Enhanced polycomb complex protein Bmi1 expression in adult stem cells of the skeletal muscle leads to improved muscle function in a model of Duchenne Muscular Dystrophy via metallothionein1-mediated protection from oxidative stress.
Abstract
The Polycomb group (PcG) protein Bmi1 is an essential epigenetic regulator of stem cell function during normal development and in adult organ systems. We show that mild up-regulation of Bmi1 expression in the adult stem cells of the skeletal muscle leads to a remarkable improvement of muscle function in a mouse model of Duchenne muscular dystrophy. The molecular mechanism underlying enhanced physiological function of Bmi1 depends on the injury context and it is mediated by metallothionein 1 (MT1)–driven modulation of resistance to oxidative stress in the satellite cell population. These results lay the basis for developing Bmi1 pharmacological activators, which either alone or in combination with MT1 agonists could be a powerful novel therapeutic approach to improve regeneration in muscle wasting conditions.
Skeletal muscle is characterized by a remarkable capacity to regenerate after injury, mainly due to the function of satellite cells, the main skeletal muscle stem cells (Brack and Rando, 2012; Wang and Rudnicki, 2012). Polycomb group (PcG) proteins are essential regulators of stem cell function during normal development and in adult organs. They form multi-protein chromatin-associated complexes that play an essential role in the genome-wide epigenetic-mediated remodeling of gene expression during myogenic differentiation of satellite cells, mainly through posttranslational modifications of histones (Asp et al., 2011). Ezh2 and Bmi1 are required for adult satellite cell homeostasis and proliferation in response to muscle injury, an effect mediated at least in part by repression of the ink4a locus (Juan et al., 2011; Robson et al., 2011). Importantly, although Bmi1 is expressed in several types of cancer and its mechanism of action may be similar in a non-neoplastic and neoplastic context, its overexpression does not initiate tumorigenesis (He et al., 2009; Yadirgi et al., 2011).
An emerging role for PcG proteins is their involvement in DNA repair (Liu et al., 2009; Facchino et al., 2010; Ismail et al., 2010; Ginjala et al., 2011; Pan et al., 2011). Bmi1−/−-derived cells show significant mitochondrial dysfunction accompanied by sustained increase in reactive oxygen species (ROS) production that are sufficient to engage the DNA repair pathway (Liu et al., 2009), which is in turn impaired, thus leading to a magnified cellular damage. The balance between intracellular ROS and antioxidant molecules is vital in determining the rate of oxidative damage accumulation and the impaired function of satellite cells in aging and in myopathies, in which decreased anti-oxidative capacity has been documented (Fulle et al., 2005; Whitehead et al., 2006; Tidball and Wehling-Henricks, 2007). X-linked Duchenne muscular dystrophy (DMD) is the most common primary myopathy caused by the loss of the dystrophin protein from the plasma membrane, which causes loss of its integrity and fiber damage during repeated cycles of muscle degeneration and regeneration (Duncan, 1989). The proliferative capacity of myogenic cells was reported to be rapidly exhausted in dystrophin-deficient muscle, also because they are more sensitive to oxidative stress injury, leading to reduced and defective regeneration of the muscle as the disease progresses (Blau et al., 1983, 1985; Disatnik et al., 1998). Moreover, enzymatic adaptations to exercise-induced production of ROS and free radical damage are significantly decreased in dystrophic compared with normal muscles (Faist et al., 1998, 2001). Overall, an impaired protection against ROS in dystrophic muscle appears to contribute to disease progression as also indicated by the beneficial, albeit transient, effect of antioxidants in ameliorating the skeletal muscle pathophysiology of DMD patients (Whitehead et al., 2008).
Metallothionein 1 (MT1) and MT2 are ubiquitously expressed (Kägi and Hunziker, 1989) low molecular weight, cysteine–rich zinc binding proteins. Although the role of MT1 in promoting cell proliferation is controversial (Smith et al., 2008), studies on MT-null liver cells showed their failure to regenerate after oxidative stress injury (Oliver et al., 2006).
Here, we show that overexpression of Bmi1 in the satellite cells significantly improves muscle strength through enhanced MT1-mediated protection of these cells from oxidative stress in a mouse model of dystrophinopathies but not after acute traumatic injury.
RESULTS
Bmi1 expression in mouse models of acute traumatic and chronic degenerative skeletal muscle injuries
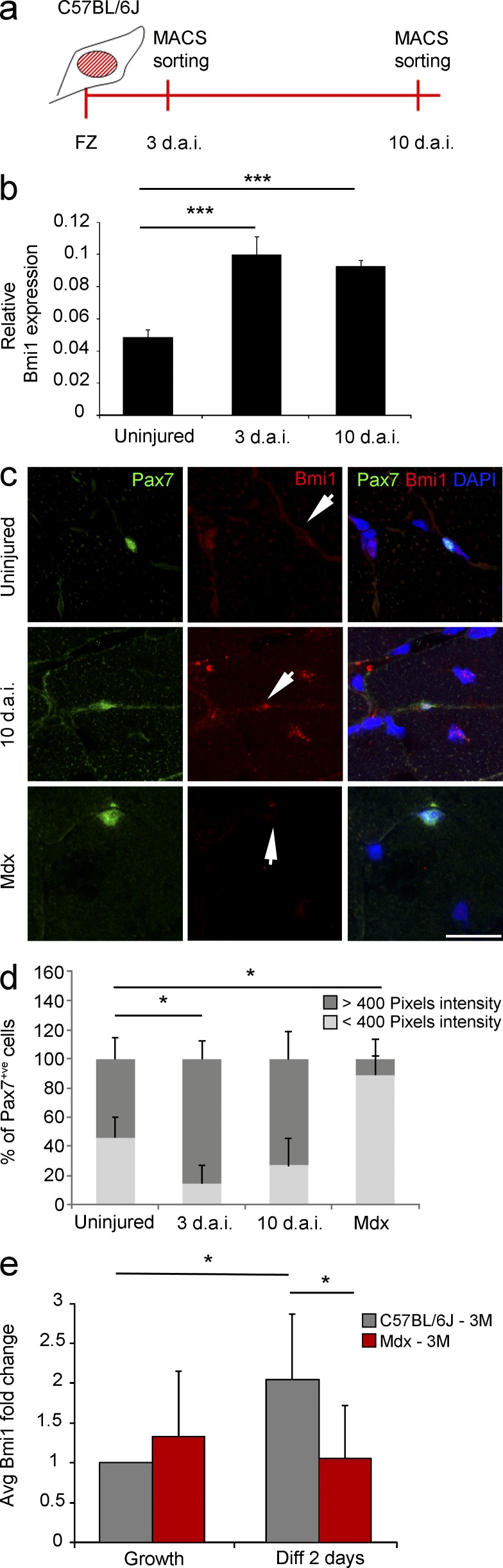
To understand the potential impact of fine tuning Bmi1 expression in muscle injury, we characterized its expression profile in satellite cells at representative time points (3 and 10 d after injury [d.a.i.]) in a well-established model of acute traumatic muscle injury: the freeze injury model (Gayraud-Morel et al., 2007). Satellite cells were isolated 3 and 10 d.a.i. by magnetic activated cell sorting using SM/C-2.6 antibody (Fukada et al., 2004; Fig. 1 a). qRT-PCR analysis revealed significant increase in the expression of Bmi1 in these cells at both time points (Fig.1 b), a result which was confirmed by an increased percentage of Pax7+ve cells expressing high level of Bmi1 in the muscle tissue, as assessed by double immunolabeling for Bmi1 and Pax7 and quantification of staining intensity (van der Laak et al., 2000; Gavet and Pines, 2010; Fig. 1, c and d [quantification]).
Figure 1.
Bmi1 expression in mouse models of acute and chronic skeletal muscle injuries. (a) Schematic representation of satellite cell isolation after freeze injury in C57BL/6J mice. (b) Relative Bmi1 expression represented as 1/dCT values at 3 d.a.i. and 10 d.a.i. (mean ± SEM from three independent experiments with n = 3 uninjured, n = 3 3 d.a.i., and n = 2 10 d.a.i.; ***, P < 0.001). (c) Immunofluorescence for Pax7 and Bmi1 on muscle sections from uninjured mice, injured mice (10 d.a.i.), and mdx mice (representative results from n = 4 uninjured, n = 2 3 d.a.i., n = 2 10 d.a.i., and n = 4 mdx). Arrows indicate Bmi1 staining intensity (expression level) in Pax7+ve satellite cells in three different conditions (uninjured, 10 d.a.i., and mdx). (d) Quantification of Bmi1 staining of Pax7+ve cells with pixel intensity above 400 at 3 and 10 d.a.i. compared with uninjured controls (mean ± SEM from two independent experiments with n = 4 uninjured, n = 2 3 d.a.i., n = 2 10 d.a.i., and n = 4 mdx; *, P < 0.05). (e) Bmi1 expression in satellite cells obtained from mdx single fibers after switching from growth to differentiation-inducing culturing conditions (48 h in differentiation conditions) compared with control satellite cells (mean ± SEM from two independent experiments with n = 3 for each condition; *, P < 0.05). Bar, 50 µm.
The mdx mouse is a well characterized model of chronic degenerative muscle pathology (Bulfield et al., 1984), which is genetically and biochemically similar to the human DMD (Collins and Morgan, 2003). We observed down-regulation of Bmi1 expression in Pax7+ve satellite cells in the muscle tissue (Fig. 1, c and d [quantification]). Interestingly, although Bmi1 expression increased significantly in control satellite cell cultures obtained from single fibers after switching from growth to differentiation-inducing conditions, no significant change was observed in mdx satellite cells (Fig. 1 e). These results show that the levels of Bmi1 expression are differentially regulated during the regenerative process in a pathological specific context.
Conditional Bmi1 expression increases the satellite cell pool and does not prevent myogenic differentiation in vitro
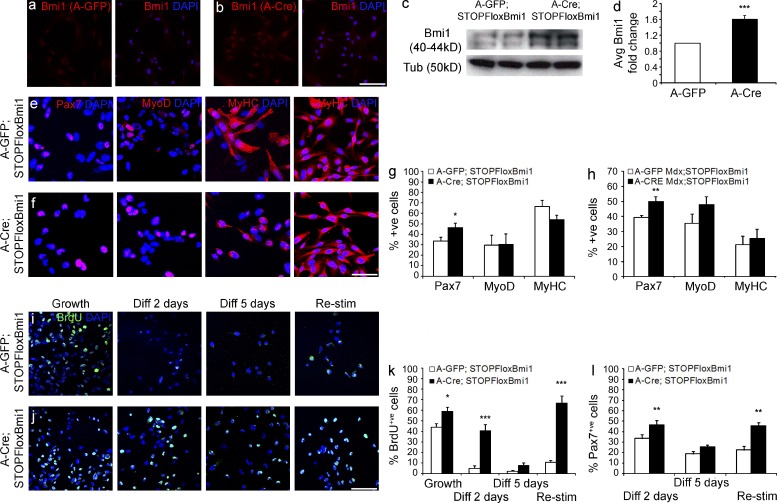
To explore the biological effect of enhancing Bmi1 expression in the satellite cells, we used a mouse line where Bmi1 expression can be activated in a cell- and time-specific fashion upon Cre-mediated recombination (Yadirgi et al., 2011). Transduction of satellite cell cultures derived from STOPFloxBmi1 muscles with Adeno-Cre (A-Cre) virus induced efficient recombination and increased Bmi1 expression at protein (Fig. 2, a–c) and RNA level (Fig. 2 d) compared with Adeno-GFP (A-GFP)–infected cells.
Figure 2.
Conditional Bmi1 activation in satellite cell cultures increases the percentage of Pax7+ve cells. (a and b) Satellite cells derived from P60 STOPFloxBmi1 single fibers, infected with A-GFP or A-Cre viruses and immunostained for Bmi1 (representative results from three independent experiments with n = 3 for each condition). (c and d) Levels of expression of Bmi1 are analyzed with WB (c; representative blot from 2 independent experiments with n = 4 biological samples) and qRT-PCR on the same samples (d; mean ± SEM from three independent experiments with n = 3 for each condition; ***, P < 0.001). (e and f) Representative images (e; 6 independent experiments with n = 8 for each condition) of satellite cell cultures infected with either A-GFP or A-Cre viruses (Bmi1Over cells; f) and stained for Pax7, MyoD, and MyHC, 2 and 5 d.a.i. in differentiation conditions. (g and h) Quantification of the findings in normal (g; mean ± SEM from six independent experiments with n = 8 for each condition; *, P < 0.05) and dystrophic context (h; mean ± SEM from 4 independent experiments with n = 4 for each condition; **, P < 0.01). (i and j) Representative images of the BrdU staining on A-GFP (i) and A-Cre–infected satellite cells (j) in four different conditions (six independent experiments with n = 8 for each condition): growth, 2 d differentiation, 5 d differentiation, and restimulation (i and j). (k) Quantification of BrdU+ve cells in growth, differentiation, and upon high serum restimulation conditions compared with A-GFP–infected satellite cells (mean ± SEM from six independent experiments with n = 8 for each condition; *, P < 0.05; ***, P < 0.001). (l) Quantification of Pax7+ve satellite cells in Bmi1Over satellite cells compared with A-GFP–infected satellite cells in growth, differentiation, and upon high serum restimulation conditions (mean ± SEM from four independent experiments with n = 4 for each condition; *, P < 0.05; **, P < 0.01). Bars, 62.5 µm. Bmi1Over: Bmi1-overexpressing cells.
Next, satellite cell cultures derived from single fibers isolated from the SOL muscle of postnatal day 60 (P60) STOPFloxBmi1 mice were infected with A-GFP or A-Cre virus and induced to differentiate by switching to low serum conditions for 2 d (Fig. 2, e–g). The number of Pax7+ve cells was increased by 14% in cultures overexpressing Bmi1 (Bmi1Over), whereas no alteration in the proportion of MyoD+ve cells (29% in both conditions) was found (Fig. 2, e, f, and g [quantification]). The higher number of Pax7+ve;MyoD-ve cells in Bmi1Over cultures suggested that the stem cell pool of satellite cells was increased (Fig. 2 g). Importantly, Bmi1 overexpression in satellite cells derived from P60 mice did not inhibit differentiation and the percentage of MyHC+ve cells remained constant as compared with controls (Fig. 2, e, f, and g [quantification]). Similar results were obtained also when satellite cells were isolated from mdx;STOPFloxBmi1 muscles (Fig. 2 h).
To investigate whether these results could be explained by modulation of satellite cell proliferation by Bmi1, satellite cells were treated with BrdU in four different culturing conditions: growth (proliferation medium), early differentiation (2 d in differentiation medium), late differentiation (5 d in differentiation medium), and restimulation (2 d in proliferation medium after 5 d of differentiation, Fig. 2, i and j). Under growth conditions, 59% of Bmi1Over satellite cells had incorporated BrdU compared with 44% of the controls, and in early and late differentiation, 40% as compared with 5% and 8% as compared with 2%, respectively (Fig. 2, i, j, and k [quantification]). These results indicate that Bmi1 overexpression prolongs proliferation even in differentiation-inducing conditions. When satellite cell cultures were returned to high serum conditions after 5 d of differentiation (restimulation), incorporation of BrdU was found in 12% of the cells, whereas almost 70% of the Bmi1Over satellite cells incorporated BrdU after 2 d of growth restimulation (Fig. 2, i, j, and k [quantification]). Importantly, at this time point a higher number of Pax7+ve cells was observed, in keeping with an increase in the pool of satellite cells even after a prolonged induction of differentiation (Fig. 2 l).
Conditional activation of Bmi1 expression in satellite cells improves muscle strength in dystrophic muscles but not after traumatic injury in vivo
To assess the impact of Bmi1 overexpression on muscle regeneration in vivo, we used an inducible CreER line targeting satellite cells (Pax7CreERT2; Mourikis et al., 2012). Crosses with the R26R reporter mouse (Soriano, 1999), followed by Tamoxifen-mediated activation of Cre expression in double transgenic mice and whole mount LacZ staining, revealed activation of the construct in 75% of the Pax7CreERT2;R26R injected mice (unpublished data). LacZ/Laminin double labeling confirmed the specificity of the recombination in satellite cells (unpublished data). FACS analysis revealed that at least 65% of the satellite cells isolated from Pax7CreERT2;R26R muscles by means of M-Cadherin labeling (Cooper et al., 1999) were β-galactosidase positive, hence confirming high recombination efficiency (unpublished data).
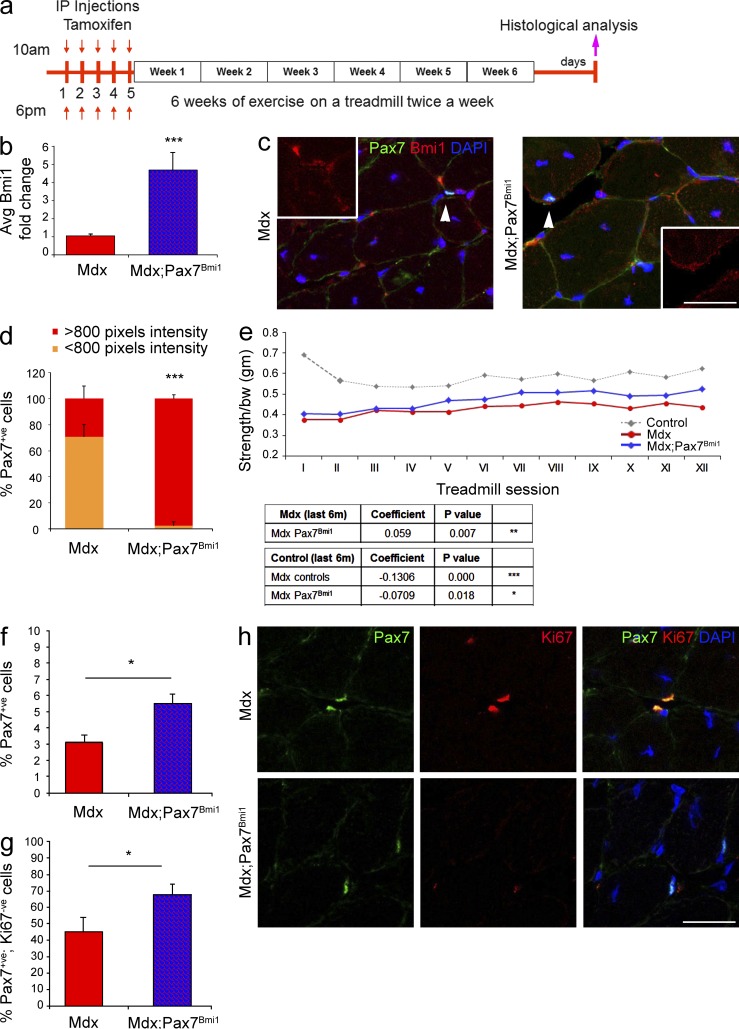
To test the hypothesis that Bmi1 overexpression in satellite cells could enhance regeneration in a chronic injury model, triple transgenic mice mdx;Pax7CreERT2;STOPFloxBmi1, here called mdx;Pax7Bmi1, were generated. Mdx mice are more susceptible to exercise-induced injury, and a cycle of forced exercise on a treadmill has often been applied to accelerate muscle pathology and to test the efficacy of novel treatments in preclinical trials (De Luca et al., 2003; Whitehead et al., 2006). Triple transgenic mice, mdx, and nontransgenic littermates were injected with Tamoxifen, followed by a 6-wk period of running on a treadmill for 30 min twice weekly (Fig. 3 a). Bmi1 overexpression was confirmed both at RNA level in whole muscle tissue (Fig. 3 b) and at protein level specifically in the targeted satellite cells by double immunolabeling for Pax7/Bmi1 (Fig. 3, c and d [quantification]). Before each treadmill session, a forelimb grip test was performed to follow the progression of the disease. The forelimb strength of mdx mice was clearly compromised compared with controls (Fig. 3 e) but the strength of both groups (mdx and control littermates) was stable throughout the duration of the experiment. Strikingly, mdx;Pax7Bmi1 mice showed a significant improvement in muscle strength from the fourth week onwards (Fig. 3 e).
Figure 3.
Conditional activation of Bmi1 expression in satellite cells improves muscle strength in dystrophic muscles in vivo. (a) Schematic temporal summary of the experiment. (b and c) Quantification of Bmi1 expression upon Tamoxifen injections in mdx;Pax7Bmi1 mice compared with controls at RNA (b, mean ± SEM from four independent experiments with n = 12 mdx;Pax7Bmi1 and n = 12 mdx; ***, P < 0.001) and at protein level (c; arrowheads indicate a Pax7+ve cell then included in the insert, representative results from two independent experiments with n = 4 mdx, n = 3 mdx;Pax7Bmi1, n = 12 fields for mdx, and n = 11 fields for mdx;Pax7Bmi1). (d) Quantification of Bmi1 pixel intensity in mdx;Pax7Bmi1 mice compared with controls (mean ± SEM from two independent experiments with n = 4 mdx, n = 3 mdx;Pax7Bmi1, n = 12 fields for mdx, and n = 11 fields for mdx;Pax7Bmi1; ***, P < 0.001). (e) The progression of the muscle pathology is followed by the forelimb grip strength test before each treadmill session. The table reports the statistical significant differences between groups and the coefficient representing the improving (if positive) or worsening (if negative) of the phenotype (four independent experiments with n = 5 controls, n = 9 mdx, n = 11 mdx;Pax7Bmi1; mdx vs. mdx;Pax7Bmi1: **, P < 0.01; mdx vs. control: ***, P < 0.001; and control vs. mdx;Pax7Bmi1: *, P < 0.05). (f and g) Quantification of Pax7+ve cells in mdx;Pax7Bmi1 and mdx control (f) and quantification of double-positive cells Pax7+ve/Ki67+ve in mdx;Pax7Bmi1 and mdx control (g; mean ± SEM from n = 3 mdx, n = 7 mdx;Pax7Bmi1; *, P < 0.05). (h) Representative images of muscle sections stained for Pax7 and Ki67 in mdx;Pax7Bmi1 as compared with mdx mice (n = 3 mdx, n = 7 mdx;Pax7Bmi1; *, P < 0.05). Mdx;Pax7CreERT2;STOPFloxBmi1: mdx;Pax7Bmi1 (Bmi1 overexpression in satellite cells in a dystrophic environment). Bars: (c) 125 µm; (h) 62.5 µm.
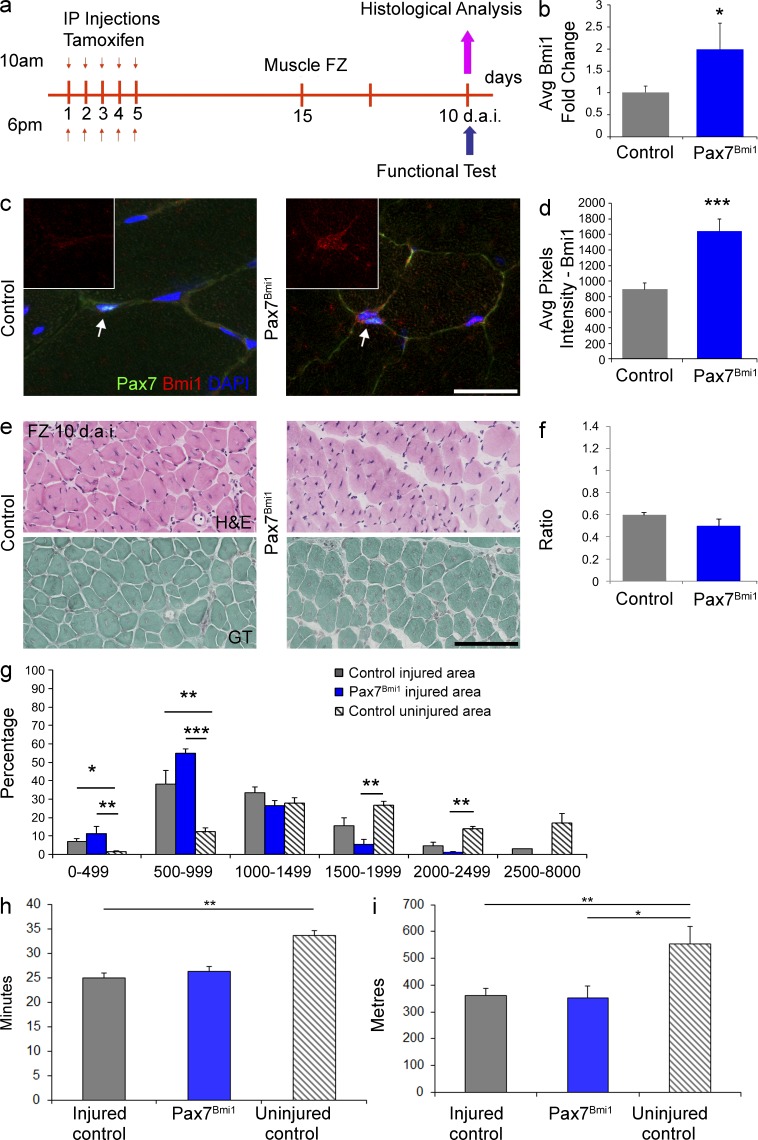
Histological examination of the muscle did not reveal morphological differences between mdx and mdx;Pax7Bmi1, in particular an impact on the degree of endomysial fibrosis or on adipose tissue dedifferentiation was excluded (unpublished data). Immunolabeling for Pax7 showed an increased percentage of Pax7+ve cells in mdx;Pax7Bmi1 mice as compared with mdx controls (Fig. 3, h and f [quantification]). Double labeling for Pax7 and the proliferation marker Ki67 revealed that it was the Ki67−ve component of the Pax7+ve population which was increased in mdx;Pax7Bmi1 (Fig. 3, h and g [quantification]), suggesting that at this time point overexpression of Bmi1 in Pax7+ve cells had increased the pool of quiescent satellite cells in a dystrophic environment. Interestingly, when a muscle freeze injury (FZ) was performed on the TA of hind limbs of Pax7CreERT2;STOPFloxBmi1 compound mutant mice—here called Pax7Bmi1—after Tamoxifen injection (Fig. 4 a) to induce Bmi1 overexpression (Fig. 4, b–d), no significant differences were found morphologically (Fig. 4, e–g) or functionally (Fig. 4, h and i) in injured Pax7Bmi1 as compared with injured controls.
Figure 4.
Conditional activation of Bmi1 expression in satellite cells does not improve skeletal muscle regeneration after traumatic injury in vivo. (a) Schematic temporal summary of the experiment. (b–d) Quantification of Bmi1 expression upon Tamoxifen injections in Pax7Bmi1 mice compared with controls at RNA (b; mean ± SEM from two independent experiments with n = 5 controls and n = 3 Pax7Bmi1; *, P < 0.05) and at protein level (via double immunolabeling for Pax7 and Bmi1; c; arrows indicate a Pax7+ve cell that is included in the insert, representative results from one experiment with n = 3 controls and n = 3 Pax7Bmi1); quantification of the average pixel intensity of Bmi1 staining in Pax7+ve cells (d; mean ± SEM with n = 3 controls and n = 3 Pax7Bmi1; ***, P < 0.001). (e) H&E staining of hind limbs 10 d.a.i. in injured controls and injured Pax7Bmi1 mice (top, representative results from two independent experiments with n = 6 controls and n = 3 Pax7Bmi1). Gomori Trichrome staining for endomysial fibrosis in the same samples of H&E (injured controls and injured Pax7Bmi1 mice; bottom; representative pictures from one experiment with n = 3 controls and n = 3 Pax7Bmi1). (f) Quantification of the ratio of regeneration in Pax7Bmi1 mice compared with controls (f, mean ± SEM from two independent experiments with n = 6 controls and n = 3 Pax7Bmi1). (g) Quantification of the CSA distribution of CNF in injured Pax7Bmi1 and injured controls: the percentage of CNF of control mice (gray), Pax7Bmi1 mice (blue), and the distribution of mature fibers (striped gray) is shown (mean ± SEM from two independent experiments and five independent experiments with n = 8 injured controls, n = 3 Pax7Bmi1, and n = 15 uninjured controls; *, P < 0.05; **, P < 0.01; ***, P < 0.001). (h) Treadmill performance in terms of the time run (h, mean ± SEM from three independent experiments with n = 12 uninjured controls, n = 7 injured controls and n = 3 Pax7Bmi1. **, P < 0.01) in injured control mice (gray) as compared with uninjured mice (striped), as well as in the distances run (i; mean ± SEM from three independent experiments with n = 7 injured controls, n = 3 Pax7Bmi1, and n = 12 uninjured controls; *, P < 0.05; **, P < 0.01). Bars, 125 µm.
Transcriptome analysis reveals increased expression of MT1 in satellite cells upon activation of Bmi1 expression
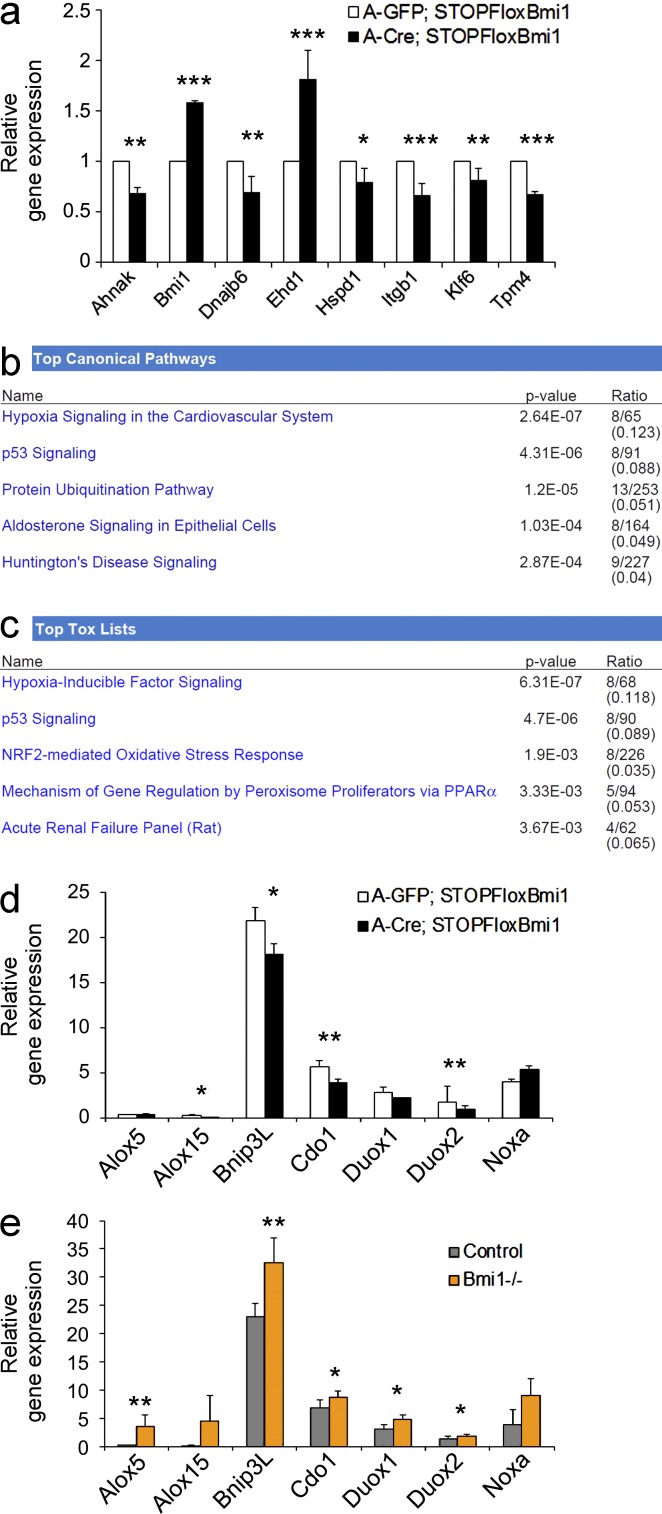
To gain insights into the downstream mechanisms mediating the observed phenotype, we first analyzed the canonical Bmi1 downstream target genes p16ink4a, p19Arf, and p21waf1/cip1. No changes were found in the expression level of these genes in Bmi1over satellite cells cultured under differentiation-inducing conditions or upon restimulation (unpublished data). Alternative ink4a/p21-independent mechanisms were then explored by whole-genome transcriptome analysis on the same cultures (Illumina platform mouse v2). A set of 254 genes was identified (Table S1), the expression of which was deregulated in satellite cell cultures overexpressing Bmi1 compared with controls (P < 0.05 and absolute fold change > 1.9). Out of 10 genes (Ahnak, Bmi1, DNAjb6, Ehd1, Hspd1, Itgb1, Klf6, MT1, p21waf1/cip1, and Tpm4) selected for validation analysis, all but one (p21waf1/cip1) were confirmed to be differentially expressed (Figs. 5 a and 6 a; and not depicted). MT1 was the most significantly up-regulated gene (5.27 ± 0.99-fold) in these cultures (Fig. 6, a and b) and importantly, its expression was up-regulated (5.73 ± 2.03-fold) also in Bmi1Over cultures upon restimulation (Fig. 6 a, last two bars on the right). IPA system biology analysis on all differentially expressed genes showed hypoxia signaling in the cardiovascular system, p53 signaling, and protein ubiquitination pathway (Fig. 5 b) to be the most significantly deregulated top canonical pathways. Hypoxia-inducible factor signaling and NRF2-mediated oxidative stress response were most significantly affected among the top tox lists (Fig. 5 c). To further extend and validate these findings, the expression of a selection of genes involved in Bmi1-mediated control of redox homeostasis through generation of ROS (Liu et al., 2009) was analyzed. We found a significant repression of Alox15, Bnip3L, Cdo1, and Duox2 (Fig. 5 d). Down-regulation of MT1 expression (Fig. 6 a, first two bars; and Fig. 7 a) and increased expression of Alox5, Bnip3L, Cdo1, Duox1, and Duox2 in Bmi1−/− satellite cells confirmed the specificity of these associations (Fig. 5 e).
Figure 5.
Validation of deregulated genes and Ingenuity (IPA) system biology analysis. (a) Genes chosen for validation in Bmi1Over satellite cell cultures compared with A-GFP–infected controls (mean ± SEM from two independent experiments with n = 3 for each condition; *, P < 0.05; **, P < 0.01; ***, P < 0.001). (b and c) System biology analysis using the Ingenuity platform reporting genes in the top canonical pathways (b) and the top tox lists (c; n = 3 for each condition). (d and e) qRT-PCR analysis of expression of redox homeostasis genes in Bmi1Over satellite cell cultures compared with A-GFP–infected controls (mean ± SEM from three independent experiments with n = 5 for each condition; *, P < 0.05; **, P < 0.01) and in Bmi1−/− satellite cell cultures as compared with controls (e; mean ± SEM from two independent experiments with n = 4 for each condition; *, P < 0.05; **, P < 0.01; ***, P < 0.001).
Figure 6.
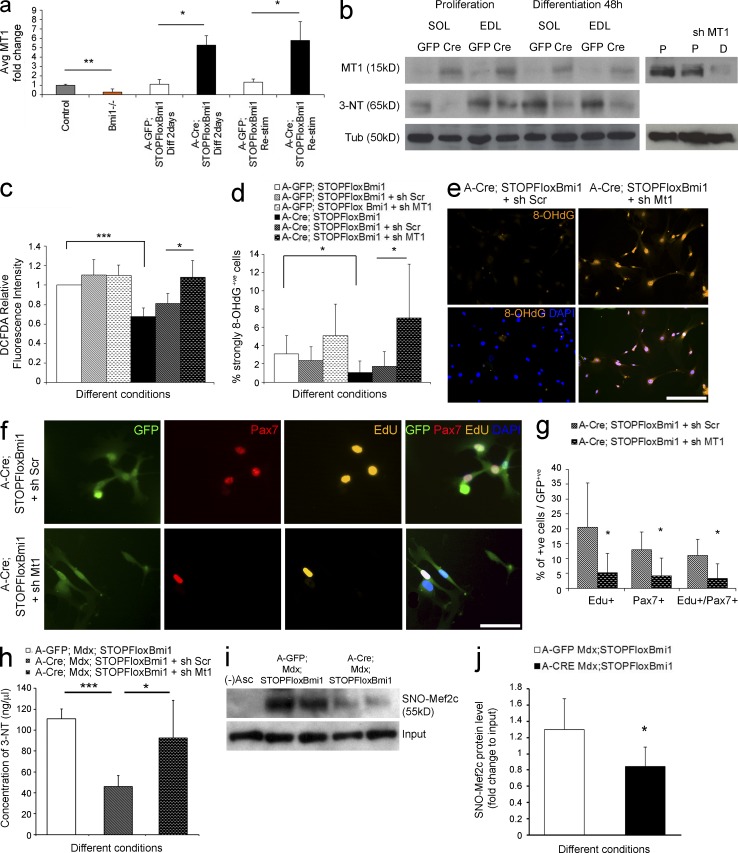
The satellite cell pool is increased by Bmi1 overexpression in vitro via up-regulation of MT1. (a) Relative expression level of MT1 in Bmi1−/−, Bmi1Over, and control satellite cells (mean ± SEM from two independent experiments with n = 3 for each condition; *, P < 0.05; **, P < 0.01). (b) Expression levels of MT1 are also analyzed at the protein level in Bmi1 Over satellite cells both in growth and differentiation-inducing conditions (left, three independent experiments with n = 3 for each condition). Efficient knock down of MT1 is also shown in proliferation (P) and differentiation (D) conditions (b, right; representative results from three independent experiments with n = 3 for each condition). WB analysis of the levels of 3-NT modification in Bmi1Over satellite cells both in proliferation and differentiation-inducing conditions (b, left; three independent experiments with n = 3 for each condition). (c) The amount of ROS production is measured via DCFDA relative fluorescence intensity in Bmi1Over satellite cells compared with controls and treated with either sh Scramble or knockdown of MT1 (mean ± SEM from two independent experiments with n = 4 for each condition; *, P < 0.05; ***, P < 0.001). (d and e) Quantification of the percentage of strongly 8OHdG+ve satellite cells in Bmi1Over cultures with and without knockdown of MT1 (mean ± SEM from two independent experiments with n = 4 for each condition; *, P < 0.05) and representative examples of immunolabeling (e) are shown. (f) Representative examples of immunolabeling for Pax7 and Edu in Bmi1Over satellite cell cultures upon MT1 knockdown (two independent experiments with n = 4 for each condition). (g) Quantification of the percentage of Edu+ve or Pax7+ve or Edu+ve/Pax7+ve cells is shown (mean ± SEM from two independent experiments with n = 4 for each condition; *, P < 0.05). (h) Total concentration of nitrosylated proteins (ng/µl) in Bmi1Over mdx;Bmi1Over satellite cells compared with A-GFP–infected controls, with and without Mt1 knockdown (mean ± SEM from two independent experiments with n = 4; ***, P < 0.001; *, P < 0.05). (i and j) The level of SNO-Mef2c is shown in mdx;Bmi1Over satellite cells compared with controls A-GFP–infected satellite cells as assessed by a modified biotin switch assay, followed by immunoblot (i) and quantification (j; mean ± SEM from two independent experiments with n = 4; *, P < 0.05). MT1: Metallothionein1; 8OHdG: 8-HydroxydeoxyGuanosine; MT1 up: MT1 up-regulation; MT1 nc: MT1 not changed. Bars: (e) 125 µm; (f) 62.5 µm.
Figure 7.
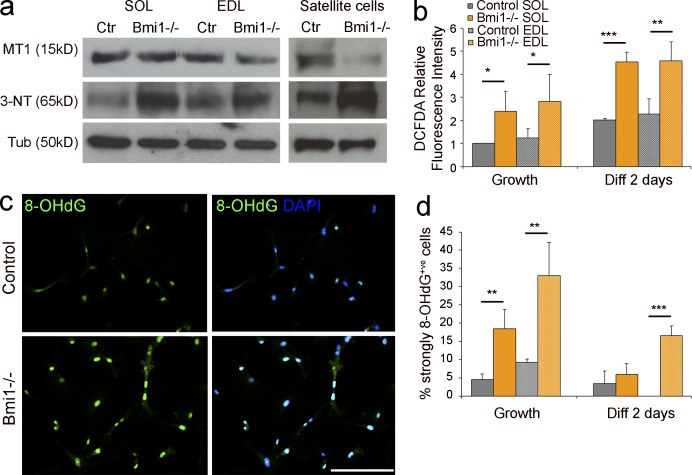
Increased ROS production and oxidative-stress induced DNA and protein damage in Bmi1−/− cells is accompanied by MT1 down-regulation. (a) Protein analysis of MT1 and 3-NT levels in Bmi1−/− muscle tissue and satellite cells cultures (representative results from three independent experiments with n = 3 for each condition). (b) The amount of ROS production is measured via DCFDA relative fluorescence intensity in Bmi1−/− satellite cells compared with controls isolated from both SOL and EDL and cultured either in growth or differentiation conditions (mean ± SEM from two independent experiments with n = 4 for each condition; *, P < 0.05; **, P < 0.01; ***, P < 0.001). (c) Immunolabeling for 8-OHdG in Bmi1−/− satellite cell cultures and controls (representative results from two independent experiments with n = 4 for each condition). (d) Quantification of 8-OHdG+ve cells in Bmi1−/− satellite cell cultures and controls in both growth and differentiation conditions (mean ± SD from two independent experiments with n = 4 for each condition; **, P < 0.01; ***, P < 0.001). Bar, 125 µm.
MT1 is a crucial mediator of the enhanced satellite cell pool elicited by Bmi1 overexpression in vitro
Because the DMD pathology is linked to accumulation of ROS in satellite cells with a consequent increase in the oxidative stress induced cellular damage (Rando et al., 1998; Zhuang et al., 2001; Wozniak and Anderson, 2007), we analyzed whether Bmi1 overexpression would exert a protective role against oxidative stress-induced cellular damage in satellite cells. We observed a decrease in ROS production in Bmi1Over satellite cell cultures incubated with the fluorescent dye H2DCFDA, followed by flow cytometry (Fig. 6 c, white and black bars), which led to decreased oxidative damage to the cells as assessed by 8-hydroxydeoxyguanosine (8-OHdG) staining, a marker of oxidative stress-dependent DNA damage (Won et al., 1999; Fig. 6, e and d [quantification]).
These findings suggest that the amount of oxidative damage incurred by satellite cells in vitro depends on Bmi1 expression levels and raise the possibility that this effect is mediated by MT1. To test this hypothesis functionally, we set out to knock down MT1 expression (MT1kd) in both Bmi1Over and mdx;Bmi1Over cultures and we analyzed ROS production, the percentage of 8-OHdG+ve cells, and the number of proliferating satellite cells and/or Pax7+ve cells. Western blot analysis confirmed efficient knockdown of MT1 in mdx;Bmi1Over cultures in growth and in differentiation conditions (Fig. 6 b). A significant increase in ROS production was observed upon MT1kd in Bmi1Over and in mdx;Bmi1Over satellite cells (Fig. 6 c and not depicted) with concomitant increase in the percentage of 8-OHdG+ve cells (Fig. 6, d and e) and significantly fewer EdU+ve, Pax7+ve and EdU+ve;Pax7+ve cells (Fig. 6, f and g [quantification]), compared with controls treated with scrambled sequences.
Under stress conditions, cytotoxicity is also caused by excessive production of peroxynitrite anions, which react with tyrosine in proteins generating 3-nytrotyrosine (3-NT) residues, an irreversible oxidative modification (Vasilaki et al., 2007). 3-NT is regarded as a sensitive measure of oxidative damage to myofibrillar proteins and results in impaired contractile function of the muscle in addition to contributing to the impairment of the satellite cell function (Vasilaki et al., 2007; Sakellariou et al., 2011). We found that the levels of 3-NT were significantly decreased in satellite cells upon Cre-mediated Bmi1 overexpression (Fig. 6, b [WB analysis] and h [ELISA]), an effect which was dependent on MT1 up-regulation, as shown by the reversal of the effect upon MT1kd in mdx;Bmi1Over satellite cells (Fig. 6 h). Importantly, reduced nitrosylation upon Bmi1 overexpression was confirmed also on a specific protein, Mef2c (Fig. 6, i and j [quantification]), known to play an essential role in the muscle regenerative process (Liu et al., 2014; Panda et al., 2014) and to be inhibited by nitrosylation in the context of oxidative stress-induced protein damage (Ryan et al., 2013). Experiments performed on satellite cells isolated from Bmi1−/− mice showed increased ROS production (Fig. 7 b), increased number of 8-OHdG+ve cells (Fig. 7, c and d [quantification]), and increased levels of 3-NT in Bmi1−/− muscles and satellite cell cultures (Fig. 7 a), hence supporting the interpretation that the observed protection from oxidative stress is a Bmi1-dependent effect.
Bmi1-driven improved muscle strength in dystrophic mice is mediated by MT1 up-regulation
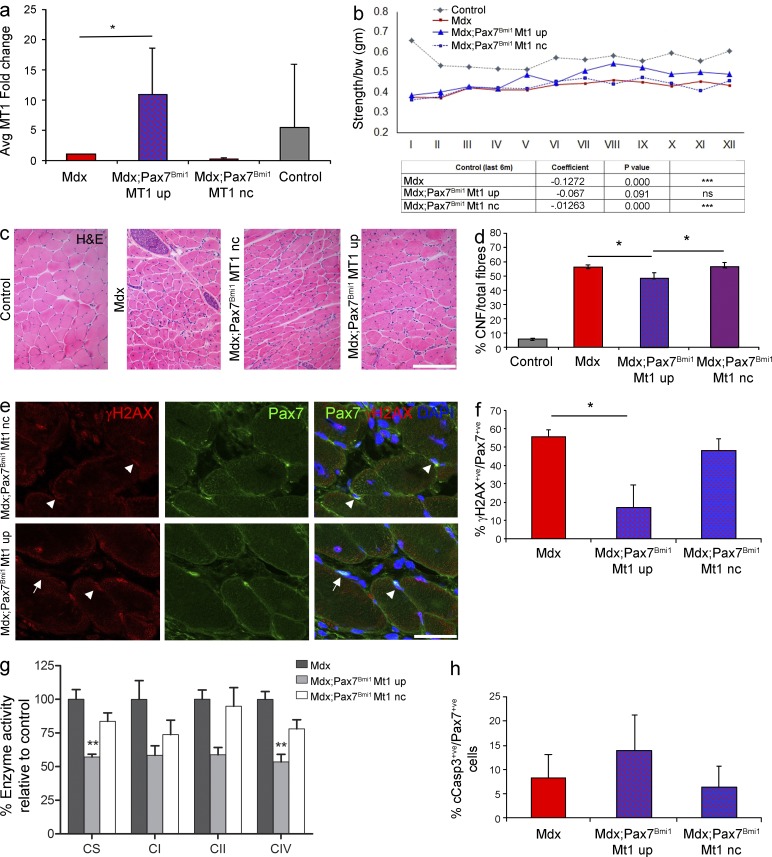
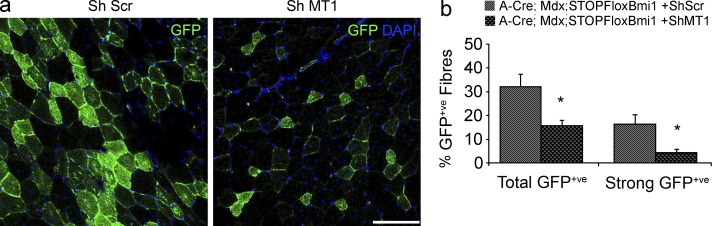
Next, we assessed whether up-regulation of MT1 mediates the improved muscle strength observed in the mdx model upon Bmi1 overexpression in satellite cells. MT1 up-regulation was detected in 33% of mdx;Pax7Bmi1 (Fig. 8 a). Importantly, no MT1 up-regulation was ever detected in mdx control mice (Fig. 8 a), in keeping with a Bmi1-induced effect. Re-evaluation of the muscle strength data with focus on MT1 expression revealed that mdx;Pax7Bmi1 mice with MT1 up-regulation were the animals with the best improvement in forelimb grip strength (Fig. 8 b) as compared with mdx control mice. mdx;Pax7Bmi1 mice up-regulating MT1 showed a behavior similar to that of unaffected control mice, whereas mdx;Pax7Bmi1 with no MT1 up-regulation showed a performance comparable to mdx control mice (Fig. 8 b). These findings show that Bmi1-induced MT1 up-regulation molecularly mediates the impact of Bmi1 overexpression on muscle strength. Furthermore, centrally nucleated fiber (CNF) percentage was significantly reduced in mdx;Pax7Bmi1 with MT1 up-regulation compared with mdx control mice and to mdx;Pax7Bmi1 mice without MT1 up-regulation (Fig. 8, c and d [quantification]), suggesting that muscle fibers were more resistant to exercise-induced damage in mdx;Pax7Bmi1 with MT1 up-regulation. The reduced number of γH2AX+ve (a marker of DNA double-strand break) satellite cells detected in these mice (double immunofluorescence for Pax7 and γH2AX; Fig. 8, e and f [quantification]) is in keeping with the protective effect having been exerted specifically on the satellite cells. We did not observe differences in the number of apoptotic satellite cells or in the total number of apoptotic nuclei (double immunolabeling for Pax7 and cCaspase3; Fig. 8 h and not depicted), or in the percentage of necrotic fibers (not depicted), but a generalized decrease in mitochondrial oxidative enzyme activities was detected by means of in vitro biochemical analysis in mdx;Pax7Bmi1 with MT1 up-regulation (Fig. 8 g). To functionally validate these findings, we transplanted satellite cells isolated from mdx;STOPFloxBmi1 mice induced to overexpress Bmi1 in vitro and either up-regulating MT1 (Bmi1-induced) or not (ShRNA-MT1 mediated knockdown) into the TA muscle of young mdx mice. We show that the contribution of the cells to the regenerative process was more efficient when MT1 was up-regulated, as demonstrated by the reduced number of donor satellite cell–derived GFP-positive fibers upon MT1 knockdown (Fig. 9, a and b [quantification]).
Figure 8.
Bmi1-driven improved muscle strength in dystrophic mice is mediated by concomitant up-regulation of MT1. (a) Relative expression of MT1 in mdx;Pax7Bmi1 compared with mdx control mice (mean ± SEM from three independent experiments with n = 19 mdx, n = 4 mdx;Pax7Bmi1 Mt1 UP, n = 6 mdx;Pax7Bmi1 Mt1 nc, and n = 5 controls; *, P < 0.05). (b) mdx;Pax7Bmi1 forelimb grip strength test is reanalyzed taking into account MT1 up-regulation (mean from four independent experiments with n = 19 mdx, n = 4 mdx;Pax7Bmi1 Mt1 UP, n = 6 mdx;Pax7Bmi1 Mt1 nc, n = 5 controls; control vs mdx: ***, P < 0.001; control vs. mdx;Pax7Bmi1 Mt1 UP: control vs. mdx;Pax7Bmi1 Mt1 nc: ***, P < 0.001). (c) H&E staining of representative fields of forelimbs of controls, mdx, and mdx;Pax7Bmi1 with and without MT1 up-regulation (representative results from four independent experiments with n = 6 controls, n = 19 mdx, n = 4 mdx;Pax7Bmi1 Mt1 UP, and n = 6 mdx;Pax7Bmi1 Mt1 nc). (d) Quantification of CNF of mdx;Pax7Bmi1 with and without MT1 up-regulation in mdx;Pax7Bmi1 MT1 up mice, mdx;Pax7Bmi1 MT1 nc, and mdx mice (mean ± SEM from four independent experiments with n = 6 controls, n = 19 mdx, n = 4 mdx;Pax7Bmi1 Mt1 UP, and n = 6 mdx;Pax7Bmi1 Mt1 nc; mdx vs. mdx;Pax7Bmi1 Mt1 UP: *, P < 0.05; mdx vs mdx;Pax7Bmi1 nc UP: *, P < 0.05). (e and f) Immunolabeling for Pax7 and γH2AX (e, representative results from n = 5 mdx, n = 4 mdx;Pax7Bmi1 Mt1 UP, and n = 6 mdx;Pax7Bmi1 Mt1 nc; arrowhead: γH2AX+ve;Pax+ve; arrow: Pax7+ve) in mdx;Pax7Bmi1 MT1 UP compared with mdx controls and mdx;Pax7Bmi1 MT1 nc (f [quantification]; mean ± SEM from n = 5 mdx, n = 4 mdx;Pax7Bmi1 Mt1 UP, and n = 6 mdx;Pax7Bmi1 Mt1 nc; *, P > 0.05). (g) The activities of mitochondrial respiratory chain enzymes including complex I (NADH: ubiquinone oxidoreductase (CI)), complex II (succinate:ubiquinone oxidoreductase (CII)), and complex IV (cytochrome c oxidase (CIV)) and the matrix marker citrate synthase (CS) are determined in post–600 g supernatants prepared from frozen gastrocnemius muscle samples from mdx and mdx;Pax7Bmi1 mice either up-regulating MT1 or not. Activities (calculated relative to mitochondrial protein) are shown relative to those observed in mdx mice (denoted 100% activity; mean ± SEM from one experiment with n = 5 mdx, n = 4 mdx;Pax7Bmi1 MT1 UP, and n = 6 mdx;Pax7Bmi1 MT1 nc; **, P < 0.01). (h) Quantification of double staining for Pax7 and Cleaved Caspase3 (cCasp3) on the forelimb of mdx and mdx;Pax7Bmi1 mice with up-regulation of MT1 and mdx;Pax7Bmi1 mice with MT1 nc after 6 wk of treadmill exercise (mean ± SEM from one experiment with n = 5 mdx, n = 4 mdx;Pax7Bmi1 MT1 UP, and n = 5 mdx;Pax7Bmi1 MT1 nc). Bars, 62.5 µm.
Figure 9.
MT1 up-regulation is essential for the efficient contribution of Bmi1Over satellite cells to muscle regeneration upon transplantation in the mdx mouse. (a) Satellite cells are isolated from mdx;STOPFloxBmi1 mice, cultured for 14 d, infected with A-Cre virus to activate Bmi1 expression, and treated either with Sh scramble and Mt1 knockdown, both viruses expressing GFP in infected cells. 6 d after treatment, the cells are transplanted in the TA muscles of mdx recipient mice. The transplanted muscles are collected after 27 d. Representative pictures of mdx muscles transplanted with A-Cre; mdx;STOPFloxBmi1 + Sh Scr satellite cells (a, left) or A-Cre; mdx;STOPFloxBmi1 + Sh MT1 satellite cells (a, right) are shown (a, representative results from n = 4 for each condition). (b) Quantification of GFP+ve fibers in A-Cre; mdx;STOPFloxBmi1 + Sh MT1 transplanted satellite as compared with A-Cre; mdx;STOPFloxBmi1 + Sh MT1 transplanted satellite (mean ± SEM from one experiment with n = 4 A-Cre;Mdx;STOPFloxBmi1+Sh Scr and n = 4 Mt1 A-Cre;Mdx;STOPFloxBmi1+ShMt1; *, P < 0.05). Bar, 125 µm.
Bmi1 expression is reduced in patients affected by DMD
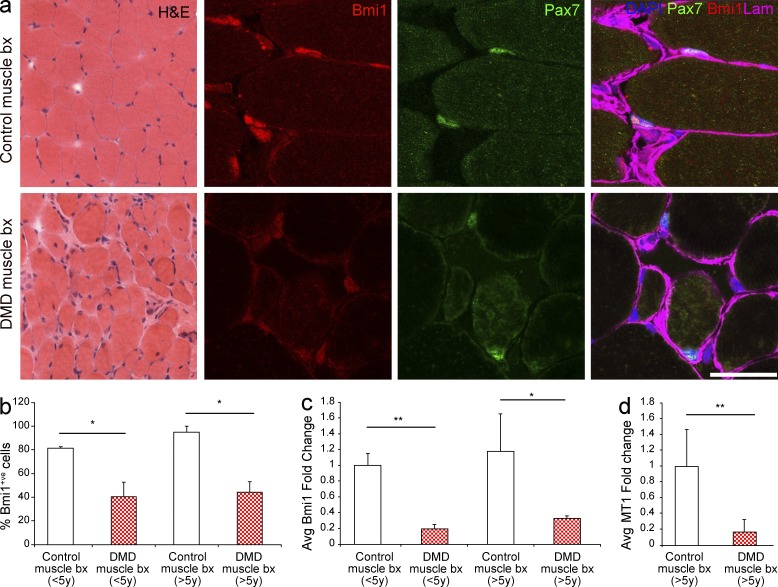
To translate the significance of these findings to human pathology, we analyzed the expression of Bmi1 in DMD muscles and in morphologically normal controls. Four DMD patients aged 2.2, 3.2, 6.1, and 9.5 yr and six age-matched controls were used. Reduction of Bmi1 expression was found in DMD muscle biopsies at all ages (Fig. 10 a). Immunolabeling for Bmi1 performed on adjacent sections confirmed a reduction in the number of Bmi1+ve cells in DMD cases (Fig. 10, a and b) and double labeling for Bmi1 and Pax7 revealed lower levels of Bmi1 expression in Pax7+ve satellite cells in DMD patients (Fig. 10 a). Importantly, reduced expression of Bmi1 (Fig. 10 c) and MT1 (Fig. 10 d) was detected also in DMD patients at the RNA level.
Figure 10.
Bmi1 expression is reduced in the muscle of human DMD cases. (a) H&E staining of a DMD case as compared with an age-matched morphologically normal control. The same muscles are double stained for Bmi1 and Pax7 (representative results from two different cases). (b) Quantification of Bmi1+ve cells in DMD muscle biopsies and age matched controls (mean ± SEM from two independent experiments with n = 2 control and DMD in both age groups, five high power fields/case; *, P < 0.05; **, P < 0.01). (c) Levels of expression of Bmi1 are analyzed by qRT-PCR in the muscle biopsies of DMD patients as compared with age matched unaffected muscles (mean ± SEM from two independent experiments, n = 3 control and n = 2 DMD in both age groups, two technical replicas for each case; *, P < 0.05; **, P < 0.01). (d) Expression levels of MT1 are measured by qRT-PCR in the muscle biopsies of DMD patients >5 yr compared with age-matched unaffected muscles (mean ± SEM from two independent experiments with n = 3 control and n = 2 DMD in both age groups, two technical replicas for each case; *, P < 0.05; **, P < 0.01). Bar: (H&E) 62.5 µm; (immunofluorescence) 31 µm.
DISCUSSION
Here, we show for the first time that mild overexpression of a chromatin modifier gene has a functionally relevant impact on muscle regeneration. Bmi1 overexpression in satellite cells confers enhanced protection from exercise-induced injury and improves muscle strength in a mouse model of DMD. We also demonstrate that the mechanism underlying the effect of enhanced physiological function of Bmi1 in the satellite cells is mediated by MT1-driven modulation of resistance against oxidative stress.
Skeletal muscle is an important and highly relevant platform to assess how manipulation of stem cell properties may enhance a functionally significant regenerative process in vivo. We show that the expression level of Bmi1 is modulated in response to pathological conditions in the satellite cells. Indeed, Bmi1 is up-regulated upon freeze injury in wild-type mice, whereas reduction of the proportion of Pax7+ve cells with high Bmi1 expression is found in the mdx mouse, with the latter finding corroborated by the lack of Bmi1 up-regulation in mdx satellite cell cultures upon induction of differentiation. In keeping with these findings, overexpression of Bmi1 in satellite cells has a beneficial effect solely in the mdx mouse, where down-regulation of its expression was observed.
The best characterized molecular mechanisms mediating Bmi1 function relies on transcriptional repression of the ink4a locus, encoding for p16ink4a and p19arf (Jacobs et al., 1999), as well as of p21waf1/cip1. However, we found no change in the expression levels of these genes upon activation of Bmi1 expression raising the intriguing possibility that a novel mechanism may mediate the observed phenotype. The generation of free radicals and ROS/reactive nitrogen species (RNS) is a physiological, continuous process in aerobic organisms. When the production of ROS/RNS exceeds the endogenous antioxidant buffering capacity, oxidative stress-induced cellular damage occurs. There is increasing evidence that oxidative stress level is elevated in diseases such as DMD and in normal aging (Fulle et al., 2004; Whitehead et al., 2006; Jackson, 2009; Cozzoli et al., 2011; Spassov et al., 2011). Bmi1 has an important role in mitochondrial function and redox homeostasis in thymocytes and in neurons (Liu et al., 2009; Abdouh et al., 2012); however, the mechanisms mediating this role of Bmi1 are currently unclear. Up-regulation of Bmi1 expression in Pax7+ve satellite cells increases the pool of quiescent Ki67−ve cells in vivo after 6 wk of training on a treadmill, indicating that Bmi1 is involved in the regulation of self-renewal of these cells. To explain these results, two scenarios can be envisaged: it is possible that a phase of increased proliferation has also occurred in vivo, perhaps immediately after induction of Bmi1 overexpression in a similar fashion as we observed in vitro. Alternatively, satellite cells overexpressing Bmi1 are better able to reenter quiescence, hence preventing the typical exhaustion of the satellite cell pool seen in dystrophic muscle conditions after repeated degeneration/regeneration cycles. Genome-wide transcriptome analysis of satellite cells overexpressing Bmi1 revealed significantly increased expression of MT1, which has been shown to be necessary to maintain proliferation and normal cellular functions under stress conditions, including when cells reenter the cell cycle from a quiescent state (Zbinden et al., 2010). In keeping with this notion, improvement of muscle strength is seen only in those mdx mice in which up-regulation of MT1 occurred concomitantly with overexpression of Bmi1 in satellite cells. The reduced contribution to regeneration after transplantation in a dystrophic environment of Bmi1Over satellite cells upon MT1 knockdown lends additional support to this conclusion. At present it is unclear why up-regulation of MT1 occurs only in a proportion of mdx mice overexpressing Bmi1 and it will be important to investigate this fully in future studies. However, the correlation of muscle strength and MT1 up-regulation suggests that Bmi1 exerts its effect through protection against oxidative stress-induced cellular damage in vitro and in vivo, hence delaying muscle wasting through reduction of ROS-induced oxidative stress. The reduction of γH2AX+ve satellite cells observed in the mdx;Pax7Bmi1 with concomitant MT1 up-regulation is in keeping with this conclusion, as it is the MT1-dependent reduction of 3-NT levels in mdx;Pax7Bmi1 satellite cell cultures and the reduced levels of nitrosylated Mef2c, a transcription factor with an essential role in muscle regeneration (Liu et al., 2014; Panda et al., 2014). We also observed a decrease in metabolic and mitochondrial oxidative enzyme activities in these mice, raising the possibility that because satellite cells are better protected against exercise-induced oxidative stress in a dystrophic environment, metabolic adaptation is less prominent. Although additional studies will be required to fully elucidate the impact of Bmi1 overexpression on metabolism, it is intriguing that decreased mitochondrial oxidative phosphorylation activities have been shown to increase resistance to metabolic diseases through prevention of ROS accumulation (Pospisilik et al., 2007) and to be associated with better tolerance to oxidative stress (Patel et al., 2013). It is currently unclear what mediates Bmi1-induced up-regulation of MT1 expression in the skeletal muscle. As the main mode of function of PcG genes is through transcriptional repression, it is possible to speculate that a MT1 repressor may be involved in this process.
Finally, we show here an important correspondence of Bmi1 expression between mice and humans, including its depletion in human DMD muscles. Interestingly there is evidence that MT1 expression increases in human skeletal muscle 24 h after intense exercise (Urso et al., 2006); here, we show decreased expression of MT1 in human DMD muscle, raising the possibility that manipulation of Bmi1 expression could be a useful approach to enhance satellite cell–driven regeneration also in human primary myopathies. Identifying novel pharmacological agents to enhance and sustain muscle regeneration in DMD is highly desirable as they could have a tremendous impact on DMD therapy either alone or, most importantly, in combination with genetic therapies aiming at reconstituting dystrophin expression.
MATERIALS AND METHODS
Animals.
The following transgenic lines were used: C57BL/6J-BL/10, STOPFloxBmi1 (Yadirgi et al., 2011), Bmi1−/− (van der Lugt et al., 1994), R26R (Soriano, 1999), mdx (Bulfield et al., 1984), and Tg:Pax7CreERT2 (Mourikis et al., 2012). Animals were genotyped with PCR on gDNA extracted from ear notches according to standard protocols. Published primer sequences were used. All procedures had Home Office approval (Animals Scientific Procedures Act 1986, PPL 70/7275).
Satellite cell culture.
Single fibers from the soleus (SOL) and the extensor digitorum longus (EDL) muscles of adult Bmi1−/−, STOPFloxBmi1, mdx;STOPFloxBmi1, and nontransgenic littermates were isolated and cultured to obtain pure satellite cell cultures according to standard protocols. In brief, the muscles were dissected from P60 mice being careful not to stretch the muscles. Tendon and other connective tissues were carefully removed and the muscles incubated in 0.2% Collagenase type 1 (Worthington Biochemical Corporation) at 37°C for 2 h. Fibers were liberated by gentle trituration in DMEM. Single fibers were placed in a 24-well plate (BD) coated with 1mg/ml Geltrex Reduced Growth Factor Basement Membrane Matrix in DMEM (Invitrogen) and allowed to adhere for 5 min at 37°C before 500 µl of plating medium was added (DMEM supplemented with 10% horse serum [PAA] and 0.5% chick embryo extract [Gibco] with antibiotics) and incubated at 37°C in 5% CO2. After 3 d, the media was changed to proliferation media (DMEM supplemented with 20% FCS [PAA] and 2% chick embryo extract [Gibco] plus antibiotics). To induce differentiation, satellite cells were plated at 2,000 cells per well in an 8-well chambered slides (BD) and cultured in differentiation media (DMEM supplemented with 2% horse serum and antibiotics) for 2 or 5 d. Satellite cells derived from the same fiber of STOPFloxBmi1 and mdx;STOPFloxBmi1 mice were set up in parallel wells and treated with either A-GFP or A-Cre (Akagi et al., 1997) at the time of plating in the differentiating media. To control for nonspecific effects of viral treatment and overexpression of any exogenous proteins, infection with A-GFP was used as control for all experiments where A-Cre infection was performed.
Magnetic activated satellite cells sorting.
TA muscles were isolated from C57BL/6J mice at time zero (uninjured controls), and 3 and 10 d after freeze injury. The muscles were dissected and finely chopped with scissors and incubated with 0.2% Collagenase in DMEM for 30 min at 37°C while gently shaking (250 rpm). This step of the procedure was performed twice. The cells were then filtered with 30-µm filters (Miltenyi Biotec). 1 ml of serum was added and the suspension was centrifuged for 20 min at 1,200 rpm at 4°C. The supernatant was decanted and the staining was performed with the SM/C-2.6 antibody (Fukada et al., 2004; 1:200 for 40 min on ice) and with anti-Biotin MicroBeads (Miltenyi Biotec) as secondary antibody according to manufacturer’s protocol. After magnetic sorting the cells were centrifuged 20 min at 2,500 rpm and the pellet was snap frozen on dry ice.
Immunohistochemistry.
All myoblast cultures and muscle sections were fixed with 4% PFA for 4 and 10 min, respectively. For immunolabeling, primary antibodies were applied overnight at room temperature; appropriate fluorescent secondary antibodies were used and the myogenic cultures or sections were mounted using VECTASHIELD mounting medium with DAPI (Vector Laboratory). The following primary antibodies were used: chicken anti–β-galactosidase (1:1,000; Abcam), mouse anti-Bmi1 (1:500; Millipore), goat anti-Bmi1 (1:100; Santa Cruz Biotechnology, Inc.), Click-iT TM EdU Imaging kits (Invitrogen), rabbit anti-Laminin (1:1,000; Sigma-Aldrich), mouse monoclonal to Metallothionein 1 (1:50; Abcam), rabbit anti-Myf5 (C-20; 1:100; Santa Cruz Biotechnology, Inc.), mouse anti-MyoD (5.8A; 1:100; Novacastra), rabbit anti-MyoD (1:100; Santa Cruz Biotechnology, Inc.), mouse anti-pan myosin heavy chain (A4.1025; 1:10; Developmental Studies Hybridoma Bank), mouse anti-Pax7 (1:1; Developmental Studies Hybridoma Bank), rabbit anti-Ki67 (1:300; Novacastra), rabbit anti-γH2AX (phosphor S139)–DNA double-strand break marker (1:200; Abcam), and rabbit anti-cleaved Caspase3 (cCasp3; 1:200; Cell Signaling Technology). The following secondary antibodies were used: donkey anti–mouse IgG Alexa Fluor 488 (1:500), goat anti–mouse IgG Alexa Fluor 488 (1:1,000), goat anti–mouse IgG1 Alexa Fluor 546 (1:1,000), donkey anti–rabbit IgG Alexa Fluor 488 (1:500), donkey anti–rabbit IgG Alexa Fluor 647 (1:500), goat anti–rabbit IgG Alexa Fluor 546 (1:1,000), goat anti–rabbit IgG Alexa Fluor 488 (1:1,000), donkey anti–goat Alexa Fluor 568 (1:500), rabbit anti–goat Alexa Fluor 488 (1:1,000), goat anti–mouse IgG2a Alexa Fluor 488 (1:1,000), goat anti–chicken Alexa Fluor 594 (1:1,000), goat anti–rat Alexa 488 (1:1,000; all from Invitrogen), and biotinylated goat anti–mouse IgG (H+L) (ready to use; Abcam).
Microscopy and quantification.
Fluorescent and bright field image capture was performed using an epifluorescent (Leica) or confocal (Meta 510 LSM or LSM 710; Zeiss) microscope. In myoblast cultures, all DAPI-positive cells and antigen-positive cells were counted and the percentage of Antigen+ve cells was calculated. Data from three or more cultures were pooled to give a population mean ± SEM.
For the quantification of the intensity staining, all samples were stained simultaneously and the pictures were acquired with an LSM 710 confocal microscope at the same ratio of emissions. For the analysis with ImageJ software (National Institutes of Health), the shape of each Pax7+ve cell was drawn around the nucleus and the intensity of Bmi1 staining calculated for each cell as mean of integrated density (mean intensity × area). The mean integrated density was calculated among all cells per each field acquired. The percentage of cells above or below certain pixel intensity (determined by the mean integrated density between all measured cells) was calculated out of the total number of measured cells per each muscle section analyzed. The method was also validated using Definiens Tissue Studio (IF) 2.1.1, action library 3.6.1, and Developer XD 2.1.1 (Definiens AG). In particular for the validation on sections stained for Pax7 and Bmi1, an initial analysis was performed using Tissue Studio to identify the nuclei based on DAPI stain and Pax7. Nuclei were identified using a stain threshold of 1.62 (RGB space converted to HSD model to give a range of 0–2; van der Laak et al., 2000) and typical size of 18 µm2. Pax7 was processed twice using thresholds of 0 and 25 (RGB space) to classify all nonnuclear tissue (Marker 1) and areas with significant Pax7 staining (Marker 2), respectively. These are overlaid to give classifications of marker 1 and markers 1 and 2. Developer was used to perform post-processing on the classification resultant from Tissue Studio. Before classification of nuclei as positive or negative, nuclei with area <5 µm2 were merged with connecting nuclei. In cases where these small nuclei did not border another nucleus, no change occurred. In cases where a small nucleus borders multiple nuclei, the merge resulting in the lowest elliptical fit was selected. Several dynamic thresholds were calculated for each image: the mean Pax7 stain intensity for each area classified as marker 1 was used to give Pax7 Background; areas of marker 1 <10 µm2 were excluded from this calculation to prevent skewing of resultant threshold toward the upper limit of 24 because these are generally isolated areas within the intensely stained regions (markers 1 and 2) just below the threshold of 25. The threshold for classifying nuclei as positive Pax7 threshold was calculated as: maximum Pax7 pixel value − (2 × Pax7 Background). A second set of thresholds were calculated based on the Bmi1 stain to segment positive nuclei staining as below mean, low intensity, and high intensity. The mean Bmi1 stain intensity in all pixels with intensity <50 (RGB space) gives mean Bmi1, and the range between mean Bmi1 and the maximum Bmi1 value was divided by two to give low Bmi1 and high Bmi1. Nuclei with more than five pixels with Pax7 greater than Pax7 threshold were classified as positive nuclei. Positive nuclei were then segmented based on BMI1 stain as below mean, low intensity, and high intensity. To compare this analysis with the manual analysis with ImageJ, the mean Bmi1 intensity for each single slice of the stack where Bmi1 was on focus was multiplied to the area (in pixel) so to obtain integrated density values. The percentage of cells above or below certain pixel intensity (determined by the mean integrated density between all measured cells) was calculated out of the total number of measured cells per each muscle section analyzed.
For the analysis of satellite cells in vitro, satellite cells were stained for Bmi1 and counterstained with DAPI. Immunofluorescent images taken at 16-bit were analyzed using ImageJ and Developer XD (2.1.1; Definiens AG). The analysis in ImageJ was conducted using the same settings as above. For the analysis in Definiens, pixels with intensity above 30 (RGB space) were classified as nucleus, those below 30 remained unclassified; and small unclassified areas bordering only nucleus were reclassified as nucleus and merged in. The distance of each pixel classified as nucleus to unclassified was then calculated to facilitate the splitting of connected nuclei. Nuclei with irregular shapes (roundness >0.5; roundness is based on the difference between the largest enclosing ellipse and the smallest enclosed ellipse) were then segmented based on these distances; local minima provide seeds for nuclei. These seeds were then grown to fill the space originally classified as nuclei, with separate nuclei growing to share a border rather than joining, thus splitting connected nuclei. Nuclei with area < 100 µm2 were then removed. For each nucleus, the sum of the Bmi1 staining for each pixel was exported and this value was multiplied for the area of each single nucleus. For both the analyses, the fold change of increased Bmi1 staining was calculated using the average of Bmi1 staining of A-GFP–infected cells as one per each coverslip analyzed.
RNA extraction and qPCR analysis.
Satellite cell cultures were harvested after 48 h of either differentiation or restimulation. The cell pellets were lysed in RLT buffer from RNeasy Micro purification kit (QIAGEN). Total RNA was isolated using the same kit. Digestion with DNase (on column) was performed to remove genomic DNA. cDNA synthesis was performed with SuperScript III Reverse transcription kit (Invitrogen). For the sorted satellite cells, the retrotranscription was performed with QuantiTect Whole Transcriptome kit (QIAGEN) according to manufacturer’s protocols. Analysis of gene expression was performed with a Rotor-Gene apparatus (Corbett Life Science) using SYBR Green JumpStart TaqReadyMix (Sigma-Aldrich) according to the manufacturer’s instruction. A list of the primers used is reported. The Ct values of all the genes analyzed were normalized with GAPDH and fold changes were calculated. qRT-PCR analysis for Bmi1 was performed with TaqMan assays (Applied Biosystems) with FAM-labeled probes in 96-well plates using the 7500 RT-PCR machine (Applied Biosystems) according to the manufacturer’s instructions (Bmi1: Mm 00776122_gH). The cDNA content was normalized against the expression of the housekeeping gene β-actin (Mm 00607939_s1). Technical duplicates for each culture or muscle tissue and three or more independently derived cultures or biological samples were analyzed. The following primers were used in qPCR experiments: Alox5 sense, 5′-CATGTTACCGCTGGATCA-3′, and antisense, 5′-CCACTCCATCCATCTATACTG-3′; Alox15 sense, 5′-GACTTGGCTGAGCGAGGACT-3′, and antisense, 5′-CTTGACACCAGCTCTGCA-3′; Bmi1 sense, 5′-GTTCACAAGACCAGACCACTAC-3′, and antisense, 5′-CTGCAACCTCTCCTCTATCTTC-3′; Bnip3L sense, 5′-CTGAGCACACCTTCTGCCAGC-3′, and antisense, 5′-TAGGGATCATGCTTACAATAGGTC-3′; Cdo1 sense, 5′-GTGGATCAAGGAAATGGGA-3′, and antisense, 5′-CTTGATCATCTCGTTGGA-3′; Dnajb6 sense, 5′-AAGTTGCTCCATGGCTACTAC-3′, and antisense, 5′-AGCTACAGCCTTCACTTGACT-3′; Duox1 sense, 5′-CTGTACCTCGATGGACCGTTTGGAGA-3′, and antisense, 5′-AGTCCTTGTCACCCAGATGAAGTAGA-3′; Duox2 sense, 5′-TCCATCCTCAAAGACCTGGTCTTCA-3′, and antisense, 5′-CTCAGCCAGCTGAGTAATGTAGATGT-3′; Ehd1 sense, 5′-GGTGCTCCTTTATCACCTTC-3′, and antisense: 5′-GACTAGTTCGAGACAGAGCAAG-3′; GAPDH sense, 5′-GCACAGTCAAGGCCGAGAAT-3′, and antisense, 5′-GCCTTCTCCATGGTGGTGAA-3′; Hspd1 sense, 5′-ACTTCAGAGAAGTCACCTGGA-3′, and antisense, 5′-GAACAGTAGAGATGCCTCAGTG-3′; Itgb1 sense, 5′-GGTGCTCCTTTATCACCTTC-3′, and antisense, 5′-GACTAGTTCGAGACAGAGCAAG-3′; Klf6 sense, 5′-TGGTTCCTAACCTTCTGAGAG-3′, and antisense, 5′-CCTTGGTATACTCCTCACACAC-3′; Mt1 sense, 5′-AAGTGCACCTCCTGCAA-3′, and antisense, 5′-GGGTGGAACTGTATAGGAAGAC-3′; Noxa sense, 5′-GAACGCGCCAGTGAACCCAA-3′, and antisense, 5′-CTTTGTCTCCAATCCTCCGG-3′; P16Ink4a sense, 5′-CCAAGAGCAGAGCTAAATCC-3′, and antisense, 5′-CCTTCGCTCAGTTTCTCAT-3′; P19Arf sense, 5′-CCCACTCCAAGAGAGGGTTT-3′, and antisense, 5′-TCTGCACCGTAGTTGAGCAG-3′; P21 sense, 5′-GACAAGAGGCCCAGTACTTC-3′, and antisense, 5′-CTCTTGCAGAAGACCAATCTG-3′; Tpm4 sense, 5′-CCGGAGGTATCTGAACTAAAG-3′, and antisense, 5′-CAGGTCATCGATTGTCTTCTC-3′.
Western blot.
EDL and SOL muscles were mechanically chopped and lysed for 1 h on ice with RIPA buffer (1% Igepal CA-630, 2 M TrisHCl, pH 8.0, 0.5% Na-deoxycholate, 0.1% SDS, 2 mM EDTA, and 150 mM NaCl including proteinase inhibitor complete mini) followed by max speed centrifugation (10 min). The supernatants were collected and the protein concentration was measured using BCA Protein Assay kit (Thermo Fisher Scientific). 90 µg of protein per lane were separated on a NuPage 4–12% Bis-Tris Gel (Invitrogen) for 1 h at 200 V and electroblotted onto nitrocellulose PROTRAN (GE Healthcare) for 2 h at 35 V. After blocking overnight (ON) in TBST buffer (25 mM Tris HCl, 137 mM NaCl, and 0.1% Tween 20, pH 7.5) containing 5% skimmed milk, immunodetection of proteins was performed with the following antibodies: mouse anti–3-NT antibody (1:200, ON at 4°C; Cayman), anti-MT1 antibody (1:500, ON at 4°C; Abcam), mouse anti-Bmi1 (Millipore), mouse anti-Mef2C (1:1,000; Cell Signaling Technology), followed by HRP-conjugated anti–mouse IgG secondary antibody (1:5,000 for 1 h; GE Healthcare) at room temperature. Enhanced chemoluminescence (ECL Plus; GE Healthcare) was used for detection of the bands. α-Tubulin (1:5,000; Sigma-Aldrich) was used as a control for gel loading followed by anti–mouse IgG HRP (1:5,000; GE Healthcare).
When satellite cells were analyzed, cell pellets were collected from different culture conditions and lysed with RIPA buffer as mentioned above. 40 µg of protein extracted from cell samples was loaded per lane on the NuPage 4–12% Bis-Tris gel.
Production of shRNA-Mt1 lentiviral vector.
A pGIPZ-lentiviral shRNAmir vector containing a hairpin sequence targeting MT1 is commercially available from Open Biosystems. The shRNA-containing lentiviral vector was cotransfected with lentiviral packaging, using the calcium phosphate method into 293T cells. The medium was refreshed and viruses were harvested after 48 h of transfection, passed through 0.45-µm filters, concentrated by PEG precipitation, and stored at −80°C. The infectious titer was determined by FACS analysis of GFP-positive 293T cells.
Lentiviral-mediated Mt1 knockdown.
Satellite cells were seeded in multiple-well plates and transduced with concentrated lentivirus containing shRNA that targets MT1, along with A-Cre (to induce Bmi1 overexpression) in proliferation medium. The medium was replaced on the second day. 96 h after transduction, cells were switched to differentiation medium for 48 h and the transduction efficiency was analyzed. Cells were collected for Western blot, immunocytochemistry, or ROS assay.
ROS assay.
Satellite cells were treated with 40 µM H2DCFDA in differentiation medium for 1 h at 37°C before being harvested and resuspended in 500 µl of PBS. Cells were then immediately processed for flow cytometry analysis using an LSRII instrument (BD) and the relative fluorescence intensity was calculated.
ELISA for 3-NT detection.
Satellite cells prepared from mdx;STOPFloxBmi1 EDL muscles were transfected with A-Cre to overexpress Bmi1 and a proportion then treated with LV-shMt1. Cells infected with A-GFP were used as controls. Proteins were extracted from the cells and concentration was measured by BCA protein assay kit (Thermo Fisher Scientific). The amount of 3-NT protein in the lysates was determined by the Nitrotyrosine ELISA kit (Abcam) according to the manufacturer’s protocol. The OD was recorded at 450 nm and the data were analyzed with the program XLfit 5.3.1.3 (IBDS).
Modified biotin switch assay to detect SNO-MEF2c.
The biotin-switch assay (BST) is a modified immunoblot analysis used to detect the levels of S-nitrosylated proteins. Here, we used a modified version of this assay. In brief, cell lysates were prepared from A-Cre– or A-GFP–treated mdx;STOPFloxBmi1 satellite cell cultures in HENS Buffer (Pierce S-Nitrosylation Western Blot kit; Thermo Fisher Scientific) and the BCA Protein Assay was used (Thermo Fisher Scientific) to determine the protein concentration. 300 µg of protein from each sample was mixed with 20 mM methyl methane thiosulfonate (MMTS) for 30 min at room temperature to block free thiol groups. After removing excess MMTS by acetone precipitation (1 h at −20°C), S-nitrosothiols were reduced to thiols with 20 mM sodium ascorbate. In the negative control, ultrapure water was used instead of sodium ascorbate. Labeling reagent containing tandem mass tag (TMT) was added to label the newly formed thiols. After acetone precipitation, samples were resuspended into HENS buffer. TMT labeled proteins were loaded onto spin columns with pre-added immobilized anti-TMT resin (Thermo Fisher Scientific) and eluted. The eluted proteins were concentrated using Pierce Concentrator (Thermo Fisher Scientific) and further analyzed by immunoblotting. Input was used for the control blot.
Pax7CreERT2T2 tamoxifen injection.
Tg:Pax7CreERT2 mice were crossed with R26R to optimize the injections protocol. 2-mo-old littermate Pax7CreERT2;R26R and R26R (as controls) were injected with Tamoxifen (Sigma-Aldrich) 20 mg/ml suspended in corn oil (Sigma-Aldrich; 4.5 mg/40 g of body weight) twice a day for five consecutive days. 5 d after the last injection the mice were culled and the efficiency of recombination was assessed. Pax7CreERT2 mice were bred with the STOPFloxBmi1 mice to overexpress Bmi1 in a cell type–specific fashion and the mdx mice for the experiments in a dystrophic model. In particular, the following compound mutant mice were generated: Pax7CreERT2;STOPFloxBmi1 (Pax7Bmi1) and mdx;Pax7CreERT2;STOPFloxBmi1 (mdx;Pax7Bmi1).
Acute muscle injury and histological analysis.
Pax7Bmi1 and littermate control mice (2–4 mo) were used in this experiment. Mice were injected with Tamoxifen (as described above) to induce Bmi1 overexpression in satellite cells. 10 d after the last injection, muscle freeze injury (FZ) was performed on these mice. 2% isoflurane inhalation was used to anesthetize the mice. The left hind limb was shaved and then wet with 70% ethanol to remove hair. To induce the damage to the Tibialis Anterior (TA) muscle, an incision of 5 mm was done in the skin above the muscle; then, a dry ice precooled probe was applied on the muscle for 10 s. The incision was closed by veterinary glue (VETBOND Tissue Adhesive). The mice were either sacrificed or used in a test for treadmill performance 10 d.a.i. Both the injured and contralateral hind limbs were collected. The injured leg was embedded in OCT and frozen in liquid nitrogen-precooled isopentane, whereas the contralateral muscles were snap-frozen in liquid nitrogen and processed for RNA extraction to check Bmi1 expression levels.
The frozen muscles were serially sectioned at 8 µm (with a step of 120 µm) and processed for H&E staining and/or immunohistochemistry. The percentage of CNF was counted in the injured area of three different sections along the length of the leg for each animal. In the same sections, the cross-sectional areas (CSAs) of the fibers in regenerating and uninjured areas (as control) were measured using the MetaMorph software (Life Sciences Research Imaging Systems). The researcher who performed all the analysis was blinded to the animal’s genotype.
Treadmill performance test after FZ injury.
Tamoxifen-injected Pax7Bmi1 and littermate controls mice (2–4 mo) were used in this experiment. 10 d after the muscle freeze injury mice performed a functional test running on a treadmill (LE8710M, Panlab Harvard Apparatus). 3 d before the test, mice were trained for each day for 5 min, at the speed of 10 m/min, with an inclination of 5°. The day of the test, the mice started running for 4 min (speed 10 m/min) and then every 4 min the speed was increased of 2 m/min until the mice were tired. Tiredness was defined as the inability of the mouse to go back to the treadmill belt despite five mechanical stimulations (De Luca, 2008a). The researcher who performed all the analysis was blinded to the animal’s genotype.
Enforced treadmill exercise and histological analysis.
The dystrophic animals mdx;Pax7Bmi1, mdx control littermates and C57BL/6J-BL/10 mice (2–4 mo) were used in this experiment. Mice were injected with Tamoxifen. Starting from 3 d after the last injection, one mouse was put in each lane of the treadmill and forced to run at the speed of 12 m/min for 30 min, twice a week for 6 wk. This protocol was not exhausting and generally all the mice were able to perform the exercise. Whenever an avoiding behavior was observed, the mice were stimulated to run by a gentle touch of the tail or with the presence of a soft barrier at the end of the treadmill lane (De Luca, 2008b). The mice were sacrificed 6 d after the last treadmill session. One forelimb was embedded in OCT and frozen in isopentane cooled at the liquid nitrogen temperature. The other forelimb was dissected and the muscles snap-frozen in liquid nitrogen to perform RNA extraction and to check Bmi1 expression levels.
The frozen muscles were serially sectioned at 8 µm and processed for H&E staining and/or immunohistochemistry. The percentage of CNF was counted in the injured area of three different sections along the length of the leg for the same animal. The researcher who performed all the analysis was blinded to the animal’s genotype.
Assessment of muscle strength.
The dystrophic animals mdx;Pax7Bmi1, mdx control littermates and C57BL/6J-BL/10 mice (2–4 mo) performing the enforced treadmill exercise were used in this experiment. Before each treadmill session, a forelimb grip test was performed (De Luca, 2008c) to assess the course of the disease. The test was performed using a horizontal grip strength meter (Linton). In brief, each mouse was lifted by the tail toward the metal pull bar. Once the mouse had grasped the bar with the forelimbs, the mouse was pulled back in a horizontal plane. The protocol was repeated three consecutive times within 30 s. The grip strength was normalized against the body weight.
Cell transplantation experiments.
Satellite cells isolated from mdx;STOPFloxBmi1 were treated as for the MT1 kd experiments. 6 d later, cells were harvested and 20,000 cells were transplanted into surgically exposed TA muscles of 3-wk-old mdx mice. After 27 d, the muscle tissues were fixed in 4% PFA for 6 h, left on in 30% sucrose, and embedded in OCT.
Serial sections (8 µm thick) were cut, mounted, and analyzed at the microscope for GFP positivity (two sections/mouse; three fields/section). The same settings were applied to take pictures of MT1 kd and MT1 SCr transplanted mice. For the analysis of the strong GFP positivity a defined threshold was applied to all pictures and the GFP+ve fibers were counted out of the total number of fibers/microscopic field. The percentage of strong GFP+ve fibers was calculated out of the total number of GFP+ve fibers/field. The researcher who performed all the analysis was blinded to the different treatments of the transplanted satellite cells.
Microarray analysis.
Three independently prepared satellite cell cultures isolated from STOPFloxBmi1 mice, treated with A-Cre infection and induced to differentiate for 2 d were analyzed by whole-genome Illumina platform mouse v2 as compared with the same cells treated with A-GFP. Integrity of starting total RNA was evaluated using capillary electrophoresis (Bioanalyzer 2100; Agilent Technologies) and quantified using a NanoDrop 1000 (NanoDrop). Aliquots of RNA (250 ng) samples were amplified according to the specifications of the Illumina TotalPrep RNA Amplification kit (Ambion). The cRNA samples obtained were applied to the MouseWG-6 v2.0 Expression BeadChip arrays (Illumina) and hybridized according to the manufacturer’s specification. Each array on the BeadChip targets >45,200 transcripts using 3-µm beads bearing covalently attached 50-base oligonucleotide probes. The BeadChips were scanned with the BeadSrray system scanner (Illumina). The hybridization images signal intensity was extracted and background subtracted using BeadStudio software (version 3.3.7 Illumina). The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession no. GSE41878.
All the data produced were quality checked with array QualityMetrics package under R version 2.15.1 Bioconductor software to detect possible outliers. To identify differentially expressed genes, based on a moderate t test, the Limma package was used and genes were selected based on a p-value cut-off <0.05 and a fold change threshold of 1.9. The resultant list of differentially expressed probes was then analyzed using IPA software (Ingenuity) to detect the enrichment of biofunctions and networks Core according to IPA analysis settings: reference set, MouseWG-6 v2.0; relationship to include, direct and indirect; includes endogenous chemicals; filter summary, consider only molecules and/or relationships where (species = rat OR mouse OR uncategorized (e.g., chemicals)) AND (confidence = high (predicted) OR experimentally observed) AND (data sources = BIND OR BIOGRID OR ClinicalTrials.gov OR Cognia OR DIP OR Gene Ontology (GO) OR GVK Biosciences OR HumanCyc OR Ingenuity Expert Findings OR Ingenuity ExpertAssist Findings OR INTACT OR Interactome studies OR Kyoto Encyclopedia of Genes and Genomes (KEGG) OR MINT OR MIPS OR miRBase OR Mouse Genome Database (MGD) OR Obesity Gene Map Database).
Statistical analysis.
For the histological analysis on muscle sections, the number of fibers in each range of areas (µm2) was counted and the percentage of fibers in each range was calculated. Percentages from different animals were pooled and the mean calculated. For the ratio of regeneration, the mean CSA of CNF was divided by the mean CSA of mature fibers. Unless specified in the text, all the graphs represent population mean ± SEM. Statistical analysis was performed using Prism statistical analysis software (GraphPad), and tests applied included Student’s t test for single comparisons and ANOVA, Kruskal-Wallis, or Student’s t test with Bonferroni correction for multiple comparison. Significance is indicated as ***, P < 0.001; **, P < 0.01; *, P < 0.05. For behavioral tests, each point of forelimb grip strength represents the mean of the normalized strength between all animals per genotype at that specific time point. Mixed effects linear regression was used to model the change in forelimb grip strength over the last six attempts and to compare results between groups of mice. Likelihood ratio tests were performed to determine whether the inclusion of random intercepts and slopes significantly improved the models. All analyses were performed in Stata (version 10; StatCorp).
Respiratory chain enzyme activity measurements.
The activities of mitochondrial respiratory chain (RC) enzymes and the mitochondrial matrix marker citrate synthase were determined in mouse muscle (Gastrocnemius) after 600 g supernatants as described by Kirby et al. (2007). Statistical analyses (one-way ANOVA with Tukey’s post-test) of activities between different groups were performed using Prism version 5.00 for Windows (GraphPad Software). Histochemical demonstration of mitochondrial oxidative enzyme activities and determination of muscle fiber type were undertaken using standard protocols.
Online supplemental material.
Table S1 lists differentially expressed genes in satellite cell cultures upon Bmi1 overexpression. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20140317/DC1.
Supplementary Material
Acknowledgments
We thank S. Brandner for critically reading this article and for many useful suggestions on the Definiens software; P. Pallier, J. Bestwick, and Paul Creeke for advice on the behavioral experiments and on statistics, respectively; all members of the Marino Lab for helpful discussions; and S. Fukada and S. Takeda for the gift of the SM/C-2.6 antibody. We are grateful to the BSU staff for help in the daily care of our mouse colony.
This work is supported by grants of the Medical Research Council UK (G0802546/1) and Muscular Dystrophy Campaign (RA4/788/4) to S. Marino and L.G. Robson. F. Muntoni is supported by the Great Ormond Street Children’s Charity. R.W. Taylor is funded by the Wellcome Trust (096919/Z/11/Z) and the Medical Research Council UK Centre for Neuromuscular Diseases (G0601943). The MRC Neuromuscular Centre biobank is also gratefully acknowledged.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- A-Cre
- Adeno-Cre
- A-GFP
- Adeno-GFP
- CNF
- centrally nucleated fiber
- CSA
- cross-sectional area
- d.a.i.
- days after injury
- DMD
- Duchenne muscular dystrophy
- MT1
- metallothionein 1
- PcG
- Polycomb group
- ROS
- reactive oxygen species
References
- Abdouh M., Chatoo W., El Hajjar J., David J., Ferreira J., and Bernier G.. 2012. Bmi1 is down-regulated in the aging brain and displays antioxidant and protective activities in neurons. PLoS ONE. 7:e31870 10.1371/journal.pone.0031870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi K., Sandig V., Vooijs M., Van der Valk M., Giovannini M., Strauss M., and Berns A.. 1997. Cre-mediated somatic site-specific recombination in mice. Nucleic Acids Res. 25:1766–1773 10.1093/nar/25.9.1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asp P., Blum R., Vethantham V., Parisi F., Micsinai M., Cheng J., Bowman C., Kluger Y., and Dynlacht B.D.. 2011. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc. Natl. Acad. Sci. USA. 108:E149–E158 10.1073/pnas.1102223108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau H.M., Webster C., Chiu C.P., Guttman S., and Chandler F.. 1983. Differentiation properties of pure populations of human dystrophic muscle cells. Exp. Cell Res. 144:495–503 10.1016/0014-4827(83)90431-7 [DOI] [PubMed] [Google Scholar]
- Blau H.M., Webster C., Pavlath G.K., and Chiu C.P.. 1985. Evidence for defective myoblasts in Duchenne muscular dystrophy. Adv. Exp. Med. Biol. 182:85–110 10.1007/978-1-4684-4907-5_7 [DOI] [PubMed] [Google Scholar]
- Brack A.S., and Rando T.A.. 2012. Tissue-specific stem cells: lessons from the skeletal muscle satellite cell. Cell Stem Cell. 10:504–514 10.1016/j.stem.2012.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulfield G., Siller W.G., Wight P.A., and Moore K.J.. 1984. X chromosome-linked muscular dystrophy (mdx) in the mouse. Proc. Natl. Acad. Sci. USA. 81:1189–1192 10.1073/pnas.81.4.1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C.A., and Morgan J.E.. 2003. Duchenne’s muscular dystrophy: animal models used to investigate pathogenesis and develop therapeutic strategies. Int. J. Exp. Pathol. 84:165–172 10.1046/j.1365-2613.2003.00354.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper R.N., Tajbakhsh S., Mouly V., Cossu G., Buckingham M., and Butler-Browne G.S.. 1999. In vivo satellite cell activation via Myf5 and MyoD in regenerating mouse skeletal muscle. J. Cell Sci. 112:2895–2901. [DOI] [PubMed] [Google Scholar]
- Cozzoli A., Nico B., Sblendorio V.T., Capogrosso R.F., Dinardo M.M., Longo V., Gagliardi S., Montagnani M., and De Luca A.. 2011. Enalapril treatment discloses an early role of angiotensin II in inflammation- and oxidative stress-related muscle damage in dystrophic mdx mice. Pharmacol. Res. 64:482–492 10.1016/j.phrs.2011.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca A.2008a. Use of treadmill and wheel exercise to assess dystrophic state. SOP Number: DMD_M.2.1.003. Treat-NMD Neuromuscular Network. Wellstone Muscular Dystrophy Center; Washington, DC: http://www.treat-nmd.eu/downloads/file/sops/dmd/MDX/DMD_M.2.1.003.pdf (accessed November 17, 2014). [Google Scholar]
- De Luca A.2008b. Use of treadmill and wheel exercise for impact on mdx mice phenotype. SOP Number: DMD_M.2.1.001. Treat-NMD Neuromuscular Network. Wellstone Muscular Dystrophy Center; Washington, DC: http://www.treat-nmd.eu/downloads/file/sops/dmd/MDX/DMD_M.2.1.001.pdf (accessed November 13, 2014). [Google Scholar]
- De Luca A.2008c. Use of grip strength meter to assess the limb strength of mdx mice. SOP Number: DMD_M.2.2.001. Treat-NMD Neuromuscular Network. Wellstone Muscular Dystrophy Center; Washington, DC: http://www.treat-nmd.eu/downloads/file/sops/dmd/MDX/DMD_M.2.2.001.pdf (accessed November 14, 2014). [Google Scholar]
- De Luca A., Pierno S., Liantonio A., Cetrone M., Camerino C., Fraysse B., Mirabella M., Servidei S., Rüegg U.T., and Conte Camerino D.. 2003. Enhanced dystrophic progression in mdx mice by exercise and beneficial effects of taurine and insulin-like growth factor-1. J. Pharmacol. Exp. Ther. 304:453–463 10.1124/jpet.102.041343 [DOI] [PubMed] [Google Scholar]
- Disatnik M.H., Dhawan J., Yu Y., Beal M.F., Whirl M.M., Franco A.A., and Rando T.A.. 1998. Evidence of oxidative stress in mdx mouse muscle: studies of the pre-necrotic state. J. Neurol. Sci. 161:77–84 10.1016/S0022-510X(98)00258-5 [DOI] [PubMed] [Google Scholar]
- Duncan C.J.1989. Dystrophin and the integrity of the sarcolemma in Duchenne muscular dystrophy. Experientia. 45:175–177 10.1007/BF01954866 [DOI] [PubMed] [Google Scholar]
- Facchino S., Abdouh M., Chatoo W., and Bernier G.. 2010. BMI1 confers radioresistance to normal and cancerous neural stem cells through recruitment of the DNA damage response machinery. J. Neurosci. 30:10096–10111 10.1523/JNEUROSCI.1634-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faist V., Koenig J., Hoeger H., and Elmadfa I.. 1998. Mitochondrial oxygen consumption, lipid peroxidation and antioxidant enzyme systems in skeletal muscle of senile dystrophic mice. Pflugers Arch. 437:168–171 10.1007/s004240050764 [DOI] [PubMed] [Google Scholar]
- Faist V., König J., Höger H., and Elmadfa I.. 2001. Decreased mitochondrial oxygen consumption and antioxidant enzyme activities in skeletal muscle of dystrophic mice after low-intensity exercise. Ann. Nutr. Metab. 45:58–66 10.1159/000046707 [DOI] [PubMed] [Google Scholar]
- Fukada S., Higuchi S., Segawa M., Koda K., Yamamoto Y., Tsujikawa K., Kohama Y., Uezumi A., Imamura M., Miyagoe-Suzuki Y., et al. . 2004. Purification and cell-surface marker characterization of quiescent satellite cells from murine skeletal muscle by a novel monoclonal antibody. Exp. Cell Res. 296:245–255 10.1016/j.yexcr.2004.02.018 [DOI] [PubMed] [Google Scholar]
- Fulle S., Protasi F., Di Tano G., Pietrangelo T., Beltramin A., Boncompagni S., Vecchiet L., and Fanò G.. 2004. The contribution of reactive oxygen species to sarcopenia and muscle ageing. Exp. Gerontol. 39:17–24 10.1016/j.exger.2003.09.012 [DOI] [PubMed] [Google Scholar]
- Fulle S., Di Donna S., Puglielli C., Pietrangelo T., Beccafico S., Bellomo R., Protasi F., and Fanò G.. 2005. Age-dependent imbalance of the antioxidative system in human satellite cells. Exp. Gerontol. 40:189–197 10.1016/j.exger.2004.11.006 [DOI] [PubMed] [Google Scholar]
- Gavet O., and Pines J.. 2010. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev. Cell. 18:533–543 10.1016/j.devcel.2010.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayraud-Morel B., Chrétien F., Flamant P., Gomès D., Zammit P.S., and Tajbakhsh S.. 2007. A role for the myogenic determination gene Myf5 in adult regenerative myogenesis. Dev. Biol. 312:13–28 10.1016/j.ydbio.2007.08.059 [DOI] [PubMed] [Google Scholar]
- Ginjala V., Nacerddine K., Kulkarni A., Oza J., Hill S.J., Yao M., Citterio E., van Lohuizen M., and Ganesan S.. 2011. BMI1 is recruited to DNA breaks and contributes to DNA damage-induced H2A ubiquitination and repair. Mol. Cell. Biol. 31:1972–1982 10.1128/MCB.00981-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S., Iwashita T., Buchstaller J., Molofsky A.V., Thomas D., and Morrison S.J.. 2009. Bmi-1 over-expression in neural stem/progenitor cells increases proliferation and neurogenesis in culture but has little effect on these functions in vivo. Dev. Biol. 328:257–272 10.1016/j.ydbio.2009.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail I.H., Andrin C., McDonald D., and Hendzel M.J.. 2010. BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J. Cell Biol. 191:45–60 10.1083/jcb.201003034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M.J.2009. Strategies for reducing oxidative damage in ageing skeletal muscle. Adv. Drug Deliv. Rev. 61:1363–1368 10.1016/j.addr.2009.07.018 [DOI] [PubMed] [Google Scholar]
- Jacobs J.J., Kieboom K., Marino S., DePinho R.A., and van Lohuizen M.. 1999. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 397:164–168 10.1038/16476 [DOI] [PubMed] [Google Scholar]
- Juan A.H., Derfoul A., Feng X., Ryall J.G., Dell’Orso S., Pasut A., Zare H., Simone J.M., Rudnicki M.A., and Sartorelli V.. 2011. Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes Dev. 25:789–794 10.1101/gad.2027911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kägi J.H., and Hunziker P.. 1989. Mammalian metallothionein. Biol. Trace Elem. Res. 21:111–118 10.1007/BF02917243 [DOI] [PubMed] [Google Scholar]
- Kirby D.M., Thorburn D.R., Turnbull D.M., and Taylor R.W.. 2007. Biochemical assays of respiratory chain complex activity. Methods Cell Biol. 80:93–119 10.1016/S0091-679X(06)80004-X [DOI] [PubMed] [Google Scholar]
- Liu J., Cao L., Chen J., Song S., Lee I.H., Quijano C., Liu H., Keyvanfar K., Chen H., Cao L.Y., et al. . 2009. Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature. 459:387–392 10.1038/nature08040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Nelson B.R., Bezprozvannaya S., Shelton J.M., Richardson J.A., Bassel-Duby R., and Olson E.N.. 2014. Requirement of MEF2A, C, and D for skeletal muscle regeneration. Proc. Natl. Acad. Sci. USA. 111:4109–4114 10.1073/pnas.1401732111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourikis P., Sambasivan R., Castel D., Rocheteau P., Bizzarro V., and Tajbakhsh S.. 2012. A critical requirement for notch signaling in maintenance of the quiescent skeletal muscle stem cell state. Stem Cells. 30:243–252 10.1002/stem.775 [DOI] [PubMed] [Google Scholar]
- Oliver J.R., Jiang S., and Cherian M.G.. 2006. Augmented hepatic injury followed by impaired regeneration in metallothionein-I/II knockout mice after treatment with thioacetamide. Toxicol. Appl. Pharmacol. 210:190–199 10.1016/j.taap.2005.05.007 [DOI] [PubMed] [Google Scholar]
- Pan M.R., Peng G., Hung W.C., and Lin S.Y.. 2011. Monoubiquitination of H2AX protein regulates DNA damage response signaling. J. Biol. Chem. 286:28599–28607 10.1074/jbc.M111.256297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda A.C., Abdelmohsen K., Yoon J.H., Martindale J.L., Yang X., Curtis J., Mercken E.M., Chenette D.M., Zhang Y., Schneider R.J., et al. . 2014. RNA-binding protein AUF1 promotes myogenesis by regulating MEF2C expression levels. Mol. Cell. Biol. 34:3106–3119 10.1128/MCB.00423-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel H., Chen J., Das K.C., and Kavdia M.. 2013. Hyperglycemia induces differential change in oxidative stress at gene expression and functional levels in HUVEC and HMVEC. Cardiovasc. Diabetol. 12:142 10.1186/1475-2840-12-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospisilik J.A., Knauf C., Joza N., Benit P., Orthofer M., Cani P.D., Ebersberger I., Nakashima T., Sarao R., Neely G., et al. . 2007. Targeted deletion of AIF decreases mitochondrial oxidative phosphorylation and protects from obesity and diabetes. Cell. 131:476–491 10.1016/j.cell.2007.08.047 [DOI] [PubMed] [Google Scholar]
- Rando T.A., Disatnik M.H., Yu Y., and Franco A.. 1998. Muscle cells from mdx mice have an increased susceptibility to oxidative stress. Neuromuscul. Disord. 8:14–21 10.1016/S0960-8966(97)00124-7 [DOI] [PubMed] [Google Scholar]
- Robson L.G., Di Foggia V., Radunovic A., Bird K., Zhang X., and Marino S.. 2011. Bmi1 is expressed in postnatal myogenic satellite cells, controls their maintenance and plays an essential role in repeated muscle regeneration. PLoS ONE. 6:e27116 10.1371/journal.pone.0027116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan S.D., Dolatabadi N., Chan S.F., Zhang X., Akhtar M.W., Parker J., Soldner F., Sunico C.R., Nagar S., Talantova M., et al. . 2013. Isogenic human iPSC Parkinson’s model shows nitrosative stress-induced dysfunction in MEF2-PGC1α transcription. Cell. 155:1351–1364 10.1016/j.cell.2013.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakellariou G.K., Pye D., Vasilaki A., Zibrik L., Palomero J., Kabayo T., McArdle F., Van Remmen H., Richardson A., Tidball J.G., et al. . 2011. Role of superoxide-nitric oxide interactions in the accelerated age-related loss of muscle mass in mice lacking Cu,Zn superoxide dismutase. Aging Cell. 10:749–760 10.1111/j.1474-9726.2011.00709.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P.J., Wiltshire M., Furon E., Beattie J.H., and Errington R.J.. 2008. Impact of overexpression of metallothionein-1 on cell cycle progression and zinc toxicity. Am. J. Physiol. Cell Physiol. 295:C1399–C1408 10.1152/ajpcell.00342.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P.1999. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21:70–71 10.1038/5007 [DOI] [PubMed] [Google Scholar]
- Spassov A., Gredes T., Gedrange T., Pavlovic D., Lupp A., and Kunert-Keil C.. 2011. Increased oxidative stress in dystrophin deficient (mdx) mice masticatory muscles. Exp. Toxicol. Pathol. 63:549–552 10.1016/j.etp.2010.04.006 [DOI] [PubMed] [Google Scholar]
- Tidball J.G., and Wehling-Henricks M.. 2007. The role of free radicals in the pathophysiology of muscular dystrophy. J. Appl. Physiol. 102:1677–1686 10.1152/japplphysiol.01145.2006 [DOI] [PubMed] [Google Scholar]
- Urso M.L., Scrimgeour A.G., Chen Y.W., Thompson P.D., and Clarkson P.M.. 2006. Analysis of human skeletal muscle after 48 h immobilization reveals alterations in mRNA and protein for extracellular matrix components. J. Appl. Physiol. 101:1136–1148 10.1152/japplphysiol.00180.2006 [DOI] [PubMed] [Google Scholar]
- van der Laak J.A., Pahlplatz M.M., Hanselaar A.G., and de Wilde P.C.. 2000. Hue-saturation-density (HSD) model for stain recognition in digital images from transmitted light microscopy. Cytometry. 39:275–284 [DOI] [PubMed] [Google Scholar]
- van der Lugt N.M., Domen J., Linders K., van Roon M., Robanus-Maandag E., te Riele H., van der Valk M., Deschamps J., Sofroniew M., van Lohuizen M., et al. . 1994. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 8:757–769 10.1101/gad.8.7.757 [DOI] [PubMed] [Google Scholar]
- Vasilaki A., Simpson D., McArdle F., McLean L., Beynon R.J., Van Remmen H., Richardson A.G., McArdle A., Faulkner J.A., and Jackson M.J.. 2007. Formation of 3-nitrotyrosines in carbonic anhydrase III is a sensitive marker of oxidative stress in skeletal muscle. Proteomics Clin. Appl. 1:362–372 10.1002/prca.200600702 [DOI] [PubMed] [Google Scholar]
- Wang Y.X., and Rudnicki M.A.. 2012. Satellite cells, the engines of muscle repair. Nat. Rev. Mol. Cell Biol. 13:127–133. [DOI] [PubMed] [Google Scholar]
- Whitehead N.P., Yeung E.W., and Allen D.G.. 2006. Muscle damage in mdx (dystrophic) mice: role of calcium and reactive oxygen species. Clin. Exp. Pharmacol. Physiol. 33:657–662 10.1111/j.1440-1681.2006.04394.x [DOI] [PubMed] [Google Scholar]
- Whitehead N.P., Pham C., Gervasio O.L., and Allen D.G.. 2008. N-Acetylcysteine ameliorates skeletal muscle pathophysiology in mdx mice. J. Physiol. 586:2003–2014 10.1113/jphysiol.2007.148338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won M.H., Kang T.C., Jeon G.S., Lee J.C., Kim D.Y., Choi E.M., Lee K.H., Choi C.D., Chung M.H., and Cho S.S.. 1999. Immunohistochemical detection of oxidative DNA damage induced by ischemia-reperfusion insults in gerbil hippocampus in vivo. Brain Res. 836:70–78 10.1016/S0006-8993(99)01611-X [DOI] [PubMed] [Google Scholar]
- Wozniak A.C., and Anderson J.E.. 2007. Nitric oxide-dependence of satellite stem cell activation and quiescence on normal skeletal muscle fibers. Dev. Dyn. 236:240–250 10.1002/dvdy.21012 [DOI] [PubMed] [Google Scholar]
- Yadirgi G., Leinster V., Acquati S., Bhagat H., Shakhova O., and Marino S.. 2011. Conditional activation of Bmi1 expression regulates self-renewal, apoptosis, and differentiation of neural stem/progenitor cells in vitro and in vivo. Stem Cells. 29:700–712 10.1002/stem.614 [DOI] [PubMed] [Google Scholar]
- Zbinden S., Wang J., Adenika R., Schmidt M., Tilan J.U., Najafi A.H., Peng X., Lassance-Soares R.M., Iantorno M., Morsli H., et al. . 2010. Metallothionein enhances angiogenesis and arteriogenesis by modulating smooth muscle cell and macrophage function. Arterioscler. Thromb. Vasc. Biol. 30:477–482 10.1161/ATVBAHA.109.200949 [DOI] [PubMed] [Google Scholar]
- Zhuang W., Eby J.C., Cheong M., Mohapatra P.K., Bredt D.S., Disatnik M.H., and Rando T.A.. 2001. The susceptibility of muscle cells to oxidative stress is independent of nitric oxide synthase expression. Muscle Nerve. 24:502–511 10.1002/mus.1033 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.