Abstract
Uniparental transcripts during embryogenesis may arise due to gamete delivery during fertilization or genomic imprinting. Such transcripts have been found in a number of plant species and appear critical for the early development of embryo or endosperm in seeds. Although the regulatory expression mechanism and function of these genes in embryogenesis require further elucidation, recent studies suggest stage-specific and highly dynamic features that might be essential for critical developmental events such as zygotic division and cell fate determination during embryogenesis. Here, we summarize the current work in this field and discuss future research directions.
Keywords: uniparental transcripts, gamete-delivered transcript, maternal control, paternal allele, genomic imprinting, embryo
INTRODUCTION
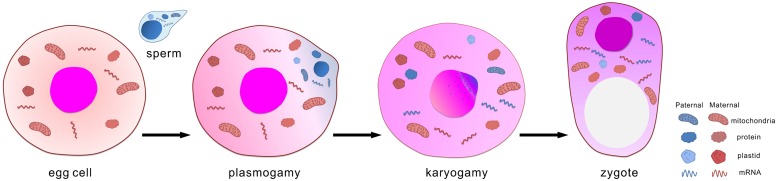
During sexual reproduction of flowering plants, male and female gametes are formed in the haploid gametophytic generation (Walbot and Evans, 2003; Chang et al., 2011). In angiosperm, the typical female gametophyte contains two kinds of female gametes, a haploid egg cell and a diploid central cell with two identical copies of the maternal genome. The male gametophyte is found in pollen, which carries one generative cell or two sperm cells. During pollen generation, sperm cells are transported though the pollen tube to the female gametophyte. Upon double fertilization, two sperm cells enter the embryo sac. One sperm cell fuses with the egg cell, and the other fuses with the central cell. This integration of the two gamete genomes results in the formation of a diploid embryo and a triploid endosperm, respectively. After fertilization, the embryo forms basic morphological and physiological structures (Le et al., 2007), during which the endosperm plays a nutritive role, similar to the placenta of mammals, to support embryonic development (Lopes and Larkins, 1993; Olsen, 2004). During this process, both paternal and maternal genetic information may contribute to the fertilization and development of the embryo, which leads to generation of the sporophyte. These parental information includes RNA that transcribed in sperm and/or egg cells, proteins that synthesized and deposited in gametes, paternal or maternal genome, and mitochondria and plastid genome. After fertilization they are brought into and integrated in zygote (Figure 1). Due to technical limitations the contribution of gamete-delivered proteins, mitochondria and plastid genome to zygote development and early embryogenesis are hardly investigated. Current studies as pioneer works mainly focus on de novo expression of imprinted genes and gamete-delivered transcripts.
FIGURE 1.
Fertilization integrates the maternal and paternal genetic information.
The molecular mechanisms of fertilization and early embryogenesis, especially the role of parent-of-origin genes, have been well studied in animals. However, little is known about these processes in plants due to technical limitations. Gametogenesis, fertilization and embryogenesis occur deep in the plant saprophytic tissues, thus rendering it difficult to observe the developmental events and investigate the molecular mechanisms of these processes directly. Modern technological advances have allowed the isolation and analysis of gametes, zygotes, and early embryos in a wide variety of plants including maize, tobacco, Arabidopsis, rice, and wheat (Engel et al., 2003; Zhao et al., 2011; Nodine and Bartel, 2012; Anderson et al., 2013; Domoki et al., 2013). Therefore, great advances have been made toward understanding the role of uniparental transcripts in plant embryogenesis.
In animals, maternal allele products synthesized during gametogenesis exert control in all aspects of embryonic development prior to the global activation of the zygotic genome (Tadros and Lipshitz, 2009). However, in plants, the parental contribution in early embryogenesis has not yet been fully understood. Early reports indicated that the transcripts in early embryos were mainly originated from the maternally inherited alleles and the transcription of paternal alleles was delayed (Vielle-Calzada et al., 2000; Baroux et al., 2001; Golden et al., 2002; Grimanelli et al., 2005). Baroux et al. (2008) further suggested that early embryogenesis in plants was maternally controlled similar to that in animals, as early studies indicated that maternal transcripts could support embryonic development until the proembryo stage. At the same time, some other researches presented evidences of early activated paternal genome (Weijers et al., 2001; Scholten et al., 2002; Lukowitz et al., 2004; Sheldon et al., 2008). Recently, paternal transcripts were proved to be critical for the normal development of early embryo (Ueda et al., 2011; Babu et al., 2013). More impressively, interleukin-1 receptor-associated kinase (IRAK)/Pelle-like kinase gene, SHORT SUSPENSOR (SSP) transcripts were found to be produced in mature pollen and were believed to be carried into the egg cell via fertilization in Arabidopsis thaliana (Bayer et al., 2009). SSP functioned during the asymmetric first division in the zygote, indicating that the paternal transcripts from sperm cells may be involved in many aspects of zygotic development and early embryogenesis in plants. Our previous work also confirmed that paternal transcripts in sperm cells could be found in zygotes soon after fertilization (Xin et al., 2011), suggesting the possibility that sperm-delivered paternal transcripts may be involved in zygotic development.
Meyer and Scholten (2007) reported the relative expression levels of parental transcripts in zygotes, suggesting equivalent parental contribution in maize zygotic development. Maternally expressed in embryo 1 (mee1) in maize was the first reported imprinted gene in a plant embryo, although its function is unclear (Jahnke and Scholten, 2009). Using deep sequencing in a genome-wide analysis, Autran et al. (2011) assessed the parental contributions in early embryogenesis and found that the maternal transcripts predominated at early embryonic stages in Arabidopsis. With development, the relative paternal contribution arose due to the gradual activation of the embryonic genome. Subsequently, Nodine and Bartel (2012) found that a majority of genes were expressed equally from both parents at the beginning of embryogenesis in Arabidopsis. Interestingly, some of these works focused on the quantitative ratio of maternal and paternal transcripts, some mainly analyzed the regulatory roles of these genes. It is not surprised to see various conclusions. Even more, a latest report indicated that the different results might be due to the different material (e.g., ecotypes) they used in their experiments (Del Toro-De León et al., 2014). Despite all these discussions, it is believed that some transcripts are derived primarily from one parent or from imprinted genes in embryos soon after fertilization (Autran et al., 2011; Nodine and Bartel, 2012). These studies indicate that the parent-of-origin gene transcripts indeed exist in the zygote or early embryo. Such transcripts could arise from both the gamete-delivered and de novo expression of imprinted genes. Each type of uniparental transcript may play specific roles in plant development, since they are regulated by different molecular mechanisms. This review highlights the characteristics of uniparental transcripts during early embryogenesis.
GAMETE-CARRIED MATERNAL OR PATERNAL TRANSCRIPTS INVOLVED IN EARLY EMBRYOGENESIS
MATERNAL TRANSCRIPTS
The embryo originates from a fertilized egg cell, termed a zygote. Two sequential events occur during the integration of a sperm and an egg cell: plasmogamy and karyogamy. Not only do the two genomes integrate, but also various components of the cytoplasm mix during the fertilization process. For example, sperm mitochondria could be found in fertilized egg cells of tobacco (Yu and Russell, 1994) although mitochondria is usually inherited maternally.
During early embryogenesis in most animal species, maternal transcripts deposited in the egg cells are involved in various developmental processes before activation of the zygotic genome, such as formation of embryonic axes, cell differentiation, and morphogenesis (Johnston, 1995; Wylie et al., 1996; Nishida, 1997; Angerer and Angerer, 2000; Mohr et al., 2001; Pellettieri and Seydoux, 2002). Although various experimental data support the hypothesis that maternal control may also exist during early embryogenesis in plants (Baroux et al., 2008), little is known about transcripts stored in egg cells and their role in early embryogenesis (Xin et al., 2012).
Using microdissection, Sprunck et al. (2005) constructed a cDNA library from wheat egg cells, and a total of 404 clusters were found to function in metabolic activity, mRNA translation and protein turnover. Subsequently, another 226 expressed sequence tags (ESTs) were studied in wheat egg cells (Domoki et al., 2013). In a similar analysis carried out in tobacco, thousands of ESTs were detected, which may be involved in a variety of developmental processes (Ning et al., 2006; Zhao et al., 2011). In addition, microarray technology combined with laser-assisted microdissection (LAM) was used to analyze the expression profile in Arabidopsis egg cells (Wuest et al., 2010). Transcriptomic analysis of egg cells isolated by manual manipulation was performed in rice (Ohnishi et al., 2011; Abiko et al., 2013), and genome-wide deep sequencing was used to characterize the gene expression profile in rice egg cells (Anderson et al., 2013). The functional categories of approximately 27,000 genes detected proved to be comprehensive. However, a comparison of the egg-specific expression of transcriptomes in rice and Arabidopsis revealed relatively different sets of genes in egg cells of rice and Arabidopsis (Ohnishi et al., 2011).
The role of mRNA stored in egg cells has been investigated. Although downregulation of RNA polymerase II by RNA interference (RNAi) impeded de novo transcription, the development of Arabidopsis embryos continued until the preglobular stage (Pillot et al., 2010). In tobacco, zygotic development continued without de novo transcription until 72 h after pollination (HAP). The cytological observation of developmental events in transcriptionally inhibited zygotes showed that maternal transcripts stored in egg cells were functionally competent in gamete fusion, zygote volume reduction, complete cell wall formation, large vacuole disappearance, and limited cell enlargement during early developmental stages. However, de novo transcripts would then seize control of embryogenesis to trigger subsequent developmental processes (Zhao et al., 2011).
Interestingly, small RNA-mediated transposon silencing is thought to be an essential regulatory mechanism in male and female gametes (Slotkin et al., 2009; Martínez and Slotkin, 2012). Anderson et al. (2013) evaluated the expression of genes involved in the miRNA and siRNA pathways in transcriptomes of rice gametes and showed that all important components involved in these pathways were active in egg cells rather than in sperm cells. Thus, transposon silencing is mediated by small RNAs produced in egg cells; moreover, it is regulated in the zygote by small RNAs inherited from the egg cells (Anderson et al., 2013).
Currently, the roles of female gamete transcripts in zygotes and early embryogenesis are unclear. Although de novo transcription in the zygotic genome is activated within hours after fertilization in maize, tobacco and Arabidopsis, the maternal transcripts deposited in the egg cells still play a key role in the initial stages of zygotic development (Meyer and Scholten, 2007; Zhao et al., 2011; Nodine and Bartel, 2012).
PATERNAL TRANSCRIPTS
The sperm cell, the other contributor to zygote, has a simple structure including the karyoplasm and very little cytoplasm. Due to the condensed chromatin observed in sperm cells, it was generally thought that inactive male transcription made no contribution to early embryogenesis prior to zygotic genome activation. This view might be supported in animals, since almost all mRNAs in zygotes are inherited from egg cells (Ostermeier et al., 2004; Krawetz, 2005). However, the cytoplasm of sperm cells may play an important role during early embryogenesis after fertilization in plants, as the extracted sperm nuclei in maize was insufficient to achieve successful fertilization in vitro (Matthys-Rochon et al., 1994).
Recently, increasing evidence has confirmed the presence of a number of transcripts in the sperm cell, refuting the hypothesis that highly condensed chromatin in sperm cells impede activation of transcription. Various cDNA libraries have been constructed based on isolated sperm cells from rice (Gou et al., 2001), tobacco (Xin et al., 2011), maize (Zea mays; Engel et al., 2003), and Plumbago dimorphic (Gou et al., 2009). Additionally, genome-wide expression has been detected in different plants. Using microarray analysis, the transcriptomic profile in sperm cells was investigated in rice and Arabidopsis (Borges et al., 2008; Russell et al., 2012; Abiko et al., 2013). The transcriptomes of rice sperm cells were studied by deep sequencing (Anderson et al., 2013), and ~25000 genes were analyzed. These studies revealed a diverse and broad constitution of mRNAs in sperm cells. Subsequently, sperm transcription profiles were compared among different plant species. Only 35 genes were found in common among 1,048 ESTs in tobacco (Xin et al., 2011), 5,829 genes in Arabidopsis (Borges et al., 2008), and 5,174 ESTs in maize (Engel et al., 2003). Analysis of these 35 genes suggested that active transcription in sperm cells is involved in many basic pathways and processes such as metabolism, transcription, translation, signal transduction and intercellular trafficking (Xin et al., 2011).
For years, plant scientists have questioned whether male transcripts are delivered to the zygote during the fertilization process, and if so, whether these sperm-carrying transcripts have a role in zygote development or early embryogenesis. In our previous work, we identified sperm-specific transcripts in zygotes at 96 HAP (Ning et al., 2006). Subsequently, two kinds of sperm transcripts with unknown function were revealed in zygotes ~10 h after fertilization (HAF). These results strongly suggested that paternal transcripts could be delivered into zygotes, where they might play a role in zygote activation and/or early embryogenesis (Xin et al., 2011). Similarly, Ohnishi et al. (2014) found abundant expression of the Os07g0182900 rice gene in sperm cells (Abiko et al., 2013), but not in unfertilized egg cells. The fact that its transcripts could be detected in the zygote ~10–20 min after fertilization indicated that transcripts in plant zygotes could be delivered from the sperm cells by plasmogamy. The Os07g0182900 gene encoding cytosine-5 DNA methyltransferase 1 (MET1) may be involved in the transition from the zygote to two-celled pre-embryo stage, as the process could be partially inhibited by a specific inhibitor of MET1 (Abiko et al., 2013). In Arabidopsis, the polarity of elongated zygotes contributed substantially to regular embryonic development. Corrected asymmetric cell division led to normal formation of the initial apical–basal axis and the embryo and suspensor ancestors in plants (Jeong et al., 2011; Zhang and Laux, 2011; Ueda and Laux, 2012). Sperm transcripts are now believed to be essential in this critical developmental process. Bayer et al. (2009) reported that transcripts of the IRAK/Pelle-like kinase gene, SSP, were produced in mature sperm cells and translated in zygotes after fertilization. Defective SSP influenced the elongation of the zygote and the formation of suspensor through the YODA-dependent MAPKKK signaling pathway. These examples suggest that sperm mRNAs might have vital functions in normal developmental embryogenesis.
IMPRINTED GENES IN EARLY EMBRYOGENESIS
In mammals and flowering plants, genomic imprinting is a general epigenetic mechanism associated with the differential expression of parental alleles (Feil and Berger, 2007). The differential de novo transcription of parental alleles is caused by different epigenetic influences established in the germ line, rather than the nucleotide changes or uniparental transcripts caused by gamete delivery. Maternally expressed imprinted genes (MEGs) are expressed maternally but silenced paternally, whereas maternally expressed imprinted genes (PEGs) are expressed paternally but silenced maternally.
IMPRINTED GENES IN PLANT EMBRYOS
Imprinting is another cause of unequal contributions from parental transcripts in the early embryo. A minority of imprinted genes has been identified in endosperm using conventional methods, such as sequence homologies, small-scale transcriptional surveys, assays for reduced DNA methylation and mutant identification. The mee1 gene in maize provided the evidence confirming the presence of imprinted genes in embryos (Raissig et al., 2011). The differential methylation status between paternal and maternal alleles regulates the maternal expression of mee1 in the embryo and endosperm. Dynamic expression of mee1 was found in the early embryo, but its function remains unclear (Jahnke and Scholten, 2009).
Recently, genome-wide approaches have been used to identify imprinted genes in Arabidopsis, maize and rice (Gehring et al., 2011; Hsieh et al., 2011; Luo et al., 2011; Waters et al., 2011; Wolff et al., 2011; Zhang et al., 2011). Several 100 endosperm-specific imprinted genes were newly detected in these species. However, the presence of imprinted genes in the embryo remains controversial. For example, Hsieh et al. (2011) identified 116 MEGs and 10 PEGs in Arabidopsis endosperm 7–8 days after pollination (DAP), while 37 MEGs and one PEG were found in the embryo during the same period. However, the imprinted genes in the embryo were considered to be false positives due to contamination with endosperm or maternal tissue (Hsieh et al., 2011). Similarly, Gehring et al. (2011) identified 165 MEGs and 43 PEGs in Arabidopsis endosperm at 6–7 DAP; additionally, 17 MEGs and one PEG were found in embryos during the same period. However, the imprinted genes in the embryo could have been due to endosperm contamination or biased expression dependent on an unchangeable allele (Gehring et al., 2011). In monocots, Luo et al. (2011) found 262 imprinted loci in rice endosperm at 5 DAF. An imprinted gene was detected in both the embryo and endosperm; however, this candidate requires further confirmation by confirming its expression in gametes (Luo et al., 2011). Waters et al. (2011) found 54 MEGs and 46 PEGs in maize endosperm at 14 DAP, with 29 MEGs and nine PEGs in embryos during the same period. However, these imprinted genes in embryos might be due to contamination, trafficking of transcripts produced in the endosperm to the embryo, or relatively stable transcripts inherited from the gametes (Waters et al., 2011).
Currently, genomic imprinting in Arabidopsis embryos has not been validated conclusively. Raissig et al. (2013) constructed cDNA libraries using 2 to 4-cells embryos and globular embryos isolated from the reciprocal cross of the Col-0 and the Ler accessions. Imprinted gene candidates were then chosen, and their relative expression levels between parental alleles were assessed by reverse transcription polymerase chain reaction (RT-PCR) and Sanger sequencing (Raissig et al., 2013). A total of 11 MEGs were expressed at the 2 to 4-cells and globular embryo stages, and one PEG was expressed at the 2 to 4-cells embryo stage. No transcripts in the one PEG or in nine of the MEGs were detected in the gametes, indicating that their imprinted expression in the embryo was derived from de novo transcription and was reliable. To avoid contamination, strict procedures were adopted in constructing the cDNA libraries. In addition, an independent assay was used to confirm the genomic imprinting in embryos by fusing the promoters of seven MEGs and one PEG with the reporter gene β-glucuronidase (GUS). Promoter-GUS reporter lines (Col-0 background) were crossed reciprocally with wild-type plants (Col-0), and the analysis of stained F1 embryos showed that six MEG reporter lines were either imprinted fully or showed a strong bias for maternal expression (Raissig et al., 2013). Furthermore, Raissig et al. (2013) detected imprinted expression of all embryonic MEGs and the PEG in other samples, as early Col-0 × Cvi embryos (different accession, similar stage; Nodine and Bartel, 2012) and late torpedo-stage Col-0 × Ler embryos (same accessions, but later stage; Gehring et al., 2011). The results confirmed that the expression of most imprinted genes during early embryogenesis was maintained regardless of the different accessions or later developmental stage (Raissig et al., 2013). Therefore, these results indicated that genomic imprinting may not be restricted to the endosperm and may be more extensive in embryos than thought previously.
FUNCTION OF IMPRINTED GENES INVOLVED IN EMBRYOGENESIS
In mammals, 100s of imprinted genes have been identified that are connected to the location of nutrient transfer from mother to offspring, embryogenesis, and postnatal development (Constância et al., 2004; Gregg et al., 2010). Abnormal imprinting can harm fetal growth, hormone systems after birth, and adult brain function. Whereas genome-wide approaches have revealed many imprinted genes involved in transcriptional regulation, chromatin modification, hormone signaling, ubiquitin degradation, small RNA pathways and metabolism (Gehring et al., 2011; Hsieh et al., 2011; Luo et al., 2011; Raissig et al., 2011, 2013) in plants, little is known regarding the involvement of imprinted genes in plant development (Table 1).
Table 1.
Uniparental genes in sexual plant reproduction.
| Genes | Epigenetic mark | Ecotype/inbred line for detection | Period of detection | Mutant/RNAi phenotype | Reference | |
|---|---|---|---|---|---|---|
| Endosperm | ||||||
| Arabidopsis | MEA (MEG)1,2 | H3K27me3 DNA-me |
Ler&RLD | 6,7,8 DAP | Seed abortion |
Kinoshita et al. (1999), Vielle-Calzada et al. (1999) |
| FWA (MEG) | DNA-me | Col-0, Ler, WS | 6,7,8 DAP | — | Kinoshita et al. (2004), | |
| Jullien et al. (2006) | ||||||
| FIS2 (MEG)1 | DNA-me | C24&Col | 0.5-5 DAP | Seed abortion | Jullien et al. (2006, 2008), | |
| Luo et al. (2000) | ||||||
| MPC (MEG)1 | DNA-me | Col&Ler | 3,5,7 DAP | Abnormal seed | Tiwari et al. (2008) | |
| PHE1 (PEG)1,2 | H3K27me3 | Col&C24 | 1–4 DAP | — | Köhler et al. (2003, 2005), | |
| DNA-me | Makarevich et al. (2008) | |||||
| FH5 (MEG)1,2 | H3K27me3 | Ler&C24 | 5 DAP | Endosperm defects |
Ingouff et al. (2005), Fitz Gerald et al. (2009) |
|
| AGL36 (MEG)1 | H3K27me3 DNA-me |
Col&Ler | 3 DAP | — |
Shirzadi et al. (2011), Wuest et al. (2010) |
|
| 3 MEGs&2 PEGs | DNA-me | Col-gl&Ler | Torpedo-stage | — | Gehring et al. (2009) | |
| 116 MEGs&10 PEGs | — | Col&Ler | 7–8 DAP | — | Hsieh et al. (2011) | |
| 165 MEGs&43 PEGs | — | Col-0&Ler | 6 or 7 DAP | — | Gehring et al. (2011) | |
| 39 MEGs&27 PEGs | — | Col-0&Bur-0 | 4DAP | — | Wolff et al. (2011) | |
| Maize | Fie1 (MEG)2 | DNA-me, | B73&Mo17; | 2-15 DAP | — | Danilevskaya et al. (2003), |
| H3K27me3, | SSS1&NSS1 | Gutiérrez-Marcos et al. (2006), | ||||
| H3/H4-Ac | Hermon et al. (2007) | |||||
| Fie2 (MEG) | DNA-me | B73&Mo17 | 2,5 DAP | — | Danilevskaya et al. (2003), | |
| Gutiérrez-Marcos et al. (2006), | ||||||
| Hermon et al. (2007), | ||||||
| Nrp1 (MEG)2 | DNA-me, | B73&Mo17; | 10,14,21 DAP | — | Guo et al. (2003), | |
| H3K27me3, | SSS1&NSS1 | Haun and Springer (2008) | ||||
| H3/H4-Ac | ||||||
| Peg1 (PEG) | — | W22&Tx303 | 12 DAP | — | Gutiérrez-Marcos et al. (2003) | |
| Meg1 (MEG)1,2 | DNA-me | F2, A69Y, W23 | 4 DAP | Reduced-size seeds | Gutiérrez-Marcos et al. (2004), Costa et al. (2012) | |
| Mez1 (MEG) | DNA-me, | B73&Mo17 | 8-27 DAP | — | Haun et al. (2007) | |
| H3K27me3, | Haun and Springer (2008) | |||||
| H3/H4-Ac | ||||||
| Mee1 (MEG) | DNA-me | UH005&UH301 | 6 DAP | — | Jahnke and Scholten (2009) | |
| α-Tubulin (MEG) | DNA-me | W64A&A69Y | 20 DAP | — | Lund et al. (1995b) | |
| Zein (MEG) | DNA-me | W64A&A69Y | 19 DAP | — | Lund et al. (1995a) | |
| R gene (MEG) | — | — | — | — | Kermicle (1970), Ludwig et al. (1989) | |
| Dzr-1 (MEG) | — | BSSS53&Mo17 | 15-27 DAP | — | Chaudhuri and Messing (1994) | |
| 54 MEGs&46 PEGs | — | B73&Mo17 | 14 DAP | — | Waters et al. (2011) | |
| 93 MNCs&124 PNCs3 | — | B73&Mo17 | 10DAP | — | Zhang et al. (2011) | |
| Rice | 177 MEGs&85 PEGs | — | Nip&93-11 | 5 DAF | — | Luo et al. (2011) |
| Embryo | ||||||
| Arabidopsis | 11 MEGs&1 PEGs1,2 | H3K27me34 | Col-0&Ler | 2.5,4 DAP | — | Raissig et al. (2013) |
| Maize | Mee1 (MEG)2 | DNA-me | UH005&UH301 | 3,6,8 DAP | — | Jahnke and Scholten (2009) |
| Gamete-carried transcripts | Origin of transcripts | Defective in embryogenesis | Reference | |||
| Zygote | ||||||
| Arabidopsis | SSP | Sperm-specific | Abnormal division of zygote | Bayer et al. (2009) | ||
| Rice | Os07g0182900 | Sperm-specific | Abnormal division of zygote | Ohnishi et al. (2014) | ||
| Tobacco | Ntsp0002&Ntsp0003 | Sperm-specific | — | Xin et al. (2011) | ||
The table lists the known information of imprinted and potentially imprinted genes discovered in plants to date. Phenotypes in seed development are also displayed. These data are partially adapted from Raissig et al. (2011). —: not shown or not known.
1Reporter activity of parent-of-origin–expression was identified (part of imprinted genes in Arabidopsis embryo).
2De novo transcription was identified (part of imprinted genes in Arabidopsis embryo).
3MNCs, maternal expressed transcripts; PNCs, paternal expressed transcripts. MNCs and PNCs include protein-coding genes and long non-coding RNAs.
4Epigenetic marks of a part of imprinted genes in Arabidopsis embryo.
To date, only four imprinted genes in endosperm are known to be involved in embryogenesis (Raissig et al., 2011; Costa et al., 2012). In Arabidopsis, the FERTILIZATION-INDEPENDENT SEED (FIS) genes MEA (FIS1) and FIS2 belong to the Polycomb group family (PcG). MEDEA (MEA) is expressed in both the embryo and endosperm; however, maternal imprinting has been confirmed only in the latter, and it remains to be determined whether MEA is imprinted in the embryo (Raissig et al., 2013). FIS2 is a maternally imprinted gene, and its expression was detected both in the central cell before fertilization and endosperm after fertilization (Luo et al., 2000). Double fertilization products that contained maternal alleles of mea and fis2 resulted in failure of endosperm cellularization. Moreover, embryogenesis ceased at the heart/torpedo stage, resulting in seed abortion (Luo et al., 2000). Another novel maternal imprinted gene MPC was found to be active in the central cell before fertilization and in the endosperm from fertilization to 4 DAP. Knockdown of MPC through RNAi resulted in defective seed development, with delayed embryogenesis and abnormal embryo and endosperm morphology (Tiwari et al., 2008). In maize, maternally expressed gene 1 (Meg1) encodes a new kind of signaling peptide located in endosperm nutrient transfer cells, where it regulates their establishment and differentiation. Meg1 is the first identified imprinted gene in plants that participates in nutrient distribution to the embryo. Interestingly, in contrast to imprinted genes in mammals, Meg1 promotes rather than restricts the transfer of nutrient flow from the mother to fetus (Costa et al., 2012).
The imprinted genes in the endosperm mentioned above have defined roles in the endosperm; however, the role of the imprinted genes in the embryo remain unknown. To identify the contribution of imprinted genes in the embryo during embryogenesis, T-DNA gene insertions were used to search for deviant phenotypes relative to embryonic development, but no obvious phenotypes were observed (Raissig et al., 2013). Interestingly, all the maternally imprinted genes in the Arabidopsis embryo were expressed in the seed coat, and some even showed a slightly biased expression toward the basal embryo and the suspensor (Raissig et al., 2013). Notably, some maternally imprinted genes were involved in metabolism (Raissig et al., 2013). Therefore, maternally imprinted genes in the embryo might function at the interface between the embryo and maternal tissue, possibly by linking seed coat metabolism and embryo metabolism, and rendering the genes in the embryo under maternal control (Raissig et al., 2013). This result may support the maternal–offspring coadaptation theory, which posits that maternally imprinted genes are critical for the events during mother–offspring interactions (Bateson, 1994; Wolf and Hager, 2006). Further research on the roles of imprinted genes in the embryo will lead to a better understanding of the function and evolution of genomic imprinting in plants.
REGULATION OF IMPRINTED GENES IN EMBRYOS
DNA methylation and histone modification are two distinct epigenetic mechanisms involved in the regulation of genomic imprinting in plants. The differential DNA methylation status of parental alleles in the endosperm is due mainly to genome-wide hypomethylation of maternal alleles in the central cell (Gehring et al., 2009, 2011). DNA glycosylase DEMETER (DME) with 5-methylcytosine excising activity (Kinoshita et al., 2004; Gehring et al., 2006) and the repression of MET1 involved in maintaining DNA methylation (Jullien et al., 2008; Hsieh et al., 2011) are responsible for the DNA demethylation at CG sites. However, sometimes DNA methylation alone is not sufficient to establish different imprinting markers of some genes, and the Polycomb repressive complex 2 (PRC2) that catalyzes the trimethylation of histone H3 on lysine 27 (H3K27me3) is required (Baroux et al., 2006; Makarevich et al., 2008; Hennig and Derkacheva, 2009).
To investigate the epigenetic mechanism of genomic imprinting in the embryo, the fertilization-independent endosperm (fie) mutant was crossed reciprocally with wild-type plants, and met1-3 mutants were used to pollinate wild-type plants (Raissig et al., 2013). F1 hybrid embryos were isolated, and mutant embryonic cDNA libraries were created. The detection of the allele-specific expression pattern of 11 embryonic MEGs in Arabidopsis demonstrated that imprinted expression of MEGs in embryos are not influenced by the paternal met1-3 allele. However, disruption of the maternal FIE function changed the monoallelic expression of two MEGs and one PEG. Thus, the function of PRC2 may be comprehensive in regulating imprinted expression in both the embryo and endosperm (Raissig et al., 2013). Furthermore, the role of asymmetric DNA methylation in the CHG context was negated in the establishment of imprinting in the embryo. Finally, Raissig et al. (2013) indicated that PRC2, but not MET1, played a role in regulating the imprinted genes in the embryo. This is consist with a previous conjecture that DNA methylation is unlikely to be a primary imprinting mark in maize embryos (Gutiérrez-Marcos et al., 2006; Jahnke and Scholten, 2009). Other undiscovered mechanisms may be involved in the establishment of genomic imprinting in the embryo.
PERSPECTIVES
Technological advances in genome-wide sequencing technology and acquisition of gametes, zygotes and early embryos will be important for elucidating the role of parent-of-origin genes during plant early embryogenesis. Such technological progress may lead to the identification of more uniparental transcripts in early embryos, whether gamete-delivered or imprinted gene-derived. Currently, in both gametes and early zygotes, it remains technically difficult to identify the origin of transcripts, which may be gamete-delivered during fertilization or transcribed de novo after fertilization. In addition, little is known about the functions of these parent-of-origin genes during fertilization and early embryogenesis. Obviously, no matter how parental transcripts may contribute to the transcriptome of early embryo, the function analysis of the parent-of-origin genes in specific developmental events at specific developmental stage will surely provide imperative knowledge to understand the parental effect in early seed formation.
Recently, the identification of imprinted genes in early embryos in Arabidopsis has questioned the concept that imprinted genes are restricted mainly to the endosperm. In light of so many candidate imprinted genes in the embryos of dicots and monocots, optimized methods are required to avoid contamination of maternal tissues and false positives or negatives in data collection and reliable analysis. Determination of the relevant functions of imprinted genes should shed light on our understanding of epigenetic mechanisms in promoting embryogenesis, embryo pattern formation, and cell fate determination during embryogenesis. With further technological advancement, the role of methylation in gene imprinting during embryogenesis might be further elucidated.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mr. Ce Shi for figure preparation. This project was supported by the “973” Project (2013CB126900; 2014CB943400) and National Natural Science Fund of China (31170297; 31070284).
REFERENCES
- Abiko M., Maeda H., Tamura K., Hara-Nishimura I., Okamoto T. (2013). Gene expression profiles in rice gametes and zygotes: identification of gamete-enriched genes and up- or down-regulated genes in zygotes after fertilization. J. Exp. Bot. 64 1927–1940 10.1093/jxb/ert054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S. N., Johnson C. S., Jones D. S., Conrad L. J., Gou X., Russell S. D., et al. (2013). Transcriptomes of isolated Oryza sativa gametes characterized by deep sequencing: evidence for distinct sex-dependent chromatin and epigenetic states before fertilization. Plant J. 76 729–741 10.1111/tpj.12336 [DOI] [PubMed] [Google Scholar]
- Angerer L. M., Angerer R. C. (2000). Animal-vegetal axis patterning mechanisms in the early sea urchin embryo. Dev. Biol. 218 1–12 10.1006/dbio.1999.9553 [DOI] [PubMed] [Google Scholar]
- Autran D., Baroux C., Raissig M. T., Lenormand T., Wittig M., Grob S., et al. (2011). Maternal epigenetic pathways control parental contributions to Arabidopsis early embryogenesis. Cell 145 707–719 10.1016/j.cell.2011.04.014 [DOI] [PubMed] [Google Scholar]
- Babu Y., Musielak T., Henschen A., Bayer M. (2013). Suspensor length determines developmental progression of the embryo in Arabidopsis. Plant Physiol. 162 1448–1458 10.1104/pp.113.217166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroux C., Autran D., Gillmor C. S., Grimanelli D., Grossniklaus U. (2008). The maternal to zygotic transition in animals and plants. Cold Spring Harb. Symp. Quant. Biol. 73 89–100 10.1101/sqb.2008.73.053 [DOI] [PubMed] [Google Scholar]
- Baroux C., Blanvillain R., Gallois P. (2001). Paternally inherited transgenes are down-regulated but retain low activity during early embryogenesis in Arabidopsis. FEBS Lett. 509 11–16 10.1016/S0014-5793(01)03097-6 [DOI] [PubMed] [Google Scholar]
- Baroux C., Gagliardini V., Page D. R., Grossniklaus U. (2006). Dynamic regulatory interactions of Polycomb group genes: MEDEA autoregulation is required for imprinted gene expression in Arabidopsis. Genes Dev. 20 1081–1086 10.1101/gad.378106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateson P. (1994). The dynamics of parent-offspring relationships in mammals. Trends Ecol. Evol. 9 399–403 10.1016/0169-5347(94)90066-90063 [DOI] [PubMed] [Google Scholar]
- Bayer M., Nawy T., Giglione C., Galli M., Meinnel T., Lukowitz W. (2009). Paternal control of embryonic patterning in Arabidopsis thaliana. Science 323 1485–1488 10.1126/science.1167784 [DOI] [PubMed] [Google Scholar]
- Borges F., Gomes G., Gardner R., Moreno N., McCormick S., Feijó J. A., et al. (2008). Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiol. 148 1168–1181 10.1104/pp.108.125229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F., Wang Y., Wang S., Ma H. (2011). Molecular control of microsporogenesis in Arabidopsis. Curr. Opin. Plant Biol. 14 66–73 10.1016/j.pbi.2010.11.001 [DOI] [PubMed] [Google Scholar]
- Chaudhuri S., Messing J. (1994). Allele-specific parental imprinting of dzr1, a posttranscriptional regulator of zein accumulation. Proc. Natl. Acad. Sci. U.S.A. 91 4867–4871 10.1073/pnas.91.11.4867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constância M., Kelsey G., Reik W. (2004). Resourceful imprinting. Nature 432 53–57 10.1038/432053a [DOI] [PubMed] [Google Scholar]
- Costa L. M., Yuan J., Rouster J., Paul W., Dickinson H., Gutierrez-Marcos J. F. (2012). Maternal control of nutrient allocation in plant seeds by genomic imprinting. Curr. Biol. 22 160–165 10.1016/j.cub.2011.11.059 [DOI] [PubMed] [Google Scholar]
- Danilevskaya O. N., Hermon P., Hantke S., Muszynski M. G., Kollipara K., Ananiev E. V. (2003). Duplicated fie genes in maize: expression pattern and imprinting suggest distinct functions. Plant Cell 15 425–438 10.1105/tpc.006759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Toro-De León G., García-Aguilar M., Gillmor C. S. (2014). Non-equivalent contributions of maternal and paternal genomes to early plant embryogenesis. Nature 514 624–627 10.1038/nature13620 [DOI] [PubMed] [Google Scholar]
- Domoki M., Szűcs A., Jäger K., Bottka S., Barnabás B., Fehér A. (2013). Identification of genes preferentially expressed in wheat egg cells and zygotes. Plant Cell Rep. 32 339–348 10.1007/s00299-012-1367-0 [DOI] [PubMed] [Google Scholar]
- Engel M. L., Chaboud A., Dumas C., McCormick S. (2003). Sperm cells of Zea mays have a complex complement of mRNAs. Plant J. 34 697–707 10.1046/j.1365-313X.2003.01761.x [DOI] [PubMed] [Google Scholar]
- Feil R., Berger F. (2007). Convergent evolution of genomic imprinting in plants and mammals. Trends Genet. 23 192–199 10.1016/j.tig.2007.02.004 [DOI] [PubMed] [Google Scholar]
- Fitz Gerald J. N., Hui P. S., Berger F. (2009). Polycomb group-dependent imprinting of the actin regulator AtFH5 regulates morphogenesis in Arabidopsis thaliana. Development 136 3399–3404 10.1242/dev.036921 [DOI] [PubMed] [Google Scholar]
- Gehring M., Bubb K. L., Henikoff S. (2009). Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324 1447–1451 10.1126/science.1171609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M., Huh J. H., Hsieh T. F., Penterman J., Choi Y., Harada J. J., et al. (2006). DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124 495–506 10.1016/j.cell.2005.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M., Missirian V., Henikoff S. (2011). Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds. PLoS ONE 6:e23687 10.1371/journal.pone.0023687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden T. A., Schauer S. E., Lang J. D., Pien S., Mushegian A. R., Grossniklaus U., et al. (2002). SHORT INTEGUMENTS1/SUSPENSOR1/CARPEL FACTORY, a Dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis. Plant Physiol. 130 808–822 10.1104/pp.003491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou X. P., Tang L., Yan F., Chen F. (2001). Representative cDNA library from isolated rice sperm cells. Acta Bot. Sin. 43 1093–1096. [Google Scholar]
- Gou X., Yuan T., Wei X., Russell S. D. (2009). Gene expression in the dimorphic sperm cells of Plumbago zeylanica: transcript profiling, diversity, and relationship to cell type. Plant J. 60 33–47 10.1111/j.1365-313X.2009.03934.x [DOI] [PubMed] [Google Scholar]
- Gregg C., Zhang J., Weissbourd B., Luo S., Schroth G. P., Haig D., et al. (2010). High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science 329 643–648 10.1126/science.1190830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimanelli D., Perotti E., Ramirez J., Leblanc O. (2005). Timing of the maternal-to-zygotic transition during early seed development in maize. Plant Cell 17 1061–1072 10.1105/tpc.104.029819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Rupe M. A., Danilevskaya O. N., Yang X., Hu Z. (2003). Genome-wide mRNA profiling reveals heterochronic allelic variation and a new imprinted gene in hybrid maize endosperm. Plant J. 36 30–44 10.1046/j.1365-313X.2003.01852.x [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Marcos J. F., Costa L. M., Biderre-Petit C., Khbaya B., O’Sullivan D. M., Wormald M., et al. (2004). Maternally expressed gene1 is a novel maize endosperm transfer cell-specific gene with a maternal parent-of-origin pattern of expression. Plant Cell 16 1288–1301 10.1105/tpc.019778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Marcos J. F., Costa L. M., Dal Prà M., Scholten S., Kranz E., Perez P., et al. (2006). Epigenetic asymmetry of imprinted genes in plant gametes. Nat. Genet. 38 876–878 10.1038/ng1828 [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Marcos J. F., Pennington P. D., Costa L. M., Dickinson H. G. (2003). Imprinting in the endosperm: a possible role in preventing wide hybridization. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358 1105–1111 10.1098/rstb.2003.1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haun W. J., Laoueillé-Duprat S., O’connell M. J., Spillane C., Grossniklaus U., Phillips A. R., et al. (2007). Genomic imprinting, methylation and molecular evolution of maize Enhancer of zeste (Mez) homologs. Plant J. 49 325–337 10.1111/j.1365-313X.2006.02965.x [DOI] [PubMed] [Google Scholar]
- Haun W. J., Springer N. M. (2008). Maternal and paternal alleles exhibit differential histone methylation and acetylation at maize imprinted genes. Plant J. 56 903–912 10.1111/j.1365-313X.2008.03649.x [DOI] [PubMed] [Google Scholar]
- Hennig L., Derkacheva M. (2009). Diversity of Polycomb group complexes in plants: same rules, different players? Trends Genet. 25 414–423 10.1016/j.tig.2009.07.002 [DOI] [PubMed] [Google Scholar]
- Hermon P., Srilunchang K. O., Zou J., Dresselhaus T., Danilevskaya O. N. (2007). Activation of the imprinted Polycomb Group Fie1 gene in maize endosperm requires demethylation of the maternal allele. Plant Mol. Biol. 64 387–395 10.1007/s11103-007-9160-0 [DOI] [PubMed] [Google Scholar]
- Hsieh T. F., Shin J., Uzawa R., Silva P., Cohen S., Bauer M. J., et al. (2011). Regulation of imprinted gene expression in Arabidopsis endosperm. Proc. Natl. Acad. Sci. U.S.A. 108 1755–1762 10.1073/pnas.1019273108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingouff M., Fitz Gerald J. N., Guérin C., Robert H., Sørensen M. B., Van Damme D., et al. (2005). Plant formin AtFH5 is an evolutionarily conserved actin nucleator involved in cytokinesis. Nat. Cell Biol. 7 374–380 10.1038/ncb1238 [DOI] [PubMed] [Google Scholar]
- Jahnke S., Scholten S. (2009). Epigenetic resetting of a gene imprinted in plant embryos. Curr. Biol. 19 1677–1681 10.1016/j.cub.2009.08.053 [DOI] [PubMed] [Google Scholar]
- Jeong S., Bayer M., Lukowitz W. (2011). Taking the very first steps: from polarity to axial domains in the early Arabidopsis embryo. J. Exp. Bot. 62 1687–1697 10.1093/jxb/erq398 [DOI] [PubMed] [Google Scholar]
- Johnston S. D. (1995). The intracellular localization of messenger RNAs. Cell 81 161–170 10.1016/0092-8674(95)90324-0 [DOI] [PubMed] [Google Scholar]
- Jullien P. E., Kinoshita T., Ohad N., Berger F. (2006). Maintenance of DNA methylation during the Arabidopsis life cycle is essential for parental imprinting. Plant Cell 18 1360–1372 10.1105/tpc.106.041178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien P. E., Mosquna A., Ingouff M., Sakata T., Ohad N., Berger F. (2008). Retinoblastoma and its binding partner MSI1 control imprinting in Arabidopsis. PLoS Biol. 6:e194 10.1371/journal.pbio.0060194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermicle J. L. (1970). Dependence of the R-mottled aleurone phenotype in maize on mode of sexual transmission. Genetics 66 69–85 10.3410/f.718486711.793497084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T., Miura A., Choi Y., Kinoshita Y., Cao X., Jacobsen S. E., et al. (2004). One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303 521–523 10.1126/science.1089835 [DOI] [PubMed] [Google Scholar]
- Kinoshita T., Yadegari R., Harada J. J., Goldberg R. B., Fischer R. L. (1999). Imprinting of the MEDEA polycomb gene in the Arabidopsis endosperm. Plant Cell 11 1945–1952 10.1105/tpc.11.10.1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C., Hennig L., Spillane C., Pien S., Gruissem W., Grossniklaus U. (2003). The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev. 17 1540–1553 10.1101/gad.257403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C., Page D. R., Gagliardini V., Grossniklaus U. (2005). The Arabidopsis thaliana MEDEA Polycomb group protein controls expression of PHERES1 by parental imprinting. Nat. Genet. 37 28–30. [DOI] [PubMed] [Google Scholar]
- Krawetz S. A. (2005). Paternal contribution: new insights and future challenges. Nat. Rev. Genet. 6 633–642 10.1038/nrg1654 [DOI] [PubMed] [Google Scholar]
- Le B. H., Wagmaister J. A., Kawashima T., Bui A. Q., Harada J. J., Goldberg R. B. (2007). Using genomics to study legume seed development. Plant Physiol. 144 562–574 10.1104/pp.107.100362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M. A., Larkins B. A. (1993). Endosperm origin, development and function. Plant Cell 5 1383–1399 10.1105/tpc.5.10.1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig S. R., Habera L. F., Dellaporta S. L., Wessler S. R. (1989). Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc. Natl. Acad. Sci. U.S.A. 86 7092–7096 10.1073/pnas.86.18.7092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund G., Ciceri P., Viotti A. (1995a). Maternal-specific demethylation and expression of specific alleles of zein genes in the endosperm of Zea mays L. Plant J. 8 571–581 10.1046/j.1365-313X.1995.8040571.x [DOI] [PubMed] [Google Scholar]
- Lund G., Messing J., Viotti A. (1995b). Endosperm-specific demethylation and activation of specific alleles of alpha-tubulin genes of Zea mays L. Mol. Gen. Genet. 246 716–722 10.1007/BF00290717 [DOI] [PubMed] [Google Scholar]
- Luo M., Bilodeau P., Dennis E. S., Peacock W. J., Chaudhury A. (2000). Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc. Natl. Acad. Sci. U.S.A. 97 10637–10642 10.1073/pnas.170292997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Taylor J. M., Spriggs A., Zhang H., Wu X., Russell S., et al. (2011). A genome-wide survey of imprinted genes in rice seeds reveals imprinting primarily occurs in the endosperm. PLoS Genet. 7:e1002125 10.1371/journal.pgen.1002125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W., Roeder A., Parmenter D., Somerville C. (2004). A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell 116 109–119 10.1016/S0092-8674(03)01067-5 [DOI] [PubMed] [Google Scholar]
- Makarevich G., Villar C. B., Erilova A., Köhler C. (2008). Mechanism of PHERES1 imprinting in Arabidopsis. J. Cell Sci. 121(Pt 6) 906–912 10.1242/jcs.023077 [DOI] [PubMed] [Google Scholar]
- Martínez G., Slotkin R. K. (2012). Developmental relaxation of transposable element silencing in plants: functional or byproduct? Curr. Opin. Plant Biol. 15 496–502 10.1016/j.pbi.2012.09.001 [DOI] [PubMed] [Google Scholar]
- Matthys-Rochon E., Mòl R., Heizmann P., Dumas C. (1994). Isolation and microinjection of active sperm nuclei into egg cells and central cells of isolated maize embryo sacs. Zygote 2 29–35 10.1017/S0967199400001738 [DOI] [PubMed] [Google Scholar]
- Meyer S., Scholten S. (2007). Equivalent parental contribution to early plant zygotic development. Curr. Biol. 17 1686–1691 10.1016/j.cub.2007.08.046 [DOI] [PubMed] [Google Scholar]
- Mohr S. E., Dillon S. T., Boswell R. E. (2001). The RNA-binding protein Tsunagi interacts with Mago Nashi to establish polarity and localize oskar mRNA during Drosophila oogenesis. Genes Dev. 15 2886–2899 10.1101/gad.927001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning J., Peng X. B., Qu L. H., Xin H. P., Yan T. T., Sun M.-X. (2006). Differential gene expression in egg cells and zygotes suggests that the transcriptome is restructed before the first zygotic division in tobacco. FEBS Lett. 580 1747–1752 10.1016/j.febslet.2006.02.028 [DOI] [PubMed] [Google Scholar]
- Nishida H. (1997). Cell lineage and timing of fate restriction, determination and gene expression in ascidian embryos. Semin. Cell Dev. Biol. 8 359–365 10.1006/scdb.1997.0160 [DOI] [PubMed] [Google Scholar]
- Nodine M. D., Bartel D. P. (2012). Maternal and paternal genomes contribute equally to the transcriptome of early plant embryos. Nature 482 94–97 10.1038/nature10756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T., Takanashi H., Mogi M., Takahashi H., Kikuchi S., Yano K., et al. (2011). Distinct gene expression profiles in egg and synergid cells of rice as revealed by cell type-specific microarrays. Plant Physiol. 155 881–891 10.1104/pp.110.167502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi Y., Hoshino R., Okamoto T. (2014). Dynamics of male and female chromatin during karyogamy in rice zygotes. Plant Physiol. 165 1533–1543 10.1104/pp.114.236059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen O. A. (2004). Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell 16(Suppl.) S214–S227 10.1105/tpc.017111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermeier G. C., Miller D., Huntriss J. D., Diamond M. P., Krawetz S. A. (2004). Reproductive biology: delivering spermatozoan RNA to the oocyte. Nature 429:154 10.1038/429154a [DOI] [PubMed] [Google Scholar]
- Pellettieri J., Seydoux G. (2002). Anterior-posterior polarity in C. elegans and Drosophila-PARallels and differences. Science 298 1946–1950 10.1126/science.1072162 [DOI] [PubMed] [Google Scholar]
- Pillot M., Baroux C., Vazquez M. A., Autran D., Leblanc O., Vielle-Calzada J. P., et al. (2010). Embryo and endosperm inherit distinct chromatin and transcriptional states from the female gametes in Arabidopsis. Plant Cell 22 307–320 10.1105/tpc.109.071647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raissig M. T., Baroux C., Grossniklaus U. (2011). Regulation and flexibility of genomic imprinting during seed development. Plant Cell 23 16–26 10.1105/tpc.110.081018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raissig M. T., Bemer M., Baroux C., Grossniklaus U. (2013). Genomic imprinting in the Arabidopsis embryo is partly regulated by PRC2. PLoS Genet. 9:e1003862 10.1371/journal.pgen.1003862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell S. D., Gou X., Wong C. E., Wang X., Yuan T., Wei X. (2012). Genomic profiling of rice sperm cell transcripts reveals conserved and distinct elements in the flowering plant male germ lineage. New Phytol. 195 560–573 10.1111/j.1469-8137.2012.04199.x [DOI] [PubMed] [Google Scholar]
- Scholten S., Lörz H., Kranz E. (2002). Paternal mRNA and protein synthesis coincides with male chromatin decondensation in maize zygotes. Plant J. 32 221–231 10.1046/j.1365-313X.2002.01418.x [DOI] [PubMed] [Google Scholar]
- Sheldon C. C., Hills M. J., Lister C., Dean C., Dennis E. S., Peacock W. J. (2008). Resetting of FLOWERING LOCUS C expression after epigenetic repression by vernalization. Proc. Natl. Acad. Sci. U.S.A. 105 2214–2219 10.1073/pnas.0711453105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirzadi R., Andersen E. D., Bjerkan K. N., Gloeckle B. M., Heese M., Ungru A., et al. (2011). Genome-wide transcript profiling of endosperm without paternal contribution identifies parent-of-origin-dependent regulation of AGAMOUS-LIKE36. PLoS Genet. 7:e1001303 10.1371/journal.pgen.1001303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin R. K., Vaughn M., Borges F., Tanurdzić M., Becker J. D., Feijó J. A., et al. (2009). Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136 461–472 10.1016/j.cell.2008.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprunck S., Baumann U., Edwards K., Langridge P., Dresselhaus T. (2005). The transcript composition of egg cells changes significantly following fertilization in wheat (Triticum aestivum L.). Plant J. 41 660–672 10.1111/j.1365-313X.2005.02332.x [DOI] [PubMed] [Google Scholar]
- Tadros W., Lipshitz H. D. (2009). The maternal-to-zygotic transition: a play in two acts. Development 136 3033–3042 10.1242/dev.033183 [DOI] [PubMed] [Google Scholar]
- Tiwari S., Schulz R., Ikeda Y., Dytham L., Bravo J., Mathers L., et al. (2008). MATERNALLY EXPRESSED PAB C-TERMINAL, a novel imprinted gene in Arabidopsis, encodes the conserved C-terminal domain of polyadenylate binding proteins. Plant Cell 20 2387–2398 10.1105/tpc.108.061929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda M., Laux T. (2012). The origin of the plant body axis. Curr. Opin. Plant Biol. 15 578–584 10.1016/j.pbi.2012.08.001 [DOI] [PubMed] [Google Scholar]
- Ueda M., Zhang Z., Laux T. (2011). Transcriptional activation of Arabidopsis axis patterning genes WOX8/9 links zygote polarity to embryo development. Dev. Cell 20 264–270 10.1016/j.devcel.2011.01.009 [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada J. P., Baskar R., Grossniklaus U. (2000). Delayed activation of the paternal genome during seed of development. Nature 404 91–94 10.1038/35003595 [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada J. P., Thomas J., Spillane C., Coluccio A., Hoeppner M. A., Grossniklaus U. (1999). Maintenance of genomic imprinting at the Arabidopsis medea locus requires zygotic DDM1 activity. Genes Dev. 13 2971–2982 10.1101/gad.13.22.2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbot V., Evans M. M. (2003). Unique features of the plant life cycle and their consequences. Nat. Rev. Genet. 4 369–379 10.1038/nrg1064 [DOI] [PubMed] [Google Scholar]
- Waters A. J., Makarevitch I., Eichten S. R., Swanson-Wagner R. A., Yeh C. T., Xu W., et al. (2011). Parent-of-origin effects on gene expression and DNA methylation in the maize endosperm. Plant Cell 23 4221–4233 10.1105/tpc.111.092668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D., Geldner N., Offringa R., Jurgens G. (2001). Seed development: early paternal gene activity in Arabidopsis. Nature 414 709–710 10.1038/414709a [DOI] [PubMed] [Google Scholar]
- Wolf J. B., Hager R. (2006). A maternal-offspring coadaptation theory for the evolution of genomic imprinting. PLoS Biol. 4:e380 10.1371/journal.pbio.0040380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff P., Weinhofer I., Seguin J., Roszak P., Beisel C., Donoghue M. T., et al. (2011). High-resolution analysis of parent-of-origin allelic expression in the Arabidopsis endosperm. PLoS Genet. 7:e1002126 10.1371/journal.pgen.1002126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuest S. E., Vijverberg K., Schmidt A., Weiss M., Gheyselinck J., Lohr M., et al. (2010). Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr. Biol. 20 506–512 10.1016/j.cub.2010.01.051 [DOI] [PubMed] [Google Scholar]
- Wylie C., Kofron M., Payne C., Anderson R., Hosobuchi M., Joseph E., et al. (1996). Maternal beta-catenin establishes a ’dorsal signal’ in early Xenopus embryos. Development 122 2987–2996. [DOI] [PubMed] [Google Scholar]
- Xin H. P., Peng X. B., Ning J., Yan T. T., Ma L. G., Sun M. X. (2011). Expressed sequence-tag analysis of tobacco sperm cells reveals a unique transcriptional profile and selective persistence of paternal transcripts after fertilization. Sex. Plant Reprod. 24 37–46 10.1007/s00497-010-0151-y [DOI] [PubMed] [Google Scholar]
- Xin H. P., Zhao J., Sun M. X. (2012). The maternal-to-zygotic transition in higher plants. J. Integr. Plant Biol. 54 610–615 10.1111/j.1744-7909.2012.01138.x [DOI] [PubMed] [Google Scholar]
- Yu H. S., Russell S. D. (1994). Occurrence of mitochondria in the nuclei of tobacco sperm cells. Plant Cell 6 1477–1484 10.1105/tpc.6.10.1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Zhao H., Xie S., Chen J., Xu Y., Wang K., et al. (2011). Extensive, clustered parental imprinting of protein-coding and noncoding RNAs in developing maize endosperm. Proc. Natl. Acad. Sci. U.S.A. 108 20042–20047 10.1073/pnas.1112186108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Laux T. (2011). The asymmetric division of the Arabidopsis zygote: from cell polarity to an embryo axis. Sex. Plant Reprod. 24 161–169 10.1007/s00497-010-0160-x [DOI] [PubMed] [Google Scholar]
- Zhao J., Xin H., Qu L., Ning J., Peng X., Yan T., et al. (2011). Dynamic changes of transcript profiles after fertilization are associated with de novo transcription and maternal elimination in tobacco zygote, and mark the onset of the maternal-to-zygotic transition. Plant J. 65 131–145 10.1111/j.1365-313X.2010.04403.x [DOI] [PubMed] [Google Scholar]