Abstract
Several studies have investigated the association between abnormal microRNA-10b expression and the risk of various developing cancers, but the results are inconsistent. We searched all publications addressing the level of microRNA-10b expression in cancer cases and noncancerous controls (Accessed: August 2014). Thirty-six studies on 14 types of cancer were included. Among them, 25 studies were subjected to the meta-analysis with a vote-counting strategy, 13 studies were estimated using odds ratio (OR) and diagnostic accuracy, and 2 studies were assessed by both methods. It was found that vestibular schwannomas ranked first among the reported cancer types with up-regulated microRNA-10b expression; melanoma ranked first among the reported cancer types with down-regulated microRNA-10b expression; while breast cancer and hepatocellular cancer presented inconsistent microRNA-10b regulation. Of 13 included studies calculated for OR and diagnostic accuracy, it was shown that high-expression of microRNA-10b could be significantly associated with cancer risk (OR = 32.80, 95% CI: 11.90–90.37, P<0.0001), and the area under the summary receiver operating characteristic (SROC) curve for microRNA-10b high-expression in the diagnosis of cancer is 0.81, which suggested that high-expression of microRNA-10b can predict worse outcomes in some types of cancer and the regular monitoring of miR-10b expression might be useful in the clinical practice.
MicroRNAs (miRNAs) are short (average of 22 nucleotides), noncoding single-stranded RNAs that play an important role in many physiological processes through the post-transcriptional regulation of protein coding genes by binding to the 3′UTR of target mRNAs1. Meanwhile, deregulated miRNAs have been found in different types of human diseases and cancers. In particular, recent evidence has shown that altered expression of miRNA species is well established in a variety of pathological processes and cancers and that they are involved in tumor development, progression, and metastasis by targeting the mRNA of oncogenes or cancer suppressor genes. In fact, abnormal expression of specific miRNAs has been shown to correlate with a variety of cancers. Typically, miR-21 expression is up-regulated in glioblastoma, hepatocellular carcinoma, leukemia, ovarian cancer, breast cancer, colon cancer, lung cancer, pancreas cancer, prostate cancer and stomach cancer2,3,4. Moreover, miRNA expression level and function depend highly on the cellular context in which they are studied, including tissue type. For example, miR-183 family members were found to be consistently either up- or down-regulated depending on the type of cancer5. The published studies analyzed here suggested that miR-10b was abnormally expressed through either up- or down-regulation in various cancers, depending on their target genes. Several studies have demonstrated that miR-10b was highly expressed in esophageal cancer, oral cancer, lung cancer, vestibular schwannoma, pituitary adenoma, prostate cancer and glioma cancer6,7,8,9,10,11,12,13,14,15. However, down-regulation of miR-10b expression was observed in clear cell renal cell cancer (ccRCC), renal cancer, colon cancer, endometrioid endometrial carcinoma (EEC) and melanoma16,17,18,19,20,21. Other studies have reported altered miR-10b expression levels in breast cancer and hepatocellular cancer (HCC)9,22,23,24,25,26,27.
Because the oncogenic or tumor suppressive properties of miR-10b are inconsistent and often ambiguous, we conducted a systematic review and meta-analysis from all eligible studies to evaluate a more precise association between abnormal miR-10b expression and the risk of developing multiple, independent types of cancer. In the future, we want to comprehensively and quantitatively summarize evidence for the use of miR-10b as a biomarker of different clinical characteristics of cancer.
Results
Eligible studies and study characteristics
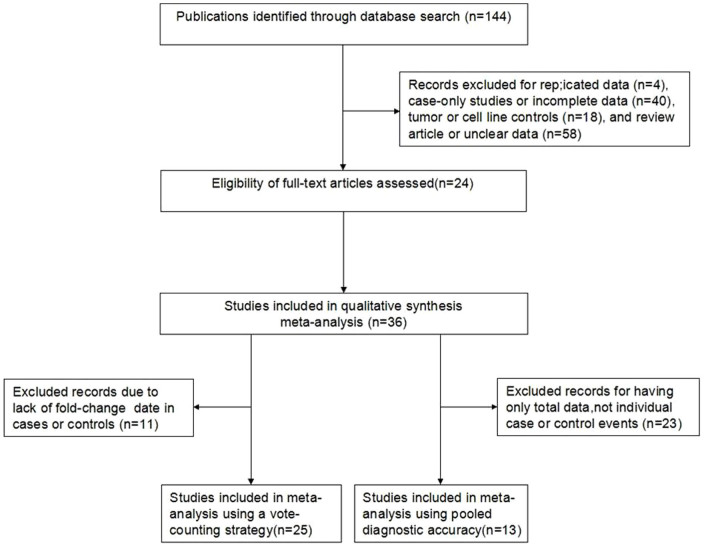
Based on the search criteria, the initial search returned a total of 144 publications, 120 of which were excluded due to ineligibility criteria (4 had multiple publication data, 40 were case-only studies or had incomplete data, 18 were controls from tumor or cell lines, and 58 had unclear data or were reviews). The 24 selected articles (Table S1) included 36 studies with 1460 patients from the United States, Germany, Ireland, Switzerland, China, Japan, Singapore, Spain, Greece, Italy and Chinese Taiwan. 25 of the studies were subjected to the meta-analysis with the vote-counting strategy6,9,11,13,14,15,16,17,18,19,20,21,22,23,24,25,28,29. Thirteen of the studies were estimated using odds ratio and diagnostic accuracy6,7,9,11,12,26,27,29. Two of the studies were subjected to both methods6,29. A flow diagram of the study selection process is shown in Figure 1. The publication dates of the included articles ranged from 2005 to 2014, and they were all retrospective in design. The types of cancer in these studies included colon, lung, breast, oral, glioma, esophageal, colorectal, HCC, vestibular schwannoma, pituitary adenoma, prostate, renal, EEC and melanoma. Thirty studies used quantitative real-time polymerase chain reaction (qRT-PCR) assays to measure the expression level of miR-10b, and 9 studies used microarray assays. Two articles investigated 5 independent studies as a sample/control set9,16, and 4 articles investigated 2 independent studies as a sample/control set10,14,17,28 (Table 1).
Figure 1. Flow chart depicting the study selection process.
Table 1. Thirty-six human cancer microRNA-10b expression studies (cancer cases versus noncancerous controls).
| Cancer type | Reference | Region | Samples (cancer/noncancerous) | Validation | Regulated features |
|---|---|---|---|---|---|
| HCC | Li QJ, 2012 | China | 41(34/7) | qRT-PCR | Up |
| HCC | Shen J, 2012 | China Taiwan | 74(37/37) | qRT-PCR | Up |
| Oral cancer | Lu YC, 2012 | China Taiwan | 97(54/43) | qRT-PCR | Up |
| Breast cancer | Ma L, 2007 | USA | 24(23/1) | qRT-PCR | Up |
| Breast cancer | Zhao FL, 2012) | China | 181(122/59) | qRT-PCR | Up |
| Breast cancer brain metastasis | Teplyuk NM, 2012 | USA | 31(16/15) | qRT-PCR | Up |
| Breast cancer leptomeningeal metastasis | Teplyuk NM, 2012 | USA | 41(26/15) | qRT-PCR | Up |
| Breast cancer | Chan M, 2013 | Singapore | 54(32/22) | qRT-PCR | Up |
| Lung cancer brain metastasis | Teplyuk NM, 2012 | USA | 43(28/15) | qRT-PCR | Up |
| Lung cancer leptomeningeal metastasis | Teplyuk NM, 2012 | USA | 19(4/15) | qRT-PCR | Up |
| NSCLC | Roth C, 2011 | Germany | 47(19/28) | qRT-PCR | Up |
| SCLC | Roth C, 2011 | Germany | 36(8/28) | qRT-PCR | Up |
| NSCLC | Cui EH, 2013 | China | 520(260/260) | qRT-PCR | Up |
| NSCLC with EGFR mutation | Shen Y, 2013 | China | 92(60/32) | qRT-PCR | Up |
| NSCLC without EGFR mutation | Shen Y, 2013 | China | 100(68/32) | qRT-PCR | Up |
| Esophageal cancer | Xie Z, 2013 | China | 58(39/19) | qRT-PCR +microarray | Up |
| Esophageal cancer | Tian YY, 2010 | China | 80(40/40) | qRT-PCR | Up |
| Glioblastoma multiforme | Teplyuk NM, 2012 | USA | 34(19/15) | qRT-PCR | Up |
| GBM | Guessous F, 2013 | USA | 25(20/5) | qRT-PCR | Up |
| Glioma | Sasayama T, 2009 | Japan | 49(43/6) | qRT-PCR | Up |
| Vestibular schwannomas | Torres-Martin M, 2013 | Spain | 10(7/3) | qRT-PCR +microarray | Up |
| Non-functioning pituitary adenomas | Liang S, 2013 | China | 12(10/2) | microarray | Up |
| Gonadotropin-secreting pituitary adenomas | Liang S, 2013 | China | 12(10/2) | microarray | Up |
| Prostate cancer | Walter BA,2013 | USA | 50(40/10) | microarray | Up |
| Prostate cancer | Heneghan HM, 2010 | Ireland | 83(20/63) | qRT-PCR | Normal range |
| Breast cancer | Heneghan HM, 2010 | Ireland | 146(83/63) | qRT-PCR | Normal range |
| HCC | Zaravinos A, 2012 | Greece | 77(56/21) | qRT-PCR | Down |
| EEC | Tsukamoto O, 2014 | Japan | 42(28/14) | qRT-PCR | Down |
| Colon cancer | Pizzini S, 2013 | Italy | 54(31/23) | microarray | Down |
| Colon cancer liver metastases | Pizzini S, 2013 | Italy | 47(24/23) | microarray | Down |
| Colon cancer | Heneghan HM, 2010 | Ireland | 93(30/63) | qRT-PCR | Down |
| Melanoma | Heneghan HM, 2010 | Ireland | 73(10/63) | qRT-PCR | Down |
| Renal cancer | Heneghan HM, 2010 | Ireland | 83(20/63) | qRT-PCR | Down |
| Breast cancer | Iorio MV, 2005 | Italy | 86(76/10) | qRT-PCR | Down |
| ccRCC | Wotschofsky Z, 2012 | Germany | 57(35/22) | qRT-PCR +microarray | Down |
| ccRCC | Wu X, 2012 | USA | 38(28/10) | microarray | Down |
Abbreviations: HCC: hepatocellular carcinoma; NSCLC: non-small cell lung cancer; SCLC: small cell lung cancer; ccRCC: clear cell renal cell carcinoma; GBM: glioblastoma; EEC: endometrioid endometrial carcinoma.
Differentially expressed miR-10b in human cancers
In a panel of miR-10b expression analyses, miR-10b expression was reported to be consistently up-regulated among ten types of cancer. Vestibular schwannomas were reported to rank first in one study with an average fold-change (FC) of 269.19. Esophageal cancer, HCC, prostate cancer, oral cancer and breast cancer were also reported in one study, with average FCs of 58.70, 17.10, 13.88, 4, and 1.17, respectively. In two studies, glioma and pituitary adenomas were reported to have average FCs of 33.86 and 27.24. In five studies, lung cancer was reported to have an average FC of 3.64. Ten studies showed that miR-10b was down-regulated among seven cancers. Melanoma and HCC were reported to rank first in one study with an average FC of -3.13. Renal cancer, EEC and breast cancer were also mentioned in one study to have an average FC of -1.53, -1.27, and -1.53, respectively. CcRCC was reported in two studies to have an average FC of -2.08. Colon cancer, reported in three studies, had an average FC of -1.94. As mentioned above, miR-10b was inconsistently expressed in breast cancer or HCC cases when compared to noncancerous/normal controls (Table 2).
Table 2. Vote-counting strategy of abnormal miR-10b expression based on tumor type.
| Subset of studies with fold-change | |||||||
|---|---|---|---|---|---|---|---|
| Direction of expression | Cancer type | No. of studies (reference) | Total sample size | No. of studies | Total sample size | Mean fold-change | Range |
| Up-regulated | HCC | 2 (Shen J, 2012;Li QJ, 2012) | 115 | 1 | 74 | 17.10 | - |
| Esophageal cancer | 2 (Tian YY, 2010;Xie Z, 2013) | 138 | 1 | 58 | 58.70 | - | |
| Oral cancer | 1(Lu YC, 2012) | 97 | 1 | 97 | 4 | - | |
| Breast cancer | 5(Ma L, 2007; Zhao FL, 2012; Teplyuk NM, 2012; Chan M, 2013) | 316 | 1 | 54 | 1.77 | - | |
| Lung cancer | 7 (Teplyuk NM, 2012; Roth C, 2011; Cui EH, 2013; Shen Y, 2013) | 805 | 5 | 735 | 3.64 | 0.77:6.50 | |
| Glioma | 3 (Sasayama T, 2009; Teplyuk NM,2012; Guessous F, 2013) | 108 | 2 | 74 | 33.86 | 0.08:364 | |
| Vestibular schwannomas | 1(Torres-Martin M, 2013) | 10 | 1 | 10 | 269.19 | - | |
| Pituitary adenomas | 2(Liang S, 2013) | 22 | 2 | 22 | 27.24 | 5.74:48.73 | |
| Prostate Cancer | 1(Walter BA,2013) | 50 | 1 | 50 | 13.88 | - | |
| Down-regulated | Renal cancer | 1 (Heneghan HM, 2010) | 83 | 1 | 83 | −1.53 | - |
| Colon cancer | 3 (Heneghan HM, 2010, Pizzini S, 2013) | 194 | 3 | 194 | −1.94 | −1.64: −2.81 | |
| Melanoma | 1 (Heneghan HM, 2010) | 73 | 1 | 73 | −3.13 | - | |
| Breast cancer | 1 (Iorio MV, 2005) | 86 | 1 | 86 | −1.53 | - | |
| HCC | 1 (Zaravinos A, 2012) | 77 | 1 | 77 | −3.13 | - | |
| EEC | 1 (Tsukamoto O, 2014) | 42 | 1 | 42 | −1.27 | - | |
| ccRCC | 2 (Wotschofsky Z, 2012; Wu X,2012) | 95 | 2 | 95 | −2.08 | −1.19: −5.55 | |
Abbreviations: HCC: hepatocellular carcinoma; ccRCC: clear cell renal cell carcinomas; NSCLC: non-small cell lung cancer; EEC: endometrioid endometrial carcinoma.
Correlation between miR-10b expression and ORs
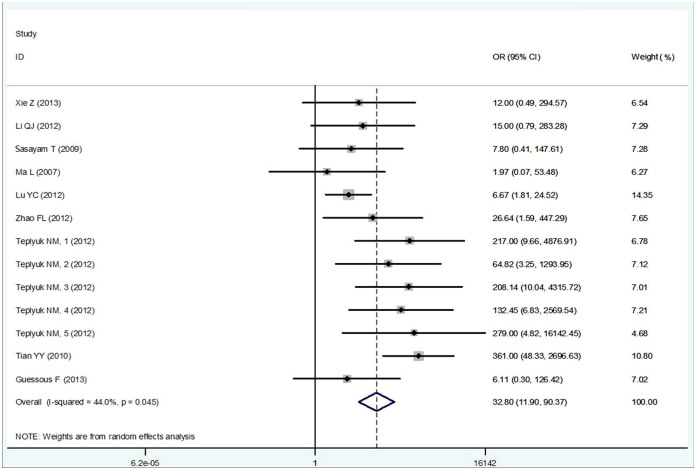
The primary results of this meta-analysis are shown in Table 3. We performed an overall analysis of the data from studies containing high-expression of miR-10b and ORs from a variety of cancers. The studies were found to have moderate heterogeneity (I2 = 44.0%, P = 0.045), so a random effects model was applied to calculate a pooled OR and its 95% confidence interval (CI) (32.80, 95% CI: 11.90–90.37, P<0.0001), which was statistically significant (Figure 2). Then, subgroup analysis by cancer type showed significant association between the high-expression of miR-10b and various types of cancer. Low heterogeneities were found among nor-digestive system cancer (I2 = 18.7%, P = 0.276); thus, a fixed effects model was applied to calculate OR (33.97, 95% CI: 11.98–96.32, P = 0.000). High heterogeneity was observed in digestive system cancer (I2 = 72.3%, P = 0.013), and so a random effects model was applied to calculate the OR (26.37, 95% CI: 3.21–216.41, P = 0.002). No significant heterogeneity existed among the studies evaluating OR for miR-10b. The result of subgroup analysis by sample source was also significant. Low and moderate heterogeneities were found from circulating based (I2 = 0%, P = 0.633) and tissue based (I2 = 37.6%, P = 0.108) miRNAs, respectively. Thus, a fixed effects model was applied to calculate ORs (circulating based: OR = 10.711, 95% CI: 3.484–32.929, P<0.0001; tissue based: OR = 45.263, 95% CI: 17.490–117.136, P<0.0001). The pooled OR being greater than 1 indicates that high-expression of miR-10b may be significantly associated with the risk of cancer (Table 3).
Table 3. Pooled diagnostic accuracy.
| Cancer vs. Non-cancer | Tissue-based miRNA | Nor-tissue-based miRNA | Digestive system cancer | Nor-digestive system cancer | |
|---|---|---|---|---|---|
| No. of studies | 13 | 10 | 3 | 4 | 9 |
| Cancer Group (No./total No.) | 211/436 | 165/253 | 46/183 | 79/135 | 132/301 |
| Control Group (No./total No.) | 6/179 | 2/74 | 4/105 | 6/93 | 0/86 |
| OR(95% CI) | 32.80 (11.90–90.37)a | 10.71 (3.48–32.93) | 8.86 (2.93–26.88) | 26.37 (3.21– 216.41)a | 33.97 (11.98– 96.32) |
| Heterogeneityb (p) | 21.41 (0.05) | 14.43 (0.11) | 0.91 (0.63) | 10.82 (0.01) | 9.84 (0.28) |
| AUC (95% CI) | 0.98 (0.96–0.99) | 0.99 (0.98– 1.00) | 0.50 (0.09–0.91) | 0.93 (0.90– 0.95) | 0.50 (0.09– 0.91) |
| SEN (95% CI) | 0.98 (0.89–0.99) | 0.99 (0.76– 1.00) | - | 0.93 (0.84– 0.97) | - |
| Heterogeneityb (p) | 33.16 (0.00) | 16.29 (0.06) | - | 4.15 (0.26) | - |
| SPE (95% CI) | 0.62 (0.39–0.82) | 0.655 (0.36– 0.87) | - | 0.67 (0.30– 0.90) | - |
| Heterogeneityb (p) | 145.83 (0.00) | 113.01 (0.00) | - | 33.5 (0.00) | - |
| PLR(95% CI) | 2.63 (1.44–4.82) | 2.891 (1.29– 6.49) | - | 2.77 (1.00– 7.70) | - |
| Heterogeneityb (p) | 240.04 (0.00) | 242.68 (0.00) | 0.00 (1.0) | 34.61 (0.00) | 0.00 (1.0) |
| NLR(95% CI) | 0.02 (0.002–0.172) | 0.003 (0.00– 0.42) | - | 0.11 (0.04– 0.29) | - |
| Heterogeneityb (p) | 32.93 (0.00) | 35.66 (0.00) | 0.00 (1.0) | 4.61 (0.20) | 0.00 (1.0) |
| DOR(95% CI) | 133.145 (13.21–1341.87) | 1035.07 (7.52–1.4e+05) | - | 25.54 (4.15–157.01) | - |
| Heterogeneityb (p) | 14.537 (0.000) | 14.249 (0.00) | - | 6.24 (0.02) | - |
aSignificant heterogeneity: the random-effects model was chosen to summarize the result; bQ-value.
OR-odds ratio; AUC-area under the curve; SEN-sensitivity; SPE-specificity; PLR-positive likelihood ratio; NLR-negative likelihood ratio; DOR-diagnostic odds ratio.
Figure 2. Meta-analysis of the miR-10b high-expression odds ratio (OR) between cancer and noncancerous groups using the random-effects model.
Bars are the 95% CI of OR in patients versus controls. The areas of the squares are proportional to the weights used for combining the data. The center of the lozenge gives the combined OR. The OR was considered statistically significant if the 95% CI for the overall OR did not cross the value 1.
Meta-analysis was not performed on the cancer cases and noncancerous controls that had low-expression of miR-10b due to insufficient data from the searched studies.
Diagnostic accuracy
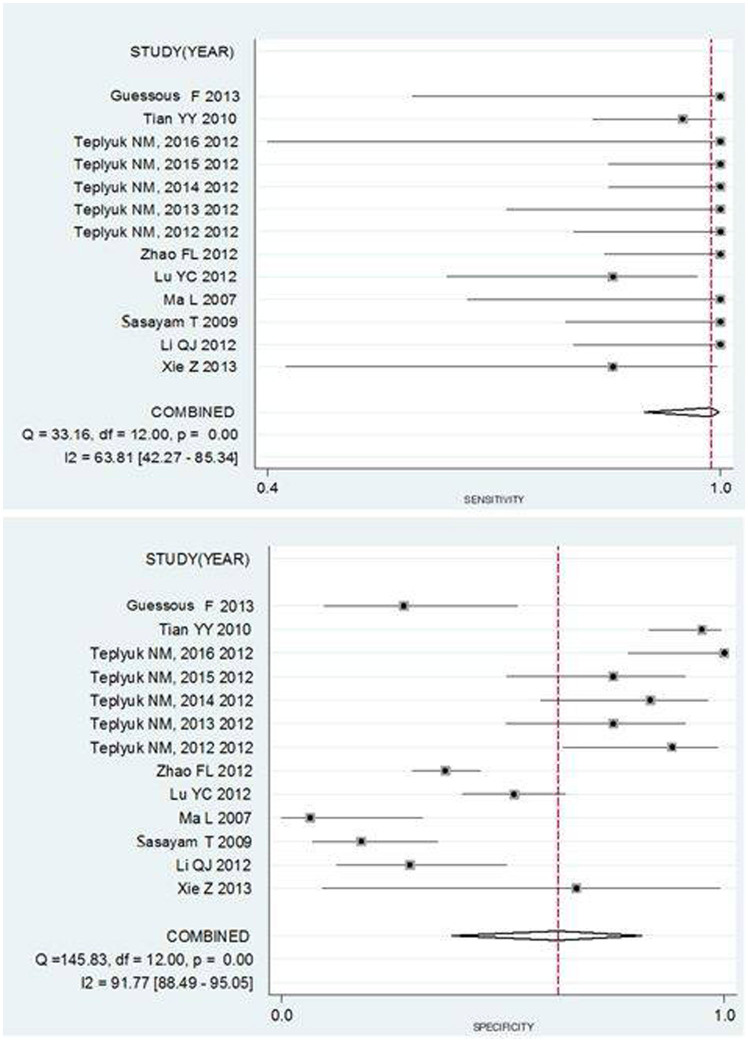
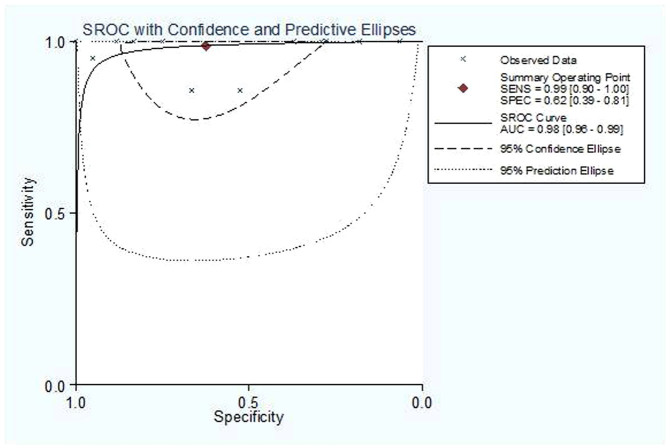
A graph shows a forest plot for the sensitivity and specificity of miR-10b assays in the diagnosis of cancer for 13 studies (Figure 3). Pooled results for the diagnostic accuracy are listed in Table 3. The sensitivity (SEN) was 0.988 (95% CI: 0.899–0.999), and the specificity (SPE) was 0.624 (95% CI: 0.386–0.815). The pooled positive likelihood ratio (PLR) was 2.630 (95% CI: 1.436–4.815), the negative likelihood ratio (NLR) was 0.020 (95% CI: 0.002–0.172), the diagnostic odds ratio (DOR) was 133.145 (95% CI: 13.211–1341.874) and the area under the SROC curve (AUC) value was 0.98 (95% CI: 0.96–0.99). These results indicate that the miR-10b assay could differentiate affected individuals from those without cancer. Chi-squared values of SEN 33.16 (p = 0.00), SPE 145.83 (p = 0.00), PLR 240.04 (p = 0.00), NLR 32.93 (p = 0.00) and DOR 14.537 (p = 0.000) all indicate that significant heterogeneity exists between studies. Our data also showed that the SROC curve is positioned near the desirable upper left corner of the graph; the red point shows the maximum joint sensitivity and specificity. The area under the curve was 0.98, indicating a high level of overall accuracy (Figure 4).
Figure 3. Forest plot estimating the sensitivity and specificity of miR-10b assays in the diagnosis of cancer from 13 studies.
The estimates of sensitivity and specificity from each study are shown as solid points. Error bars are 95% confidence intervals. Numbers indicate the referenced study listed in Table S.
Figure 4. Summary receiver operating characteristic (SROC) curve for miR-10b assays in the diagnosis of different types of cancer from the 13 included studies.
Solid circles represent each study included in the meta-analysis. The size of each study is indicated by the size of the solid circle. The regression SROC curve summarizes the overall diagnostic accuracy.
Subgroup analyses were also performed. Table 3 shows the pooled results for diagnostic accuracy in the different subgroups. The results indicate that the different sources of control subgroups and the different types of cancer subgroups were significantly divergent. Comparing the source of the control, circulating-based miRNA assays had a DOR of 8.86 (95% CI: 2.93–26.88) and an AUC of 0.50 (95% CI: 0.09–0.91), indicating lower accuracy, whereas tissue-based miRNA assays had a SEN of 0.99 (95% CI: 0.76–1.00), a SPE of 0.66 (95% CI: 0.36–0.87), a PLR of 2.89 (95% CI: 1.29–6.49), an NLR of 0.003 (95% CI: 0.01–0.43), a DOR of 1035.07 (95% CI: 7.52–1.4E+05), and an AUC of 0.99 (95% CI: 0.98–1.00), which demonstrates a higher level of accuracy. Comparing the assays studying different types of cancer, the digestive system cancer group had a higher level of accuracy: the SEN was 0.93 (95% CI: 0.84–0.97), the SPE was 0.67 (95% CI: 0.30–0.90), the PLR was 2.77 (95% CI: 1.00–7.70), the NLR was 0.11 (95% CI: 0.04–0.29), the DOR was 25.54 (95% CI: 4.15–157.01), and the AUC was 0.93 (95% CI: 0.90–0.95), indicating that miRNA-10b was more accurate at distinguishing patients with digestive system cancer from healthy people than patients with nor-digestive system cancer, which had an AUC of 0.50 (95% CI: 0.09–0.91). For meta-analysis of digestive system cancer, the chi-squared value of SEN was 4.15 (p = 0.26), SPE was 33.5 (p = 0.00), PLR was 34.61 (p = 0.00), NLR was 4.61 (p = 0.20) and DOR was 6.24 (p = 0.02), p>0.05 indicating that low significant heterogeneity exists between the studies.
To identify the accuracy of miR-10b for nor-digestive system cancer samples, we ran the algorithms using 447 digestive system cancer samples from the Sasayama, Ma, Zhao, Teplyuk, Guessous data set and obtained different source of samples, tissues or serum. The results suggested that the signatures had a high reproducibility. By detection of miR-10b up-regulate, we were able to stratify the digestive system cancer samples into low-and high-risk groups. We tested the predictive performance of miR-10b up-regulate in the five testing cohorts, which showed that 100% accuracy for high-risk groups in the testing sets containing 447 samples (Table 4). To identify the accuracy of miR-10b for digestive system cancer samples, we ran the same algorithms using the 228 digestive system cancer samples from the Xie, Li, Lu, Tian data set and obtained three source of samples, tissues, serum and saliva. The results suggested that the signatures still had a high reproducibility. By detection of miR-10b up-regulation, the digestive system cancer samples were stratified into low-and high-risk groups. The predictive performance of miR-10b up-regulation was evaluated in the four testing cohorts. Similar to nor-digestive system cancer samples, miR-10b also well performed in digestive system cancer samples, that is, 85.71–100% accuracy for high-risk groups in the testing sets containing 228 samples (Table 5).
Table 4. Accuracy of miR-10b up-regulated detection for nor-digestive system cancer samples.
| Studies | Number of samples | Low-risk group* | High-risk group** |
|---|---|---|---|
| Sasayama T | 49 | 18.18% | 100.00% |
| Ma L | 24 | 6.67% | 100.00% |
| Zhao FL | 181 | 37.11% | 100.00% |
| Teplyuk NM (1) | 34 | 39.58% | 100.00% |
| Teplyuk NM (2) | 31 | 49.04% | 100.00% |
| Teplyuk NM (3) | 41 | 58.50% | 100.00% |
| Teplyuk NM (4) | 43 | 67.96% | 100.00% |
| Teplyuk NM (5) | 19 | 77.43% | 100.00% |
| Fadila Guessous | 25 | 86.89% | 100.00% |
*Percentage of actual ‘good’ samples in the predicted low-risk group;
**Percentage of actual ‘bad’ samples in the predicted high-risk group.
Table 5. Accuracy of miR-10b up-regulated detection for digestive system cancer samples.
| Studies | Number of samples | Low-risk group* | High-risk group** |
|---|---|---|---|
| Xie Z | 10 | 66.67% | 85.71% |
| Li QJ | 41 | 29.17% | 100.00% |
| Lu YC | 97 | 52.63% | 85.71% |
| Tian YY | 80 | 95.00% | 95.00% |
*Percentage of actual ‘good’ samples in the predicted low-risk group;
**Percentage of actual ‘bad’ samples in the predicted high-risk group.
Assessment of publication bias and sensitivity analysis
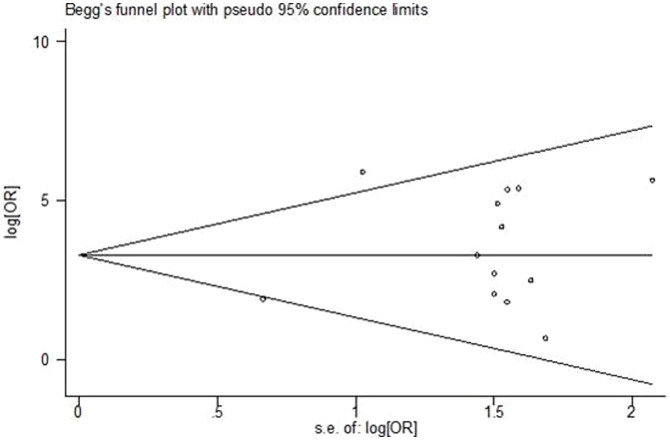
To assess publication bias in this study, the included studies were evaluated using Begg's funnel plots and the Egger's test. As shown in Figure 5, the Begg's funnel plots were almost symmetric, and the Egger's regression intercept was 0.205. Thus, there was no evidence for significant publication bias in this meta-analysis.
Figure 5. Begg's funnel plots of publication bias for studies evaluating OR of miR-10b expression in cancer.
Each point represents a separate study for the indicated association. Log [OR], natural logarithm of OR. The horizontal line represents the magnitude of the effect.
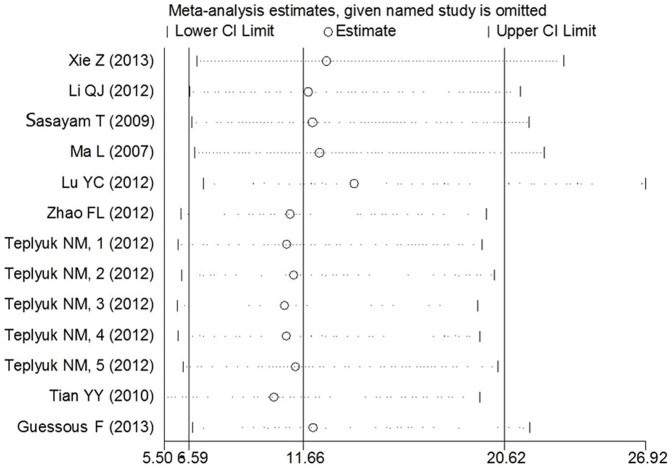
Sensitivity analysis was performed by omitting one study at a time to measure its individual effect on the pooled OR. As presented in Figure 6, no individual study dominantly influenced the overall OR.
Figure 6. Sensitivity analysis assessing the influence of individual studies on the pooled abnormal miR-10b expression OR in the cancer and noncancerous groups.
The results were computed by omitting each study in turn. Meta-analysis random-effects estimates (exponential form) were used. The two ends of the dotted lines represent the 95% CI.
Discussion
In the past decade, increasing evidence has demonstrated that aberrant expression of several miRNAs correlates with certain oncogenes or cancer suppressors30,31. miRNAs are also known to play important roles in the pathogenesis of cancer; up-regulation of oncogenic miRNAs or down-regulation of cancer suppressive miRNAs can contribute to tumorigenesis by altering many pathways, including cell cycle, angiogenesis, invasion and metastasis32,33.
To our knowledge, no meta-analysis has investigated the association between various cancers and their miR-10b expression with a consistent, statistically significant frequency in their expression level. Recent studies have shown that miR-10b is present in a variety of cancer types. Notably, miR-10b expression is elevated in glioma, liver cancer, pancreatic cancer, breast cancer, lung cancer, and esophageal cancer, among others6,7,8,9,10,11,12,13,14,15,23,24,25,26,27,28,29. Interestingly, except for its function as a cancer suppressor, miR-10b may act as a micro-oncogene in carcinogenesis to regulate cancer development and progression16,17,18,19,20,21,22. This study summarizes frequently reported cancer types with consistent or inconsistent miR-10b expression, and combines them by averaging the fold-change between independent studies, which may provide evidence for future research on the underlying mechanisms of tumorigenesis by miR-10b in specific types of cancer.
Our meta-analysis showed that vestibular schwannomas ranked at the top among consistently reported cancer types with up-regulated miR-10b (average FC: 269.19). miR-10b is also significantly up-regulated in HCC, esophageal cancer, oral cancer, glioma, pituitary adenomas and prostate cancer (average FC more than 4-fold (4 to 58.70)). The present study suggests that miR-10b is targeted to different genes in different carcinomas, such as NF1 in neurofibromatosis type 1, CDKN1A/CDKN2A in glioblastoma and KLF4 in esophageal squamous cell carcinoma34. These genes are involved in promoting the aggressive growth of carcinoma cells. High-expression of miR-10b had also been frequently reported in breast cancer, but the average fold-change was not significantly up-regulated (average FC: 1.77). Robert Weinberg's group, along with several others, have demonstrated that miR-10b is up-regulated in breast cancer and affects invasion and metastasis by targeting HOXD10 or TIAM124,35. Alternatively, melanoma ranked as the most consistently reported cancer type with down-regulated miR-10b. However, based on this study, the down-regulation of miR-10b expression in a variety of cancers is not significant (average FC less than 4-fold (-1.27 to -3.13)). Although miR-10b up- or down-regulation is inconsistent in this study, the meta-analysis using the vote-counting strategy clearly demonstrates that miR-10b had high-expression in a number of cancer samples.
The pooled odds ratio and 95% confidence intervals in this meta-analysis also confirmed the conclusion of the above-referenced vote-counting analysis. Thirteen studies from 8 articles on miR-10b expression indicated that miR-10b was significantly associated with cancer risk, included breast cancer, HCC, lung cancer, esophageal cancer and glioma (OR = 32.80, 95% CI: 11.90–90.37, P<0.0001). Low genetic effect heterogeneity was found in these cancers, even though they are from different genetic backgrounds, populations and cancers. Subgroup analysis of cancer types showed significant association between the expression of miR-10b and the increased relative risk among digestive system cancer (OR = 26.37, 95% CI: 3.21–216.41, P = 0.02), and nor-digestive system cancer (OR = 33.97, 95% CI: 11.98–96.32, P = 0.000) (Table 3).
In this study, the pooled SEN was 0.98 (95% CI: 0.89–0.99), the SPE was 0.62 (95% CI: 0.39–0.82) and the AUC was 0.98 (95% CI: 0.96–0.99), indicating a relatively high level of overall accuracy. The DOR is a single indicator of test accuracy that combines the data from sensitivity and SPE into a single number. In this meta-analysis, the pooled DOR was 133.145 (95% CI: 13.211–1341.874), indicating a good discriminatory test performance. The SROC curve and the DOR are not easy to interpret and use in clinical practice, but the likelihood ratios (PLR and NLR) are more clinically meaningful for measures of diagnostic accuracy. The PLR value of 2.63 (95% CI: 1.44–4.82) suggests that patients with cancer have an approximately 2.63-fold higher chance of being miRNA-10b assay-positive compared to control patients without cancer. The NLR value of 0.02 (95% CI: 0.00–0.17) means that the probability of the person having cancer is 2% if the miR-10b assay is negative, which is low enough to rule out cancer.
Heterogeneity is a potential problem in interpreting the results of any meta-analysis; our meta-analysis was interpreted within the context of its limitations. Subgrouping of the meta-analysis also allowed for the analysis of the different types of cancer and source of samples. The AUC value of 0.5 for the non-tissue group is too low, which means the combined SEN, SPE, PLR, NLR and DOR values cannot be determined. The PLR value of the tissue-based subgroup of the meta-analysis was 2.89, suggesting that patients with cancer have an approximately 2.89-fold higher chance of being miRNA-10b tissue-based assay-positive than control patients, and the NLR value of 0.003 in the present meta-analysis means that the probability of seeing cancer is 0.3% if the tissue-based miRNA-10b assay is negative, which is low enough to rule out cancer, including HCC, esophageal cancer, oral cancer, lung cancer, glioma and breast cancer. Our results indicate that the tissue-based level of miR-10b can distinguish cancer with more significant sensitivity and specificity than non-tissue based miR-10b. Similarly, the AUC value for the diagnostic accuracy of the meta-analysis of nor-digestive system cancer was lower (0.5) than in digestive system cancer (0.93). Only in the digestive system cancer subgroup can the value of the SEN, SPE, PLR, NLR and DOR be calculated. The pooled SEN was 0.93 (95% CI: 0.84–0.97), the SPE was 0.67 (95% CI: 0.30–0.90) and the AUC was 0.93 (95% CI: 0.90–0.95), which is high enough to consider for clinical practice. The PLR value of 2.77 suggests that patients with digestive system cancer have approximately 2.77-fold higher chance of being miRNA-10b assay-positive compared to control patients without digestive system cancer. The NLR value of 0.11 means that the probability of a patient having esophageal cancer is 11% if the miRNA-10b assay is negative, which is not low enough to rule out digestive system cancer.
As Li J et al. mentioned, interpatient and intratumour heterogeneity had an important role in affecting the robustness of gene signatures36. In this study, we screened the data from randomized source of samples to calculate the accuracy of miR-10b up-regulation detected on digestive or nor-digestive system cancers. The results showed that this strategy was able to significantly increase the predictive accuracy especially in high-risk groups (100%, 85.71–100% for nor-digestive and digestive patients, respectively).
This meta-analysis has several limitations. First, meta-analysis results often depend on the control selection procedures. This may influence heterogeneity insomuch that population-based controls should be representative of the general population to reduce the natural heterogeneity present in such genetic association studies. Second, only articles written in English were included in this meta-analysis, and articles written in other languages, unpublished data and ongoing studies were not included, which may contribute to publication bias in our meta-analysis. Third, this study combined data from microarray analysis and real time quantitative PCR (qRT-PCR), which means these data have an inherent heterogeneity based on the methodology used to determine their parameters. Finally, a lack of access to the original data from the reviewed studies limited our ability to perform meta-analysis on down-regulated miR-10b OR and its 95% CI. It was expected that a meta-analysis of low miR-10b expressing studies containing pooled ORs and 95% CIs would be included in this study; they will be analyzed in the future when sufficient data can be collected.
In conclusion, we provide evidence that high-expression of miR-10b is significantly associated with cancer risk via a systematic meta-analysis. The vote-counting results demonstrate that miR-10b up-regulation may play an important role in vestibular schwannomas, HCC, esophageal cancer, oral cancer, glioma, pituitary adenomas and prostate cancer. Furthermore, miR-10b may be useful as a new molecular target to identify some types of cancer, especially the digestive system cancer.
Methods
Literature search
We conducted a literature search using the following electronic databases: PubMed, EmBase, Web of Science and Science Direct. The following keywords and medical subheadings were used simultaneously in each set: “miR-10b or microRNA-10b” and “cancer or carcinoma or tumor or neoplasms”. Alternative spellings were also considered. Original articles published in English up to August 10, 2014 were screened. The authors were contacted via email to obtain relevant articles and data needed for the meta-analysis. The literature search was performed by two independent researchers.
Inclusion criteria
The articles collected were considered eligible if they met the following criteria: (1) they were published in English; (2) they measured miR-10b expression level in cancer patients; (3) they used tissue, saliva or blood samples obtained from surgically resected cancer cases and noncancerous/normal controls from different patients for comparison; (4) the validation method and validation sample set were reported; and (5) the studies utilized a case-control design and contained sufficient published data to allow the use of the vote-counting strategy or an estimation of the odds ratio (OR) within a 95% CI or calculate the diagnostic accuracy. Additionally, articles were excluded if they were (1) not published in English, (2) review articles, laboratory articles or letters, (3) duplicates or continued work of previous publications, (4) unqualified data, or (5) an investigation of a set of miRNAs, not miR-10b alone.
Data extraction
Key components of a qualified study were recorded: study population, type of carcinoma, study design, outcome assessment, and miR-10b measurements. Studies lacking any of these components were excluded to increase the reliability of the meta-analysis. A flowchart describing the identifying process of qualifying studies is presented in Figure 1. The following information was extracted from the full text of eligible articles: the first author's surname, year of publication, study designation, origin country, subject ethnicity, case and control sources, number of subjects, types of cancer, and assessment methods.
Quality assessment
To ensure the quality of the meta-analysis, all selected articles were scored and categorized according to the Newcastle-Ottawa Scale (NOS). Studies with a score of five or higher were considered to be of high quality (Table S1). Two authors (Y.J.L. and J.N.Y.) reached a consensus on study eligibility, and any disagreements were resolved through a discussion. If no agreement could be reached, three other authors (J.Y., Q.J.W. and X.C.) checked the extracted studies to determine their inclusion.
Statistical analysis
The vote-counting strategy of meta-analysis based on ranking potential molecular determinants has been previously described by Griffith et al.37. Each included study compared miR-10b expression between cancer cases and noncancerous (normal or benign) controls in different types of cancer. Reported miR-10b expression levels were ranked according to the following order of importance: (1) listing miR-10b expression as differentially expressed with a consistent direction of change; (2) calculating average FC of miR-10b expression in a consistent cancer type (in the studies with available fold change information); and (3) comparing the total number of samples in the studies that were in agreement. Total sample size was considered more important than average FC because many studies did not report a FC. Therefore, average FC values were based on the subset of studies with these data available. The ranking was performed using RevMan 5.1 (Cochrane collaboration, Oxford, UK).
To statistically evaluate the level of miRNA-10b expression and OR in a variety of cancers, ORs and 95% CIs were calculated to estimate each study. The statistical significance of the summary ORs was determined by Z test (P<0.05 was considered statistically significant). The subgroup pooled ORs were calculated and used for comparisons of miR-10b between different type of cancers and different assessment methods for diagnosing the cancer. Inter-study heterogeneity was tested with the chi-square based Q-test. If an absence of heterogeneity across studies was identified (P>0.05), the fixed-effects model (Mantel-Haenszel method) was used. Otherwise, the random-effects model (DerSimonian and Laird method) was performed. The I2 (I2 = 100% × (Q-df)/Q) statistic was then used to quantitatively estimate heterogeneity, where I2<25%, 25–75%, and >75% represent low, moderate and high inconsistency, respectively38. Then, the true positive rate (TPR) and false positive rate (FPR) of each study were converted by constructing a 2×2 contingency table, and the patient numbers were used to calculate the overall diagnostic accuracy. The following indexes of test accuracy were computed for each study: SEN, SPE, PLR, NLR, and DOR. Based on the measures mentioned above, SROC curves were calculated to assess diagnostic accuracy11. The AUC was also calculated. An AUC value close to 1.0 signifies that the test has almost perfect discrimination, and an AUC value close to 0.5 suggests poor discrimination. The value of a DOR ranges from 0 to infinity, with higher values indicating better discriminatory test performance39. To evaluate whether use of the miR-10b would result in a better prediction of patient outcome from various types of cancer than single type of cancer, we applied the algorithms of miR-10b up-regulating detection in two ways: (1) we divided different types of cancer into nor-digestive and digestive system cancer; (2) we collected the data random from different sources to perform the values of miR-10b up-regulation between the “good” and “bad” samples. The pooled sensitivity, specificity and other related indexes across studies were calculated using a random-effects model. The chi-square test was used to detect statistically significant heterogeneity across studies. Additionally, sensitivity analysis was performed by omitting each study independently to reflect the influence of an individual study's data on the summary ORs. Finally, evidence of publication bias was analyzed by the Begg's funnel plot and Egger's test (P<0.05 was considered a significant publication bias)40. All calculations were performed using RevMan 5.1 (Cochrane collaboration, Oxford, UK) and STATA 11.2 (STATA Corporation, College Station, TX).
Author Contributions
Conceived and designed the experiments: Y.J.L., J.N.Y. and X.C. Analyzed the data: Y.J.L., J.Y. and Q.J.W. Contributed reagents/materials/analysis tools: Y.J.L., J.N.Y., J.Y., Q.J.W. and X.C. Wrote the first draft of the manuscript: Y.J.L., J.Y. and X.C. Reviewed, edited and approved the manuscript: Y.J.L., J.Y., J.N.Y., Q.J.W. and X.C.
Supplementary Material
Supplementary Information
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grant number: 81172142).
References
- Bartel D. P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky A. M. & Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med 13, 39–53 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S. et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 103, 2257–2261 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X. et al. Prognostic role of microRNA-21 in various carcinomas: a systematic review and meta-analysis. Eur J Clin Invest 41, 1245–1253 (2011). [DOI] [PubMed] [Google Scholar]
- Lin S. et al. Prognostic Role of MicroRNA-181a/b in Hematological Malignancies: A Meta-Analysis. PLoS One 8, e59532 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasayama T., Nishihara M., Kondoh T., Hosoda K. & Kohmura E. MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC. Int J Cancer 125, 1407–1413 (2009). [DOI] [PubMed] [Google Scholar]
- Lu Y. C. et al. Oncogenic function and early detection potential of miRNA-10b in oral cancer as identified by microRNA profiling. Cancer Prev Res 5, 665–674 (2012). [DOI] [PubMed] [Google Scholar]
- Cui E. H. et al. Serum microRNA 125b as a diagnostic or prognostic biomarker for advanced NSCLC patients receiving cisplatinbased chemotherapy. Acta Pharmacol Sin 34, 309–313 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplyuk N. M. et al. MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro Oncol 14, 689–700 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth C., Kasimir-Bauer S., Pantel K. & Schwarzenbach H. Screening for circulating nucleic acids and caspase activity in the peripheral blood as potential diagnostic tools in lung cancer. Mol Oncol 5, 281–291 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z. et al. Salivary microRNAs as promising biomarkers for detection of esophageal cancer. PLoS One 8, e57502 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y. Y. et al. MicroRNA-10b Promotes Migration and Invasion through KLF4 in Human Esophageal Cancer Cell Lines. J Biol Chem 285, 7986–7994 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Martin M. et al. Global profiling in vestibular schwannomas shows critical deregulation of microRNAs and upregulation in those included in chromosomal region 14q32. PLoS One 8, e65868 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S., Chen L., Huang H. & Zhi D. The experimental study of miRNA in pituitary adenomas. Turk Neurosurg 23, 721–727 (2013). [DOI] [PubMed] [Google Scholar]
- Walter B. A., Valera V. A., Pinto P. A. & Merino M. J. Comprehensive microRNA Profiling of Prostate Cancer. J Cancer 4, 350–357 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneghan H. M., Miller N., Kelly R., Newell J. & Kerin M. J. Systemic miRNA-195 differentiates breast cancer from other malignancies and is a potential biomarker for detecting noninvasive and early stage disease. The Oncologist 15, 673–682 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzini S. et al. Impact of microRNAs on regulatory networks and pathways in human colorectal carcinogenesis and development of metastasis. BMC Genomics 14, 589 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. et al. De novo sequencing of circulating miRNAs identifies novel markers predicting clinical outcome of locally advanced breast cancer. J Transl Med. 10, 42 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaravinos A. et al. Expression of miRNAs involved in Angiogenesis, tumer cell proliferation, tumer suppressor inhibithon, epithelial-mesenchymal transition and activation of metastasis in bladder cancer. J Urol 188, 615–623 (2012). [DOI] [PubMed] [Google Scholar]
- Tsukamoto O. et al. Identification of endometrioid endometrial carcinoma-associated microRNAs in tissue and plasma. Gynecol Oncol 132, 715–721 (2014). [DOI] [PubMed] [Google Scholar]
- Wotschofsky Z. et al. Identification of metastamirs as metastasis-associated MicroRNAs in clear cell renal cell carcinomas. Int J Biol Sci 8, 1363–1374 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio M. V. et al. MicroRNA gene expressionderegulation in humanbreast cancer. Cancer Res 65, 7065–7070 (2005). [DOI] [PubMed] [Google Scholar]
- Chan M. et al. Identification of circulating microRNA signatures for breast cancer detection. Clin Cancer Res 19, 4477–4487 (2013). [DOI] [PubMed] [Google Scholar]
- Ma L., Teruya-Feldstein J. & Weinberg R. A. Tumor invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449, 682–688 (2007). [DOI] [PubMed] [Google Scholar]
- Shen J. et al. Genome-wide aberrant DNA methylation of microRNA host genes in hepatocellular carcinoma. Epigenetics 7, 1230–1237 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. J. et al. MicroRNA-10b promotes migration and invasion through CADM1 in human hepatocellular carcinoma cells. Tumor Biol 33, 1455–1465 (2012). [DOI] [PubMed] [Google Scholar]
- Zhao F. L. et al. Serum overexpression of MicroRNA-10b in patients with bone metastatic primary breast cancer. J Int Med Res 40, 859–866 (2012). [DOI] [PubMed] [Google Scholar]
- Shen Y. et al. microRNA expression profiles associated with survival, disease progression, and response to gefitinib in completely resected non-small-cell lung cancer with EGFR mutation. Med Oncol 30, 750 (2013). [DOI] [PubMed] [Google Scholar]
- Guessous F. et al. Oncogenic effects of miR-10b in glioblastoma stem cells. J Neurooncol 112, 153–163 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S. M. et al. RAS is regulated by the let-7 microRNA family. Cell 120, 635–647 (2005). [DOI] [PubMed] [Google Scholar]
- Deng S., Calin G. A., Croce C. M., Coukos G. & Zhang L. Mechanisms of microRNA deregulation in human cancer. Cell Cycle 7, 2643–2646 (2008). [DOI] [PubMed] [Google Scholar]
- Hong L. et al. The malignant phenotype-associated microRNA in gastroenteric, hepatobiliary and pancreatic carcinomas. Expert Opin Biol Ther 10, 1693–1701 (2010). [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. MicroRNA-143 targets MACC1 to inhibit cell invasion and migration in colorectal cancer. Mol Cancer 11, 23 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriely G., Teplyuk N. M. & Krichevsky A. M. Context effect: microRNA-10b in cancer cell proliferation, spread and death. Autophagy 7, 1384–1386 (2011). [DOI] [PubMed] [Google Scholar]
- Ma L. et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol 28, 341–347 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. et al. Identification of high-quality cancer prognostic markers and metastasis network modules. Nat Commun 1, 34 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith O. L., Melck A., Jones S. J. & Wiseman S. M. Meta-analysis and meta-review of thyroid cancer gene expression profiling studies identifies important diagnostic biomarkers. J Clin Oncol 24, 5043–5501 (2006). [DOI] [PubMed] [Google Scholar]
- Higgins J. P., Thompson S. G., Deeks J. J. & Altman D. G. Measuring inconsistency in meta-analyses. BMJ 327, 557–560 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res 2, 807–813 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg C. B. & Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics 50, 1088–1101 (1994). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information