Gao et al. show that E-box proteins dampen the generation and function of Foxp3+ regulatory T cells in part by inhibiting IL-2Rα expression and IL-2 responsiveness.
Abstract
E-proteins are TCR-sensitive transcription factors essential for intrathymic T cell transitions. Here, we show that deletion of E-proteins leads to both enhanced peripheral TGF-β–induced regulatory T (iT reg) cell and thymic naturally arising T reg cell (nT reg cell) differentiation. In contrast, deletion of Id proteins results in reduced nT reg cell differentiation. Mechanistic analysis indicated that decreased E-protein activity leads to de-repression of signaling pathways that are essential to Foxp3 expression. Decreased E-protein binding to an IL-2Rα enhancer locus facilitated TCR-induced IL-2Rα expression. Similarly, decreased E-protein activity facilitated TCR-induced NF-κB activation and generation of c-Rel. Consistent with this, microarray analysis indicated that cells with E-protein depletion that are not yet expressing Foxp3 exhibit activation of the IL-2 and NF-κB signaling pathways as well as enhanced expression of many of the genes associated with Foxp3 induction. Finally, studies using Nur77-GFP mice to monitor TCR signaling showed that TCR signaling strength sufficient to induce Foxp3 differentiation is accompanied by down-regulation of E-protein levels. Collectively, these data suggest that TCR stimulation acts in part through down-regulation of E-protein activity to induce T reg cell lineage development.
Naturally arising T regulatory cells (nT reg cells) undergo a differentiation program in the thymus during which they acquire Foxp3 expression. Recent studies suggests that T reg cell differentiation is a process involving, first, TCR stimulation with a signal intensity that enables the nT reg cell precursor to respond to IL-2R signaling and, second, signaling via the latter to activate STAT5 (Burchill et al., 2007; Burchill et al., 2008). The most important cytokine mediating such signaling during intrathymic T reg cell differentiation is IL-2, as shown by the fact that mice lacking IL-2 or its receptor subunits, IL-2Rα (CD25) and IL-2Rβ (CD122), have major deficits in numbers of CD4+CD25+ T reg cells and develop autoimmune disease similar to that observed in Foxp3−/− mice (Bayer et al., 2007; Burchill et al., 2007; Malek, 2008; Cheng et al., 2011).
The major outcome of IL-2R signaling relative to Foxp3 expression is the generation of activated STAT5, a key regulatory element controlling Foxp3 expression (Yao et al., 2007; Burchill et al., 2008). TCR signaling, however, is not only important to nT reg cell differentiation because of its effect on STAT5 activation, but also because it results in NF-κB activation. This was shown in studies of mice bearing a transgene expressing a constitutively active mutant form of Iκκ-β kinase (IKKEE) that exhibits enhanced NF-κB activity associated with marked increases in Foxp3+ thymocytes (Long et al., 2009). It should be noted, however, that the mechanism of TCR stimulation of NF-κB activation appears to be quite separate from TCR-mediated effects on IL-2R signaling and STAT5 activation because the latter was not enhanced in IKKEE transgenic mice (Long et al., 2009).
Thymocyte differentiation has been shown to be regulated by members of E-protein family of transcription factors (Engel et al., 2001; Jones and Zhuang, 2007); it is therefore possible that these factors could also exert an influence on T reg cell development. E-proteins consist of a family of four proteins: the E2A proteins, E12 and E47 (TCF3) that are alternatively spliced forms of the same gene, as well as HEB (TCF12) and E2-2 (TCF4; Murre, 2005; Kee, 2009). Although E-proteins are necessary for early thymocyte development preceding T-lineage commitment, they later exert an inhibitory effect on DN to DP transitions and DP to SP transitions that must be overcome by down-regulation of E-protein activity mediated by preTCR or TCR signaling (Engel et al., 2001; Jones and Zhuang, 2007). Given the fact that, as indicated above, TCR stimulation of thymocytes initiates T reg cell development, such TCR-mediated down-regulation of E-protein may define the possible area of E-protein influence on T reg cell development. An example of how this could occur comes from studies showing that inhibition of E-proteins by transgenes that express E-protein inhibitors (Id1 and Tal1) leads to NF-κB activation, and thus possible effects of NF-κB transcription factors on Foxp3+ T reg cell induction (Kim et al., 2002).
In this study, we investigated the effect of E-proteins on T reg cell development in E2A/HEB (E-protein) conditional KO mice. We found that E-protein depletion leads to a markedly increased Foxp3+ induced T reg (iT reg) cell and nT reg cell development, whereas increased E-protein activity in Id2−/−Id3−/− mice leads to a striking reduction of Fox3+ nT reg cells. In subsequent studies, we found that decreased E-protein activity impacted on two important processes associated with T reg cell development: (1) it increased IL-2Rα+ (CD25) cell and STAT5 phosphorylation through direct de-suppression of CD25 transcription and (2) it enhanced expression of c-Rel due to an effect of E-protein activity on NF-κB activation. Given previous studies establishing a relation between TCR signaling and E-protein activity, these data suggest that E-protein activity plays an essential role in setting the threshold for the TCR-induced NF-κB activation and STAT5 phosphorylation that accompany T reg cell development.
RESULTS
E-protein regulates iT reg cell differentiation
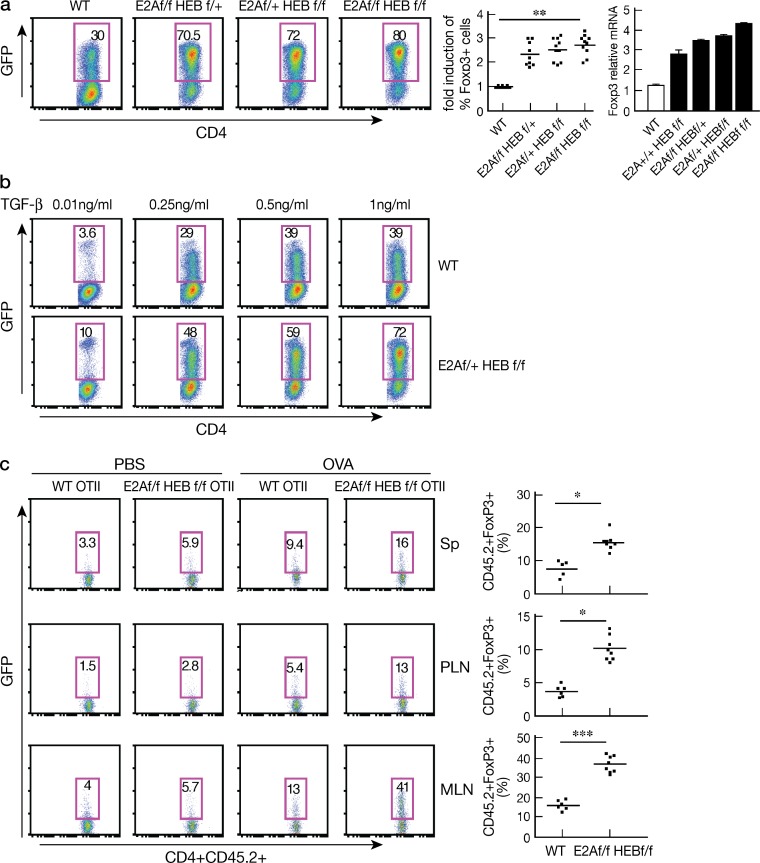
To investigate the role of the E-protein in the regulation of Foxp3+T reg cell development, we used inducible E-protein conditional KO mice (E2Af/fHEBf/f Rosa26-ER-Cre; Jones et al., 2009). In initial studies focused on iT reg cell development we obtained (∼99% pure) purified naive CD4+ T cells from WT and E2Af/f/HEBf/f ER-Cre or from WT Foxp3-GFP-KI mice and E2Af/f/HEBf/f ER-Cre/Foxp3-GFP-KI and then cultured the cells in the presence of TGF-β plus 4-hydroxytamoxifen (4-OHT) for 3 d. We found that naive cells from E-protein conditional KO mice undergo significantly greater induction into Foxp3+ cells and significantly greater up-regulation of Foxp3 mRNA compared with WT mice (Fig. 1 a). In addition, such increased iT reg cell differentiation occurred in a TGF-β dose-dependent manner (Fig. 1 b).
Figure 1.
E-proteins regulate iT reg cell differentiation. (a) CD4+ cells obtained from WT Foxp3-GFP-KI mice or floxed E2A/HEB ER-Cre/Foxp3KI mice were activated with anti-CD3/CD28 and cultured in the presence of TGF-β and 2 µM 4-OHT for 3 d. (left) Foxp3+ cells were identified by GFP; numbers in box indicate percentages; middle, fold-induction of Foxp3+ cells in each group of floxed mice; each dot represents data from 1 mouse (7–9 mice total for each group); data are pooled from at least three independent experiments; means are shown. **, P = 0.012. (right) Cells cultured as in (a) were subjected to RNA extraction and Foxp3 expression was analyzed by real-time RT-PCR; data are representative of two independent experiments, at least two mice for genotype were included in each experiment. (b) CD4+ cells obtained from WT Foxp3-GFP-KI mice and E2A/HEBf/f ER-Cre/Foxp3-GFP-KI mice were cultured in the presence of different amounts of TGF-β (0.01–1 ng/ml) and 4-OHT for 3 d. Foxp3 (GFP)+ cells were analyzed by flow cytometry; data are representative of two independent experiments. At least two mice for each genotype were included in each experiment. (c) Naive CD4+ (∼99%) cells purified from spleens of WT Foxp3-GFP-KI OTII mice and E2Af/f/HEBf/f ER-Cre/Foxp3-GFP-KI OTII (CD45.2) mice were transferred (2 × 106 cells) into congenic WT (CD45.1) mice, which were then administered tamoxifen. 2 d after final tamoxifen administration, the mice were subcutaneously and orally administered with either PBS or OVA peptide (0.4 mg/mouse). (left) Analysis of Foxp3+(GFP+) cells in spleen, peripheral and mesenteric LN 2 d after PBS or OVA administration. (right) Frequency of CD4+Foxp3+(GFP)+ cells in various tissues after OVA administration; each dot represents data from 1 mouse (6–8 mice total in each group). Data are pooled from two independent experiments. Means are shown. *, P < 0.05, ***, P < 0.001.
To determine if the E-protein affects iT reg cell differentiation in vivo, we transferred (∼99% pure) purified naive CD45.2+CD4+CD62Lhi cells from WT Foxp3-GFP-KI/OT-II or E2Af/fHEBf/f Er-Cre/Foxp3-GFP-KI/OT-II mice into congenic WT CD45.1 mice (2 × 106 cells/mouse), and then treated the mice with tamoxifen three times; we then administered OVA peptide or PBS by both subcutaneous and oral routes and, 2 d after completion of such administration, determined Foxp3+ cells in various tissues. We found that the percentage of Foxp3+ cells was greatly increased in spleen, peripheral LNs, and mesenteric LNs from mice transferred E2Af/fHEBf/fEr-Cre/Foxp3-GFP-KI/OT-II CD4+ T cells compared with mice transferred WT Foxp3-GFP-KI/OT-II cells or administered PBS (Fig. 1 c). These data strongly suggested that E-protein activity regulates iT reg cell differentiation both in vivo and in vitro.
E-protein regulates Foxp3+ nT reg cell development in the thymus
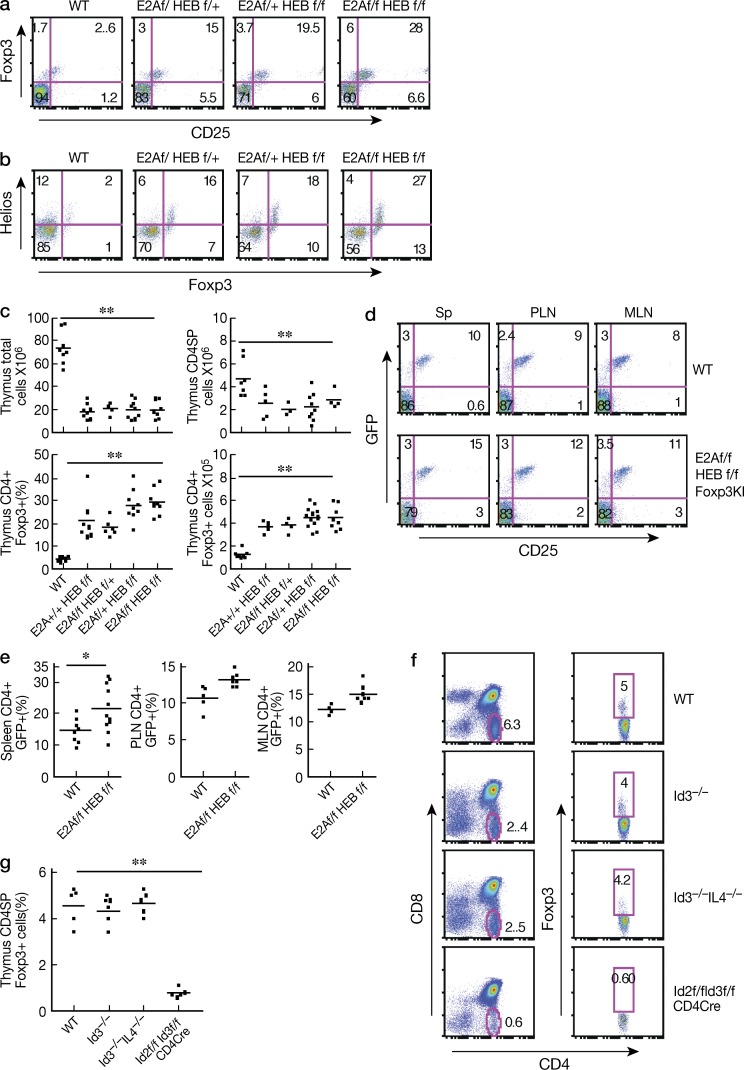
In additional studies, we investigated whether E-protein activity also regulates Foxp3+ nT reg cell development. In this case, we used E-protein conditional KO mice with either four allele deletion (f/f f/f, double KO mice) or three allele deletion of E2A/HEB (f/f f/+ or f/+ f/f) in which E-protein had been deleted by tamoxifen injection (Jones et al., 2009). As expected, we found that such tamoxifen-treated E-protein KO mice display increased percentages of both CD4SP (single-positive) and CD8SP cells (unpublished data), which is consistent with previous findings that E-proteins provide a checkpoint for SP thymocyte development (Engel et al., 2001). In addition, we found that the percentage of the Foxp3+ cells was greatly increased (∼15–30% in KO mice compared with 3–5% in WT mice; Fig. 2 a) and most of these Foxp3+ thymocytes were also positive for CD25 and Helios (Fig. 2, a and b). Importantly, the absolute number of Foxp3+ cells was increased approximately twofold in KO mice compared with WT mice in spite of the fact that both the total number of thymocytes and the total number of CD4SP thymocytes were significantly decreased (Fig. 2 c). Similar data were obtained from E2A+/+HEBf/f Cre, E2Af/fHEB+/+ Cre mice (i.e., single KO E-protein KO mice), and E2Af/fHEBf/f Er-cre/Foxp3-GFP-Knock-In mice treated with tamoxifen (unpublished data). Collectively, these observations indicate that the E-protein activity regulates nT reg cell development in the thymus.
Figure 2.
E-proteins regulate thymic foxp3+ T reg cell development. (a and b) Foxp3+ and CD25+ or HelioE + thymocytes from tamoxifen-treated WT Cre+ (WT), E2Af/fHEBf/+ER-Cre (f/f f/+); E2Af/+HEBf/f ER-Cre (f/+f/f); E2Af/fHEBf/f ER-Cre(f/f f/f) mice analyzed by flow cytometry. Numbers in each quadrant indicate percentage. Data are representative of at least three independent experiments. (c) The total number of thymocytes, total number of CD4+ cells, the percentage and absolute number of Foxp3+ CD4SP thymocytes from each tamoxifen-treated genotyped group of mice were calculated from flow cytometry data. Each dot represents data from one mouse (7–10 mice total in each group); means are shown. **, P < 0.01. Data are pooled from at least three independent experiments. (d) Tamoxifen-treated 3–4-mo-old WT Cre and E2Af/fHEBf/f ER-Cre Foxp3-GFP-KI mice were analyzed for Foxp3+ cells in spleen, pLN, and mLN by flow cytometry; numbers in each quadrant indicate percentages. (e) The percentage of Foxp3+ cells among CD4+ cells in spleen, pLN, and mLN. Each dot represents data from 1 mouse (6–9 mice total in each group); graph shows means. *, P < 0.05. Data are representative of at three independent experiments (f and g) Thymocytes from age-matched (6–8 wk) WT Id3−/−, Id3−/−IL-4−/−, Id2f/fId3f/f CD4-Cre mice were analyzed by flow cytometry. (f) Representative flow cytometry plots showing the percentage of CD4+SPFoxp3+ cells in one mouse. (g) Combined data; each dot represents data from one mouse (5 mice total in each group). **, P < 0.01. Data are pooled from three independent experiments.
The finding that E-protein depletion is inversely related to thymic T reg cell differentiation prompted us to investigate if E-proteins affect cellular homeostasis in peripheral lymphoid organs. Whereas there is a mild increase in the percentage of CD4+ and CD8+ cells in the spleens of E-protein KO mice when compared with WT mice (unpublished data), the total cell numbers in the spleen or in the mesenteric and peripheral LNs did not change (unpublished data). In addition, T cells in E-protein KO mice did not exhibit increased activation as evaluated by CD44 and CD62L expression (unpublished data). However, E-protein KO mice exhibited a significantly increase of CD4+Foxp3+ cells in the spleen compared with WT mice, but not in the peripheral or mesenteric nodes (Fig. 2, d and e).
nT reg cell differentiation in Id protein-deficient mice
To further investigate the effect of E-proteins on T reg cell development, we examined Foxp3+ cell development in conditional Id protein-deficient mice, because, as mentioned, Id proteins bind to E-proteins and inhibit the latter’s capacity to bind to DNA, mice with Id protein deficiency provide a setting in which E-protein activity is increased rather than decreased.
We first determined the percentage of Foxp3+ thymocytes in Id3−/− and Id3−/−IL-4−/− mice, including the latter in this study because Id3−/− mice have been reported to produce increased amount of IL-4 (Verykokakis et al., 2010), a cytokine that has been shown to have an inhibitory effect on Foxp3 expression (Maruyama et al., 2011). We found that, contrary to a previous study (Maruyama et al., 2011), the percentage of Foxp3+ thymocytes in the thymuses of Id3−/− and Id3−/−IL-4−/− mice were not significantly different from that of WT mice (Fig. 2 f and Fig. 2 g). We realized, however, that this result could be due to the fact that various members of the Id protein family (mainly Id2 and Id3 in lymphocytes) have redundant capacities to inhibit E-protein activity, and thus Id2 might inhibit E-protein activity in the absence of Id3. To examine this possibility, we determined Foxp3+ cell development in Id2f/f/Id3f/f CD4-Cre mice, i.e., mice known to have high levels of E-protein expression (Zhang et al., 2014). As observed here and as shown previously, such mice have greatly reduced thymic cellularity and reduced numbers of CD4- or CD8-SP thymocytes compared with WT mice (Fig. 2 f, left). Nevertheless, the percentage of Foxp3+ cells in the CD4-SP cell population still present was significantly decreased (Fig. 2 f, right; and Fig. 2 g). These results thus provide confirmation that E-proteins control Foxp3+ T cell development.
E-protein KO nT reg cell maintain regulatory function
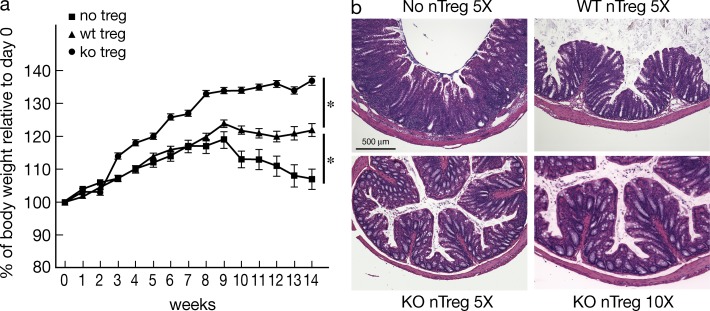
To address the functional capacity of T reg cells from E-protein KO mice, we next assessed their ability to ameliorate cell transfer colitis. Accordingly, WT naive CD45RBhiCD4+ T cells alone or in combination with CD4+GFP+(Foxp3+) thymocytes obtained from tamoxifen-treated E2Af/fHEBf/f Er-cre/Foxp3-GFP-KI mice (KO nT reg cells) or from WT Foxp3-GFP-KI mice (WT nT reg cells) were transferred into RAG2−/− recipient mice, and colitis development was assessed by body weight loss and colon tissue histology. RAG2−/− mice that had been transferred naive CD4+ T cells along with WT T reg cells developed significantly less colitis than mice that had been transferred naive CD4+ T cells alone. Somewhat unexpectedly, however, mice co-transferred T reg cells from E-protein KO mice (KO T reg cell) exhibited significantly less colitis than mice co-transferred WT T reg cells (Fig. 3, a and b). These findings thus suggest that nT reg cells from E-protein KO mice exhibit normal or even increased regulatory function.
Figure 3.
Suppressive activity of nT reg cells from E-Protein KO mice. Rag2−/− recipient mice were adoptively transferred naive CD4+ T cells alone or in combination of T reg cells from tamoxifen-treated WT Foxp3-GFP-KI mice (WT T reg cells) or floxed E2A/HEB ER-Cre/Foxp3-GFP-KI mice (KO T reg cells), and then monitored for development of colitis. (a) Changes in body weight after cell transfer. (b) Hematoxylin and eosin staining of colon sections 14 wk after cell transfer in (a). Data are representative of two independent experiments. *, P < 0.05 (10 mice total in each group).
E-protein regulation of Foxp3+ cell development is cell intrinsic
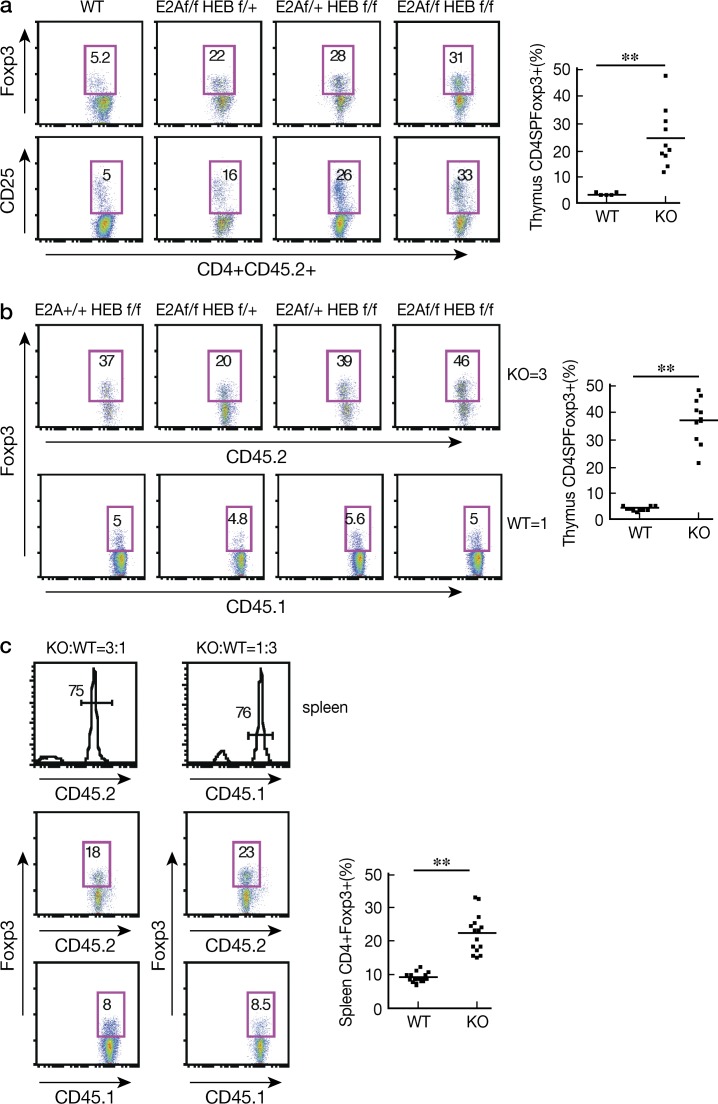
To investigate whether the E-protein effect on T reg cell development is cell intrinsic, we first generated BM chimeric mice by transferring BM cells from CD45.2+ E-protein KO or CD45.1 WT mice into sublethally irradiated RAG2−/− mice and, 5 wk later, administered tamoxifen to both groups of mice. As expected, chimeric mice that had been transferred BM cells from E-protein KO mice exhibited a higher percentage of CD4+ Foxp3+ and CD4+CD25+ cells compared with mice transferred BM from WT mice (Fig. 4 a). In further studies, we generated mixed BM chimeric mice by transferring BM cells from CD45.2 E2Af/fHEBf/f Er-cre and WT mice or E2Af/fHEBf/f Er-cre Foxp3-GFP-KI and CD45.1 Foxp3-GFP-KI WT mice mixed at different ratios (3:1, 1:1, and 1:3) into sublethally irradiated RAG2−/− mice as above. At each of the ratios, E-protein KO cells exhibited a higher percentage of Foxp3+ cells compared with cells from WT mice (Fig. 4 b and not depicted). Furthermore, this increase in Foxp3+ cells was also evident in the E-protein KO cells repopulating the spleen of the BM chimeric mice (Fig. 4 c). Collectively, these observations indicated that E-proteins regulate T reg cell development in a cell-intrinsic manner.
Figure 4.
E-proteins regulate nT reg cell development in a cell-intrinsic manner. (a) Rag2−/− recipient mice were sublethally irradiated and reconstituted with CD45.2 WT or floxed E2A/HEB ER-Cre BM cells and treated with tamoxifen 5 wk after cell transfer. (b) Rag2−/− mice were reconstituted with a 3:1 mixture of BM from CD45.2-floxed E2A/HEB ER-Cre and CD45.1 WT mice. Numbers in boxes represent percentages. Plots at right show the frequency of CD4SPFoxp3+ cells among thymic CD4SP cells; each dot represents data from 1 mouse (10 mice total in each group); means are shown. (c) Tamoxifen-treated Rag2−/− recipient mice were reconstituted with a 3:1 or a 1:3 mixture of BM cells from CD45.1 WT and CD45.2 floxed E2A/HEB ER-Cre mice, and percentage of Foxp3+ cells in splenic CD4+ cells was assessed by flow cytometry. Plots at right show data from a total of 15 mice; each dot represents data from one mouse; means are shown. **, P < 0.01. The data are representative of two independent experiments.
E-proteins are differentially regulated in WT CD4+Foxp3+ and CD4+Foxp3− cells
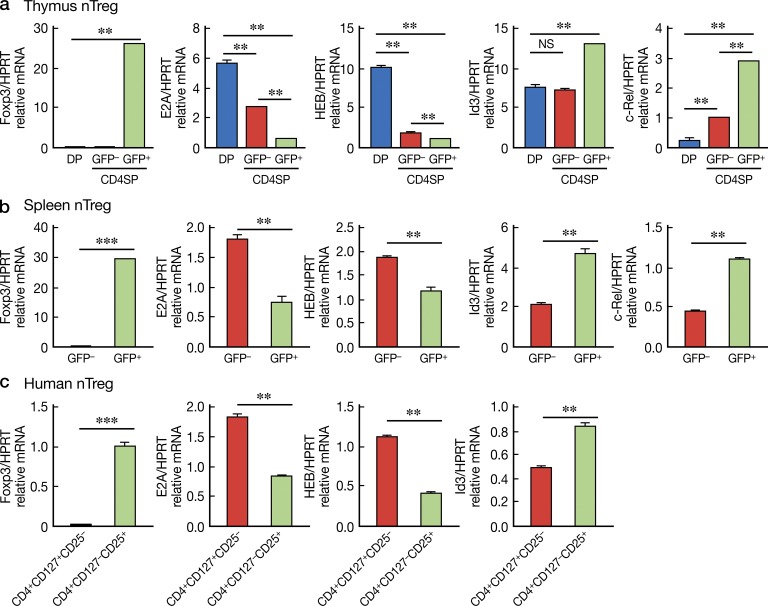
The aforementioned findings suggested that down-regulation of E-protein activity (i.e., E-protein function determined by the E-protein/Id protein ratio) in CD4+ T cells is a necessary precondition for T reg cell development; this, in turn, implies that E-protein levels may be differentially regulated in WT CD4+Foxp3+ and CD4+Foxp3− cells. To investigate these possibilities, CD4SPCD25+Foxp3+, CD4SPCD25−Foxp3−, and DP cells were sorted from thymus or spleen of Foxp3-GFP-KI WT mice and selected gene expression profiles were evaluated by quantitative RT-PCR. We found that both E2A and HEB expression were down-regulated during the DP-to-CD4SP transition, and, importantly, E-protein expression were further down-regulated in Foxp3+(GFP+) CD4SP cells compared with Fox3−(GFP−) CD4SP cells (Fig. 5 a). In contrast, Id3 expression, as mentioned above, an inhibitor of E-protein DNA binding activity, is up-regulated in CD4+Foxp3+ cells, but not in CD+Foxp3− cells. Similar findings were obtained upon examination of cells from spleens of Foxp3+ and Foxp3− mice (Fig. 5 b). The decreased E-proteins levels combined with the increased Id3 protein levels indicate that the total E-protein activity in Foxp3+ cells is much lower than that of conventional CD4+ T cells. Finally, similar studies were performed with sorted human CD4+CD25+CD127− cells (nT reg cells) and CD4+CD25−CD127+ cells (conventional T cells), and we found that the gene expression patterns of human nT reg cells are very similar to that of mice (Fig. 5 c). Collectively, these findings suggest that, as in the case of E-protein KO mice, dynamic changes in E-protein activity accompany T reg cell development in WT mice.
Figure 5.
Decreased E-protein activity is associated with Foxp3 expression. (a) DP, CD4+SPGFP+, CD4+SPGFP− cells were purified from thymus of Foxp3−GFP-KI WT mice by cell sorting and then subjected to real-time RT-PCR to assess the expression of the indicated genes. Gene expression was normalized by hprt. Data are representative of three independent experiments. (b) CD4+Foxp3+(GFP+) and CD4+Foxp3−(GFP−) cells were purified from the spleen of Foxp3-GFP-KI WT mice by cell sorting and then subjected to real-time RT-PCR as in (a). Data are representative of three independent experiments. **, P < 0.01; ***, P < 0.001. (c) Human CD4+CD127+CD25− and CD4+CD127−CD25+ (nT reg) cells were purified from human blood by cell sorting and then subjected to real-time RT-PCR as in (a). Data are representative of two independent experiments. **, P < 0.01; ***, P < 0.001.
Deletion of E-proteins is associated with enhanced IL-2 receptor–mediated signaling
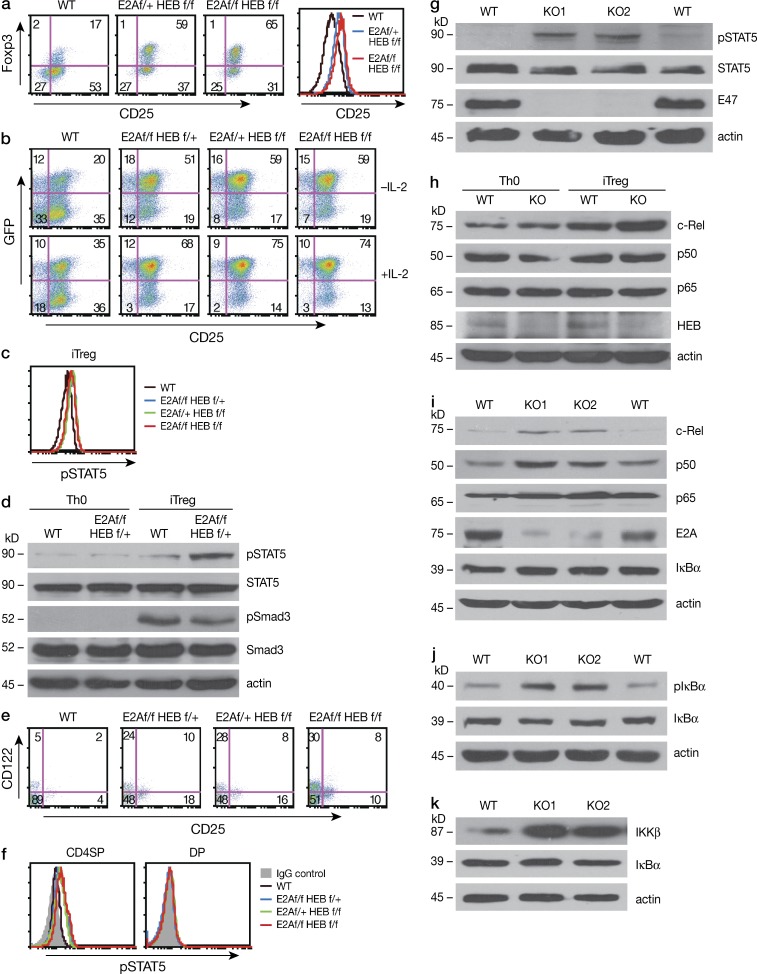
Common γ chain cytokines, primarily IL-2 (but also IL-7 and IL-15), have been shown to play an important role in iT reg cell induction in the periphery or in nT reg cell development in the thymus (Vang et. al.,2008). Therefore, deletion of E-protein may promote the generation of increased numbers of Foxp3+ cells by increasing IL-2R signaling. To investigate this possibility, we initially determined if E-protein deletion affects the expression and function of IL-2Rα (CD25). We found that purified naive CD4+ T cells from E2Af/fHEBf/f/Er-cre mice E-protein-KO mice cultured in the presence of 4-OHT under iT reg cell conditions express higher amount of IL-2Rα (CD25; Fig. 6 a) and were more responsive to IL-2 than CD4+ T cells from WT mice cultured under similar conditions (Fig. 6 b). In accord with these findings, STAT5 was more highly phosphorylated in iT reg cells from the E-protein KO mice than WT cells as assessed by flow cytometry (Fig. 6 c) and immunoblot (Fig. 6 d). It should be noted that Smad3 phosphorylation, one of the most important events occurring during TGF-β induced iT reg cells, was equivalent in E-protein KO and WT iT reg cells (Fig. 6 d).
Figure 6.
IL-2-STAT5 signaling and NF-κB activation in E-protein KO mice. (a) CD4+ cells were purified from WT and floxed E2A/HEB ER-Cre mice and were cultured in the presence of TGF-β and 2 µM 4-OHT for 3 d. Foxp3 (GFP)+ cell and CD25 expression was evaluated by flow cytometry. Data are representative of three independent experiments. (b) CD4+ cells purified from WT Foxp3KI and floxed E2A/HEB Er-Cre/Foxp3-GFP-KI mice cultured as in (a) with or without IL-2. Foxp3 (GFP)+ cells and CD25 expression were evaluated by flow cytometry; histogram on the right shows CD25 expression. Data are representative of two independent experiments. (c) CD4+ cells were purified from WT and floxed E2AHEB ER-Cre mice and cultured as described in (a), and pSTAT5 was evaluated by flow cytometric analysis. Data are representative of three independent experiments. (d) CD4+ cells were purified from WT and floxed E2AHEB ER-Cre mice and cultured under Th0 (no cytokine) or iT reg cell conditions in the presence of 2 µM 4-OHT for 3 d; Expression of pSTAT5, STAT5, pSmad3, and Smad3 was assessed by immunoblot. Data are representative of three independent experiments. (e) Frequency of CD25 (IL-2Rα)+/CD122 (IL-2β)+ cells among CD4SP thymocytes from tamoxifen-treated WT and floxed E2A/HEB ER-Cre mice was assessed by flow cytometry. Numbers in quadrants indicate percentage. Data shown are representative at least three independent experiments. (f) STAT5 phosphorylation in CD4SP and DP thymocytes from tamoxifen-treated WT and floxed E2A/HEB ER-Cre mice was assessed by flow cytometry. Data shown are representative of three independent experiments (g) Whole-cell extracts were prepared from thymocytes of tamoxifen-treated WT and E2Af/fHEBf/f ER-Cre (KO) mice, and expression of pSTAT5, STAT5, E47 and actin was assessed by immunoblot. Data shown are representative of three independent experiments. (h) CD4+ cells were purified from WT and floxed E2AHEB ER-Cre mice and cultured under Th0 (no cytokine) or iT reg cell conditions in the presence of 2 µM 4-OHT for 3 d. Expression of c-Rel, p50, p65, HEB, and actin was assessed by immunoblot. Data shown are representative three independent experiments. (i) Whole-cell extracts prepared as in (g). c-Rel, p50, p65, E2A, IκBα, and actin were assessed by immunoblot. Data shown are representative of three independent experiments. (j) Whole-cell extracts prepared as in (g). p-IκBα, IκBα, and actin were assessed by immunoblot. Data shown are representative of three independent experiments. (k) Whole-cell extracts prepared as in (g). Iκκβ, IκBα and actin were assessed by immunoblot. Data shown are representative of three independent experiments.
We next determined if the observed up-regulation of IL-2 signaling and increased phosphorylation of STAT5 was due to increased IL-2 production in E-protein KO iT reg cells. Accordingly, we determined IL-2 production in purified naive CD4+ T cells from WT and E-protein KO mice cultured in the presence of 4-OHT under Th0 (no cytokine) or iT reg cell conditions. We found that although E-proteins were greatly diminished in iT reg cells from the E-protein KO mice, these cells did not produce more IL-2 than iT reg cell WT cells (unpublished data). Thus, the increased phosphorylation of STAT5 and Foxp3 expression in E-protein KO iT reg cells was not caused by effects on IL-2 production, but more likely by the effects of responsiveness to IL-2 resulting from increased IL-2R expression. It should be noted, however, that the increased responsiveness did not promote the expansion of E-protein KO iT reg cells, as the proliferation of naive CD4+ cells labeled with CFSE from E-protein KO mice and from WT mice cultured under iT reg cell conditions as above was similar (unpublished data).
In parallel studies to investigate IL-2 signaling in developing nT reg cells, we determined the percentage of CD25+ T cells among nT reg cells in the thymus of E-protein KO and WT mice. We found that CD25+ and CD122+ T cells were markedly increased within the CD4SP cell population from E-protein KO mice as compared with those from WT mice (Fig. 6 e). In initial studies (using an anti–CD25-APC antibody), this increase in CD25+ T cells paralleled that of Foxp3+ cells (Fig. 2 a); however, in a subsequent study (using an anti–CD25-PE antibody), the increased IL-2Rα expression was observed in both Foxp3+ and Foxp3− cells (see Fig. 9 and further discussion below). These changes in nT reg cell T cells (as in the case iT reg cell T cells) were accompanied by equivalent increases STAT5 phosphorylation by flow cytometry (Fig. 6 f) and by immunoblot when compared with WT thymocytes (Fig. 6 g). In addition, STAT5 increases were also observed in both Foxp3+ and Foxp3− cells (unpublished data).
Figure 9.
Microarray analysis of E-protein effect versus Foxp3+ differentiation. (a) Venn diagram showing the top 332 genes each for E-protein effect, Foxp3+ differentiation, and T reg cell signature (Hill, et al., 2007). Top genes were selected based on magnitude of log ratios after prefiltering to include only genes assayed in both studies. (b) Heat map showing relative expression differences associated with E-protein effect, Foxp3+ differentiation, and T reg cell signature genes; color coding reflects the log ratios for each of these sets of genes (left to right) for the 118 genes that compose the overlap of Foxp3+ differentiation and T reg cell signature genes (see text). Ratio values range from +5.5 (red) to −5.5 (green) on the log base 2 scale. (c) CD4+GFP−CD25− cells were sorted from tamoxifen-treated WT Er-Cre and E2Af/fHEBf/f Foxp3KI/ER-Cre mice and were analyzed by real-time RT-PCR for IL-2Rα (CD25) and E2A. Data are representative of two independent experiments. (d) Thymocytes were isolated from tamoxifen-treated WT Er-Cre or E2Af/fHEBf/f Foxp3KI/ER-Cre mice and CD25+Foxp3+ and CD25+Foxp3−, and then analyzed by flow cytometry, gating on CD4SP thymocytes. Data are representative of three independent experiments. (e) Thymocytes were isolated as in (d). CD103 and CXCR5 neuropillin expression in CD4SPCD25−GFP− cells were analyzed by flow cytometry. Data are representative of three independent experiments. (f) Thymocytes were isolated as in (d), CD122+ cells were analyzed by flow cytometry. CDSP thymocytes were gated. Data are representative of three independent experiments.
In view of the above effects of E-protein deletion on IL-2 signaling, we next performed BrdU incorporation assays and Annexin V staining studies to determine if the increased Foxp3+ cells in E-protein KO mice was due to possible IL-2/STAT5 effects on thymocyte proliferation and survival. We did not observe either excessive proliferation or reduced apoptosis of E-protein KO CD4+Foxp3+ and CD4+Foxp3− cells (unpublished data), and thus the increased number of Foxp3+ cells observed in E-protein KO mice was not attributable to increased cell proliferation or survival.
Overall, these findings suggest that E-protein deletion is accompanied by the appearance of cells subject to increased IL-2R signaling, an essential component of both iT reg cell and nT reg cell development. (Yao et al., 2007).
Deletion of E-proteins results in activation of the NF-κB pathway
The NF-κB pathway has been found to be involved in the induction of IL-2 receptor component chains and c-Rel has been shown to be associated with increased Foxp3 expression both in the thymus and in the peripheral lymphoid tissue (Bellavia et al., 2000; Isomura et al., 2009; Ruan et al., 2009; Vang et al., 2010; Zheng et al., 2010). Therefore, the increased IL-2R and Foxp3+ cells in iT reg cells and nT reg cells of mice with E-protein deletion may be the result of increased NF-κB activation.
To test this possibility in iT reg cells, we determined expression of NF-κB activation components by immunoblot analysis and found that c-Rel, but not p50 or p65, was increased in TGF-β–induced iT reg cells from E-protein KO mice as compared with iT reg cells from WT mice (Fig. 6 k). Next, in similar studies of nT reg cells, we determined the level of NF-κB activation components by immunoblot analysis of whole-cell extracts of thymocytes from E-protein KO and WT mice. We found that c-Rel and p50 levels were greatly increased in E-protein KO thymocytes compared with WT thymocytes, whereas the p65 level was unchanged (Fig. 6 d). In addition, these increases were accompanied by increased phosphorylation of IκBα (Fig. 6 e), but not total IκBα (the latter perhaps due to its continuous degradation and resynthesis), as well as increased expression of Iκκ-β (Fig. 6 f). These findings thus indicate that decreased E-protein activity leads to activation of the NF-κB signal pathway and, in particular, the production of c-Rel, a factor having a direct effect on Foxp3 transcription. Parenthetically, we also observed that c-Rel expression was inversely correlated with that of E-protein in Foxp3+ cells, suggesting that NF-κB activation and Foxp3 expression occurs in cells with decreased E-protein activity (Fig. 5, a and b).
E-protein levels decrease in association with TCR stimulation accompanying expression of Foxp3+ T cells
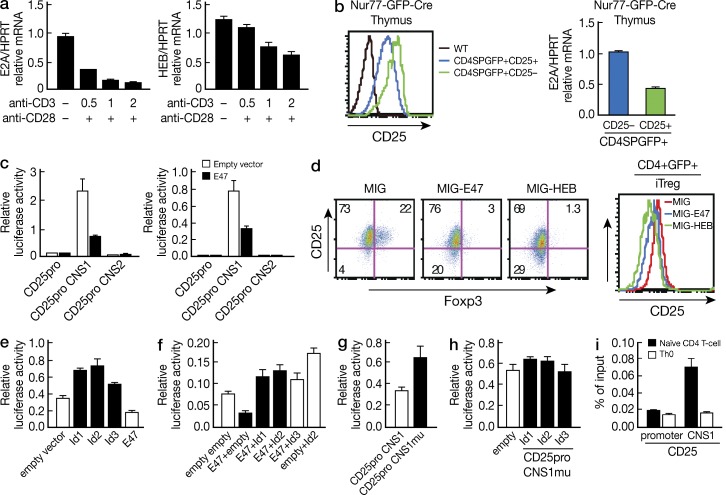
We next focused on the molecular mechanisms that underlie the influence of E-protein on both nT reg cell and iT reg cell development. In initial studies focused on the relation of TCR stimulation to E-protein expression, we determined if the intensity of TCR stimulation, a factor that regulates Foxp3 expression in the normal thymus, also regulates E-protein expression. Consistent with previous findings, we found that TCR stimulation led to dose-dependent E-protein down-regulation (Fig. 7 a). These in vitro results established that TCR signaling causes E-protein down-regulation, and thus that such signaling has the potential to regulate T reg cell differentiation via effects on E-protein levels; however, they did not address whether such regulation does in fact occur in vivo in association with the level of TCR signaling that is required for physiological nT reg cell differentiation in the thymus.
Figure 7.
E-protein activity controls CD25 expression. (a) Naive CD4+ T cells were purified from WT mice and activated with 0.5, 1, or 2 µg/ml anti-CD3ε and 1 µg/ml anti-CD28. E2A or HEB expression was analyzed by real-time RT-PCR 6 h after cell activation. Data are representative of three independent experiments. (b) Thymocytes from Nur77GFP mice were analyzed by flow cytometry. T reg cells were defined as CD4SPGFP+CD25+. (left) Flow cytometric analysis of GFP expression in CD4SPCD25+GFP+(T reg cell, green) and CD4SPCD25-GFP+ (blue) cells from the thymus of Nur77-GFP-Cre or WT (black) mice. (right) Real time RT-PCR assessment of E2A expression in the cell groups defined in Fig. 7 b. Data are representative of two independent experiments. (c) Primary T cell (middle) or EL4 cells were transfected with a CD25 luciferase reporter and empty vector or E47 expressing plasmid as well as a Renilla plasmid and then stimulated with anti-CD3 and anti-CD28; resultant luciferase activity was measured and normalized by Renilla luciferase activity. Data are representative of three independent experiments. (d) Purified CD4+ cells were stimulated by anti-CD3/anti-CD28, transduced with the indicated retroviral vectors, and then cultured under iT reg cell conditions (with TGF-β) for an additional 3 d; (left) flow cytometric plot showing CD25 and Foxp3 expression; right panel shows expression of CD25 in each group of GFP+ cells. Data are representative of three independent experiments. (e and f) EL4 cells were transfected with a reporter plasmid expressing the CD25 promoter linked to CNS1 (CD25pro+CNS1) and Id protein alone (e), or Id protein in combination with E47 (f), and then stimulated with anti-CD3 and anti-CD28; resultant luciferase activity was measured and normalized as above. Data are representative at least three independent studies. (g) EL4 cells were transfected with a reporter plasmid expressing CD25pro+CNS1 plasmid or a CD25pro+CNS1Mu plasmid with mutated E2A binding site at CNS1 and then stimulated with anti-CD3/28; resultant luciferase were measured and normalized as above. Data are representative of three independent experiments. (h) EL4 cells were transfected with a reporter plasmid expressing CD25pro+CNS1 with mutated E2A binding site at CNS1 and Id protein, and then stimulated with anti-CD3/28; resultant luciferase activity was measured and normalized as above. Data are representative of three independent experiments. (i) E-protein binding to the CD25 CNS1 region. Chromatin from purified naive CD4+ T cells and Th0 cells were subjected to immunoprecipitation with anti-E47. Binding of E-proteins to CD25 was analyzed by quantitative PCR. Results are representative of three independent experiments. Data shown are mean ± SD of duplicate PCRs.
To address this latter question, we next determined E-protein levels in Nr4a1(Nur77)-GFP transgenic mice in which it has been shown that the TCR-induced expression of Nur77 (GFP) corresponds to the strength of TCR stimulation and that SP Foxp3+ thymocytes express higher levels of Nur77 than SP Foxp3− thymocytes, indicating that differentiation into Foxp3+ cells is accompanied by more intense TCR stimulation (Moran et al., 2011). These mice thus offered a way of monitoring E-protein expression in cells before and after the physiological TCR stimulation that leads to T reg cell differentiation.
Accordingly, we determined Nur77 (GFP) expression in CD25+CD4SP and CD25−CD4SP thymocytes (i.e., cells equivalent to Foxp3+ and Foxp3− thymocytes, respectively) and showed that whereas CD25+ cells had higher Nur77-GFP than CD25− cells (Fig. 7 b, left), they expressed less E2A mRNA than CD25− cells as determined by quantitative RT-PCR (Fig. 7 b, right). Thus, there was in fact a direct relation between strength of the TCR signal and the level of E-protein expression; in addition, the level of E-protein was decreased in association with the greater strength of the TCR stimulation occurring in association with differentiation of T reg cells. These findings are consistent with the cell sorting studies shown in Fig. 5 in which we demonstrated that in WT mice E-protein levels are decreased in Foxp3+ cells relative to that in Foxp3− cells.
E-protein activity regulates IL-2Rα gene transcription and NF-κB activation
A possible mechanism by which E-proteins could be influencing the development of Foxp3+ T cells is by an effect on the molecular regulation of a key component of Foxp3 expression, the IL-2Rα gene. To investigate this possibility, we next determined if E-protein directly regulates IL-2Rα gene transcription. First, we identified two evolutionarily conserved E2A-binding sites (CANNTG) in each of two conserved noncoding sequences (termed CNS1 and CNS2) located in the intron region of the CD25 gene using the rVISTA tool (unpublished data). We then constructed promotor reporter plasmids containing CNS1 or CNS2 linked to a 0.8-kb promoter (Fig. 7 b, left) and co-transfected these plasmids into either primary CD4+ T cells (Fig. 7 b, middle) or EL4 cells with or without an E47 expression plasmid (Fig. 7 b, right) and stimulated the transfected cells with anti-CD3/CD28. Only the CNS1-containing reporter construct exhibited enhancer activity and, importantly, co-transfection of this plasmid with E2A (E47) led to greatly reduced luciferase activity (Fig. 7 b, middle and right). These data coincided with the finding that ectopic expression of E-protein in developing iT reg cells led to down-regulation of Foxp3+ cells and IL-2Rα (CD25) expression in iT reg cells (Fig. 7, c and d).
Next, we determined the effect of Id proteins (i.e., E-protein inhibitors) on CD25 transcription. We found that co-transfection of Id-protein-expressing plasmids with the CD25-promoter-CNS1 luciferase reporter into EL-4 cells greatly increased CD25 transcriptional activity (Fig. 7 e). In addition, Id protein reversed the inhibitory effect of E47 on CD25 transcription, and importantly, the luciferase activity in cells transfected with Id+E47 was higher than the basal luciferase activity of cells transfected with empty+empty plasmids (Fig. 7 f), suggesting that transfected Id proteins also counteracts the inhibitory effect of endogenous E-proteins on CD25 expression (Fig. 7 f). Confirmation of these various findings came from studies showing that mutation of the E-protein binding site in the CNS1 region of the CD25 reporter plasmid led to an increase in CD25 transcriptional activity (Fig. 7 g) and loss of Id protein augmentation of such activity (Fig. 7 h). Finally, to determine whether E-proteins bind to CD25CNS1, we performed chromatin immunoprecipitation (ChIP) assays with naive and activated CD4+ T cells. Genomic fragments containing the CD25 CNS1 but not the promoter region of CD25 were enriched for E2A binding in naive cells, but neither region was enriched for such binding in activated cells (Fig. 7 i).
The aforementioned findings, taken together with the fact that Foxp3+CD25+ cells are greatly up-regulated in mice lacking E-proteins, provide strong evidence that E-protein activity is critical in the control of CD25+ T cell development and supports the idea that one of the mechanisms by which E-proteins regulate T reg cell development is through its negative regulation of IL-2Rα expression.
NF-κB activation is increased in E-protein KO thymocytes and, in prior studies, it is also increased in thymocytes of Id1 transgenic mice (Yang et al., 2006). We therefore next investigated the effect of decreased E-protein activity on NF-κB activation. In these studies we transfected a NF-κB luciferase reporter, driven by a promoter containing five copies of an NF-κB response element (NF-κB-RE, Promega, PGL4.32), into 16610D9 cells along with Id1-, Id2-, Id3- or E47-expression constructs individually or in combination. We found that transfection of each of the of Id plasmids led to increased NF-κB reporter activity whereas, in contrast, transfection of the E-protein (E47) plasmid led to reduced NF-κB activity (Fig. 8 a). In addition, cotransfection of E47 and Id protein constructs decreased the stimulatory effect of Id protein on NF-κB luciferase activity (Fig. 8 b), suggesting that inhibition of E-protein activity by Id protein results in activation of NF-κB, whereas increased E-protein activity inhibits such activation.
Figure 8.
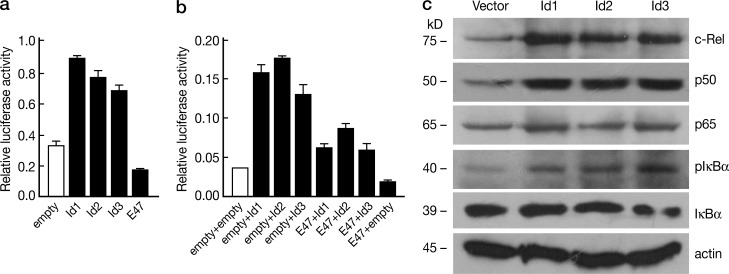
E-protein activity controls NF-κB activation. (a and b) 16610D9 cells were transiently transfected with NF-κB luciferase reporter (PGL4.32) and CMV-Renilla along with E47 and Id protein expression plasmids individually (a) or in combination (b). Transfected cells were cultured overnight, and then stimulated with 2 µg/ml anti-CD3 and 1 µg/ml anti-CD28 for 2 h. Luciferase activity was measured and normalized by Renilla luciferase activity. Data shown are derived from mean ± SD of duplicate transfections in each experiment and are representative of at least three independent experiments. (c) 16610D9 cells were retrovirally transduced with empty vector, Id1-, Id2- or Id3-expressing vectors, and were stimulated with 2 µg/ml plate-bound anti-CD3 and anti-CD28 for 2h. Whole-cell lysates obtained were immunoblotted with antibodies against the indicated proteins. Data are representative of three independent experiments.
Finally, to determine the effect of E-protein blockade on endogenous NF-κB signaling we transduced 16610D9 cells with Id protein–expressing retroviruses and then stimulated the cells with anti-CD3/anti-CD28; subsequently, cell extracts were subjected to immunoblotting with various NF-κB–related antibodies. c-Rel and p50 expression was dramatically increased in these Id-protein–transduced cells (Fig. 8 c), whereas p65 was not. As expected, this evidence of increased NF-κB activation was accompanied by elevated IκBα phosphorylation (Fig. 8 c). Thus, the effect of Id protein blockade of E-protein is consistent with the increased activation of NF-κB observed in E-protein KO cells. Collectively, these studies provide strong evidence that decreases in E-protein activity leads to NF-κB activation.
Microarray analysis reveals that E-protein deletion results in gene activation changes accompanying Foxp3 differentiation
To further identify target genes directly regulated by E-protein activity, we performed gene profiling on sorted WT CD4+Foxp3+ and CD4SPFoxp3− thymocytes and sorted CD4+Foxp3− cell from tamoxifen-treated E2Af/fHEBf/f/ER-Cre/Foxp3KI mice. With this approach, we could determine the E-protein effect distinct from the effect of Foxp3 induction by comparing gene expression profiles of CD4SPFoxp3− cells from WT and E-protein KO mice (Fig. 9 a). On the other hand, we could also determine the “Foxp3 differentiation effect” by comparing gene expression profiles of WT CD4+Foxp3− cell and WT CD4+Foxp3+ cells (Fig. 9 a). It should be noted that E-protein activity in WT Foxp3+ cells is also decreased relative to that in WT Foxp3− cells (Fig. 5).
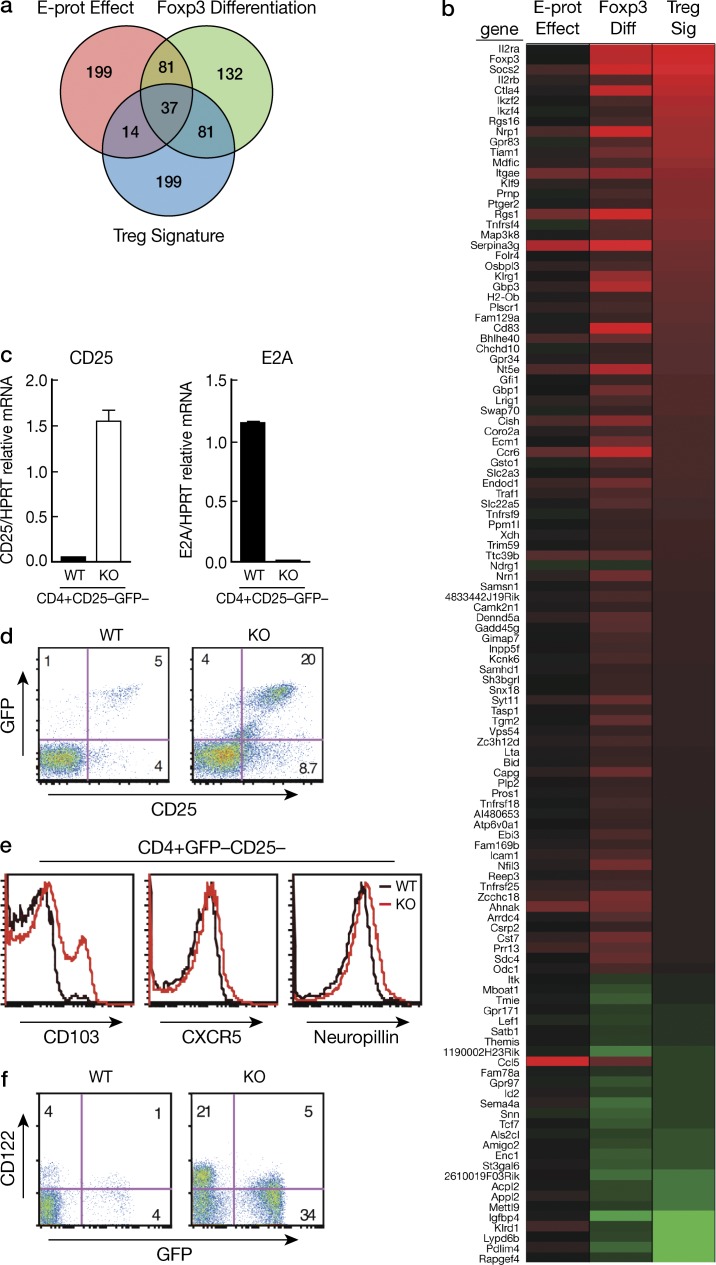
Microarray analysis to detect genes responding to the aforementioned effects indicates that ∼1,250 genes exhibited mRNA transcript changes (significant at FDR 0.05) due to the E-protein effect, including 115 genes that have linear fold changes greater than 2.0, up- or down-regulated (Table S1). Importantly, >50% (60 of 115) of these E-protein–responsive genes were also responsive to Foxp3 differentiation. Hill et al. (2007) described a set of T reg cell signature genes, 331 of which were also assayed in the present study. Of the 118 genes common to both FoxP3+ differentiation and the T reg cell signature genes, ∼30% (37/118) were also observed to be responsive to E-protein (Venn diagram, Fig. 9 a; Table S1). This latter subset of genes includes “hallmarks of T reg cells” (Hill et al., 2007) such as Itgae, Gpr83, and Nrp1. Thus, the E-protein effect (analogous to E-protein down-regulation caused by TCR signaling) accomplished many of the molecular changes accompanying and presumably necessary for Foxp3 differentiation (Fig. 9 b, heat map of 118 genes).
In an additional analysis, IPA gene annotation analysis software (from Ingenuity, Inc.) was used to search for putative modulators in the NF-κB and the IL-2 signaling pathways that could account for the observed E-protein or Foxp3 differentiation effects. For each effect, we selected 1,000 genes with the largest expression difference observed by microarray (Table S1) as input to the software, from which the program inferred the increased or decreased activity of regulatory molecules based on the known relationships (up-/down-regulated) between each cataloged regulator and its set of theoretical targets. The results indicated that the activation of regulators in the NF-κB and IL-2 pathways predicted for the E-protein effect mimicked that for Foxp3+ cell differentiation to a remarkable extent (Fig. 9 and Table S2). It should be noted, however, that this similarity of upstream signaling predictions did not extend to all signaling pathways, and, for instance, it was inferred that MAPK signaling pathways were activated by Foxp3+ differentiation but not by the E-protein effect (unpublished data). The fact that E-protein down-regulation (measured experimentally as the E-protein effect) does not capture the entire spectrum of gene changes associated with Foxp3 differentiation explains the observation that E-protein deletion does not result in a thymocyte population in which all cells express Foxp3.
As noted above, the microarray study showed that many of the Foxp3 signature genes were up-regulated in E-protein KO CD4+Foxp3− cells. However, one important signature gene that was not up-regulated was the IL-2α gene encoding CD25, despite the fact that our in vitro molecular studies showed that decreased E-protein activity leads to de-repression of this gene. To investigate this discrepancy, we obtained CD4+GFP− (Foxp3−) cells by cell sorting thymocytes from tamoxifen-treated WT Foxp3KI and E2Af/fHEBf/f Er-cre/Foxp3−GFP-KI mice and then determined the level of CD25 mRNA in the purified GFP− cells by real-time RT-PCR. We found that CD25 mRNA levels were, in fact, increased in E-protein KO CD4+Foxp3− cells (Fig. 9 c). In confirmation of this finding, we found in flow cytometric analysis of thymocytes from E-protein KO and WT mice with an anti-CD25 antibody conjugated to a different fluorochrome from that used in Fig. 2 a (anti–CD25-PE vs. the previously used anti–CD25-APC) that thymocytes from KO mice contained an increased percentage of Foxp3−CD25+ cells compared with thymocytes from WT mice (Fig. 9 d). However, the percentage of CD25+ Foxp3− cells in the E-protein KO thymus obtained in this study may underestimate the number of such cells generated in the KO thymus as these cells are subject to IL-2 signaling and thus rapid differentiation into Foxp3+ cells. Overall, these data suggest that the lack of detection of IL-2Rα gene activation in the microarray study was due to the fact that the microarray probe was relatively insensitive in detecting such activation. It should be noted, however, that they are fully consistent with the microarray IPA study that identified that the IL-2 signaling pathway is up-regulated in E-protein KO cells before Foxp3 cell development.
In further studies to verify the microarray results, we compared CD122, CD103, CXCR5, and neuropillin protein levels in CD4+Foxp3− thymocytes from WT and E-protein KO mice by flow cytometric analysis and showed that these proteins were increased in the CD4+Foxp3− cells from the KO mice, consistent with the data from the microarray study (Fig. 9, e and f).
DISCUSSION
Previous studies have shown that E-protein transcription factors in the thymus have important roles, both in the very earliest stages of thymocyte development and in the regulation of key thymocyte transitions (Murre 2005; Kee 2009; and Jones and Zhuang, 2011). In particular, they have provided convincing evidence that preTCR and TCR signaling leads to successively more profound down-regulation of E-protein levels and that such down-regulation is a precondition for DN to DP transitions and DP to SP transitions (Engel et al., 2001; Jones and Zhuang, 2007; Jones-Mason et al., 2012). Here, we used inducible E-protein KO mice (E2Af/f/HEBf/f/Er-cre mice) to study the effect of E-proteins (and the related effect of Id proteins) on the development of Foxp3+ T reg cells. We found that induced Foxp3+ T reg cell (iT reg cell) development in peripheral tissues was significantly increased both in vitro and in vivo in E-protein KO mice. In addition, this increase in iT reg cell development was accompanied by a significant increase in both the percentage of Foxp3+ cells and total number of Foxp3+ cells (nT reg cells) in the thymus of E-protein KO mice. Finally, the effects of E-protein deletion in E-protein KO mice were mimicked in CD4+ thymocytes in normal mice cells in that Foxp3+ thymocytes exhibit lower E-protein activity than CD4+Foxp3− cells. Thus, our findings provide strong evidence that E-proteins, in addition to being regulators of thymocyte transitions, also are regulators of T reg cell development.
E-protein deletion was associated with several events, both in developing iT reg cells and nT reg cells that facilitated the development of T reg cells. One of these was the up-regulation of IL-2 receptor chains that, in turn, led to increased activation of STAT5, a critical regulator of Foxp3 transcription (Yao et al., 2007; Burchill et al., 2008). Thus, the effect of E-protein deletion on Foxp3 expression is in accord with the “two-step model” of T reg cell development that holds that TCR signaling leading to induction of Foxp3 acts first through its up-regulation of proximal IL-2 signaling components (IL-2Rα) and second through facilitation of cytokine-mediated (i.e., Stat5-mediated) induction of Foxp3 (Lio and Hsieh, 2008). Analysis of the mechanism of E-protein function led to the important finding that TCR signaling causes down-regulation of E-protein expression and that E-protein is a negative regulator of IL-2Rα transcription that acts via binding to a conserved CD25 intronic enhancer. Thus, the picture that emerges is that TCR-induced down-regulation of E-protein facilitates CD25 expression by relieving the negative effect of E-protein on IL-2Rα transcription. This conclusion is consistent with our finding that E-protein deletion was accompanied by increased IL-2R signaling, and a previous finding that decreased E47 expression is associated with increased IL-2Rα transcription (Schwartz et al., 2006).
A second consequence of E-protein deletion that relates to its effect on T reg cell development is our observation that such deletion was associated with greatly increased NF-κB activation in both developing nT reg cells and iT reg cells. This finding was presaged by previous studies that showed that in mice with deletion of signaling molecules linked to TCR-induced NF-κB activation, nT reg cell development is severely impaired (Gupta et al., 2008; Medoff et al., 2009). In addition, it had been shown that mice bearing transgenes that inhibit E-protein activity (Id1 and Tal1) exhibit increased NF-κB activity in thymocytes (Kim et al., 2002). In our studies, both p50 and c-Rel in E-protein KO mice displayed increases in p50 and c-Rel but not in p65. These findings are in accord with previous studies showing that c-Rel is critically important to Foxp3 transcription (Isomura et al., 2009; Ruan et al., 2009) as well as studies showing that while p50 (NF-kB1) is not important in the development of nT reg cell progenitors, it does have a role in the development of mature T reg cells (Vang et al., 2010). It should be noted, however, that it is not likely that the up-regulation of CD25 expression in E-protein KO mice is due to an direct effect of simultaneous NF-κB up-regulation, because in previous studies of thymocytes bearing a transgene expressing Iκκ-β and thus constitutive NF-κB activation, the cells did not express increased CD25 (Long et al., 2009).
In studies of the mechanism of E-protein on NF-κB activation, we used an NF-κB reporter assay and retroviral transduction system to show that Id proteins have a direct enhancing effect on NF-κB activation, whereas E-protein inhibits such activation. These findings are consistent with previous studies that have shown that mice bearing an Id1 transgene exhibit increased basal NF-κB activity that is further increased by TCR stimulation (Yang et al., 2006). Overall, these findings suggest that changes in E-protein activity (i.e., the ratio of E protein/Id protein) during TCR stimulation is a critical regulator of NF-κB activation in developing T reg cells, and thus in the generation of key transcription factors necessary for T reg cell development. However, the precise mechanism by which E-protein affects initiation of the NF-κB activation cascade remains an area of further investigation.
In a previous study of mice with Id3 deletion, it was shown that Foxp3+ cell development, both in vivo and in vitro (after TGF-β induction), was impaired in cells mice with such deletion due to the fact that, in the absence of Id3, IL-4 production is greatly increased and this leads to increased expression of GATA3, a factor that has a negative effect on Foxp3 transcription (Maruyama et al., 2011). However, we found normal thymic Foxp3+ cells in 6–8-wk-old Id3−/− mice or in Id3−/−IL-4−/− mice. It was only in Id2−/−Id3−/− mice that we observed decreased Foxp3+ cells, presumably because Id2 and Id3 are functionally redundant. It should be noted, however, that these studies were conducted with relatively young mice (6–8 wk of age), and we did observe that Foxp3+ cells decreased somewhat in 4–6-mo-old Id3−/− mice (unpublished data), possibly due to the fact that as mice age, IL-4 production increases (unpublished data). The same study also found, mainly in in vitro studies (in this case, of EL-4 cells or TGF-β–induced CD4+ cells), that E-proteins bind to the Foxp3 promoter and, in doing so, promote Foxp3 expression. These data are directly contrary to our extensive in vivo and in vitro data showing that, on the one hand, E-protein deletion enhances T reg cell development, and on the other hand, combined deletion of inhibitors of E-protein, Id3, and Id2, suppress such development.
In another previous study of mice relating to the effect of deletion of Id2 and/or Id3, the effect of deletion of these proteins on Foxp3+ cells specifically were examined (Miyazaki et al., 2014). In broad agreement with the studies reported here, although the number of Foxp3+ cells were not decreased in peripheral tissues (the thymus was not examined), the expression of Foxp3 in individual cells was diminished and the percentage of CD25+/helios+ cells were decreased. In addition, Id2/Id3 deletion led to widespread Th2-mediated inflammation, suggesting the presence of diminished T reg cell function. Finally, Id protein deletion was associated with multiple changes in the phenotype of T reg cells and to functional changes in the maintenance and homing of these cells. In comparing the results of the present studies with those evaluating the effect of Id protein deletion, it should be noted that the effects of changes in E-protein and Id protein levels cannot assumed to be strictly reciprocal because Id proteins have variable effects on E-protein levels and, in addition, Id proteins affect the function of other transcription factors.
There is now reasonable consensus that TCR signaling strength plays a key role in nT reg cell development (Moran et al., 2011); thus, although low-affinity TCR engagement leads to positive selection and high-affinity TCR engagement triggers negative selection, intermediate-affinity TCR signaling leads to nT reg cell development (Hsieh et al., 2012). However, it is still not clear how these different signal strengths operate at the molecular level to induce T reg cells. One possibility is that signal strength regulates Foxp3+ T reg cell development via a direct effect on NF-κB activity. Support for this view comes from a recent study showing that mice bearing transgenes that lead to constitutive NF-κB activation exhibit markedly increased nT reg cell induction (Long et al., 2009). In these studies, NF-κB overexpression was associated with nT reg cell induction in cells bearing a TCR-recognizing a conventional antigen; such overexpression appeared to be able to lower the level of TCR affinity necessary for induction of Foxp3+ T cells. Thus, these data suggest that TCR-induced activation of NF-κB sets the threshold for nT reg cell development.
This NF-κB–centric model of nT reg cell induction is, however, likely to be incomplete because it fails to incorporate the effect of TCR-induced down-regulation of E-proteins. The significance of the latter is shown most clearly in the microarray study in which E-protein deletion itself, even in Foxp3-negative cells, was shown to lead to activation of the NF-κB and IL-2 signaling pathways. This implies that in developing WT nT reg cells where E-protein down-regulation is occurring at the same time as Foxp3 induction, the activation of NF-κB is most likely an indirect effect of TCR signaling acting via down-regulation of E-protein activity. In addition, in view of the microarray finding that E-protein deletion led to up-regulation of many Foxp3 signature genes, it is likely that down-regulation of E-protein affects many aspects of Foxp3+ T cell development in addition to NF-κB activation or CD25 up-regulation. On this basis, we favor a model of TCR-induced Foxp3 induction in which one of the key initiating events is down-regulation of E-protein followed by de-repression of the transcription of many of the genes critically involved in Foxp3 expression such as the CD25 gene and an upstream NF-κB gene. This, in turn, is followed by the activation of the NF-κB and IL-2 signaling pathways and the elaboration of factors that underpin T reg cell development. Thus, in this view, modulation of E-protein activity is setting the threshold for TCR induction of Foxp3.
EXPERIMENTAL PROCEDURES
Mice.
OT-II mice and IL-4−/− and WT CD45.1 and Nur77-GFP-Cre mice were obtained from Jackson Laboratories. Tcf3 (E2A)f/fTcf12(HEB)f/f Rosa-26-ER-Cre mice, Id2f/fId3f/f mice, Id3−/− mice. and Foxp3-GFP-KI mice were gifts from Y. Zhuang (Duke University, Durham, NC) and M. Oukka (Brigham and Woman’s Hospital, Boston, MA) respectively. Strain-matched WT-Cre mice or floxed E2A/HEB/ER-Cre− mice served as controls. E2Af/fHEBf/f ER-cre mice were crossed with Foxp3-GFP-KI mice to generate E2Af/fHEBf/f/Cre/Foxp3-GFP-KI mice. To induce E-protein deletion in vivo, WT or floxed mice were orally administered tamoxifen (Sigma-Aldrich) dissolved in corn oil 3 times every other day (3 mg/mice) over a period of 8 d. Analysis of cells in the tamoxifen-treated mice was conducted 2 d after the last tamoxifen administration. All animal experimentation was approved by and performed according to guidelines from the Institutional Animal Care and Use Committee at Chinese Academy of Science and National Institutes of Health.
Flow cytometry and cell sorting.
Flow cytometry analyses were performed by staining cells with antibodies (all from BD) as previously described (Vang et al., 2008). Flow cytometric analysis of Foxp3 (eBioscience) and phospho-STAT5 (BD) was performed according to the manufacturer’s protocols. Stained cell were analyzed on a FACSCalibur flow cytometer (BD). Cell sorting was performed using a FACSAria (BD).
Immunoblotting.
Thymocytes from tamoxifen-treated WT, E2A/HEB/ER-Cre/Foxp3-GFP-KI mice or cultured cells were lysed in RIPA buffer after which whole-cell extracts were collected and analyzed by SDS-PAGE. Antibodies against c-Rel, p50, p65, and IkBa were obtained from Santa Cruz Biotechnology, Inc.; Iκκ-β, p-IκBα, STAT5, pSTAT5, Smad3, and pSmad3 were obtained from Cell Signaling Technology.
Retroviral transduction.
Retroviral transduction was performed as previously described (Zhang et al., 2008). In brief, plated E cells were transfected with retroviral constructs expressing E47, HEB or Id1, Id2, and Id3 by Lipofectamine 2000 (Invitrogen), and retroviral supernatant was collected and transduced with activated CD4 T cells or 16610 D9 cells. Transduced cells were cultured under Th0 conditions (no cytokine added) or T reg cell conditions (TGF-β added) for an additional 3 d. Foxp3 and CD25 expression were assessed by flow cytometry. 16610 D9 cells were directly transduced with viral supernatant as above. Whole-cell lysates were analyzed by immunoblotting with the antibodies indicated.
Quantitative PCR.
RNA was purified from thymus or from sorted DP, CD4+GFP+ cells, or CD4+GFP− cells. cDNA was obtain by using a High Capacity cDNA Reverse Transcription kit (Applied Biosystems). Real-time PCR was performed using the ABI PRISM 7700 Real-time PCR system by using gene-specific primers and probes (Applied Biosystems). An internal control (Hprt) was used to normalize the expression value of the target gene. The primers and probes are as follows: Hprt, Mm00446968_m1; E2A, Mm01175588_m1; HEB, Mm00441699_m1; Id3, Mm00492575_m1; Foxp3, Mm00475162_m1; c-Rel, Mm01239661_m1. For human probe: ID3, Hs00954037_g1; E2A, Hs00413032_m1; HEB, Hs00918966_m1; Foxp3, Hs01085834_m1; Hprt, Hs02800695_m1. CD25 primer and probe were purchased from Roche. CD25 Probe1# from Universal Probelibrary, primer sequence: forward 5′-TGGTCTATATGCGTTGCTTAGG-3′, reverse 5′-AACTTGCTTTCTCGATTTGTCA-3′.
Transfer model of colitis.
Purified CD4+CD45RBhi WT cells, CD4+CD25+GFP+ cells from Tamoxifen-treated WT Foxp3-GFP-KI Cre mice (WT T reg cells) or from floxed E2A/HEB ER-Cre/Foxp3-GFP-KI mice (KO T reg cells) were obtained by sorting on a FACSAria cell sorter. 3 × 105 CD4+CD45RBhi WT cells were injected intraperitoneally alone, in combination with 2 × 105 WT T reg cells, or 2 × 105 KO T reg cells into Rag2−/− mice. Mice were weighed and assessed for clinical signs of colitis weekly. Colon pathology was assessed by microscopy of H&E stained tissues.
Generation of BM chimeras.
BM cells were isolated from the femurs and tibias of both WT or floxed E2A/HEB ER-Cre mice, BM chimeras were generated by intravenous transfer of donor BM cells separately or after mixture at different ratios (5 × 106 BM cells total) into sublethally irradiated (600 Rad) Rag2−/− mice. Tamoxifen was orally administered 5 wk after cell transfer. Foxp3 expression was analyzed 2 d after the administration of the final tamoxifen.
In vitro induction of T reg cell differentiation.
Naive CD4+ cells were purified from WT and floxed E2A/HEB ER-Cre or E2A/HEB ERCre/Foxp3-GFP-KI mice using anti–mouse CD4 beads (Miltenyi Biotec) and were activated with anti-CD3/CD28, cultured in the presence of TGF-β (2 ng/ml) plus 4-hydoxytamoxifen (4-OHT, 2 µM final concentration) for 3 d. In some of experiments, the cells were cultured in the presence of TGE-β and IL-2 as indicated. Foxp3 and CD25 were assessed by flow cytometry.
Induction of iT reg cells in vivo.
Naive CD4+CD62Lhi T cells (CD45.2) were purified from spleens of WT Foxp3-GFP-KI OT-II or E2A/HEB ERCre/Foxp3-GFP-KI OT-II mice using a Naive CD4 T cell kit (Miltenyi Biotec), and then adoptively transferred into CD45.1 mice by intravenous injection (2 × 106 cells/mouse). After cell transfer, tamoxifen was orally administered every other day for three times. 2 d after the final tamoxifen administration, the recipient mice were immunized subcutaneously twice with soluble chicken OVA323-339 peptide (OVA peptide in PBS, 0.4 mg/mouse) and orally once (0.4 mg/mouse) between the first and last subcutaneous injection. 2 d after the final injection, cells were collected from peripheral lymphoid tissues and analyzed.
Reporter assay.
CD25 promoter was obtained by PCR using mouse genomic DNA as a template. PCR product was cloned into the pGL4.10 luciferase vector (Promega) using the NheI and BglII entry sites. CNS1 and CNS2 were cloned downstream of CD25 promoter. Mutations in CNS1 were introduced by using the QuickChange II site-directed mutagenesis kit (Stratagene). PCR primer for mutation in CD25 CNS1 is 5′-GCCAAGCCCCACTCCGCAGAGCATCAGCATG-3′. All constructs were verified by sequencing. Primary T cell or EL4 cells were transfected using a Nucleofector II machine (Amaxa). 2 × 106 cells in 100 µl Amaxa solution were transfected with 3 µg of luciferase reporter construct and 0.1 µg pRL-TK plasmid along with 6 µg of plasmid encoding E protein, Id protein, or control plasmid. Transfected cells were rested for 4 h, stimulated with anti-CD3/28. Luciferase activity was measured by using a dual luciferase assay system (Promega). Transfections were performed in duplicate in each experiment.
ChIP assay.
Chromatin for ChIP assays were obtained by using naive CD4 T cell and activated T cells and were performed as previously described (Kanno et al., 2008). In brief, the chromatin was incubated with anti-E2A (Santa Cruz Biotechnology, Inc.), or normal rabbit IgG antibody coupled Dynabeads Protein A at 4°C over-night. The beads were washed extensively with wash buffer, and beads–protein–DNA complex were then subjected to proteinase K digestion at 65°C for 4 h and purified. Finally, the DNA was subjected to real-time PCR using primer pairs and probe CD25 promoter: forward, 5′-CTGAAAGGCCTCCTGGGAGAAGC-3′, reverse 5′-CGGTATTGGTTCACTCCATGCACGGTGG-3′, probe 5′-FAM-GTGAATCCCACACCCATGGAAC-TAMRA-3′; CD25CNS1 forward 5′-GACACTTGCTGACTGTGGGCAGG-3′, reverse 5′-GGTCTGAACAGAGAGGTGTCACTAGC-3′, probe 5′-FAM-CACTCAGCTGAGCATCAGCATGG-TAMRA-3′.
Microarray hybridization.
RNA quality was verified by Bioanalyzer with RNA Integrity Number (RIN) greater than 9.0 for all samples. Amplification and labeling of the RNA samples were performed using the Illumina TotalPrep RNA Amplification (Applied Biosystems) and an input of 500 ng of total RNA per sample. Biotinylated aRNA was hybridized to Illumina MouseRef-8 v2.0 Expression BeadChip (Gene Expression Omnibus accession no. GPL6885) having 25,697 unique probes, using reagents provided, and imaged using the Illumina HiScan-SQ.
Microarray expression analysis.
Signal data were extracted from the image files with the Gene Expression module (v. 1.9.0) of the GenomeStudio software (v. 2011.1) from Illumina, Inc. Signal intensities were converted to log2 scale. Calculation of detection P values is described in the GenomeStudio Gene Expression Module User Guide. Data for array probes with insufficient signal (at least two arrays must have detection P value < 0.1) were removed from the dataset. After dropping nonperforming probes, quantile normalization was applied across all arrays. ANOVA was performed on the normalized log2 expression estimates to test for mRNA expression differences by strain (WT [Verykokakis et al, 2010] or knockout [Yang et al., 2006] of E-proteins) and sort fraction (FoxP3+ or FoxP3−). A P value of 0.05 was used for the statistical significance cutoff, after adjusting for the familywize error rate (FWER) using Benjamini–Hochberg method to account for multiple testing. Statistical analysis was performed using JMP/Genomics software version 6.0 (SAS Institute Inc.).
Comparison of microarray results to previously defined “T reg cell signature” genes.
Hill et al. (2007) performed a metaanalysis to define a set of T reg cell genes based on consistent expression differences (at least 1.5 fold-change) across several microarray datasets. This “T reg cell signature” set comprised 603 probes from the Affymetrix mouse M430v2 platform. We assigned Entrez gene identifiers to these Affymetrix probes and to the Illumina Mouse Ref-8 v2.0 probes using microarray platform annotation available from the mAdb database. Microarray probes were assigned NCBI refSeq transcript IDs based on 99% sequence identity using BLAST search. Each refSeq transcript has an associated gene (Entrez gene ID) that served as the basis for our cross-indexing. Only data that could be resolved by matching gene IDs across the two platforms was used. Because the T reg cell signature probes mapped to 331 gene identifiers common to both platforms, we tested for overlaps with the top 331 genes (based on largest absolute log ratios) from each of our contrasts. This approach was used to generate an equitable representation of high scoring genes, though it is not exhaustive in terms of expression difference detection for all genes (Table S1). In general, the behavior of the T reg cell signature genes and the FoxP3 differentiation genes were well correlated.
Pathway analysis.
Upstream regulator predictions for the E-protein effect (KO, FoxP3− vs. WT, FoxP3−) and FoxP3+ differentiation (WT, FoxP3+ vs. WT, FoxP3−) were computed in IPA software (Ingenuity). As input for the search, for each contrast, we took 1000 genes having the largest expression difference. The search parameter settings were as follows: reference set = Illumina Ref-8 v2.0; species = Mouse; relationships = relaxed; confidence = Experimentally Observed or High Confidence prediction; data source = Ingenuity Expert/ExpertAssist Findings or Gene Ontology. This analysis infers the increased or decreased activity of regulatory molecules based on the behavior of their downstream target genes (expression observed by microarray) and the known relationship (as encapsulated in the IPA database) between the regulator and target. Upstream Regulator analysis indicates significance by (1) an overlap P value that tests for statistical enrichment of genes in the input set against the set of known target genes for each regulator, and (2) an activation Z-score, which tests the concordance of predicted up/down expression with those observed by microarray. The approach is described in a whitepaper titled “Ingenuity Upstream Regulator Analysis.”
Upstream regulator predictions for the E-protein effect or FoxP3+ differentiation were selected based on overlap P value < 10−4, and their activation Z-score values were mapped onto elements of the IPA canonical pathways “IL-2 Signaling” and “Regulation of IL-2 Expression in Activated T Lymphocytes” or “Regulation of NF-κB Expression in Activated T-Lymphocytes.” In contrast, regulators were predicted for FoxP3+ differentiation but not for the E-protein effect in many pathways featuring MAPK signaling cascades.
Online supplemental material.
Table S1 shows statistical analysis of genes for E-protein effect or Foxp3+differentiation. Table S2 shows predicted upstream regulatory effects based on microarray expression differences. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20132681/DC1.
Supplementary Material
Acknowledgments
The authors wish to thank Dr. Yuan Zhuang for providing mice and John O’Shea and Xuyu Zhou for critical reading of the manuscript.
This work was supported by National Natural science Foundation of China (31270933, F. Zhang), Ministry of Health of China (2013ZX10004608, F. Zhang), and Intramural Research Program of National Institute of Allergy and Infectious Diseases (W. Strober).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- iT reg cell
- induced T reg cell
- nT reg cell
- naturally arising T reg cell, regulatory T cell
References
- Bayer A.L., Yu A., and Malek T.R.. 2007. Function of the IL-2R for thymic and peripheral CD4+CD25+ Foxp3+ T regulatory cells. J. Immunol. 178:4062–4071 10.4049/jimmunol.178.7.4062 [DOI] [PubMed] [Google Scholar]
- Bellavia D., Campese A.F., Alesse E., Vacca A., Felli M.P., Balestri A., Stoppacciaro A., Tiveron C., Tatangelo L., Giovarelli M., et al. . 2000. Constitutive activation of NF-kappaB and T-cell leukemia/lymphoma in Notch3 transgenic mice. EMBO J. 19:3337–3348 10.1093/emboj/19.13.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchill M.A., Yang J., Vogtenhuber C., Blazar B.R., and Farrar M.A.. 2007. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J. Immunol. 178:280–290 10.4049/jimmunol.178.1.280 [DOI] [PubMed] [Google Scholar]
- Burchill M.A., Yang J., Vang K.B., Moon J.J., Chu H.H., Lio C.W., Vegoe A.L., Hsieh C.S., Jenkins M.K., and Farrar M.A.. 2008. Linked T cell receptor and cytokine signaling govern the development of the regulatory T cell repertoire. Immunity. 28:112–121 10.1016/j.immuni.2007.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G., Yu A., and Malek T.R.. 2011. T-cell tolerance and the multi-functional role of IL-2R signaling in T-regulatory cells. Immunol. Rev. 241:63–76 10.1111/j.1600-065X.2011.01004.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel I., Johns C., Bain G., Rivera R.R., and Murre C.. 2001. Early thymocyte development is regulated by modulation of E2A protein activity. J. Exp. Med. 194:733–745 10.1084/jem.194.6.733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Manicassamy S., Vasu C., Kumar A., Shang W., and Sun Z.. 2008. Differential requirement of PKC-theta in the development and function of natural regulatory T cells. Mol. Immunol. 46:213–224 10.1016/j.molimm.2008.08.275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J.A., Feuerer M., Tash K., Haxhinasto S., Perez J., Melamed R., Mathis D., and Benoist C.. 2007. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 27:786–800 10.1016/j.immuni.2007.09.010 [DOI] [PubMed] [Google Scholar]
- Hsieh C.S., Lee H.M., and Lio C.W.. 2012. Selection of regulatory T cells in the thymus. Nat. Rev. Immunol. 12:157–167. [DOI] [PubMed] [Google Scholar]
- Isomura I., Palmer S., Grumont R.J., Bunting K., Hoyne G., Wilkinson N., Banerjee A., Proietto A., Gugasyan R., Wu L., et al. . 2009. c-Rel is required for the development of thymic Foxp3+ CD4 regulatory T cells. J. Exp. Med. 206:3001–3014 10.1084/jem.20091411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.E., and Zhuang Y.. 2007. Acquisition of a functional T cell receptor during T lymphocyte development is enforced by HEB and E2A transcription factors. Immunity. 27:860–870 10.1016/j.immuni.2007.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.E., and Zhuang Y.. 2011. Stage-specific functions of E-proteins at the β-selection and T-cell receptor checkpoints during thymocyte development. Immunol. Res. 49:202–215 10.1007/s12026-010-8182-x [DOI] [PubMed] [Google Scholar]
- Jones M.E., Kondo M., and Zhuang Y.. 2009. A tamoxifen inducible knock-in allele for investigation of E2A function. BMC Dev. Biol. 9:51 10.1186/1471-213X-9-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Mason M.E., Zhao X., Kappes D., Lasorella A., Iavarone A., and Zhuang Y.. 2012. E protein transcription factors are required for the development of CD4(+) lineage T cells. Immunity. 36:348–361 10.1016/j.immuni.2012.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y., Takane Y., Takizawa Y., and Inouye Y.. 2008. Suppressive effect of aryl hydrocarbon receptor repressor on transcriptional activity of estrogen receptor alpha by protein-protein interaction in stably and transiently expressing cell lines. Mol. Cell. Endocrinol. 291:87–94 10.1016/j.mce.2008.05.004 [DOI] [PubMed] [Google Scholar]
- Kee B.L.2009. E and ID proteins branch out. Nat. Rev. Immunol. 9:175–184 10.1038/nri2507 [DOI] [PubMed] [Google Scholar]
- Kim D., Xu M., Nie L., Peng X.C., Jimi E., Voll R.E., Nguyen T., Ghosh S., and Sun X.H.. 2002. Helix-loop-helix proteins regulate pre-TCR and TCR signaling through modulation of Rel/NF-kappaB activities. Immunity. 16:9–21 10.1016/S1074-7613(02)00264-9 [DOI] [PubMed] [Google Scholar]
- Lio C.W., and Hsieh C.S.. 2008. A two-step process for thymic regulatory T cell development. Immunity. 28:100–111 10.1016/j.immuni.2007.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M., Park S.G., Strickland I., Hayden M.S., and Ghosh S.. 2009. Nuclear factor-kappaB modulates regulatory T cell development by directly regulating expression of Foxp3 transcription factor. Immunity. 31:921–931 10.1016/j.immuni.2009.09.022 [DOI] [PubMed] [Google Scholar]
- Malek T.R.2008. The biology of interleukin-2. Annu. Rev. Immunol. 26:453–479 10.1146/annurev.immunol.26.021607.090357 [DOI] [PubMed] [Google Scholar]
- Maruyama T., Li J., Vaque J.P., Konkel J.E., Wang W., Zhang B., Zhang P., Zamarron B.F., Yu D., Wu Y., et al. . 2011. Control of the differentiation of regulatory T cells and T(H)17 cells by the DNA-binding inhibitor Id3. Nat. Immunol. 12:86–95 10.1038/ni.1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medoff B.D., Sandall B.P., Landry A., Nagahama K., Mizoguchi A., Luster A.D., and Xavier R.J.. 2009. Differential requirement for CARMA1 in agonist-selected T-cell development. Eur. J. Immunol. 39:78–84 10.1002/eji.200838734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki M., Miyazaki K., Chen S., Itoi M., Miller M., Lu L.F., Varki N., Chang A.N., Broide D.H., and Murre C.. 2014. Id2 and Id3 maintain the regulatory T cell pool to suppress inflammatory disease. Nat. Immunol. 15:767–776 10.1038/ni.2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran A.E., Holzapfel K.L., Xing Y., Cunningham N.R., Maltzman J.S., Punt J., and Hogquist K.A.. 2011. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J. Exp. Med. 208:1279–1289 10.1084/jem.20110308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C.2005. Helix-loop-helix proteins and lymphocyte development. Nat. Immunol. 6:1079–1086 10.1038/ni1260 [DOI] [PubMed] [Google Scholar]
- Ruan Q., Kameswaran V., Tone Y., Li L., Liou H.C., Greene M.I., Tone M., and Chen Y.H.. 2009. Development of Foxp3(+) regulatory t cells is driven by the c-Rel enhanceosome. Immunity. 31:932–940 10.1016/j.immuni.2009.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz R., Engel I., Fallahi-Sichani M., Petrie H.T., and Murre C.. 2006. Gene expression patterns define novel roles for E47 in cell cycle progression, cytokine-mediated signaling, and T lineage development. Proc. Natl. Acad. Sci. USA. 103:9976–9981 10.1073/pnas.0603728103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vang K.B., Yang J., Mahmud S.A., Burchill M.A., Vegoe A.L., and Farrar M.A.. 2008. IL-2, -7, and -15, but not thymic stromal lymphopoeitin, redundantly govern CD4+Foxp3+ regulatory T cell development. J. Immunol. 181:3285–3290 10.4049/jimmunol.181.5.3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vang K.B., Yang J., Pagán A.J., Li L.X., Wang J., Green J.M., Beg A.A., and Farrar M.A.. 2010. Cutting edge: CD28 and c-Rel-dependent pathways initiate regulatory T cell development. J. Immunol. 184:4074–4077 10.4049/jimmunol.0903933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verykokakis M., Boos M.D., Bendelac A., and Kee B.L.. 2010. SAP protein-dependent natural killer T-like cells regulate the development of CD8(+) T cells with innate lymphocyte characteristics. Immunity. 33:203–215 10.1016/j.immuni.2010.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Liou H.C., and Sun X.H.. 2006. Id1 potentiates NF-kappaB activation upon T cell receptor signaling. J. Biol. Chem. 281:34989–34996 10.1074/jbc.M608078200 [DOI] [PubMed] [Google Scholar]
- Yao Z., Kanno Y., Kerenyi M., Stephens G., Durant L., Watford W.T., Laurence A., Robinson G.W., Shevach E.M., Moriggl R., et al. . 2007. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 109:4368–4375 10.1182/blood-2006-11-055756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Meng G., and Strober W.. 2008. Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat. Immunol. 9:1297–1306 10.1038/ni.1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Lin Y.Y., Dai M., and Zhuang Y.. 2014. Id3 and Id2 act as a dual safety mechanism in regulating the development and population size of innate-like γδ T cells. J. Immunol. 192:1055–1063 10.4049/jimmunol.1302694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Josefowicz S., Chaudhry A., Peng X.P., Forbush K., and Rudensky A.Y.. 2010. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 463:808–812 10.1038/nature08750 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.