest, results in severe developmental defects and embryonic lethality. Although Pax3 mutations produce profound phenotypes, the intrinsic transcriptional activation exhibited by Pax3 is surprisingly modest. We postulated the existence of transcriptional co-activators that function with Pax3 to mediate developmental functions. A high-throughput screen identified the Hippo effector proteins Taz and Yap65 as Pax3 co-activators. Synergistic co-activation of target genes by Pax3-Taz/Yap65 requires DNA binding by Pax3, is Tead-independent, and is regulated by Hippo kinases Mst1 and Lats2. In vivo, Pax3 and Yap65 co-localize in the nucleus of neural crest progenitors in the dorsal neural tube. Neural crest deletion of Taz and Yap65 results in embryonic lethal neural crest defects and decreased expression of the Pax3 target gene, Mitf. These results suggest that Pax3 activity is regulated by the Hippo pathway and that Pax factors are Hippo effectors.
INTRODUCTION
Neural crest is a multipotent population of cells arising from the embryonic dorsal neural tube. Following neural tube closure, neural crest cells undergo an epithelial-mesenchymal transition (EMT), migrate along defined routes and contribute to multiple organs and tissues including craniofacial cartilage, bone and the pigmented cells of the skin (melanocytes). Neural crest induction is dependent upon inductive signals from neighboring tissues. Wnt and Bmp signaling has been implicated in this process, though the precise details of how inductive signals result in EMT, migration and cell specification remain to be elucidated.
Pax3 plays a critical role in pre-migratory neural crest and is thought to orchestrate neural crest-specific gene expression (Goulding et al., 1991). Mutation of Pax3 in the mouse results in embryonic lethality and a wide array of neural crest abnormalities, including pigmentation defects and congenital heart disease. In the melanocyte lineage, Pax3 functions with Sox10 to directly activate the expression of microphthalmia-associated transcription factor (Mitf), which is required for melanogenesis. In humans, PAX3 mutations result in Waardenburg syndrome, characterized by pigmentation defects and deafness due to neural crest deficiencies.
Other members of the Pax family, consisting of nine related factors in mammals, are expressed in various organs and tissues where they play critical roles in organogenesis. Pax transcription factors are characterized by the presence of a DNA-binding domain termed the paired domain that mediates sequence-specific protein-DNA interactions (Chi and Epstein, 2002). Although Pax genes also encode a C-terminal transactivation domain, in vitro assays reveal relatively weak intrinsic activation capacity. The trans-activation potential of Pax factors may be modulated by interacting proteins. For example, a Pax3/Pax7 binding protein, Pax3/7BP, can recruit H3K4 histone methyltransferase activity to Pax target genes in C2C12 myoblasts, thus regulating cell proliferation and the expression of the Pax target genes Id3 and Cdc20 (Diao et al., 2012). Pax6 can interact with homeodomain-interacting protein kinase 2 (Hipk2). Hipk2 phosphorylates Pax6 and enhances Pax6 interaction with p300, thereby increasing transcriptional activation (Kim et al., 2006). However, the functional role of Pax interacting factors has not been examined in detail in vivo.
The Hippo signaling pathway is a kinase cascade, initially identified in Drosophila, that governs overall organ size through regulation of proliferation and apoptosis (Justice et al., 1995; Xu et al., 1995; Tapon et al., 2002; Harvey et al., 2003; Wu et al., 2003). Following the identification in Drosophila, mammalian Hippo signaling molecules have been defined. The upstream kinases Mst1 and Mst2 interact with a scaffolding protein Salvadore1. Mst1 and Mst2 phosphorylate the downstream kinases Lats1 and Lats2 along with associated scaffolding proteins Mobkl1A/Mobkl1b. Lats1 and Lats2 phosphorylate downstream effector molecules of the Hippo pathway, Taz and Yap65. When Hippo signaling is inactive, Taz and Yap65 are de-phosphorylated, located in the nucleus and act as transcriptional co-activators. Canonical transcriptional activation is achieved through association with Tead factors that serve as the DNA-binding partners for Taz and Yap65. Following activation of the Hippo pathway, Taz and Yap65 are phosphorylated and sequestered in the cytoplasm, thus abrogating transcriptional co-activation. In addition to Tead factors, Taz and Yap65 are capable of interacting in vitro with other proteins including Runx2, ErbB4, p63/p73, Tbx5 and Smads-1/2/3/7. (Zaidi et al., 2004; Komuro et al., 2003; Strano et al., 2001; Alarcon et al., 2009; Murakami et al., 2005, Varelas et al., 2008; Ferrigno et al., 2002). We and others have previously shown that Taz/Yap65, via interaction with Tead, can contribute to regulation of Pax3 gene expression in premigratory neural crest (Milewski et al., 2004; Degenhardt et al., 2010; Gee et al., 2011), and Taz was also identified as a Pax3-interacting protein in a yeast two-hybrid assay (Murakami et al., 2006). Interestingly, Hippo signaling has been shown to promote cellular proliferation and EMT (Lei et al., 2008), two processes critical for the in vivo function of neural crest.
In this manuscript, we demonstrate a potent, synergistic activation of target genes by Pax3 and Taz/Yap65. Target gene activation requires an intact Pax3 DNA-binding domain and is Tead-independent. Hippo kinases, Mst1 and Lats2, can inhibit the transcriptional activity of the complex, demonstrating a role for Hippo signaling. Pax3 and Yap65 are co-expressed in the dorsal neural tube at the time of neural crest delamination. Loss of Taz/Yap65 in these pre-migratory neural crest cells results in down-regulation of Mitf, a target of coordinated Pax-Hippo signaling. Other members of the Pax family are also co-activated by Taz/Yap65, suggesting that one output of Hippo signaling is activation of Pax targets.
RESULTS
A high-throughput screen identifies Taz and Yap65 as Pax3 co-activators
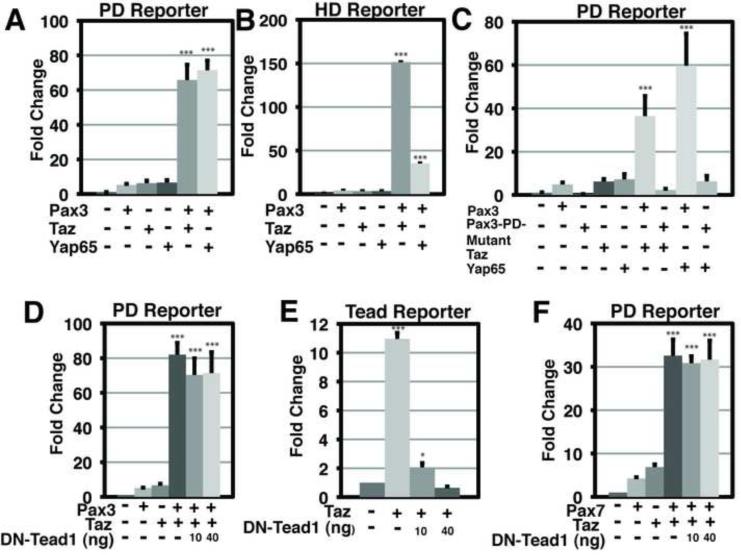
To identify potential Pax3 co-activator molecules, we conducted a high-throughput screen by co-transfecting a Pax3 expression plasmid along with a Pax3 luciferase reporter vector with each of approximately 15,000 human and mouse cDNAs arrayed in 384-well plates in HEK293T cells (Mammalian Gene Collection MGCv1 library, Open BioSystems). The Pax3 reporter contains five Pax3 paired domain DNA binding sequences (Epstein et al., 1994) upstream of a synthetic minimal promoter directing firefly luciferase expression. Potential Pax3 co-activators were identified as cDNAs that generated luciferase activity greater than that of Pax3 alone. Both Taz and Yap65 were identified among the top 5 co-activators (Table S1). We confirmed that Pax3 can interact with Yap65, like Taz (Murakami et al., 2006), using the Duolink® system which utilizes a proximity ligation assay to monitor protein-protein interactions in situ (Figure S1A). Independent validation experiments in HEK293T cells confirmed the ability of either Taz or Yap65 to induce synergistic transactivation of the synthetic Pax3 luciferase reporter when transfected with Pax3 (Figure 1A). When Pax3, Taz or Yap65 was expressed independently, each induced only ~4-5 fold activation. In contrast, when Pax3 was co-expressed with either Taz or Yap65, over 60-fold activation was induced (Figure 1A). The nine mammalian Pax genes can be divided into 4 families based upon sequence conservation, and members of each Pax sub-family can function synergistically with Taz and Yap65 (Figure S1).
Figure 1. Pax3 transcriptional activity is significantly increased in the presence of Taz or Yap65 and the activation is Tead1 independent.
(A) Results of dual luciferase reporter assays in HEK293T cells with Pax3-PairedDomain (PD)-luciferase reporter in the presence (+) or absence (−) of Pax3, Taz or Yap65. (B) Results of dual luciferase reporter assays in HEK293T cells with Pax3-HomeoDomain (HD)-luciferase reporter in the presence (+) or absence (−) of Pax3, Taz or Yap65. (C) Results of dual luciferase reporter assays in HEK293T cells with Pax3-PD-luciferase reporter in the presence (+) or absence (−) of Pax3, Pax3-PD-Mutant, Taz or Yap65. (D) Results of dual luciferase reporter assays in HEK293T cells with Pax3-PD-luciferase reporter in the presence (+) or absence (−) of Pax3, Taz or the specified nanogram (ng) amount of DN-Tead1. (E) Results of dual luciferase reporter assays in HEK293T cells with 8xGTIIC-luciferase reporter in the presence (+) or absence (−) of Taz or the specified nanogram (ng) amount of dominant negative Tead1 (DN-Tead1). (F) Results of dual luciferase reporter assays in HEK293T cells with Pax3-PD-luciferase reporter in the presence (+) or absence (−) of Pax7, Taz or the specified nanogram (ng) amount of DN-Tead1. In all experiments, luciferase activity was first normalized to the activity of a co-transfected renilla luciferase construct, then to the activity observed in the absence of specificed cDNAs (lane 1). All experiments were performed in duplicate a minimum of three individual occasions. Data depicted are the mean + standard error of the mean (SEM). Statistics were completed using an ANOVA with a Tukey-Kramer post-hoc comparison test. *** p<0.001, * p<0.05.
Several Pax family members, including Pax3, contain a homeodomain DNA binding domain in addition to the paired domain, and Pax3 can occupy both paired and homeodomain recognition motifs in vivo (Soleimani et al., 2012). To determine if the presence of Taz or Yap65 enhances Pax3 activation at homeodomain sequences, we generated a Pax-homeodomain luciferase reporter containing nine Pax homeodomain DNA binding motifs upstream of a synthetic minimal promoter directing firefly luciferase expression. Pax3 could synergize with either Taz or Yap65 to activate the synthetic homeodomain reporter (Figure 1B).
Pax3-Taz/Yap65 activation requires Pax3 DNA binding and is Tead-independent
Taz and Yap65 are generally thought to act by interacting with Tead factors, which are expressed ubiquitously, although alternative DNA binding proteins have been described to interact with Taz or Yap65 in vitro (Zhang et al., 2009; Zhao et al., 2008; Zaidi et al., 2004; Komuro et al., 2003; Strano et al., 2001; Murakami et al., 2006). To establish if DNA binding by Pax3 is required for transcriptional co-activation observed in our assays, we examined the effect of a missense mutation in the Pax3 paired-domain found in a patient with Waardenburg syndrome that impairs sequence-specific DNA interactions (Epstein et al., 1995). The Pax3 DNA binding mutant (Pax3-PD-Mutant) was unable to synergize with either Taz or Yap65 (Figure 1C). These data suggest that Pax3 must associate with DNA to mediate the robust activation observed in the presence of Taz and/or Yap65. The Pax3-PD-Mutant was able to synergize with Taz or Yap65 when a homeodomain reporter was used and protein expression was confirmed by immunoblot (Figure S2). Furthermore, inhibition of Tead activity by expression of a dominant-negative Tead mutant (DN-Tead1) consisting of a truncated protein that retains DNA-binding capacity but cannot recruit Taz (Zhang et al., 2009; Zhao et al., 2008) failed to prevent Pax3 synergy with Taz (Figure 1D), although it efficiently abolished Taz activation of a Tead-dependent reporter (Figure 1E). Pax7, which is closely related to Pax3 and is co-expressed in the dorsal neural tube, can also synergize with Taz in a Tead-independent manner (Figure 1F). Thus, we infer that Taz and Yap65 can act as direct co-activators of Pax3/7 target genes.
Pax3 and Yap65 co-localize in the dorsal neural tube
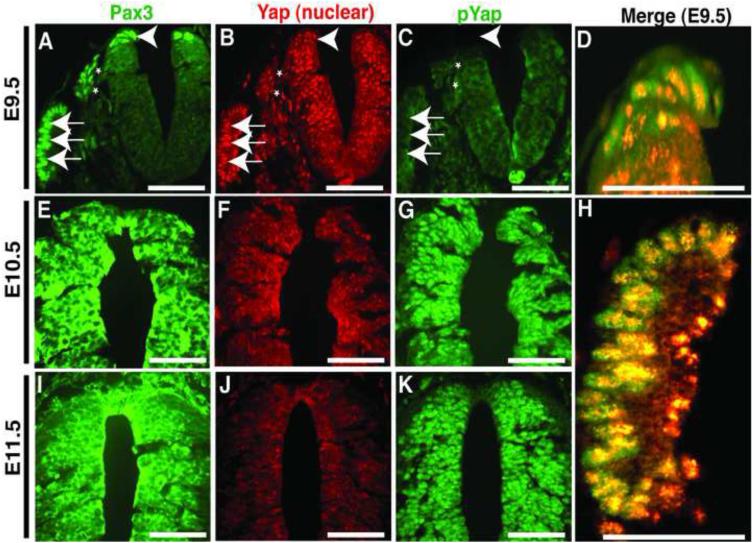
Pax3 is actively transcribed in the dorsal neural tube during early/mid gestation (~E8-10) when neural crest cells undergo an epithelial-to-mesenchymal transition, delaminate, and migrate throughout the body (Goulding et al., 1991). Yap65 is ubiquitously expressed at similar stages (Morin-Kensicki et al., 2006). At E9.5 we observed Pax3 and Yap65 nuclear co-localization in the dorsal neural tube (Figure 2), the dorsal root ganglia which is derived from neural crest (Figure 2A, asterisk) and the somites (Figure 2H). Within the dorsal neural tube, Pax3 and Yap65 are co-localized in the dorsal-most aspect, the site at which neural crest cells undergo EMT and delaminate prior to migration. At E10.5 (Figure 2E-G) and E11.5 (Figure 2I-K), phosphorylated Yap65 becomes prominent (Figure 2G,K) and nuclear Yap65 is greatly reduced (Figure 2F,J), indicating activation of Hippo signaling. Thus, Pax3 and Yap65 are co-localized in the nucleus of premigratory neural crest cells.
Figure 2. Pax3 and Yap65 are co-expressed in the dorsal neural tube at E9.5.
(A-D, H) Frontal sections of E9.5 wildtype embryos. (E-G) Frontal sections of E10.5 wildtype embryos. (I-K) Frontal sections of E11.5 wildtype embryos. Panels A, E and I display immunostaining for Pax3. Panels B, F and J depict immunostaining for Yap65. Panels C, G and K display immunostaining for phosho-Yap. At E9.5 (A and B), Pax3 and Yap are co-localized in the dorsal neural tube (arrowhead), the dorsal root ganglion (asterisks) and somites (arrows). Higher magnification Pax3/Yap merged images of the dorsal neural tube (D) and the somite (H) display the high level of overlap of Pax3 and Yap. Panels A-C, E-G and I-K scale bars: 100μm. In panels D and H, scale bars: 50μm.
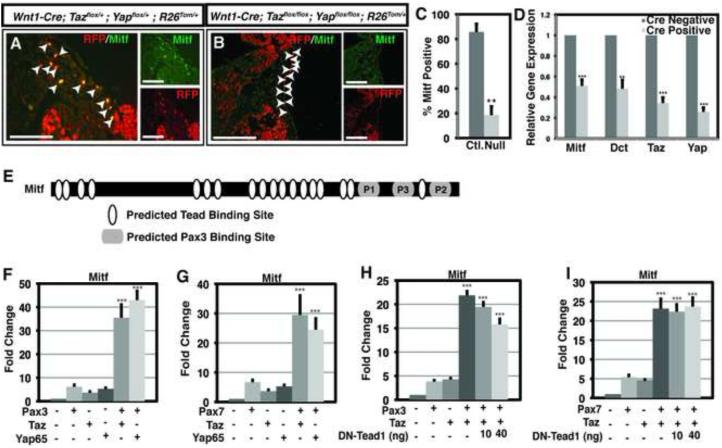
Loss of Taz and Yap in neural crest results in craniofacial defects and down-regulation of Mitf
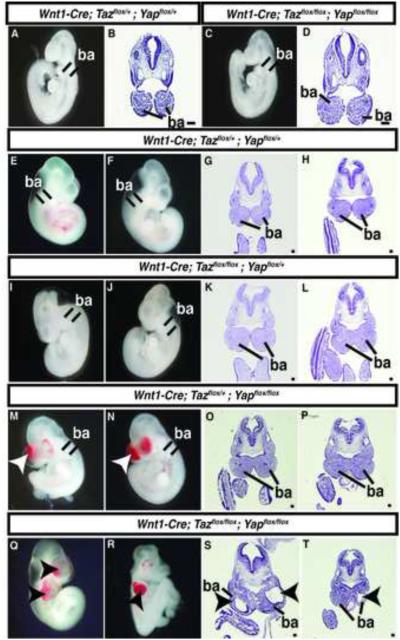
To examine the biological significance of Taz and Yap65 in neural crest, floxed alleles were utilized to delete Taz and Yap65 in premigratory neural crest using Wnt1-Cre (Xin et al., 2011; Xin et al., 2013). Genotyping of 76 P10 pups resulting from Wnt1-Cre; Tazflox/+;Yapflox/+ x Tazflox/flox;Yapflox/flox crosses identified no surviving Wnt1-Cre;Tazflox/flox;Yapflox/flox or Wnt1-Cre;Tazflox/+;Yapflox/flox mice and 5 of 9 surviving Wnt1-Cre;Tazflox/flox;Yapflox/+ mice had craniofacial defects (Table 1). Further analysis of genotype frequencies at various stages of gestation indicated that Wnt1-Cre;Tazflox/flox;Yapflox/flox embryos undergo fetal demise prior to E11.5 although they were identified at the expected frequency at E9.5 (Table 1) when they appeared grossly normal (Figure 3A-D). At E10.5, 13 of 13 Wnt1-Cre;Tazflox/flox;Yapflox/flox embryos exhibited overt craniofacial defects (Table 1 and Figure 3). Within the neural tubes of the E10.5 Yap65/Taz deletion embryos, there was no detectable change in the percentage of proliferating or apoptotic cells (Figure S3). Grossly, hemorrhages were evident in the branchial arch regions in the Yap65/Taz double null embryos (Figure 3Q,R). Histologic analysis revealed deficiencies of facial mesenchyme in the maxillary and mandibular branchial arches (Figure 3S,T), regions normally populated by neural crest. Deletion of both Yap65 alleles and a single Taz allele resulted in a similar hemorrhagic phenotype (Figure 3M,N) although facial mesenchyme appeared intact (Figure 3O,P), and mice of this genotype were not identified postnatally (Table 1). In embryos in which both Taz alleles and a single Yap65 allele were removed, the embryos were phenotypically normal at E10.5 (Figure 3I-L) and they survived postnatally (Table 1).
Table 1.
Genotyping results of Wnt1-Cre;Tazflox/+;Yapflox/+ × Tazflox/flox;Yapflox/flox Cross
| Genotype Wnt1-Cre;Tazflox/+;Yapflox/+ × Tazflox/flox;Yapflox/flox Cross |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Age |
Tazflox/+;
Yapflox/+ |
Tazflox/flox;
Yapflox/+ |
Tazflox/+;
Yapflox/flox |
Tazflox/flox;
Yapflox/flox |
Wnt1-
Cre; Tazflox/+; Yapflox/+ |
Wnt1-
Cre; Tazflox/flox; Yapflox/+ |
Wnt1-
Cre; Tazflox/+; Yapflox/flox |
Wnt1-
Cre; Tazflox/flox; Yapflox/flox |
Total | χ2 |
| E9.5 | 14 | 7 | 8 | 12 | 8 | 8 | 8 | 8 | 73 | N/S |
| E10.5 | 5 | 7 | 10 | 4 | 7 | 7 | 4 | 13a | 57 | N/S |
| E11.5 | 5 | 10 | 8 | 2 | 9 | 6 | 7 | 0 | 47 | 0.0493 |
| P10 | 12 | 8 | 14 | 18 | 15 | 9b | 0 | 0 | 76 | 0.0001 |
13/13 Craniofacial abnormalities
5/9 Craniofacial hemorrhage
Figure 3. Taz and Yap65 are required for normal neural crest development.
Whole mount bright field images (A,C,E,F,I,J,M,N,Q,R) and H&E stained transverse sections (B,D,G,H,K,L,O,P,S,T) of representative embryos of the indicated genotypes at E9.5 (A-D) and E10.5 (E-T). Arrowheads in M-N and Q-T indicate regions of abnormal facial mesenchyme. ba= branchial arches. Scale bars: 100μm.
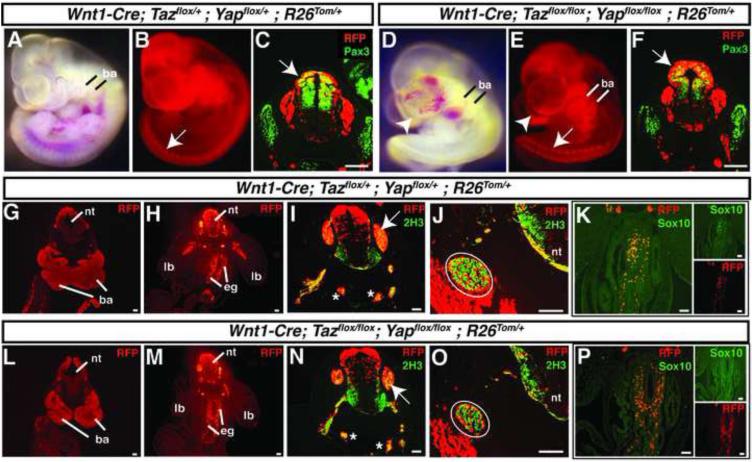
We followed the fate of neural crest cells with or without Yap65 and Taz by including a R26tdTomato (R26Tom/+) Cre reporter allele. Wnt1-derived cells can be identified in resultant embryos by expression of tdTomato and detection with an anti-RFP antibody. Examination of control and double mutant embryos at E10.5 showed that neural crest migration was grossly intact (Figure 4). Neural crest derivatives populated the branchial arches and peripheral ganglia (Figure 4B,E) and Pax3 expression was maintained in double null embryos (Figure 4C,F). Serial sections of fate-mapped E10.5 embryos revealed intact migration of neural crest throughout the embryo where they populated regions of presumptive facial mesenchyme, the peripheral nervous system and the enteric ganglia (Figure 4G,H,L,M). Neurofilament expression by neural crest populating peripheral ganglia appeared normal, as determined by co-staining of fate-mapped neural crest cells with the 2H3 anti-neurofilament antibody (Figure 4I,J,N,O). Enteric ganglia derivatives expressed Sox10 at levels indistinguishable from controls (Figure 4K,P). Thus, many aspects of neural crest migration and fate-specification appeared intact despite the loss of Taz/Yap65.
Figure 4. Characterization of embryos lacking Taz and Yap65.
(A,B) Wnt1-Cre; Taz flox/+ ;Yap flox/+; R26Tom/+ E10.5 embryo imaged in bright field (A) and fluorescence (B). (C) Transverse sections of E10.5 Wnt1-Cre; Taz flox/+ ;Yap flox/+; R26Tom/+ embryo stained tdTomato (RFP) and Pax3. (D,E) Wnt1-Cre; Taz flox/flox;Yap flox/flox; R26Tom/+ E10.5 embryo imaged in bright field (D) and fluorescence (E). (F) Transverse sections of E10.5 Wnt1-Cre; Taz flox/flox;Yap flox/flox; R26Tom/+ embryo stained for tdTomato (RFP) and Pax3. Arrows C,F designate co-staining in the dorsal neural tube. Branchial arches (ba) are invested with red-fluorescing Wnt1-derived neural crest (A,B and D,E). (G-K) Transverse sections of E10.5 Wnt1-Cre; Taz flox/+ ;Yap flox/+;R26Tom/+ embryos stained for tdTomato (RFP) (G and H), tdTomato (RFP) and neurofilament (2H3) (I and J) or tdTomato (RFP) and Sox10 (K). (L-P) Transverse sections of E10.5 Wnt1-Cre; Taz flox/flox ;Yap flox/flox;R26Tom/+ embryos stained for tdTomato (RFP) (L and M), tdTomato (RFP) and neurofilament (2H3) (N and O) or tdTomato (RFP) and Sox10 (P). Arrows in I, N mark dorsal root ganglia while asterisks demark the right and left sympathetic trunks. Circled areas in J, O denote the glossopharyngeal (IX) preganglion. ba=branchial arches, lb=limb bud, nt=neural tube, eg=enteric ganglia. Images were merged after global adjustment for brightness and were produced by combining respective red and green channels using Photoshop software (Adobe). Scale bars: 100μm.
Mitf is coordinately regulated by Pax3 and Yap/Taz
Neural crest derived cells were also observed adjacent to surface ectoderm in both control and double null embryos where migrating melanoblasts are normally found (Figure 5A,B). In control embryos, these cells expressed Mitf, a transcriptional activator of melanogenesis regulated by Pax3 (Watanabe et al., 1998) (Figure 5A). However, in double knockout embryos, Mitf expression was markedly down-regulated in these neural crest derivatives (Figure 5B). Quantitation of RFP/Mitf positive cells adjacent to and within the surface ectoderm revealed a significant decrease in the percentage of RFP+ cells that also expressed Mitf in Yap65/Taz null embryos (Figure 5C). In mouse embryonic fibroblasts derived from Tazflox/flox;Yapflox/flox embryos, treatment with a Cre virus to delete both alleles of Yap65 and Taz produced a significant decrease in Mitf expression (Figure 5D). Dopachrome tautomerase (Dct) is a direct transcriptional target of Mitf (Lang et al., 2005) and was similarly decreased following deletion of Yap65 and Taz (Figure 5D). We sought to determine if altered Mitf expression in Yap/Taz double knockout embryos was due to loss of synergy between Pax3 and Yap/Taz, or if this was a result of Tead-dependent Yap/Taz gene regulation. Regulatory elements controlling Mitf expression have been extensively characterized previously (Watanabe et al., 1998; Bondurand et al., 2000; Potterf et al., 2000). Both Pax3 and Pax7 can synergize with Taz and Yap65 to activate a Mitf-luciferase reporter construct (Figure 5F, G). Although 18 potential Tead binding sites are located in the Mitf promoter/enhancer region included in this assay (Figure 5E), DN-Tead1 failed to significantly impair synergistic Pax-Taz/Yap65 co-activation (Figure 5H,I), suggesting that Mitf activation by Pax3 and Taz is Tead-independent.
Figure 5. Mitf is coordinately regulated by Pax3 and Yap/Taz.
(A,B) Fluorescent images of transverse sections of E10.5 embryos immunostained for Mitf and tdTomato (RFP). (A) Co-labeled cells adjacent to surface ectoderm where migrating melanoblasts are normally found are indicated by arrowheads. (B) Migratory neural crest cells near the ectoderm that do not express Mitf in double mutant embryos are indicated by arrowheads. Scale bars: 100μm. (C) Quantitation of RFP/Mitf expressing cells adjacent to and in the surface ectoderm of control (Ctl.) and Yap65/Taz double null embryos (Null). Data depicted are the average percentage of RFP+ cells that also express Mitf + SEM. A minimum of 60 cells were quantified from both control and null animals encompassing sections from three individual control and null embryos. Images were merged after global adjustment for brightness and were produced by combining respective red and green channels using Photoshop software (Adobe). (D) Quantitative real-time PCR of untreated or Cre-virus treated Tazflox/flox;Yapflox/flox mouse embryonic fibroblasts for Mitf, Dct, Taz and Yap65. Data depicted in D are the mean + standard error of the mean (SEM). Statistics in C and D were completed using a Student’s t-test ** p<0.01, *** p<0.001. (E) Schematic representation of a Mitf luciferase reporter plasmid. Pax3 binding sites are denoted within gray oblong circles. Predicted TEAD binding sites are denoted with white ovals, representing the consensus sequence XDGHATXT where X = A, T, C, or G; D = A or T; and H = A, T, or C (Anbanandam et al., 2006). (F) Results of dual luciferase reporter assays in HEK293T cells with Mitf-luciferase reporter in the presence (+) or absence (−) of Pax3, Taz or Yap65. (G) Results of dual luciferase reporter assays in HEK293T cells with Mitf-luciferase reporter in the presence (+) or absence (−) of Pax7, Taz or Yap65. (H) Results of dual luciferase reporter assays in HEK293T cells with Mitf-luciferase reporter in the presence (+) or absence (−) of Pax3, Taz or the specified nanogram (ng) amount of DN-Tead1. (I) Results of dual luciferase reporter assays in HEK293T cells with Mitf-luciferase reporter in the presence (+) or absence (−) of Pax7, Taz or the specified nanogram (ng) amount of DN-Tead1. In all experiments, luciferase activity was first normalized to the activity of a co-transfected renilla luciferase construct, then to the activity observed in the absence of specificed cDNAs (lane 1). All experiments were performed in duplicate a minimum of three individual occasions. Data depicted in F-I are the mean + standard error of the mean (SEM). Statistics were completed using an ANOVA with a Tukey-Kramer post-hoc comparison test. *** p<0.001
Two Pax3 paired-domain binding sites have been described within this Mitf enhancer: site P1 at position −260 to −244 and site P2 at position −40 to −26 upstream of the transcription start site (Watanabe et al., 1998; Bondurand et al., 2000; Potterf et al., 2000). Sequence analysis utilizing the TRANSFAC database identified an additional previously uncharacterized potential Pax3 binding site, which we termed P3, located at position −160 to −143 relative to the transcription start site. A 519 base pair (bp) fragment of the Mitf enhancer, which contains the P1, P2 and P3 sites, retained the ability to be synergistically activated by Pax3 with Taz or Yap65 with activity comparable to the full length Mitf reporter (Figure S4). Each Pax3 binding site was then evaluated individually in ~100-200 bp Mitf promoter/enhancer fragments. Fragments containing P2 and P3 exhibited coactivation with Taz and Yap65, but the coactivation at site P1 was not significant in the context of the enhancer fragment. Mutation of the Pax3 binding sites within P2 and P3 led to a statistically significant decrease in the observed coactivation with Taz and Yap65. The P2 and P3 fragments each contain a single predicted Tead binding site, but DN-Tead1 did not significantly decrease Pax3-Taz or Pax3-Yap65 coactivation of the P2, P3 or 519 bp Mitf reporter (Figure S5). Taken together, we interpret these results to indicate that P2 and P3, but not P1, act as docking sites for a Pax3-Taz/Yap65 co-activation complex.
Hippo kinases Mst1 and Lats2 can inhibit Pax3-Taz/Yap65 synergistic activation
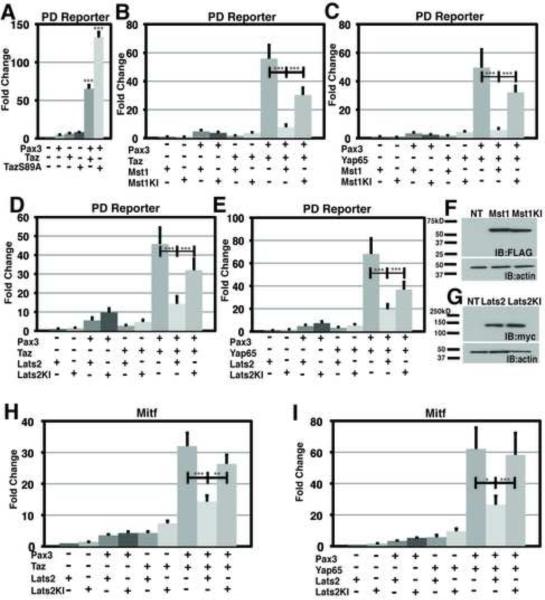
Taz and Yap65 are downstream effectors of the Hippo signaling pathway that is composed of a series of regulatory protein kinases, including Mst and Lats. Activation of the Hippo pathway culminates in phosphorylation of Taz/Yap65, which induces translocation to the cytoplasm and abrogation of nuclear functions. Serine residue 89 of the Taz protein was previously demonstrated to be a critical residue for Lats kinase phosphorylation (Lei et al., 2008). Upon mutation to alanine, TazS89A gains resistance to Lats regulation and is constitutively nuclear (Lei et al., 2008). When Pax3 was co-expressed with TazS89A there was over 130-fold activation of a Pax3 reporter (Figure 6A), consistent with constitutive activation. TazS89A was even more potent than wild type Taz, suggesting basal activity of Hippo signaling under these experimental conditions.
Figure 6. Hippo kinases Mst1 and Lats2 can inhibit the activity of the Pax3-Taz/Yap complex.
(A) Results of dual luciferase reporter assays in HEK293T cells with Pax3-PD-luciferase reporter in the presence (+) or absence (−) of Pax3, Taz or constitutively active Taz, TazS89A. (B) Results of dual luciferase reporter assays in HEK293T cells with Pax3-PD-luciferase reporter in the presence (+) or absence (−) of Pax3, Taz, Mst1 or a kinase inactive form of Mst1, Mst1KI. (C) Results of dual luciferase reporter assays in HEK293T cells with Pax3-PD-luciferase reporter in the presence (+) or absence (−) of Pax3, Yap65, Mst1 or Mst1KI. (D) Results of dual luciferase reporter assays in HEK293T cells with Pax3-PD-luciferase reporter in the presence (+) or absence (−) of Pax3, Taz, Lats2 or Lats2KI. (E) Results of dual luciferase reporter assays in HEK293T cells with Pax3-PD-luciferase reporter in the presence (+) or absence (−) of Pax3, Yap65, Lats2 or Lats2KI. (F) Anti-FLAG western blot from HEK293T cells either non-transfected (NT), transfected with a FLAG tagged Mst1 or FLAG tagged Mst1KI. Western blots of the same cellular protein lysates were also probed with anti-actin to demonstrate equivalent protein loading among samples. (G) Anti-myc western blot from HEK293T cells either non-transfected (NT), transfected with a myc tagged Lats2 or transfected with a kinase inactive form of Lats2, a myc tagged Lats2KI. Western blots of the same cellular protein lysates were also probed with anti-actin to demonstrate equivalent protein loading among samples. (H) Results of dual luciferase reporter assays in HEK293T cells with Mitf-luciferase reporter in the presence (+) or absence (−) of Pax3, Taz, Lats2 or Lats2KI. (I) Results of dual luciferase reporter assays in HEK293T cells with Mitf-luciferase reporter in the presence (+) or absence (−) of Pax3, Yap65, Lats2 or Lats2KI. In all experiments, luciferase activity was first normalized to the activity of a co-transfected renilla luciferase construct, then to the activity observed in the absence of specificed cDNAs (lane 1). All experiments were performed in duplicate a minimum of three individual occasions. Data depicted are the mean + standard error of the mean (SEM). Statistics were completed using an ANOVA with a Tukey-Kramer post-hoc comparison test. *** p<0.001, **p<0.01, * p<0.05
We probed the ability of upstream Hippo kinases Mst1 and Lats2 to modulate Pax3-Taz/Yap65 transcriptional synergy. Both Mst1 and Lats2 inhibited Pax3-Taz and Pax3-Yap65 co-activation (Figure 6B-E). Kinase inactive mutants of Mst1 or Lats2, Mst1KI (Mst1-K59R) or LATS2KI (LATS2-D809A) respectively, were relatively ineffective at inhibiting Pax3-Taz and Pax3-Yap65 co-activation (Figure 6B-E). Immunoblot analysis confirmed that the wildtype and kinase inactive forms of each Hippo kinase were expressed at equivalent protein levels (Figure 6F-G). LATS2 also significantly inhibited co-activation of the endogenous Mitf enhancer by Pax3-Taz/Yap65 (Figure 6H-I). In contrast, expression of LATS2KI did not alter Pax3-Taz or Pax3-Yap65 co-activation at the Mitf enhancer (Figure 6H-I). Thus, transcriptional synergy between Pax factors and Yap/Taz can be modulated by upstream regulators of the Hippo pathway.
We further examined the ability to Pax3 and Pax7 to co-activate another previously described Pax target gene. Myf5 is a regulator of myogenic determination that is another example of a direct transcriptional target of Pax3/7 (Bajard et al., 2006). Pax3 and Pax7 each functioned with Taz or Yap65 to co-activate a previously described Myf5 enhancer (Figure S6A-C). This activation was Tead-independent, as the presence of DN-Tead1 did not alter the co-activation (Figure S6D,E). Further Lats2, but not Lats2KI, significantly inhibited Pax3-Taz/Yap65-mediated activation of the Myf5 reporter (Figure S6F,G).
DISCUSSION
Hippo signaling is a critical regulator of proliferation and EMT during development and in cancer (Lei et al., 2008). In this study we demonstrate that the downstream effector molecules of Hippo signaling, Taz and Yap65, can function as co-activators of Pax3. This complex requires Pax3 to act as the DNA binding moiety and is regulated by upstream Hippo kinases. Unlike canonical Hippo signaling, the Pax3-Taz/Yap65 complex does not require Tead factors to mediate activity. Deletion of Taz and Yap65 in pre-migratory neural crest resulted in craniofacial defects, similar to those seen in the absence of Pax3 and Pax7 (Mansouri and Gruss, 1998), and down-regulation of the Pax3 target, Mitf. In vitro data suggests that another Pax3 target, Myf5, can also be coordinately regulated by Pax3 and Yap/Taz.
It remains unclear if all Pax targets will require Yap65 or Taz for functional activation, or if a subset of physiologic target genes are Yap/Taz independent. Clearly, many Yap/Taz targets are Pax-independent, and rely on Tead factors to mediate DNA binding. It is likely that Pax factors can activate (or repress) some downstream pathways in the absence of Yap/Taz, while a subset of targets are strongly co-activated in the presence of both Pax and Yap/Taz. Further delineation of the various classes of downstream regulated genes will be important to define the extent of Pax/Hippo coordinate regulation.
The Pax3-Taz/Yap65 complex produces potent transcriptional activation. In vivo, this complex is likely to include additional factors that may vary with context. Candidates include the previously identified Pax3/7 binding protein, Pax3/7BP which recruits an H3K4 histone methyltransferase to target gene promoters (Diao et al., 2012). Taz has been shown to interact with the histone acetyl transferases PCAF and p300 (Murakami et al., 2005). This interaction greatly enhances Taz transcriptional activity at an atrial natriuretic factor promoter reporter construct (Murakami et al., 2005). Future studies will elucidate the nature of additional Pax/Taz/Yap complex subunits.
Since Pax factors from all Pax families can utilize Taz and Yap65 as coactivators, Hippo signaling could be a common means to regulate activity of many Pax target genes in diverse developmental contexts. Pax genes function in multiple developing tissues and play key roles in organogenesis. Pax6, for example, is critical for formation of the eye and is mutated in inherited causes of blindness. Pax2 functions in early kidney formation, Pax8 in the thymus and has also been shown to co-activate with Taz, and Pax1/9 in the skeleton (Di Palma et al., 2009). Hippo signaling is known to regulate organ size. The functional interaction of Pax proteins with Yap65 and Taz may provide a physical and functional mechanism to link organogenesis with the regulation of organ size. Likewise, disruption of Pax function has been implicated in cancer, and the role of Pax/Hippo synergy in oncogenesis will be an important area of future investigation.
EXPERIMENTAL PROCEDURES
Plasmids
The firefly-luciferase reporter construct 5xPax3BS was provided by M. Buckingham (Relaix et al., 2003) and subcloned into pGL4.27 (Promega). To generate a Pax3-homeodomain reporter construct, a previously identified Pax homeodomain-binding motif (Soleimani et al., 2012), TAATTGATTA, was synthetically generated and re-iterated nine times. This sequence was then cloned upstream of pGL4.27 for use in luciferase assays (Promega). An MITF reporter previously published, containing approximately 2 kb of the 5’ proximal human promoter (chr3:69,934,337-69,936,682 (hg38), Lang et al., 2005) was subcloned into pGL4.27 (Promega). 8xGTIIC, a Tead reporter, was provided by S. Piccolo ((Dupont et al., 2011), Addgene plasmid 34615). Murine Taz and Taz-S89A expression vectors were described previously ((Kanai et al., 2000), Addgene plasmids 19025 and 19026) and were subcloned into pCMV-Sport6 (Invitrogen). Murine Yap65 was expressed in pCMV-Sport6 (Invitrogen) as described previously (Milewski et al., 2004). Dominant negative human TEAD1 (DN-TEAD1) was provided by Kun-Liang Guan (Zhang et al., 2009; Zhao et al., 2008). Murine hippo kinases Mst1 and Mst1-KI were previously described ((Lin et al., 2002), Addgene plasmids 1965 and 1966). Expression plasmids for human hippo kinases LATS2 and LATS2-KI were provided by D. Pan (Dong et al., 2007). The murine Pax3 expression construct was previously described (Lang et al., 2000) and was subcloned into pCMV-Sport6 (Invitrogen). The murine Pax3 paired domain mutant (Pax3-PD-Mutant) contains a single cytosine to thymine mutation that produces a proline to leucine change at amino acid 50 which prevents Pax3 binding to DNA sequences, was previously described (Epstein et al., 1995) and was subcloned into pCMV-Sport6 (Invitrogen). Human PAX7 was expressed in pcDNA3 (Invitrogen).
Cell Culture and Luciferase Assay
HEK293T cells were maintained at 37°C with 5% CO2 in DMEM supplemented with 10% fetal bovine serum, penicillin and streptomycin. All HEK293T cell transfections were completed using FuGene6 (Roche). Experiments utilized 250 ng of the specified firefly-luciferase reporter constructs, 80 ng Taz/Yap65, 40 ng Pax expression vector and 75 ng pGL2-Basic-renilla luciferase (Promega). In experiments with Mst1, Mst1-KI, LATS2 or LATS2-KI, 120 ng of the kinase expression vector was included. All transfections maintained an equal concentration of total DNA with the inclusion of the pCMV-Sport6 empty vector (Invitrogen). Cellular extracts were collected 48 hours post-transfection for use in a dual-luciferase assay (Promega). Twenty microliters (μl) of cellular extract was used to assess firefly and renilla luciferase activities and luciferase activity was normalized to the renilla activity. All experiments were performed in duplicate on at least three separate occasions. Statistical differences between conditions were analyzed using ANOVA, with a Tukey-Kramer post-hoc comparison test.
Western Blot
HEK293T cells were transfected with 2 ug of the specified Hippo kinase: Mst1, Mst1KI, LATS2, LATS2-KI. Mst1 and Mst1KI contain an N-terminal FLAG epitope tag while LATS2 and LATS2KI contain an N-terminal myc epitope tag. A non-transfected plate of cells was processed in parallel as a control. 48 hours post-transfection, cells were lysed in RIPA buffer (150 mm NaCl, 50 mm Tris-Base, pH 7.5, 1% IGEPAL, 0.5% sodium deoxycholate, 0.1% SDS) plus protease inhibitors (Complete Mini, Roche). All lysates were quantitated with a BCA Assay (Promega) and an equal amount of total protein was loaded per well of a 4-12% gradient gel and run at 120V for 2 hours. Blots were transferred overnight at 4°C, at 20V onto PVDF membrane (Invitrogen). Membranes were blocked in 10% non-fat dry milk/TBS-T and incubated in primary antibody overnight: M2-FLAG (mouse monoclonal, Sigma), myc (mouse monoclonal, Cell Signaling #2276), or actin (rabbit polyclonal, Cell Signaling, #4970). All blots were probed with horseradish peroxidase (HRP) conjugated secondary antibody for 1 hour with either anti-mouse secondary (Cell Signaling) or anti-rabbit secondary (Cell Signaling). Blots were developed using ECL Prime (Amersham).
Histology and Immunofluorescence
Samples were harvested, fixed overnight in 4% paraformaldehyde and dehydrated through an ethanol series. All samples were paraffin-embedded and sectioned. Antibodies used for immunofluorescence were anti-Pax3 mouse monoclonal (developed by C.P. Ordahl and obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH), anti-Yap rabbit polyclonal (Cell Signaling #4912S), anti-phosho-Yap rabbit polyclonal (Cell Signaling #4911), anti-neurofilament (2H3, developed by T. M. Jessell and J. Dodd obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH), anti-Mitf (Vector Laboratories, clone 34CA5), anti-Sox10 (Santa Cruz Biotechnology, N-20) and anti-RFP (Rockland Immunochemicals, Inc , #600-401-379). Hematoxylin and eosin (H&E) staining was completed using a standard protocol.
Melanocyte Quantitation
Transverse sections from Wnt1-Cre;Taz flox/+ ;Yap flox/+;R26Tom/+ and Wnt1-Cre;Taz flox/flox;Yap flox/flox;R26Tom/+ E10.5 embryos were co-stained for RFP and Mitf. Cells greater than 100μm from the surface ectoderm were excluded from analysis. Seven sections from three independent embryos were included in the analysis which totaled 60 RFP+ cells in control embryos and 62 RFP+ cells in null embryos. Data is reported as an average percentage of each field totaled + SEM. Statistical differences between conditions were analyzed using a Student’s t-test.
Mouse Embryo Fibroblast Preparation
Mouse Embryo Fibroblasts (MEFs) were isolated from E14.5 Tazflox/flox;Yapflox/flox embryos as described (Connor 2000). Briefly, E14.5 embryos were collected, the heads and internal organs were removed and the remains were washed twice in PBS. Embryos were then dissociated by passage through a 16-gauge needle and incubated in a trypsin solution for 5 minutes at 37°C. The resulting cells were then triturated and incubated for another 5 minutes at 37°C, an equal volume of MEF media was added and incubated for 5 minutes at RT. After centrifugation, the cell pellet was resuspended in MEF media, plated and incubated at 37°C. Cells were grown to confluence, then trypsinized and replated for treatment. In order to delete the floxed Taz and Yap65 alleles, MEFs were treated for 48 hours with an adeno-associated virus expressing a constitutively active Cre-recombinase (AAV1-CMV-Cre, Penn Vector Core, University of Pennsylvania).
RNA isolation, complementary DNA synthesis and quantitative Real-Time PCR (RT-PCR)
RNA was harvested from 1-100mm dish of untreated or AAV1-CMV-Cre treated Tazflox/flox;Yapflox/flox MEFs using the Qiagen RNeasy kit following manufacturers instructions (Qiagen). Complementary DNA (cDNA) was synthesized with the Superscript III system (Invitrogen). Quantitative RT-PCR was performed in triplicate with SYBR Green reagents (Applied Biosystems). Relative gene expression was normalized to Gapdh. Quantitative RT-PCR primer sequences are:
Mitf Forward: 5’ CGAGCTCATGGACTTTCCCTTA 3’
Mitf Reverse: 5’ CTTGATGATCCGATTCACCAAA 3’
DCT Forward: 5’ TTCGCAAAGGCTATGCGC 3’
DCT Reverse: 5’ GTTACTACCCAGGTCAGGCCAG 3’
Taz Forward: 5’ GAGAGGATTAGGATGCGTCAAG 3’
Taz Reverse: 5’ GGATCTGAGCTACTGTTGGTG 3’
Yap Forward: 5’ ACCATAAGAACAAGACCACATCC 3’
Yap Reverse: 5’ CTTCACTGGAGCACTCTGAG 3’
Gapdh Forward: 5’ CGTCCCGTAGACAAAATGGT 3’
Gapdh Reverse: 5’ GAATTTGCCGTGAGTGGAGT 3’
Mice
All mice were maintained on a mixed genetic background. Wnt1-Cre (Jiang et al., 2000), Yapflox/+ (Xin et al., 2011) and Tazflox/+ (Xin et al., 2013) alleles were genotyped as previously described. The Yapflox/+ and Tazflox/+ alleles were developed in the laboratory of Dr. Eric N. Olson at the University of Texas Southwestern Medical Center, Dallas, Texas. R26tdTomato mice (B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J) were obtained from Jackson Labs (strain number 007914). All animal protocols were approved by the University of Pennsylvania Institutional Animal Care and Use Committee.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr. James F. Martin for sharing reagents and data prior to publication. We would like to thank Jeanne Geskes for her help in performing the screen, Matthew Emmett and Feiyan Liu for technical assistance, and Min Min Lu and Lan Cheng for help with histology. This work was supported by the Spain Fund for Cardiovascular Research, the WW Smith Endowed Chair, and NIH U01 HL100405 to J.A.E. J.B.H is supported by the National Institute of Neurological Disorders and Stroke (1R01NS054794-06), the Defense Advanced Research Projects Agency (DARPA-D12AP00025, to John Harer, Duke University), and by the Penn Genome Frontiers Institute under a HRFF grant with the Pennsylvania Department of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alarcon C, Zaromytidou AI, Xi Q, Gao S, Yu J, Fujisawa S, Barlas A, Miller AN, Manova-Todorova K, Macias MJ, et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell. 2009;139:757–769. doi: 10.1016/j.cell.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anbanandam A, Albarado DC, Nguyen CT, Halder G, Gao X, Veeraraghavan S. Insights into transcription enhancer factor 1 (TEF-1) activity from the solution structure of the TEA domain. Proc Natl Acad Sci U S A. 2006;103:17225–17230. doi: 10.1073/pnas.0607171103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajard L, Relaix F, Lagha M, Rocancourt D, Daubas P, Buckingham ME. A novel genetic hierarchy functions during hypaxial myogenesis: Pax3 directly activates Myf5 in muscle progenitor cells in the limb. Genes Dev. 2006;20:2450–2464. doi: 10.1101/gad.382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondurand N, Pingault V, Goerich DE, Lemort N, Sock E, Le Caignec C, Wegner M, Goossens M. Interaction among SOX10, PAX3 and MITF, three genes altered in Waardenburg syndrome. Hum Mol Genet. 2000;9:1907–1917. doi: 10.1093/hmg/9.13.1907. [DOI] [PubMed] [Google Scholar]
- Chi N, Epstein JA. Getting your Pax straight: Pax proteins in development and disease. Trends Genet. 2002;18:41–47. doi: 10.1016/s0168-9525(01)02594-x. [DOI] [PubMed] [Google Scholar]
- Connor DA. Mouse Embryo Fibroblast (MEF) Feeder Cell Preparation. Current Protocols in Molecular Biology. 2000:23.2.1–23.2.7. doi: 10.1002/0471142727.mb2302s51. [DOI] [PubMed] [Google Scholar]
- Degenhardt KR, Milewski RC, Padmanabhan A, Miller M, Singh MK, Lang D, Engleka KA, Wu M, Li J, Zhou D, et al. Distinct enhancers at the Pax3 locus can function redundantly to regulate neural tube and neural crest expressions. Dev Biol. 2010;339:519–527. doi: 10.1016/j.ydbio.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao Y, Guo X, Li Y, Sun K, Lu L, Jiang L, Fu X, Zhu H, Sun H, Wang H, et al. Pax3/7BP is a Pax7- and Pax3-binding protein that regulates the proliferation of muscle precursor cells by an epigenetic mechanism. Cell Stem Cell. 2012;11:231–241. doi: 10.1016/j.stem.2012.05.022. [DOI] [PubMed] [Google Scholar]
- Di Palma T, D'Andrea B, Liguori GL, Liguoro A, de Cristofaro T, Del Prete D, Pappalardo A, Mascia A, Zannini M. TAZ is a coactivator for Pax8 and TTF-1, two transcription factors involved in thyroid differentiation. Exp Cell Res. 2009;315:162–175. doi: 10.1016/j.yexcr.2008.10.016. [DOI] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- Epstein J, Cai J, Glaser T, Jepeal L, Maas R. Identification of a Pax paired domain recognition sequence and evidence for DNA-dependent conformational changes. J Biol Chem. 1994;269:8355–8361. [PubMed] [Google Scholar]
- Epstein JA, Lam P, Jepeal L, Maas RL, Shapiro DN. Pax3 inhibits myogenic differentiation of cultured myoblast cells. J Biol Chem. 1995;270:11719–11722. doi: 10.1074/jbc.270.20.11719. [DOI] [PubMed] [Google Scholar]
- Ferrigno O, Lallemand F, Verrecchia F, L'Hoste S, Camonis J, Atfi A, Mauviel A. Yes-associated protein (YAP65) interacts with Smad7 and potentiates its inhibitory activity against TGF-beta/Smad signaling. Oncogene. 2002;21:4879–4884. doi: 10.1038/sj.onc.1205623. [DOI] [PubMed] [Google Scholar]
- Gee ST, Milgram SL, Kramer KL, Conlon FL, Moody SA. Yes-associated protein 65 (YAP) expands neural progenitors and regulates Pax3 expression in the neural plate border zone. PLoS One. 2011;6:e20309. doi: 10.1371/journal.pone.0020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulding MD, Chalepakis G, Deutsch U, Erselius JR, Gruss P. Pax-3, a novel murine DNA binding protein expressed during early neurogenesis. EMBO J. 1991;10:1135–1147. doi: 10.1002/j.1460-2075.1991.tb08054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- Kanai F, Marignani PA, Sarbassova D, Yagi R, Hall RA, Donowitz M, Hisaminato A, Fujiwara T, Ito Y, Cantley LC, et al. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 2000;19:6778–6791. doi: 10.1093/emboj/19.24.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EA, Noh YT, Ryu MJ, Kim HT, Lee SE, Kim CH, Lee C, Kim YH, Choi CY. Phosphorylation and transactivation of Pax6 by homeodomain-interacting protein kinase 2. J Biol Chem. 2006;281:7489–7497. doi: 10.1074/jbc.M507227200. [DOI] [PubMed] [Google Scholar]
- Komuro A, Nagai M, Navin NE, Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J Biol Chem. 2003;278:33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- Lang D, Chen F, Milewski R, Li J, Lu MM, Epstein JA. Pax3 is required for enteric ganglia formation and functions with Sox10 to modulate expression of c-ret. J Clin Invest. 2000;106:963–971. doi: 10.1172/JCI10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D, Lu MM, Huang L, Engleka KA, Zhang M, Chu EY, Lipner S, Skoultchi A, Millar SE, Epstein JA. Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature. 2005;433:884–887. doi: 10.1038/nature03292. [DOI] [PubMed] [Google Scholar]
- Lei QY, Zhang H, Zhao B, Zha ZY, Bai F, Pei XH, Zhao S, Xiong Y, Guan KL. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol Cell Biol. 2008;28:2426–2436. doi: 10.1128/MCB.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Khokhlatchev A, Figeys D, Avruch J. Death-associated protein 4 binds MST1 and augments MST1-induced apoptosis. J Biol Chem. 2002;277:47991–48001. doi: 10.1074/jbc.M202630200. [DOI] [PubMed] [Google Scholar]
- Mansouri A, Gruss P. Pax3 and Pax7 are expressed in commissural neurons and restrict ventral neuronal identity in the spinal cord. Mech Dev. 1998;78:171–178. doi: 10.1016/s0925-4773(98)00168-3. [DOI] [PubMed] [Google Scholar]
- Milewski RC, Chi NC, Li J, Brown C, Lu MM, Epstein JA. Identification of minimal enhancer elements sufficient for Pax3 expression in neural crest and implication of Tead2 as a regulator of Pax3. Development. 2004;131:829–837. doi: 10.1242/dev.00975. [DOI] [PubMed] [Google Scholar]
- Morin-Kensicki EM, Boone BN, Howell M, Stonebraker JR, Teed J, Alb JG, Magnuson TR, O'Neal W, Milgram SL. Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65. Mol Cell Biol. 2006;26:77–87. doi: 10.1128/MCB.26.1.77-87.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Nakagawa M, Olson EN, Nakagawa O. A WW domain protein TAZ is a critical coactivator for TBX5, a transcription factor implicated in Holt-Oram syndrome. Proc Natl Acad Sci U S A. 2005;102:18034–18039. doi: 10.1073/pnas.0509109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Tominaga J, Makita R, Uchijima Y, Kurihara Y, Nakagawa O, Asano T, Kurihara H. Transcriptional activity of Pax3 is co-activated by TAZ. Biochem Biophys Res Commun. 2006;339:533–539. doi: 10.1016/j.bbrc.2005.10.214. [DOI] [PubMed] [Google Scholar]
- Potterf SB, Furumura M, Dunn KJ, Arnheiter H, Pavan WJ. Transcription factor hierarchy in Waardenburg syndrome: regulation of MITF expression by SOX10 and PAX3. Hum Genet. 2000;107:1–6. doi: 10.1007/s004390000328. [DOI] [PubMed] [Google Scholar]
- Relaix F, Polimeni M, Rocancourt D, Ponzetto C, Schafer BW, Buckingham M. The transcriptional activator PAX3-FKHR rescues the defects of Pax3 mutant mice but induces a myogenic gain-of-function phenotype with ligand-independent activation of Met signaling in vivo. Genes Dev. 2003;17:2950–2965. doi: 10.1101/gad.281203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satokata I, Ma L, Ohshima H, Bei M, Woo I, Nishizawa K, Maeda T, Takano Y, Uchiyama M, Heaney S, et al. Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nat Genet. 2000;24:391–395. doi: 10.1038/74231. [DOI] [PubMed] [Google Scholar]
- Soleimani VD, Punch VG, Kawabe Y, Jones AE, Palidwor GA, Porter CJ, Cross JW, Carvajal JJ, Kockx CE, van IWF, et al. Transcriptional dominance of Pax7 in adult myogenesis is due to high-affinity recognition of homeodomain motifs. Dev Cell. 2012;22:1208–1220. doi: 10.1016/j.devcel.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strano S, Munarriz E, Rossi M, Castagnoli L, Shaul Y, Sacchi A, Oren M, Sudol M, Cesareni G, Blandino G. Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J Biol Chem. 2001;276:15164–15173. doi: 10.1074/jbc.M010484200. [DOI] [PubMed] [Google Scholar]
- Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber D, Hariharan IK. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Varelas X, Sakuma R, Samavarchi-Tehrani P, Peerani R, Rao BM, Dembowy J, Yaffe MB, Zandstra PW, Wrana JL. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat Cell Biol. 2008;10:837–848. doi: 10.1038/ncb1748. [DOI] [PubMed] [Google Scholar]
- Watanabe A, Takeda K, Ploplis B, Tachibana M. Epistatic relationship between Waardenburg syndrome genes MITF and PAX3. Nat Genet. 1998;18:283–286. doi: 10.1038/ng0398-283. [DOI] [PubMed] [Google Scholar]
- Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- Xin M, Kim Y, Sutherland LB, Murakami M, Qi X, McAnally J, Porrello ER, Mahmoud AI, Tan W, Shelton JM, et al. Hippo pathway effector Yap promotes cardiac regeneration. Proc Natl Acad Sci U S A. 2013;110:13839–13844. doi: 10.1073/pnas.1313192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Kim Y, Sutherland LB, Qi X, McAnally J, Schwartz RJ, Richardson JA, Bassel-Duby R, Olson EN. Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci Signal. 2011;4:ra70. doi: 10.1126/scisignal.2002278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Sullivan AJ, Medina R, Ito Y, van Wijnen AJ, Stein JL, Lian JB, Stein GS. Tyrosine phosphorylation controls Runx2-mediated subnuclear targeting of YAP to repress transcription. EMBO J. 2004;23:790–799. doi: 10.1038/sj.emboj.7600073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu CY, Zha ZY, Zhao B, Yao J, Zhao S, Xiong Y, Lei QY, Guan KL. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J Biol Chem. 2009;284:13355–13362. doi: 10.1074/jbc.M900843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.