Abstract
The small phenolic compound salicylic acid (SA) plays a critical role in plant defense against broad-spectrum of pathogens. The phosphate transporter gene PHT4;1 was previously shown to affect SA-mediated defense and its expression is regulated by the circadian clock. To further understand how PHT4;1 affects SA accumulation, here we analyzed the genetic interactions between the gain-of-function mutant pht4;1-1 and several known SA mutants, including sid2-1, ald1-1, eds5-3, and pad4-1. The genetic analysis was conducted in the acd6-1 background since the change of acd6-1 dwarfism can be used as a convenient readout for the change of defense levels caused by impairments in some SA genes. We found that compared with the corresponding double mutants, the triple mutants acd6-1pht4;1-1ald1-1, acd6-1pht4;1-1eds5-3, and acd6-1pht4;1-1pad4-1 accumulated lower levels of SA and PR1 transcripts, suggesting that PHT4;1 contributes to acd6-1-conferred defense phenotypes independently of these known SA regulators. Although some triple mutants had wild type (wt)-like levels of SA and PR1 transcripts, these plants were smaller than wt and displayed minor cell death, suggesting that additional regulatory pathways contribute to acd6-1-conferred dwarfism and cell death. Our data further showed that circadian expression of PHT4;1 was dependent on CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), a central oscillator component of Arabidopsis circadian clock. Recombinant CCA1 protein was demonstrated to bind to the PHT4;1 promoter in electrophoretic mobility shift assays, suggesting a direct transcriptional regulation of PHT4;1 by CCA1. Together these results indicate that PHT4;1 is a SA regulator acting independently of several known SA genes and they also implicate a role of the circadian clock mediated by CCA1 in regulating phosphate transport and/or innate immunity in Arabidopsis.
Keywords: circadian clock, defense signaling, programmed cell death, disease resistance, phosphate transporter
INTRODUCTION
Successful defense against pathogen attacks is critical to plant growth and development. In addition to pre-formed physical and chemical barriers, plants can monitor the presence of pathogens and subsequently activate defense responses to restrict further proliferation and spreading of pathogens. However, it remains challenging to identify genes that control plant defense, understand their mechanisms of action, and determine how they interact to form complex defense networks to orchestrate resistance to invaders.
The small phenolic compound salicylic acid (SA) plays a central role in plant defense signaling (Hammond-Kosack and Jones, 1996; Ryals et al., 1996; Tsuda et al., 2008). Genes that positively regulate SA-mediated defense have been identified in Arabidopsis. These genes can be grouped into three types based on their potential biochemical and molecular functions (Lu, 2009; Lu et al., 2009a). The type I SA genes encode enzymes directly involved in SA biosynthesis, which is proposed to take place in the chloroplast and cytoplasm of a cell, involving multiple pathways (Chen et al., 2009; Dempsey et al., 2011). The type I SA gene ISOCHORISMATE SYNTHASE 1 (ICS1), also called SA Induction-Deficient 2 (SID2) and ENHANCED DISEASE SUSCEPTIBILITY 16 (EDS16), contributes to the bulk SA biosynthesis (Nawrath and Metraux, 1999; Wildermuth et al., 2001; Ng et al., 2011). ICS1/SID2/EDS16 protein was shown to be chloroplast-localized (Strawn et al., 2007), indicating that the major SA biosynthetic pathway likely occurs in the chloroplast. To support this notion, the bacterial gene nahG that encodes SA hydroxylase to convert SA to the breakdown product catechol (Friedrich et al., 1995), when expressed in the chloroplast, abolishes SA accumulation in the transgenic plants challenged with pathogens or UV light (Fragniere et al., 2011).
Protein products of type II SA genes may not be directly involved in SA biosynthesis. But like SA biosynthetic enzymes (type I), they can influence SA accumulation, possibly through indirect ways, for instance, chemically modifying SA precursors, affecting availability of SA precursors and/or products, influencing expression of type I SA genes, and/or changing activities of SA biosynthetic enzymes. One example of the type II SA regulators is SID1/EDS5, which was localized to the chloroplast membrane and was proposed to transport SA from the chloroplast to the cytoplasm in a cell (Nawrath et al., 2002; Serrano et al., 2013; Yamasaki et al., 2013). The lack of such a SA-transport activity in the eds5 mutants likely leads to SA accumulation in the chloroplast that feedback-inhibits SA biosynthesis under defense conditions. Indeed, like sid2 mutants, eds5 mutants accumulate much reduced SA levels under defense conditions (Nawrath et al., 2002; Ng et al., 2011). Thus these observations further support the idea that the chloroplast is the major site for SA biosynthesis. Additional examples of type II SA regulators include Accelerated Cell Death (ACD6), AGD2-LIKE DEFENSE 1 (ALD1), EDS1, and PHYTOALEXIN DEFICIENT 4 (PAD4) (Falk et al., 1999; Jirage et al., 1999; Lu et al., 2003; Song et al., 2004). Loss of function mutations in these SA regulators often lead to EDS and partially reduced SA accumulation upon pathogen insults, compared to wild type (wt) plants. However, the mechanisms by which many of the type II SA regulators act have not been well understood.
Activation of SA signaling often leads to enhanced disease resistance in plants. The type III SA genes act downstream of SA, including SA receptors and signaling transducers. Non-expressor of PR GENES 1 (NPR1) is an example of type III SA genes that has been elegantly studied for its mechanism of action (Dong, 2004; Fu and Dong, 2013). The NPR1 protein has been shown as a key component for SA signaling, overexpression of which confers enhanced disease resistance to a range of pathogens in Arabidopsis and some crop plants (Chern et al., 2001; Ekengren et al., 2003; Fitzgerald et al., 2004; Lin et al., 2004; Makandar et al., 2006; Malnoy et al., 2007; Yuan et al., 2007; Quilis et al., 2008; Sandhu et al., 2009; Zhang et al., 2010). Two close homologs of NPR1, NPR3 and NPR4 were recently shown to be SA receptors with different binding affinities to SA (Fu et al., 2012; Fu and Dong, 2013). However, whether or not NPR1 itself is also an SA receptor remains controversial (Wu et al., 2012; Yan and Dong, 2014).
Recent studies showed that two members of a phosphate transporter family, the PHT4 family, were involved in SA regulation. The PHT4 family has six members, five of which (PHT4;1-4;5) are plastid-localized, and one (PHT4;6) is Golgi-localized (Roth et al., 2004; Guo et al., 2008a; Pavon et al., 2008; Cubero et al., 2009). Recombinant proteins of PHT4 family members were demonstrated to have phosphate transport activities (Guo et al., 2008a; Pavon et al., 2008; Cubero et al., 2009). However, only a loss of function mutation in the PHT4;6 gene but not in other five genes confers enhanced disease resistance to Pseudomonas syringae infection and high levels of SA besides reduced salt tolerance (Cubero et al., 2009; Hassler et al., 2012). These results suggest that PHT4;6 is a negative regulator of SA-mediated defense and is also involved in salt stress response.
The lack of defense and salt tolerance phenotypes in loss of function mutants of other five PHT4 members is possibly due to functional redundancy among these members. To further support roles of the PHT4 family members in defense control, we identified a gain of function mutant of the PHT4;1 gene, pht4;1-1, in a genetic screen for acd6-1 suppressors with a goal to uncover new defense genes (Wang et al., 2011b). ACD6 encodes an ankyrin repeat protein with transmembrane domain and has been shown as a major determinant of fitness in Arabidopsis ecotypes (Lu et al., 2003; Todesco et al., 2010, 2014). acd6-1 is a small gain-of-function mutant that displays extreme dwarfism, constitutive defense, and spontaneous cell death phenotypes (Rate et al., 1999; Lu et al., 2003). The small size of acd6-1 is largely in an inverse correlation with the defense level of the plant. This characteristics of acd6-1 has proven useful in genetic screens to identify novel genes critical for plant defense (Lu et al., 2009a) and in genetic analyses to interrogate interactions between known defense genes (Song et al., 2004; Ng et al., 2011; Wang et al., 2011a). The pht4;1-1 mutation suppressed high SA accumulation in acd6-1 and conferred EDS to P. syringae infection in the absence of acd6-1, which could be rescued by exogenous SA treatment (Wang et al., 2011b). This mutation was caused by a T-DNA insertion that resulted in expression of truncated PHT4;1 transcripts. Since increasing PHT4;1 expression by introducing extra copies of PHT4;1 transgene into wt also conferred EDS (Wang et al., 2011b), we conclude that pht4;1-1 is a gain of function allele and both PHT4;1 and PHT4;1-1 proteins act similarly as negative regulators of Arabidopsis defense. Genetic analysis further indicated that pht4;1-1 possibly contributed to both SID2-dependent and – independent pathways in regulating acd6-1-conferred dwarfism and cell death phenotypes. In addition, PHT4;1 expression was shown to be regulated by the circadian clock (Guo et al., 2008a; Wang et al., 2011b). Thus we propose that PHT4;1 is a type II SA regulator, the function of which implicates the circadian clock.
In this report, we further investigated the role of PHT4;1 in SA regulation and the mechanism of circadian regulation of PHT4;1. We examined genetic interactions between pht4;1-1 and several type II mutants, ald1-1, eds5-3, and pad4-1, besides the type I SA mutant sid2-1. The genetic analysis was done in the acd6-1 background because the change of acd6-1 size can be used as a convenient visual readout of functional interactions between the mutants. Our results show that pht4;1-1 acts additively with sid2-1, ald1-1, eds5-3, and pad4-1 to regulate acd6-1 dwarfism, cell death, and/or defense responses, suggesting that PHT4;1 has distinct function from these other SA regulators. To elucidate the mechanism by which PHT4;1 is circadian clock-regulated, we tested the hypothesis that PHT4;1 is a direct target of the core component of Arabidopsis circadian clock CIRCADIAN CLOCK ASSOCIATED 1 (CCA1). Our data support the hypothesis and underscore a possible role of the circadian clock mediated by CCA1 in regulating the function of PHT4;1 in phosphate transport and/or innate immunity control in Arabidopsis.
MATERIALS AND METHODS
PLANT MATERIALS
All Arabidopsis plants used in this report are in Columbia-0 background. Plants were grown in growth chambers with a 12 h light/12 h dark cycle, light intensity at 200 μmol m-2 s-1, 60% humidity, and 22°C. The triple mutant acd6-1pht4;1-1sid2-1 was previously described (Wang et al., 2011b). Additional triple mutants were made by crossing acd6-1pht4;1-1 with acd6-1ald1-1, acd6-1eds5-3, or acd6-1pad4-1 and selected for homozygotes by polymerase chain reaction (PCR) with appropriate primers (Ng et al., 2011).
RNA ANALYSIS
Whole plants of each genotype at 25-day old were harvested at ZT1 (1 h after lights on) for RNA extraction. For circadian clock-regulated gene expression, plants grown in 12 h L/12 h D were transferred to constant light (LL) and harvested starting at ZT1 at a 4 h interval for 48 h. RNA extraction and northern blotting were performed as described (Ng et al., 2011). Radioactive probes were made by PCR with antisense primers specific to individual gene fragments in the presence of [32P] dCTP. Primers used for making the PR1 probe are PR1_sense 5′ GTAGGTGCTCTTGTTCTTCCC 3′ and PR1_antisense 5′ CACATAATTCCCACGAGGATC 3′ and for making the PHT4;1 probe are PHT4;1_sense 5′ ATGAACGCGAGAGCTCTTCTTTGCTC 3′ and PHT4;1_antisense 5′ AATCGATTATCTTCTCTCCGGTTG 3′.
SA MEASUREMENT
SA was extracted from 25-day old plants and quantified by a high-performance liquid chromatography (HPLC) instrument as previously described (Ng et al., 2011; Wang et al., 2011a).
CELL DEATH STAINING
The sixth or seventh leaves of 25-day old plants were harvested for trypan blue staining as described (Ng et al., 2011). Stained leaves were washed with 50% ethanol and mounted on glass slides with cover slips for photographing with a complementary metal–oxide–semiconductor (CMOS) camera connected to a dissecting microscope (Leica M205 FA, Leica Microsystems, Germany).
PURIFICATION OF CCA1-GST RECOMBINANT PROTEIN FROM Escherichia coli
The pGEX-CCA1 construct containing CCA1-GST in the pGEX-3X vector was a kind gift from Steve Kay at University of South California. pGEX-CCA1 was transformed into the Escherichia coli strain BL21(DE3)-pLysS to express the recombinant protein. A single colony was picked for overnight culture in 5 ml LB media, which was subsequently added into 500 ml LB media for further culture. At OD600 = 0.5, the culture was treated with 0.4 mM isopropyl β-D-1-thiogalactopyranoside for 3 h followed by harvesting by centrifugation at 8000 g for 10 min at 4°C. The pellet was resuspended in 25 ml ice–cold 1 X PBS containing 1% Triton X-100 and 2x protease inhibitor (Roche, LOT# 14549800) and lysed by sonication on ice. The sonication condition was 30 s on followed by 30 s off at 30% amplitude for 20 cycles, using Virsonic Cell Disruptor (Model 16-850, The Virtis Co., New York). Cell lysates were collected by centrifugation at 8000 g for 10 min at 4°C. The supernatant was loaded onto a 2 ml glutathione spin column (Pierce, Product # 16107), incubated at 4°C for 30 min on a rocking platform. The column was washed with 10x bed volumes of equilibration/wash buffer (125 mM Tris, 150 mM NaCl, pH 8.0). The CCA1-GST recombinant proteins were eluted with elution buffer (10 mM glutathione, 125 mM Tris, 150 mM NaCl, pH 8.0), according to manufacturer’s instruction (Pierce, Product # 16107). Purified CCA1-GST protein was verified on a 6% SDS-PAGE gel and aliquoted into 30 μl per microcentrifuge tube for storage at -80°C.
ELECTROPHORETIC MOBILITY SHIFT ASSAYS
Three DNA fragments (probes) from the PHT4;1 promoter were generated by PCR amplification, purified, and used for CCA1-GST binding assays. Probe 1 (396 bp) covers from -348 to +48 bp relative to the ATG start site of the PHT4;1 promoter (primers 5′ TTGTTATTGGTATTGCCGTATTATTGTA 3′, and 5′ GTAGAGAGAGTGAATATTTGAAGA 3′). Probe 2 (118 bp) covers from -348 to -230 bp relative to the ATG start site of the PHT4;1 promoter (primers 5′ TTGTTATTGGTATTGCCGTATTATTGTA 3′, and 5′ GTTAGCTTACGAGCATAAATTGC 3′). Probe 3 (117 bp) covers from -69 to +48 bp relative to the ATG start site of the PHT4;1 promoter (primers 5′ AATCAATTCCTCTCTCTTAAAACAAA 3′, and 5′ GTAGAGAGAGTGAATATTTGAAGA 3′). The negative probe PHT4;1-NC (without CCA1 binding site) was generated by PCR amplification of the region from +134 to +668 of the PHT4;1 gene (primers 5′ CTACCCGCGAAATAGGTCCAGTG 3′, and 5′ ATCAACAAACCACTGATTCAACTACACTT 3′). Probes (60 ng each) were end-labeled with γ-[32P]-dATP, using T4 polynucleotide kinase (Thermo Scientific, product # EK0031) in the following reaction: 2 pmol DNA fragment, 2 μl 10x forward reaction buffer, 4 pmol γ-[32P]-dATP, 1 μl T4 PNK, in a total volume of 20 μl. The reaction was carried out at 37°C for 30 min, then added 1 μl of 0.5 M EDTA (pH 8.0) and incubated at 75°C for 10 min to terminate the reaction. Labeled DNA probes were purified by using a PCR purification kit (Qiagen, cat#28104) and eluted with 30 μl sterile water. Binding reactions were carried out as following: 2 μl 5X electrophoretic mobility shift assays (EMSA) buffer [125 mM HEPES-KOH (pH 7.5), 12.5 mM DTT, 5 mM PMSF, 250 mM KCl], 2 μl 50% glycerol, 1 μl 1 μg/ul poly-dIdC, 30–90 ng CCA1-GST recombinant protein, 1 μl labeled probe, in a total volume of 10 μl. For a competition assay, excessive amount of a corresponding cold probe or the negative probe PHT4;1-NC at the indicated concentrations was added to a binding reaction. Both binding and competition reactions were incubated on ice for 20 min before being immediately loaded onto a 6% non-denaturing polyacrylamide gel, prepared in 0.5X TBE buffer [40 mM Tris-Cl (pH 8.3), 45 mM boric acid, 1 mM EDTA]. Electrophoresis was conducted at 100 V for ∼1 h at room temperature to separate free probes from DNA-protein complexes. The gels were dried on a gel dryer (Hoefer, model SE1160) at 80°C for 1 h followed by exposure to X-ray film for 2–4 days.
RESULTS
PHT4;1 INTERACTS ADDITIVELY WITH MULTIPLE SA REGULATORS TO AFFECT ACD6-1 DWARFISM
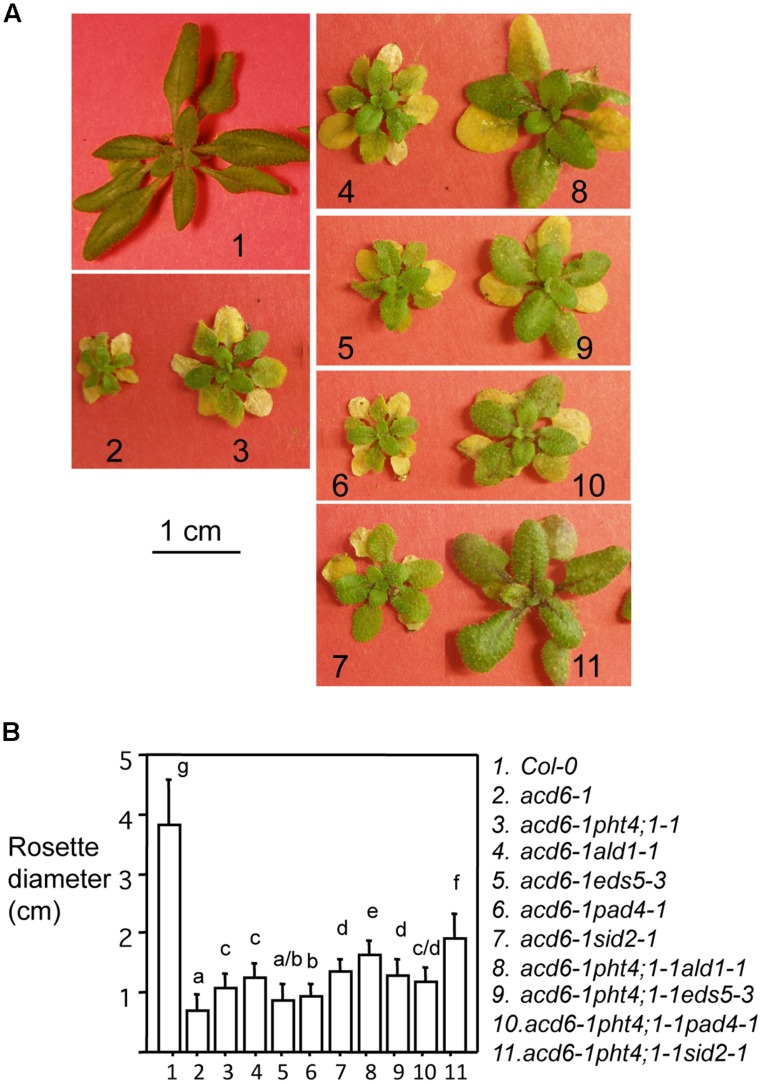
Our previous data suggest that the PHT4;1 gene acts upstream of SA to regulate SA accumulation (Wang et al., 2011b). To further investigate the role of PHT4;1 in SA regulation, we sought to examine genetic interactions between the gain of function mutant pht4;1-1 and mutants disrupting type II SA genes, ADL1, EDS5, and PAD4 (Jirage et al., 1999; Nawrath et al., 2002; Song et al., 2004). We crossed pht4;1-1 to these mutants in the acd6-1 background because the small size of acd6-1 is sensitized to the change of defense levels and thus acd6-1 can be conveniently used to dissect the functional relationship among SA genes (Song et al., 2004; Ng et al., 2011). A previous similar experiment showed that pht4;1-1 acts additively with the type I SA mutant sid2-1 in affecting acd6-1 dwarfism (Wang et al., 2011b). We found here that similar to acd6-1pht4;1-1sid2-1, the triple mutants acd6-1pht4;1-1ald1-1, acd6-1pht4;1-1eds5-3, and acd6-1pht4;1-1pad4-1 were significantly larger than their corresponding double mutants (Figures 1A,B), suggesting that PHT4;1 acts additively with multiple SA regulators in influencing acd6-1 dwarfism.
FIGURE 1.
pht4;1-1 acts additively with SA mutants to suppress acd6-1 dwarfism. (A) Pictures of 25-day old plants. The single mutants pht4;1-1, ald1-1, eds5-3, pad4-1, and sid2-1 are morphologically similar to Col-0 (not shown). The scale bar represents 1 cm and applies to all panels. (B) Plant size measurement. Plants shown in (A) were measured for their rosette diameters. Statistical analysis was performed with Student’s t-test (StatView 5.0.1). Different letters indicate significant difference among the samples (P < 0.05; n = 10).
PHT4;1 INTERACTS ADDITIVELY WITH MULTIPLE SA REGULATORS TO AFFECT DEFENSE PHENOTYPES AND CELL DEATH IN ACD6-1
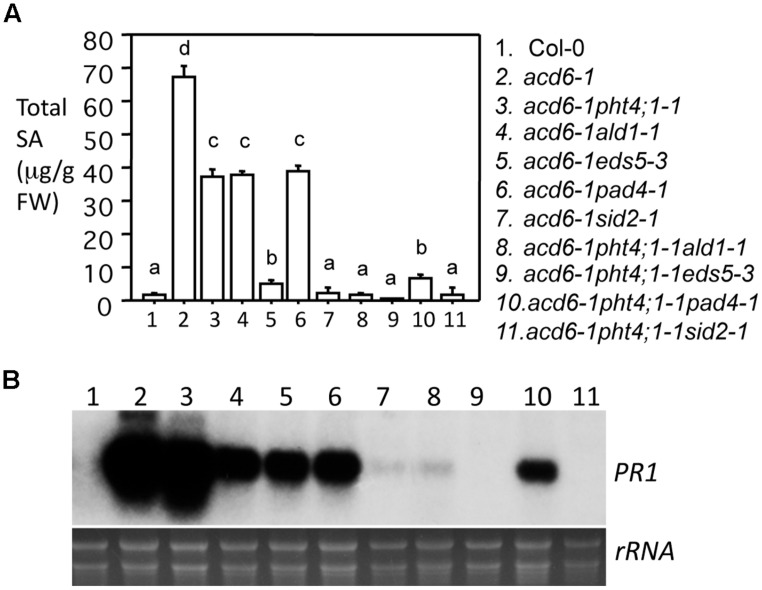
Since previous studies showed that the dwarfism of acd6-1 is grossly in reverse correlation with the defense level of the plant (Song et al., 2004; Lu et al., 2009a; Ng et al., 2011; Wang et al., 2011a), the increased size of the triple mutants shown in Figure 1 suggests reduced defense of the plants. To further test this, we measured SA levels and expression of the defense marker gene PR1 in these plants. Indeed we found that acd6-1pht4;1-1ald1-1 and acd6-1pht4;1-1eds5-3 accumulated near wt-level of SA and PR1 transcripts (Figures 2A,B). acd6-1pht4;1-1pad4-1, on the other hand, had much reduced SA level than the two parental double mutants but this level was still significantly higher than that seen in wt. Expression of PR1 was only slightly reduced in acd6-1pht4;1-1pad4-1, compared with the corresponding double mutants (Figure 2B). These results indicate that PHT4;1 has distinct function from these type II SA genes in regulating SA accumulation and PR1 expression. Consistent with a major role of SID2 in SA biosynthesis (Wildermuth et al., 2001; Ng et al., 2011), we found that SA accumulation and PR1 expression in acd6-1pht4;1-1sid2-1 were comparable to those of acd6-1sid2-1 and wt. We also noticed that the near-wt level of SA in some triple mutants (acd6-1pht4;1-1ald1-1, acd6-1pht4;1-1eds5-3, and acd6-1pht4;1-1sid2-1) was not correlated with a complete suppression of acd6-1 dwarfism, suggesting there are additional pathways independent of SA contributing to plant size regulation in acd6-1.
FIGURE 2.
pht4;1-1 acts additively with SA mutants to suppress SA accumulation and PR1 expression in acd6-1. Twenty five-day old plants were harvested for SA extraction followed by HPLC analysis and RNA preparation followed by northern blotting. (A) SA quantitation. Statistical analysis was performed with Student’s t-test (StatView 5.0.1). Different letters indicate significant difference among the samples (P < 0.05; n = 3). (B) PR1 expression. rRNA was shown as a loading control.
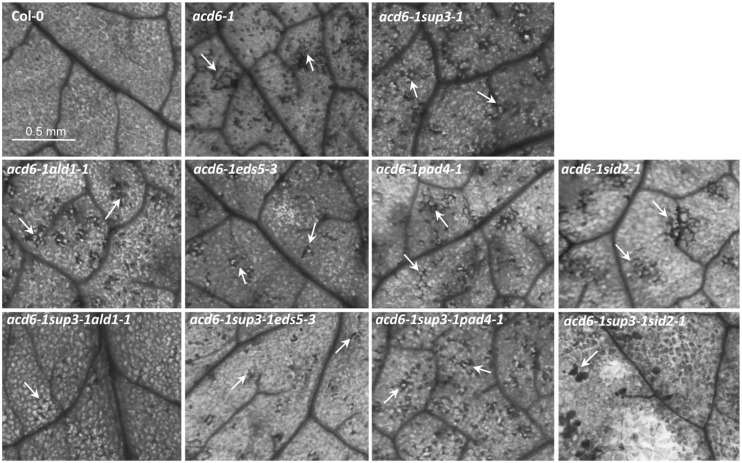
Besides dwarfism and enhanced defense phenotypes, the acd6-1 mutant displays severe cell death, even in the absence of pathogen challenge. Suppression of acd6-1-conferred dwarfism and defense phenotypes is usually associated with reduced cell death (Song et al., 2004; Lu et al., 2009a; Ng et al., 2011; Wang et al., 2011a). Consistent with this previous observation, we found that triple mutants acd6-1pht4;1-1ald1-1, acd6-1pht4;1-1eds5-3, and acd6-1pht4;1-1sid2-1 had substantially reduced but not abolished cell death on their leaves when the plants were stained with trypan blue to visualize cell death (Figure 3). Since these mutants accumulated wt-level SA, like plant-size regulation, cell death formation in these plants could be influenced by additional SA-independent pathway(s). Interestingly acd6-1pht4;1-1pad4-1 displayed similar cell death as acd6-1pht4;1-1. This could be due to the relatively high level of SA presented in the triple mutant. Alternatively PHT4;1 and PAD4 could act in the same pathway to affect cell death of acd6-1.
FIGURE 3.
pht4;1-1 acts additively with SA mutants to suppress cell death in acd6-1. The sixth or seventh leaves of 25-day old plants were harvested for trypan blue staining as described (Ng et al., 2011). Stained leaves were photographed with a CMOS camera connected to a dissecting microscope (Leica M205 FA, Leica Microsystems, Germany). Cell death is shown in the dark stained spots or patches on a leaf (arrows). At least four leaves of each genotype were stained and examined for cell death. No cell death was detected in ald1-1, eds5-3, pad4-1, sid2-1, and pht4;1-1 (data not shown). The scale bar represents 0.5 mm and applies to all panels.
CIRCADIAN EXPRESSION OF PHT4;1 IS CCA1-DEPENDENT
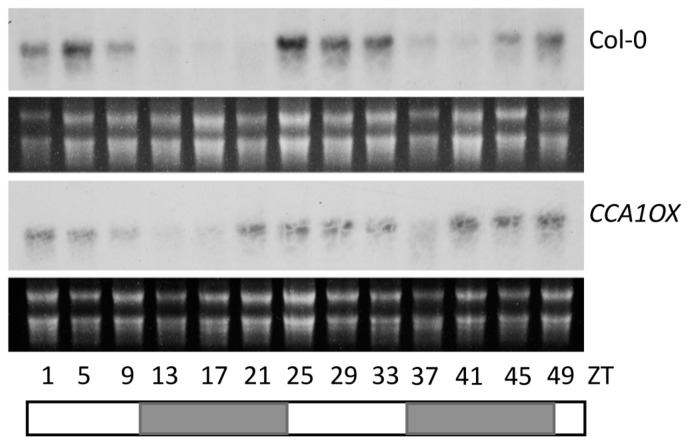
Expression of PHT4;1 was previously shown to be regulated by the circadian clock (Guo et al., 2008a; Wang et al., 2011b). Such a circadian expression pattern persisted in pht4;1-1 and acd6-1 mutants and in the presence of P. syringae challenge (Wang et al., 2011b). Consistent with being regulated by the circadian clock, the PHT4;1 promoter has two cis-elements (the CBS motifs), starting at -17 and -281 bp positions, respectively, that are putative binding sites for the core clock component CCA1 (Alabadi et al., 2002; Green and Tobin, 2002; Michael and McClung, 2002). Thus we hypothesized that CCA1 directly targets PHT4;1 promoter for expression regulation. To test this hypothesis, we first examined PHT4;1 expression in CCA1 overexpressing (CCA1ox) plants, which display arrhythmic clock activity in both constant light (LL) and light/dark (LD) conditions (Wang and Tobin, 1998; Zhang et al., 2013). We found that when the plants were transferred from LD to LL, PHT4;1 demonstrated a circadian expression pattern in wt Col-0. However, CCA1ox disrupted this expression pattern of PHT4;1 (Figure 4). This result indicates a role of CCA1 in controlling PHT4;1 expression but could not pinpoint whether such an effect is direct or indirect.
FIGURE 4.
Circadian expression of PHT4;1 is CCA1-dependent. Twenty five-day-old Col-0 and CCA1ox plants grown in a chamber with a 12 h light/12 h dark cycle and 22°C were transferred to LL at 22°C. Starting at ZT1, plants were harvested at every 4 h for 48 h for RNA extraction followed by northern blotting. White boxes indicate subjective light periods and gray boxes indicate subjective dark periods in LL. rRNA was shown as a loading control.
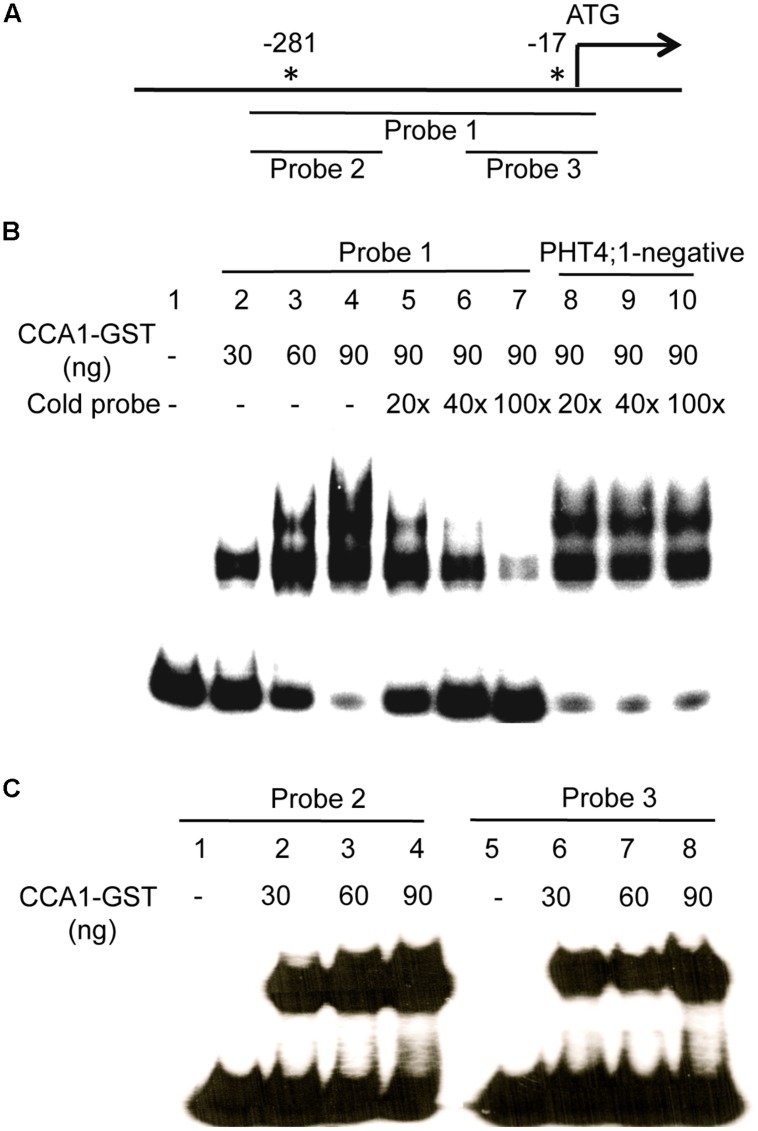
To further test if CCA1 directly binds to the PHT4;1 promoter, we conducted EMSA with CCA1-GST recombinant protein and PHT4;1 promoter fragments. The probe 1 is a PHT4;1 fragment containing two CBS motifs (Figure 5A). We found that probe 1 was bound by recombinant CCA1-GST protein, resulting in slower moving bands containing protein-DNA complexes (Figure 5B, lane 2–4). Unlabeled probe 1 could compete with isotope-labeled probe 1 for CCA1-GST binding in a dose-dependent manner (Figure 5B, lane 5–7). However, excess amount of a negative fragment (PHT4;1-negative) from PHT4;1 without a CBS motif did not compete with isotope-labeled probe 1 in CCA1-GST binding (Figure 5B, lane 8–10). These results suggest that the binding between probe 1 and CCA1-GST protein is specific. We also noticed that there were two shifted bands in most lanes from probe 1 and CCA1-GST binding reactions (Figure 5B, lane 3–5 and 8–10). We speculated that both CBS motifs in probe 1 can be bound by CCA1-GST when the protein is present in abundance. To test this, we incubated two shorter PHT4;1 promoter fragments (probe 2 and probe 3), containing only one CBS motif each, with CCA1-GST (Figure 5C). Indeed, both probe 2 and 3 were bound by CCA1-GST, forming a single DNA-protein complex that separated from the free probes. Thus these in vitro binding assays support our hypothesis that PHT4;1 is a direct target of CCA1.
FIGURE 5.
CCA1 binds to the PHT4;1 promoter in EMSA. (A) Positions of the PHT4;1 promoter fragments (probes) and the CBS motifs (asterisks). Positions are relative to the translation start site (ATG). (B) EMSA with probe 1. (C) EMSA with probe 2 and probe 3. The probes were end-labeled with γ-32P and incubated with purified recombinant CCA1-GST protein. For competition assays in (B), unlabeled fragments (probe 1 or PHT4;1-negative) at the indicated folds more than the input (isotope-labeled probe 1) were added to the binding reactions. The reactions were resolved on 6% native PAGE gels followed by gel drying and exposure to X-ray film.
DISCUSSION
In this study, we took biochemical, genetic, and molecular approaches to further investigate the function of the phosphate transporter gene PHT4;1. Our results show that PHT4;1 genetically interacts with several SA genes, including SID2, ALD1, EDS5, and PAD4, in regulating defense responses. In addition, we show that circadian expression of PHT4;1 is dependent on the circadian clock protein CCA1, which could directly bind to the PHT4;1 promoter. These results corroborate the role of PHT4;1 in defense regulation and also suggest that the circadian clock gene CCA1 regulates phosphate transport and/or defense responses, possibly through influencing PHT4;1-mediated pathway.
Our previous study indicated that PHT4;1 is a negative defense regulator acting upstream of SA (Wang et al., 2011b). Genetic analysis conducted here further showed that pht4;1-1 acts additively with SA mutants ald1-1, eds5-3, and pad4-1 to suppress high levels of SA accumulation and PR1 expression in acd6-1 (Figure 2). Thus PHT4;1 likely functions in a separate pathway from ALD1, EDS5, and PAD4 in regulating these defense outputs. Consistent with this notion, expression of ACD6, ALD1, and PAD4 are inducible by SA treatment, suggesting that these genes are involved in signal amplification loops with SA (Nawrath et al., 2002; Lu et al., 2003; Song et al., 2004). However, expression of PHT4;1 is not affected by SA treatment (data not shown), suggesting that unlike ACD6, ALD1, and PAD4, PHT4;1 is not part of SA-signal amplification loop. Together these results further support a previous notion that there are multiple pathways affecting SA-mediated defense in Arabidopsis (Song et al., 2004; Ng et al., 2011; Wang et al., 2011a). Interestingly although some triple mutants show wt-like levels of SA and PR1 expression, none of these triple mutants revert to wt-like phenotypes in terms of plant size and cell death (Figures 1 and 3). These results suggest that the regulation of plant size and cell death can be uncoupled from that of some defense phenotypes in acd6-1. Additional SA-independent pathway(s) could contribute to the regulation of plant size and cell death formation in acd6-1.
While the gain of function mutant pht4;1-1 displayed compromised defense phenotypes, the loss of function alleles of PHT4;1 did not show altered defense responses (Wang et al., 2011b). This can be explained by possible functional redundancy among some PHT4 family members. Indeed, PHT4;1 and four other members in the family share high levels of homology and are all plastid-localized. Functional redundancy among these members could prevent manifestation of defense phenotypes in single loss of function mutants. So far only one disrupted member, PHT4;2, showed small effects on plant growth (Irigoyen et al., 2011). Besides pht4;1 loss of function mutants, available single loss of function mutants of PHT4;4 and PHT4;5 are indistinguishable from wt in morphology and defense responses (data not shown).
While five plastid-localized PHT4 family members could share redundant function, the sixth member of the family, PHT4;6, might be functionally divergent from other members in the family. PHT4.6 is localized to the Golgi and was shown to have Pi transport activity in the Golgi (Guo et al., 2008a; Cubero et al., 2009). A single loss of function mutation in PHT4;6 results in enhanced disease resistance to P. syringae infection, dwarfism, and reduced salt tolerance (Cubero et al., 2009; Hassler et al., 2012). The pht4;6 mutant also accumulates modestly higher levels of SA than wt. Thus like PHT4;1, PHT4;6 is also a negative regulator of plant defense.
The involvement of two members of the PHT4 family in defense suggests a possibility that phosphate transport is critical for host-pathogen interactions. Phosphorus (P) is essential for plant growth and development. However, plants do not produce P but take up inorganic phosphate ion (Pi) from the soil to the root, reallocate Pi to different tissue and cell types, and redistribute Pi to different organelles within a cell in order to fulfill the Pi requirement for cellular functions. These processes are mediated by phosphate transporters to maintain phosphate homeostasis and the normal function of cells. At least five phosphate transporter families, PHT1, PHT2, PHT3, PHT4, and pPT, have been reported in Arabidopsis (Poirier and Bucher, 2002; Guo et al., 2008b). Among these phosphate transporter families, only mutations in some PHT1 genes and one PHT2 gene resulted in alterations in Pi concentration in planta (Versaw and Harrison, 2002; Shin et al., 2004; Gonzalez et al., 2005). The PHT1 genes encode plasma membrane-localized high affinity Pi/H+ symporters and are expressed abundantly in the root (Karthikeyan et al., 2002; Mudge et al., 2002). The PHT2 gene encodes a chloroplast-localized phosphate transporter and is highly expressed in the green tissue (Versaw and Harrison, 2002). Based on these tissue- and cell-specific expression patterns, PHT1 was proposed to acquire Pi from the root whereas PHT2 was proposed to influence the reallocation of phosphate within different tissues of a plant. PHT4 and other phosphate transporter families have not been reported to have a major effect on phosphate concentration at the whole plant level. Except two members of the PHT4 family (PHT4;1 and PHT4;6), none of the other phosphate transporter genes have been demonstrated a role in defense regulation. Therefore it is currently unknown whether perturbation of phosphate concentration in planta could result in altered defense responses. However, there is evidence to support a connection between altered phosphate signaling and defense control. One example is the SIZ1 gene encoding a SUMO E3 ligase that targets PHR1, a MYB transcriptional activator critical for phosphate response. A siz1 mutant demonstrated reduced phosphate response and enhanced disease resistance (Rubio et al., 2001; Miura et al., 2005; Lee et al., 2007; Jin et al., 2008).
Since PHT4;1 is not known to perturb phosphate concentration at the whole plant level, the defense phenotypes observed in the pht4;1-1 mutant could be caused by altered PHT4;1 transport activity at the subcellular level. PHT4;1 is mainly expressed in the shoot tissue (Guo et al., 2008a,b). Pavon et al. (2008) showed that the PHT4;1 protein was localized to the thylakoid member of the chloroplast and thus proposed that PHT4;1 transports Pi across thylakoid lumen and stroma in the chloroplast, using its Na+ and/or H+- dependent phosphate transporter activity (Guo et al., 2008b). In another study, Roth et al. (2004) localized PHT4;1 to the inner membrane of the plastid. Although the precise localization of PHT4;1 remains to be determined, these studies pointed to the connection of PHT4;1 with the chloroplast, the central organelle for photosynthesis and many secondary and primary metabolisms, including SA biosynthesis. It is conceivable that Pi transported by PHT4;1 could directly or indirectly affect SA biosynthetic pathways or proteins/processes that affect SA accumulation. Such function of PHT4;1 could be shared by other four plastid-localized PHT4 family members (PHT4;2-4;5). However, Golgi-localized PHT4;6 may influence SA accumulation and SA-mediated defense through a different mechanism from that used by PHT4;1.
The observation of circadian clock regulated PHT4;1 expression has prompted us to elucidate the role of the circadian clock in defense control (Guo et al., 2008a; Wang et al., 2011b; Zhang et al., 2013). The circadian clock is an internal time measuring machinery important for development and fitness of plants (Green et al., 2002; Michael et al., 2003; Dodd et al., 2005; Ni et al., 2009; Graf et al., 2010; Dong et al., 2011). Increasing evidence supports a role of the circadian clock in defense regulation. First, like PHT4;1, expression of some defense genes were reported to be under the circadian clock control (Wang et al., 2001, 2011b; Sauerbrunn and Schlaich, 2004; Weyman et al., 2006; Roden and Ingle, 2009). Second, wt Arabidopsis shows temporal variations in a day in its susceptibility to P. syringae infection and such variations can be disrupted by overexpression of CCA1 (Bhardwaj et al., 2011). Third, misexpression of several core clock genes, including CCA1, its close homolog LATE ELONGATED HYPOCOTYL (LHY) (Alabadi et al., 2002; Mizoguchi et al., 2002; Lu et al., 2009b), and TIME FOR COFFEE (Hall et al., 2003; Ding et al., 2007), leads to compromised resistance to the bacterial pathogen P. syringae and/or to the oomycete pathogen Hyaloperonospora arabidopsidis (Hpa) (Bhardwaj et al., 2011; Wang et al., 2011c; Shin et al., 2012; Zhang et al., 2013). Data from our study further indicate that defense activation can reciprocally regulate clock activity, suggesting crosstalk between the circadian clock and plant innate immunity (Zhang et al., 2013).
Both experimental studies and in silico analysis of circadian clock-regulated gene expression indicate that PHT4;1 is the only member in the PHT4 family that demonstrates a robust circadian expression pattern (Mockler et al., 2007; Guo et al., 2008a, and data not shown). We presented here evidence to further demonstrate that PHT4;1 could be a direct transcriptional target of the circadian clock protein CCA1 (Figures 4 and 5). Interestingly, while we show here that PHT4;1 is an SA regulator that acts independently of several known SA genes, our previous study indicated that the clock genes CCA1 and LHY acted in a SA-independent manner in defense regulation (Zhang et al., 2013). The cca1-1lhy-20 double mutations suppressed acd6-1-conferred constitutive defense but not its dwarfism and high SA accumulation. Such a discrepancy in terms of SA regulation by PHT4;1 and CCA1 suggest that CCA1-regulation of PHT4;1 might be important for phosphate transport activity of PHT4;1 but may not be directly related to the role of PHT4;1 in SA regulation. It is also possible that there are additional factor(s) affecting circadian expression of the PHT4;1 gene and/or phosphate transport activity of the PHT4;1 protein. Alternatively, CCA1 and its close homolog LHY could regulate expression of multiple defense genes, including both positive and negative SA regulators. Thus in the CCA1 and LHY loss of function background, the effect on SA accumulation could be negated by the changes of these two opposing groups of SA genes. Additional biochemical, genetic, and molecular studies are required to further elucidate the biological relevance of CCA1 binding on the PHT4;1 promoter in terms of phosphate transport and defense regulation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank members in the Lu laboratory for assistance with this work. We thank N. S. Alberola and D. Eisenmann for suggestions in the EMSA experiments. This work was partly supported by a grant from Citrus Research and Development Foundation and a grant from National Science Foundation (NSF RIG-0818651) to Hua Lu and a scholarship from China Scholarship Council to Chong Zhang.
REFERENCES
- Alabadi D., Yanovsky M. J., Mas P., Harmer S. L., Kay S. A. (2002). Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr. Biol. 12 757–761 10.1016/S0960-9822(02)00815-1 [DOI] [PubMed] [Google Scholar]
- Bhardwaj V., Meier S., Petersen L. N., Ingle R. A., Roden L. C. (2011). Defence responses of Arabidopsis thaliana to infection by Pseudomonas syringae are regulated by the circadian clock. PLoS ONE 6:e26968 10.1371/journal.pone.0026968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Zheng Z., Huang J., Lai Z., Fan B. (2009). Biosynthesis of salicylic acid in plants. Plant Signal. Behav. 4 493–496 10.4161/psb.4.6.8392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern M. S., Fitzgerald H. A., Yadav R. C., Canlas P. E., Dong X., Ronald P. C. (2001). Evidence for a disease-resistance pathway in rice similar to the NPR1-mediated signaling pathway in Arabidopsis. Plant J. 27 101–113 10.1046/j.1365-313x.2001.01070.x [DOI] [PubMed] [Google Scholar]
- Cubero B., Nakagawa Y., Jiang X., Miura K., Li F., Raghothama K. G., et al. (2009). The phosphate transporter PHT4; 6 Is a determinant of salt tolerance that is localized to the Golgi apparatus of Arabidopsis. Mol. Plant 2 535–552 10.1093/mp/ssp013 [DOI] [PubMed] [Google Scholar]
- Dempsey D. A., Vlot A. C., Wildermuth M. C., Klessig D. F. (2011). Salicylic acid biosynthesis and metabolism. Arabidopsis Book 9:e0156 10.1199/tab.0156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Millar A. J., Davis A. M., Davis S. J. (2007). TIME FOR COFFEE encodes a nuclear regulator in the Arabidopsis thaliana circadian clock. Plant Cell 19 1522–1536 10.1105/tpc.106.047241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd A. N., Salathia N., Hall A., Kevei E., Toth R., Nagy F., et al. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309 630–633 10.1126/science.1115581 [DOI] [PubMed] [Google Scholar]
- Dong M. A., Farre E. M., Thomashow M. F. (2011). Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108 7241–7246 10.1073/pnas.1103741108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X. (2004). NPR1, all things considered. Curr. Opin. Plant Biol. 7 547–552 10.1016/j.pbi.2004.07.005 [DOI] [PubMed] [Google Scholar]
- Ekengren S., Liu Y., Schiff M., Dinesh-Kumar S., Martin G. (2003). Two MAPK cascades, NPR1, and TGA transcription factors play a role in Pto-mediated disease resistance in tomato. Plant J. 36 905–917 10.1046/j.1365-313X.2003.01944.x [DOI] [PubMed] [Google Scholar]
- Falk A., Feys B. J., Frost L. N., Jones J. D., Daniels M. J., Parker J. E. (1999). EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. U.S.A. 96 3292–3297 10.1073/pnas.96.6.3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald H. A., Chern M. S., Navarre R., Ronald P. C. (2004). Overexpression of (At)NPR1 in rice leads to a BTH- and environment-induced lesion-mimic/cell death phenotype. Mol. Plant Microbe Interact. 17 140–151 10.1094/MPMI.2004.17.2.140 [DOI] [PubMed] [Google Scholar]
- Fragniere C., Serrano M., Abou-Mansour E., Metraux J. P., L’haridon F. (2011). Salicylic acid and its location in response to biotic and abiotic stress. FEBS Lett. 585 1847–1852 10.1016/j.febslet.2011.04.039 [DOI] [PubMed] [Google Scholar]
- Friedrich L., Vernooij B., Gaffney T., Morse A., Ryals J. (1995). Characterization of tobacco plants expressing a bacterial salicylate hydroxylase gene. Plant Mol. Biol. 29 959–968 10.1007/BF00014969 [DOI] [PubMed] [Google Scholar]
- Fu Z. Q., Dong X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64 839–863 10.1146/annurev-arplant-042811-105606 [DOI] [PubMed] [Google Scholar]
- Fu Z. Q., Yan S., Saleh A., Wang W., Ruble J., Oka N., et al. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486 228–232 10.1038/nature11162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez E., Solano R., Rubio V., Leyva A., Paz-Ares J. (2005). PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. Plant Cell 17 3500–3512 10.1105/tpc.105.036640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf A., Schlereth A., Stitt M., Smith A. M. (2010). Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc. Natl. Acad. Sci. U.S.A. 107 9458–9463 10.1073/pnas.0914299107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R. M., Tingay S., Wang Z. Y., Tobin E. M. (2002). Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 129 576–584 10.1104/pp.004374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R. M., Tobin E. M. (2002). The role of CCA1 and LHY in the plant circadian clock. Dev. Cell 2 516–518 10.1016/S1534-5807(02)00184-3 [DOI] [PubMed] [Google Scholar]
- Guo B., Irigoyen S., Fowler T. B., Versaw W. K. (2008a). Differential expression and phylogenetic analysis suggest specialization of plastid-localized members of the PHT4 phosphate transporter family for photosynthetic and heterotrophic tissues. Plant Signal. Behav. 3 784–790 10.4161/psb.3.10.6666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B., Jin Y., Wussler C., Blancaflor E. B., Motes C. M., Versaw W. K. (2008b). Functional analysis of the Arabidopsis PHT4 family of intracellular phosphate transporters. New Phytol. 177 889–898 10.1111/j.1469-8137.2007.02331.x [DOI] [PubMed] [Google Scholar]
- Hall A., Bastow R. M., Davis S. J., Hanano S., Mcwatters H. G., Hibberd V., et al. (2003). The TIME FOR COFFEE gene maintains the amplitude and timing of Arabidopsis circadian clocks. Plant Cell 15 2719–2729 10.1105/tpc.013730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond-Kosack K. E., Jones J. D. (1996). Resistance gene-dependent plant defense responses. Plant Cell 8 1773–1791 10.1105/tpc.8.10.1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassler S., Lemke L., Jung B., Mohlmann T., Kruger F., Schumacher K., et al. (2012). Lack of the Golgi phosphate transporter PHT4;6 causes strong developmental defects, constitutively activated disease resistance mechanisms and altered intracellular phosphate compartmentation in Arabidopsis. Plant J. 72 732–744 10.1111/j.1365-313X.2012.05106.x [DOI] [PubMed] [Google Scholar]
- Irigoyen S., Karlsson P. M., Kuruvilla J., Spetea C., Versaw W. K. (2011). The sink-specific plastidic phosphate transporter PHT4;2 influences starch accumulation and leaf size in Arabidopsis. Plant Physiol. 157 1765–1777 10.1104/pp.111.181925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J. B., Jin Y. H., Lee J., Miura K., Yoo C. Y., Kim W. Y., et al. (2008). The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J. 53 530–540 10.1111/j.1365-313X.2007.03359.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage D., Tootle T. L., Reuber T. L., Frost L. N., Feys B. J., Parker J. E., et al. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. U.S.A. 96 13583–13588 10.1073/pnas.96.23.13583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan A. S., Varadarajan D. K., Mukatira U. T., D’Urzo M. P., Damsz B., Raghothama K. G. (2002). Regulated expression of Arabidopsis phosphate transporters. Plant Physiol. 130 221–233 10.1104/pp.020007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Nam J., Park H. C., Na G., Miura K., Jin J. B., et al. (2007). Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J. 49 79–90 10.1111/j.1365-313X.2006.02947.x [DOI] [PubMed] [Google Scholar]
- Lin W. C., Lu C. F., Wu J. W., Cheng M. L., Lin Y. M., Yang N. S., et al. (2004). Transgenic tomato plants expressing the Arabidopsis NPR1 gene display enhanced resistance to a spectrum of fungal and bacterial diseases. Transgenic Res. 13 567–581 10.1007/s11248-004-2375-9 [DOI] [PubMed] [Google Scholar]
- Lu H. (2009). Dissection of salicylic acid-mediated defense signaling networks. Plant Signal. Behav. 4 713–717 10.4161/psb.4.8.9173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Rate D. N., Song J. T., Greenberg J. T. (2003). ACD6, a novel ankyrin protein, is a regulator and an effector of salicylic acid signaling in the Arabidopsis defense response. Plant Cell 15 2408–2420 10.1105/tpc.015412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Salimian S., Gamelin E., Wang G., Fedorowski J., Lacourse W., et al. (2009a). Genetic analysis of acd6-1 reveals complex defense networks and leads to identification of novel defense genes in Arabidopsis. Plant J. 58 401–412 10.1111/j.1365-313X.2009.03791.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S. X., Knowles S. M., Andronis C., Ong M. S., Tobin E. M. (2009b). CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL function synergistically in the circadian clock of Arabidopsis. Plant Physiol. 150 834–843 10.1104/pp.108.133272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makandar R., Essig J. S., Schapaugh M. A., Trick H. N., Shah J. (2006). Genetically engineered resistance to Fusarium head blight in wheat by expression of Arabidopsis NPR1. Mol. Plant Microbe Interact. 19 123–129 10.1094/MPMI-19-0123 [DOI] [PubMed] [Google Scholar]
- Malnoy M., Jin Q., Borejsza-Wysocka E. E., He S. Y., Aldwinckle H. S. (2007). Overexpression of the apple MpNPR1 gene confers increased disease resistance in Malus × domestica. Mol. Plant Microbe Interact. 20 1568–1580 10.1094/MPMI-20-12-1568 [DOI] [PubMed] [Google Scholar]
- Michael T. P., McClung C. R. (2002). Phase-specific circadian clock regulatory elements in Arabidopsis. Plant Physiol. 130 627–638 10.1104/pp.004929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael T. P., Salome P. A., Yu H. J., Spencer T. R., Sharp E. L., Mcpeek M. A. (2003). Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302 1049–1053 10.1126/science.1082971 [DOI] [PubMed] [Google Scholar]
- Miura K., Rus A., Sharkhuu A., Yokoi S., Karthikeyan A. S., Raghothama K. G., et al. (2005). The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc. Natl. Acad. Sci. U.S.A. 102 7760–7765 10.1073/pnas.0500778102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T., Wheatley K., Hanzawa Y., Wright L., Mizoguchi M., Song H. R., et al. (2002). LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2 629–641 10.1016/S1534-5807(02)00170-3 [DOI] [PubMed] [Google Scholar]
- Mockler T. C., Michael T. P., Priest H. D., Shen R., Sullivan C. M., Givan S. A., et al. (2007). The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb. Symp. Quant. Biol. 72 353–363 10.1101/sqb.2007.72.006 [DOI] [PubMed] [Google Scholar]
- Mudge S. R., Rae A. L., Diatloff E., Smith F. W. (2002). Expression analysis suggests novel roles for members of the PHT1 family of phosphate transporters in Arabidopsis. Plant J. 31 341–353 10.1046/j.1365-313X.2002.01356.x [DOI] [PubMed] [Google Scholar]
- Nawrath C., Heck S., Parinthawong N., Metraux J. P. (2002). EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14 275–286 10.1105/tpc.010376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C., Metraux J. P. (1999). Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng G., Seabolt S., Zhang C., Salimian S., Watkins T. A., Lu H. (2011). Genetic dissection of salicylic acid-mediated defense signaling networks in Arabidopsis. Genetics 189 851–859 10.1534/genetics.111.132332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z., Kim E. D., Ha M., Lackey E., Liu J., Zhang Y., et al. (2009). Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457 327–331 10.1038/nature07523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavon L. R., Lundh F., Lundin B., Mishra A., Persson B. L., Spetea C. (2008). Arabidopsis ANTR1 is a thylakoid Na+-dependent phosphate transporter: functional characterization in Escherichia coli. J. Biol. Chem. 283 13520–13527 10.1074/jbc.M709371200 [DOI] [PubMed] [Google Scholar]
- Poirier Y., Bucher M. (2002). Phosphate transport and homeostasis in Arabidopsis. Arabidopsis Book 1:e0024 10.1199/tab.0024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilis J., Penas G., Messeguer J., Brugidou C., San Segundo B. (2008). The Arabidopsis AtNPR1 inversely modulates defense responses against fungal, bacterial, or viral pathogens while conferring hypersensitivity to abiotic stresses in transgenic rice. Mol. Plant Microbe Interact. 21 1215–1231 10.1094/MPMI-21-9-1215 [DOI] [PubMed] [Google Scholar]
- Rate D. N., Cuenca J. V., Bowman G. R., Guttman D. S., Greenberg J. T. (1999). The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell 11 1695–1708 10.1105/tpc.11.9.1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden L. C., Ingle R. A. (2009). Lights, rhythms, infection: the role of light and the circadian clock in determining the outcome of plant-pathogen interactions. Plant Cell 21 2546–2552 10.1105/tpc.109.069922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth C., Menzel G., Petetot J. M., Rochat-Hacker S., Poirier Y. (2004). Characterization of a protein of the plastid inner envelope having homology to animal inorganic phosphate, chloride and organic-anion transporters. Planta 218 406–416 10.1007/s00425-003-1121-5 [DOI] [PubMed] [Google Scholar]
- Rubio V., Linhares F., Solano R., Martin A. C., Iglesias J., Leyva A., et al. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15 2122–2133 10.1101/gad.204401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals J. A., Neuenschwander U. H., Willits M. G., Molina A., Steiner H. Y., et al. (1996). Systemic acquired resistance. Plant Cell 8 1809–1819 10.1105/tpc.8.10.1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu D., Tasma I. M., Frasch R., Bhattacharyya M. K. (2009). Systemic acquired resistance in soybean is regulated by two proteins, orthologous to Arabidopsis NPR1. BMC Plant Biol. 9:105 10.1186/1471-2229-9-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauerbrunn N., Schlaich N. L. (2004). PCC1: a merging point for pathogen defence and circadian signalling in Arabidopsis. Planta 218 552–561 10.1007/s00425-003-1143-z [DOI] [PubMed] [Google Scholar]
- Serrano M., Wang B., Aryal B., Garcion C., Abou-Mansour E., Heck S., et al. (2013). Export of salicylic acid from the chloroplast requires the multidrug and toxin extrusion-like transporter EDS5. Plant Physiol. 162 1815–1821 10.1104/pp.113.218156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H., Shin H. S., Dewbre G. R., Harrison M. J. (2004). Phosphate transport in Arabidopsis: Pht1; 1 and Pht1; 4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 39 629–642 10.1111/j.1365-313X.2004.02161.x [DOI] [PubMed] [Google Scholar]
- Shin J., Heidrich K., Sanchez-Villarreal A., Parker J. E., Davis S. J. (2012). TIME FOR COFFEE represses accumulation of the MYC2 transcription factor to provide time-of-day regulation of jasmonate signaling in Arabidopsis. Plant Cell 24 2470–2482 10.1105/tpc.111.095430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. T., Lu H., Mcdowell J. M., Greenberg J. T. (2004). A key role for ALD1 in activation of local and systemic defenses in Arabidopsis. Plant J. 40 200–212 10.1111/j.1365-313X.2004.02200.x [DOI] [PubMed] [Google Scholar]
- Strawn M. A., Marr S. K., Inoue K., Inada N., Zubieta C., Wildermuth M. C. (2007). Arabidopsis isochorismate synthase functional in pathogen-induced salicylate biosynthesis exhibits properties consistent with a role in diverse stress responses. J. Biol. Chem. 282 5919–5933 10.1074/jbc.M605193200 [DOI] [PubMed] [Google Scholar]
- Todesco M., Balasubramanian S., Hu T. T., Traw M. B., Horton M., Epple P., et al. (2010). Natural allelic variation underlying a major fitness trade-off in Arabidopsis thaliana. Nature 465 632–636 10.1038/nature09083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M., Kim S. T., Chae E., Bomblies K., Zaidem M., Smith L. M., et al. (2014). Activation of the Arabidopsis thaliana immune system by combinations of common ACD6 alleles. PLoS Genet. 10:e1004459 10.1371/journal.pgen.1004459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Sato M., Glazebrook J., Cohen J. D., Katagiri F. (2008). Interplay between MAMP-triggered and SA-mediated defense responses. Plant J. 53 763–775 10.1111/j.1365-313X.2007.03369.x [DOI] [PubMed] [Google Scholar]
- Versaw W. K., Harrison M. J. (2002). A chloroplast phosphate transporter, PHT2; 1, influences allocation of phosphate within the plant and phosphate-starvation responses. Plant Cell 14 1751–1766 10.1105/tpc.002220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. F., Seabolt S., Hamdoun S., Ng G., Park J., Lu H. (2011a). Multiple roles of WIN3 in regulating disease resistance, cell death, and flowering time in Arabidopsis. Plant Physiol. 156 1508–1519 10.1104/pp.111.176776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. Y., Shi J. L., Ng G., Battle S. L., Zhang C., Lu H. (2011b). Circadian clock-regulated phosphate transporter PHT4; 1 plays an important role in Arabidopsis defense. Mol. Plant 4 516–526 10.1093/mp/ssr016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Barnaby J. Y., Tada Y., Li H., Tor M., Caldelari D., et al. (2011c). Timing of plant immune responses by a central circadian regulator. Nature 470 110–114 10.1038/nature09766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. X., Yamanouchi U., Katayose Y., Sasaki T., Yano M. (2001). Expression of the Pib rice-blast-resistance gene family is up-regulated by environmental conditions favouring infection and by chemical signals that trigger secondary plant defences. Plant Mol. Biol. 47 653–661 10.1023/A:1012457113700 [DOI] [PubMed] [Google Scholar]
- Wang Z. Y., Tobin E. M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93 1207–1217 10.1016/S0092-8674(00)81464-6 [DOI] [PubMed] [Google Scholar]
- Weyman P. D., Pan Z., Feng Q., Gilchrist D. G., Bostock R. M. (2006). A circadian rhythm-regulated tomato gene is induced by Arachidonic acid and Phythophthora infestans infection. Plant Physiol. 140 235–248 10.1104/pp.105.068874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth M. C., Dewdney J., Wu G., Ausubel F. M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414 562–565 10.1038/35107108 [DOI] [PubMed] [Google Scholar]
- Wu Y., Zhang D., Chu J. Y., Boyle P., Wang Y., Brindle I. D., et al. (2012). The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 1 639–647 10.1016/j.celrep.2012.05.008 [DOI] [PubMed] [Google Scholar]
- Yamasaki K., Motomura Y., Yagi Y., Nomura H., Kikuchi S., Nakai M., et al. (2013). Chloroplast envelope localization of EDS5, an essential factor for salicylic acid biosynthesis in Arabidopsis thaliana. Plant Signal. Behav. 8:e23603 10.4161/psb.23603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S., Dong X. (2014). Perception of the plant immune signal salicylic acid. Curr. Opin. Plant Biol. 20C, 64–68 10.1016/j.pbi.2014.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Zhong S., Li Q., Zhu Z., Lou Y., Wang L., et al. (2007). Functional analysis of rice NPR1-like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnol. J. 5 313–324 10.1111/j.1467-7652.2007.00243.x [DOI] [PubMed] [Google Scholar]
- Zhang C., Xie Q., Anderson R. G., Ng G., Seitz N. C., Peterson T., et al. (2013). Crosstalk between the circadian clock and innate immunity in Arabidopsis. PLoS Pathog. 9:e1003370 10.1371/journal.ppat.1003370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Francis M. I., Dawson W. O., Graham J. H., Orbovic V., Triplett E. W., et al. (2010). Over-expression of the Arabidopsis NPR1 gene in citrus increases resistance to citrus canker. Eur. J. Plant Pathol. 128 91–100 10.1007/s10658-010-9633-x [DOI] [Google Scholar]