Abstract
The manipulation of distinct signaling pathways and transcription factors has been shown to influence life span in a cell-non-autonomous manner in multicellular model organisms such as Caenorhabditis elegans. These data suggest that coordination of whole-organism aging involves endocrine signaling, however, the molecular identities of such signals have not yet been determined and their potential relevance in humans is unknown. Here we describe a novel metabolomic approach to identify molecules directly associated with extended life span in C. elegans that represent candidate compounds for age-related endocrine signals. To identify metabolic perturbations directly linked to longevity, we developed metabolomic software for meta-analysis that enabled intelligent comparisons of multiple different mutants. Simple pairwise comparisons of long-lived glp-1, daf-2, and isp-1 mutants to their respective controls resulted in more than 11,000 dysregulated metabolite features of statistical significance. By using meta-analysis, we were able to reduce this number to six compounds most likely to be associated with life-span extension. Mass spectrometry-based imaging studies suggested that these metabolites might be localized to C. elegans muscle. We extended the metabolomic analysis to humans by comparing quadricep muscle tissue from young and old individuals and found that two of the same compounds associated with longevity in worms were also altered in human muscle with age. These findings provide candidate compounds that may serve as age-related endocrine signals and implicate muscle as a potential tissue regulating their levels in humans.
Keywords: Aging, C. elegans, Metabolism, Metabolomics, Meta-analysis, Sarcopenia
1 Introduction
C. elegans is an attractive model organism to study aging because it is genetically tractable, the location and fate of its 959 cells is known, it has a relatively short life span, and it undergoes age-related changes such as sarcopenia that are also known to occur in mammals (Herndon et al. 2002; Kashyap et al. 2011; Kenyon 2010). As such, a growing number of technologies have been widely applied to analyze the regulation of life span in C. elegans and led to the identification and characterization of many genes influencing longevity. These tools have included RNAi, transcriptional analyses, chromatin immunoprecipitation studies, and mass spectrometry-based proteomic screens (Dong et al. 2007; Golden and Melov 2007; Panowski and Dillin 2009; Shim and Paik 2010). Results from the application of these techniques have shown that aging and metabolism, two fields that were once thought to be largely separate, are directly related. However, the focus of the majority of C. elegans aging studies has been genes, transcripts, and proteins. To date, broad surveys of metabolites related to aging have been limited in C. elegans as well as other biological systems.
Only recently have technical innovations enabled profiling of large numbers of metabolites from biological specimens, an area of study known as global metabolomics. In addition to developments in instrumentation, the emergence of metabolite databases and bioinformatic software for data analysis has been key to establishing global metabolomic methods (Smith et al. 2005, 2006; Wishart et al. 2009). By using liquid chromatography/mass spectrometry (LC/MS) to analyze metabolite extracts, thousands of so-called “metabolite features” can be detected—where a metabolite feature is defined as a data peak with a unique mass-to-charge ratio and a unique retention time. Despite the large number of metabolite features readily detected by LC/MS, structural identification of many metabolites remains challenging due to incomplete metabolite databases and biochemical pathway maps (Baker 2011). While determining the complete catalogue of metabolites present in healthy human plasma has been a focus of significant effort, many compounds are still uncharacterized and a substantial number of metabolite features detected in other organisms such as C. elegans remain unknown (Psychogios et al. 2011). With current global metabolomic methods, even identification of known compounds in metabolite databases is a time-consuming step that requires additional analyses for confirmation (Yanes et al. 2010a). Thus, new metabolomic tools and experimental methodologies to facilitate characterization of metabolites are of great utility.
Here, we describe the application of global metabolomic software to facilitate identification of features related to longevity by using meta-analysis. Mutations in glp-1, daf-2, and isp-1 that encode for a germline-proliferation signal receptor, an insulin-like hormone receptor, and an iron-sulfur protein of mitochondrial complex III, respectively, extend the lifespan of C. elegans (Feng et al. 2001; Kenyon 2005). These mutations, however, lead to a large cascade of metabolic perturbations that alters thousands of metabolite features detectable by LC/MS (see Results and Discussion section). Given that it is unlikely that all of these altered features are associated with the aging phenotype, we sought to develop a methodology to identify features that were directly related to longevity. Specifically, we were interested in metabolites that might be acting as key signaling molecules. Indeed, endocrine signaling has been suggested to modulate longevity responses in most multicellular, long-lived organisms including C. elegans (Baumeister et al. 2006; Kleemann and Murphy 2009; Rottiers et al. 2006; Russell and Kahn 2007; Tatar et al. 2003). Germ line ablation, alterations in insulin signaling, and reduced mitochondrial function are known to affect life span in a cell-non-autonomous nature, suggesting that molecular signals from specific tissues distally regulate aging of the whole organism (Arantes-Oliveira et al. 2002; Broughton et al. 2005; Durieux et al. 2011; Hwangbo et al. 2004; Libina et al. 2003; Wolkow et al. 2000). Yet, the chemical identities of such signals remain elusive and nuclear hormone receptors known to influence life span in worms are orphans for which activating ligands have not been determined (Sluder and Maina 2001). Metabolites such as steroids and fatty acids are likely candidates for endocrine signals and dafachronic acid metabolites have already been identified as DAF-12 ligands that influence life span in C. elegans (Gerisch et al. 2007; Motola et al. 2006). Our approach was to use intelligent comparisons of genetically manipulated worms to identify candidate metabolites as secondary signals that may coordinate aging in C. elegans and humans.
2 Materials and methods
2.1 Experimental procedures
2.1.1 Preparation of C. elegans for metabolomic analysis
The following organisms were used for analysis: glp-1(e2141), CF512(fer-15(b26); fem-1(hc17), daf-16(mu86); glp-1(e2141), daf-2(e1370), daf-16(mu86); daf-2(e1370), isp-1(qm150), and N2. CF512 worms served as a sterile control and N2 worms served as a wild-type control. Eggs were harvested by bleaching gravid adults and transferred to M9 medium to aerate overnight for hatching. Synchronized, arrested L1 worms were then transferred to NGM plates seeded with E. coli (strain OP50) and harvested on day 1. Each sample group contained ~1,500 worms as determined by counting of the organisms under microscope.
2.1.2 Muscle biopsies
Muscle samples were obtained from the upper-mid left quadriceps of de-identified, healthy patients by using the Bergström technique. Caucasian adults were selected for similar weight and BMI. Participants were healthy, non-obese, nonsmoking, and not on any medications affecting metabolism or body weight. All subjects had normal fasting glucose at screening and reported no first-degree family history of type 2 diabetes. Young samples were collected from patients of age 21, 22, 23, 24, 25, 27, 28, 29, 31, and 31. Elderly samples were collected from patients of age 72, 72, 74, 74, 75, 76, 77, 77, 80, 81, and 81. The study was approved by the Institutional Review Board of the Pennington Biomedical Research Center. Samples were frozen after biopsy and stored at −80 °C. Ten mg samples were used for metabolomic analysis.
2.1.3 Metabolite isolation
Metabolites were isolated from worms and muscle tissue by using cold methanol and acetone as described previously (Yanes et al. 2010b; 2011). First, a volume of 300 µL of cold (−20 °C) acetone was added to the samples, vortexed for 30 s, and the sample incubated in liquid nitrogen. The sample was thawed at room temperature and incubated in liquid nitrogen two more times prior to a 10-min sonication. After 1 h at −20 °C, the samples were centrifuged at 13,000 rpm for 15 min and the resulting supernatant was stored at −20 °C. The precipitate was then mixed with 300 µL of methanol/water/formic acid in the ratio of 86.5/12.5/1.0 and sonicated prior to a 1 h incubation at −20 °C. After centrifugation, the supernatant was collected and transferred to that which was collected after acetone extraction. The solution was dried with a vacuum concentration (SpeedVac) at room temperature and re-dissolved in 100 µL of 95 % acetonitrile/5 % water for liquid chromatography/mass spectrometry (LC/MS) analysis.
2.1.4 LC/MS analysis
Liquid chromatography was performed by using a reversed-phase C18 column (Zorbax C18, Agilent, 5 µm, 150 × 0.5 mm diameter column) with a flow rate of 20 µl/min. Samples were analyzed by using electrospray ionization time-of-flight mass spectrometry (Agilent 6520 QTOF) in positive-ion mode with water/acetonitrile as mobile-phases A/B, each containing 0.1 % formic acid. Lock mass was not used to avoid ion suppression. Standard calibrations were performed after each group comparison to adjust for instrument drift. C18 sample analyses were performed by using the following linear changes in mobile-phase B composition with time: 0 min: 10 % B, 5 min: 10 % B, 10 min: 40 % B, 50 min: 98 % B, 65 min: 98 %B. Sample groups were randomized from run to run, each separated by wash runs, to reduce error due to instrument variability and possible carryover. Data were analyzed by using the open-source software XCMS and metaXCMS (Patti et al. 2012; Smith et al. 2006; Tautenhahn et al. 2011).
2.1.5 Metabolite assignments
Metabolite assignments were made by first searching the accurate masses of metabolite features in METLIN, HMDB, KEGG, and LIPID MAPS metabolite databases. Next, the MS/MS fragmentation pattern of the metabolite of interest was compared to that of a model compound analyzed with identical conditions and collision energy (Agilent 6520 QTOF). All fragmentation data were generated by using a collision energy of 20 V.
2.1.6 Nanostructure-initiator mass spectrometry (NIMS)
A detailed description of the preparation of NIMS surfaces is described elsewhere (Northen et al. 2007; Woo et al. 2008). In brief, single-sided polished p-type (100) silicon wafers of 500 µM thickness and low resistivity were cut into 3.3 × 3.3 cm2 squares. Wafers were soaked in a 2:1 mixture of sulfuric acid and hydrogen peroxide for 30 min, rinsed with nanopure water, and blown dry with nitrogen gas. Etching was performed by clamping the chips between gold foil in a chamber filled with 25 % ethanolic hydrofluoric acid solution. A platinum loop was immersed in the hydrofluoric acid solution to serve as a cathode. Etching was performed in constant current mode at 300 mA for 30 min. The etched surfaces were rinsed with methanol and blown dry. Worms were transferred by thaw-mounting immediately prior to mass spectrometry analysis. Images were acquired by using a 5800 Applied Biosystems TOF/TOF instrument in which 20–50 laser shots were collected per spectrum at 5 µM resolution.
3 Results and discussion
3.1 Metabolomics of glp-1 worms
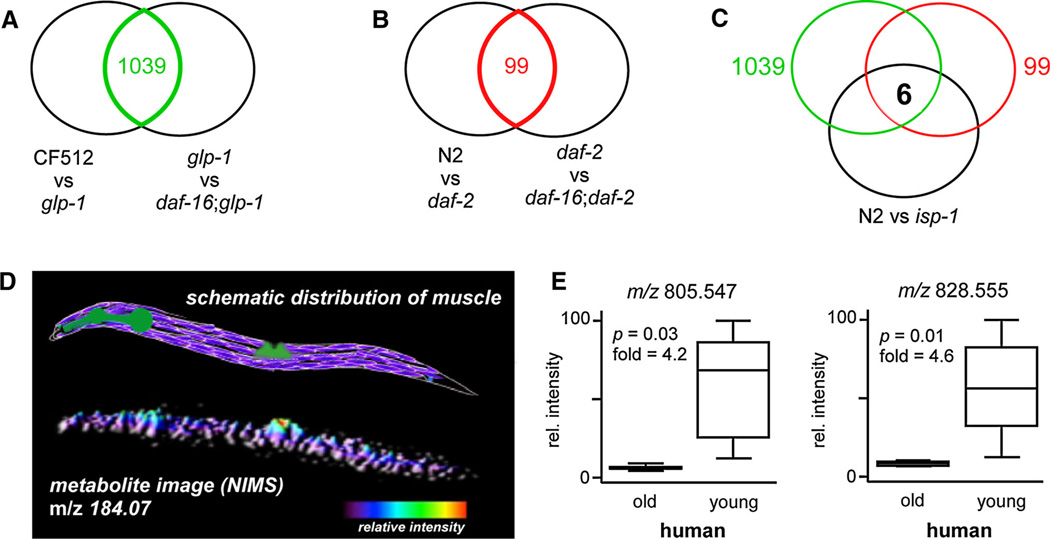
We first performed a global metabolomic comparison of sterile CF512(fer-15(b26); fem-1(hc17) control C. elegans and long-lived worms in which the germ line had been removed by glp-1(e2141) mutation. All metabolomic experiments described herein were performed on synchronized, day-1 old whole worms that were extracted and analyzed by reversed phase LC/MS in positive-ion mode as described in Materials and Methods section (n = 3 per group for all experiments with each group having ~1,500 worms). For CF512 and glp-1 organisms at 25 °C, a total of 13,639 metabolite features were detected from both groups of which 5,941 were found to be dysregulated with a p value <0.05 and a fold change >2. We have found experimentally that these are appropriate minimal thresholds for the identification of statistically significant metabolite alterations given the biological variability and analytical error associated with global metabolomic analyses (Crews et al. 2009). Here, 44 % of the detected metabolite features were altered as a result of germ line ablation. Identification of all of these metabolites is impractical with current technologies and, furthermore, it is unlikely that all of these compounds are directly related to longevity. Germ line-induced extensions in life span are dependent upon the FOXO transcription factor DAF-16 (Kenyon 2010). Double mutant daf-16; glp-1 worms are short lived and provide an organism in which metabolic perturbations caused by glp-1 mutation that are not associated with longevity can be determined. That is, metabolites uniquely associated with life-span extension should be dysregulated in long-lived glp-1 worms relative to short-lived daf-16; glp-1 worms. We therefore performed a global metabolomic comparison of long-lived glp-1 mutants and short-lived daf-16; glp-1 double mutants at 25 °C and identified 3,007 dysregulated metabolite features with a p value <0.05 and a fold change >2. By using software called metaXCMS that we developed for second-order analysis of global metabolomic data, we then compared the two datasets (glp-1 vs CF512 and glp-1 vs daf-16; glp-1) for shared and unique alterations (Tautenhahn et al. 2011). Only alterations shared in both comparisons are unique to glp-1 induced longevity. There were 1,039 features that were similarly altered between the two datasets which represented potential age-related endocrine signals (Fig. 1a). Data for these features (e.g., mass-to-charge ratios and fold changes) as well as all other features identified in our studies can be accessed online at http://metlin.scripps.edu/data/aging/.
Fig. 1.
Metabolites associated with longevity revealed by metabolomics. a metaXCMS comparison of long-lived glp-1 to both its wild-type and a short-lived double mutant reveals shared metabolites that are uniquely altered in glp-1 induced longevity. b metaXCMS comparison of long-lived daf-2 to both its wild-type and a short-lived double mutant reveals shared metabolites that are uniquely altered in daf-2 induced longevity. c Three-way comparison of metabolites altered in long-lived glp-1, long-lived daf-2, and long-lived isp-1 relative to its wild-type identifies six shared alterations as potential age-related signaling molecules. Information about the chemical identities of these molecules is listed in Table 1. d Schematic distribution of muscle in C. elegans. Body-wall muscle is purple, pharyngeal and vulval muscle is green (top). NIMS image of characteristic fragment m/z 184.07 shows a strong localization of phosphatidylcholine to muscular region of worms (bottom). e Box plots of compounds that are down-regulated in elderly human quadricep muscle relative to young. These compounds are similarly altered in long-lived worms (Table 1)
3.2 Metabolomics of daf-2 worms
We applied a similar data reduction strategy to investigate metabolic perturbations in long-lived daf-2 worms. First we compared N2 wild-type controls to daf-2 organisms. Out of the total 6,801 metabolite features detected from both groups, 2,718 features were determined to be significantly altered with p values <0.05 and fold changes >2. As in germ line-induced longevity, life-span extensions due to alterations in IGF-1/insulin signaling from daf-2 mutations are dependent upon the FOXO transcription factor DAF-16 (Kenyon 2010). Double mutant daf-16; daf-2 worms are short lived and provide a tool to identify metabolite perturbations associated with altered IGF-1/insulin signaling that do not affect longevity. That is, metabolites uniquely associated with longevity will be dysregulated in long-lived daf-2 worms relative to short-lived daf-16; daf-2 worms. A metabolomic comparison of daf-2 mutants to daf-16; daf-2 double mutants showed 248 altered features. We then compared these altered features to those dysregulated between N2 controls and daf-2 mutants to identify shared differences that were specific to daf-2 induced longevity (Fig. 1b). The meta-analysis identified 99 metabolite features as potential age-related signaling molecules.
3.3 Metabolomics of isp-1 worms
In a final profiling experiment, we compared N2 wild-type controls to long-lived isp-1 worms characterized by a mutation in iron sulfur protein of mitochondrial complex III. From the total 6,683 metabolite features detected from both groups, 2,765 were determined to be significantly altered with p values <0.05 and fold changes >2. Although each of the long-lived glp-1, daf-2, and isp-1 worms have different underlying mechanisms governing longevity, we speculated that there may be fundamental metabolic alterations shared among the organisms and that these alterations may represent converging downstream pathways intrinsic to life-span regulation or signaling molecules to coordinate aging between cells and tissues at a whole-organism level. We therefore used metaXCMS to search for shared alterations among each of the comparisons (Fig. 1c). For glp-1 and daf-2, we only considered metabolite features that were altered in glp-1 or daf-2 relative to both their respective wild-types and short-lived double mutants. Strikingly, we found that six altered features were shared in the meta-analysis as listed in Table 1. It should be noted that in searching for shared and unique alterations between the groups, we did not impose the direction of dysregulation as a restriction. Our logic is that alterations in the level of metabolite, whether up or down, correspond to dysregulation of associated metabolic pathways. For global metabolomic experiments, up- and down-regulation can correspond to either increased or decreased flux.
Table 1.
Altered metabolites shared between long-lived C. elegans
| m/z | Retention time (min) | Identification | Chemical formula | Adduct observed |
|---|---|---|---|---|
| 124.040 | 1.5 | Niacin | C6H5NO2 | [M + H]+ |
| 383.389 | 62.9 | Unknown | Unknown | Unknown |
| 742.538 | 64.1 | Phosphatidylcholine | C41H76NO8P | [M + H]+ |
| 792.545 | 58.9 | Phosphatidylcholine | C45H78NO8P | [M + H]+ |
| 805.546 | 58.8 | Unknown | Unknown | Unknown |
| 828.555 | 60.3 | Phosphatidylcholine | C48H78NO8P | [M + H]+ |
We searched the accurate mass of each of the six features in metabolite databases such as METLIN, HMDB, KEGG, and LIPID MAPS. Putative assignments made on the basis of accurate mass were supported by comparing tandem mass spectral data from each of the detected compounds in worms to that of model standards. One of the features was confirmed to be niacin. Three features were confirmed to be phosphatidylcholines whose mass and molecular formula are listed in Table 1. Although several database hits were returned for the remaining two features, tandem mass spectral data for the features were inconsistent with that from model compounds and the features were thus classified as unknowns.
3.4 Mass-based imaging of metabolites in C. elegans by NIMS
Given that we analyzed whole C. elegans worms, we could not determine the anatomical location of these altered metabolites within the organism from our global metabolomic data. We therefore applied a novel mass spectrometry-based imaging approach called nanostructure-initiator mass spectrometry (NIMS), as described in Materials and Methods section, to gain insight with respect to their spatial distribution (Northen et al. 2007; Patti et al. 2010). While we were not able to observe the intact metabolites listed in Table 1 directly, we were able to localize the characteristic fragment of phosphatidylcholine metabolites (m/z 184. 073) to muscular regions of the worm as shown in Fig. 1d. These data suggested that the phosphatidylcholine alterations shared among the longevity models might be a result of metabolic changes in C. elegans muscle. Importantly, mutations increasing life span are known to slow the progression of age-related muscle atrophy in worms (Kashyap et al. 2011). Additionally, in mammals, age-related changes in skeletal muscle have been shown to affect physiological functions of distal organ systems other than skeletal muscle itself such as cardiovascular oxygenation and nerve conduction (Imagita et al. 2009).
3.5 Metabolomics of human skeletal muscle
Next, we were interested in whether alterations in these small molecules were characteristic of aging muscle and if alterations of the same molecules could be detected in mammals. We extended our studies to humans by performing global metabolomics on muscle biopsies from the quadriceps of young and elderly adults ranging from 21–32 and 72–81 years of age, respectively (n = 10 per group, ages are included in Materials and Methods section). We identified a number of metabolite features that were down-regulated in elderly individuals more than two fold with p values < 0.05 which had accurate masses consistent with phosphatidylcholine assignments. In particular, we found that two of the same compounds determined to be similarly altered in long-lived worms as listed in Table 1 were also dysregulated in human muscle (Fig. 1e). The assignment of one of these features as a phosphatidylcholine was supported by MS/MS data. The other similarly altered feature remains unknown. Other metabolites listed in Table 1 were not identified in the untargeted screen to be altered in human muscle with age.
4 Concluding remarks
Metabolomic profiling has the power to survey large numbers of small molecules within biological systems whose levels may be altered in response to metabolic perturbations. Some metabolic perturbations, such as those introduced to extend the life span of C. elegans, can lead to a statistically significant difference in the intensities of many compounds detected. Here, determining the most biologically relevant differences to study and ultimately characterize can be a major challenge. In this work, we show the application of two metabolomic approaches that complement untargeted profiling studies: meta-analysis and mass-based imaging. By applying these strategies to study long-lived mutants of C. elegans, we identified two small molecules that were similarly altered in worms and aging human muscle. Further investigation is needed to determine the molecular mechanism underlying the alteration of these metabolites and the resulting phenotypic effect. However, it is intriguing to consider the potential role of these compounds as age-related signals originating from muscle. These results may have implications for the role of healthy muscle in whole-organism aging.
Acknowledgments
This work was supported by the California Institute of Regenerative Medicine Grant TR1-01219 (GS), the National Institutes of Health grants R24 EY017540-04 (GS), P30 MH062261-10 (GS), P01 DA026146-02 (GS), R01 ES022181 (GJP), and L30 AG0 038036 (GJP). Financial support was also received from the Department of Energy grants FG02-07ER64325 (GS) and DE-AC0205CH11231 (GS).
Contributor Information
Gary J. Patti, Email: gjpattij@wustl.edu, Departments of Chemistry, Genetics, and Medicine, Washington University School of Medicine, 660 S Euclid Ave, Campus Box 8232, St. Louis, MO 63110, USA.
Ralf Tautenhahn, Departments of Chemistry and Molecular Biology, The Center for Mass Spectrometry and Metabolomics, The Scripps Research Institute, 10550 N Torrey Pines Road, La Jolla, CA 92037, USA.
Darcy Johannsen, Human Physiology, The Pennington Biomedical Research Center, Baton Rouge, LA, USA.
Ewa Kalisiak, Departments of Chemistry and Molecular Biology, The Center for Mass Spectrometry and Metabolomics, The Scripps Research Institute, 10550 N Torrey Pines Road, La Jolla, CA 92037, USA.
Eric Ravussin, Human Physiology, The Pennington Biomedical Research Center, Baton Rouge, LA, USA.
Jens C. Brüning, Department of Mouse Genetics and Metabolism, Cologne Excellence Cluster on Cellular Stress Responses in Aging Associated Diseases (CECAD), Institute for Genetics and Center for Molecular Medicine, University of Cologne (CMMC), Zülpicher Str. 47, 50674 Cologne, Germany Max-Planck-Institute for Neurological Research, Gleueler Str. 50a, 50931 Cologne, Germany.
Andrew Dillin, Howard Hughes Medical Institute, Glenn Center for Aging Research, The Salk Institute for Biological Studies, La Jolla, CA, USA.
Gary Siuzdak, Email: siuzdak@scripps.edu, Departments of Chemistry and Molecular Biology, The Center for Mass Spectrometry and Metabolomics, The Scripps Research Institute, 10550 N Torrey Pines Road, La Jolla, CA 92037, USA.
References
- Arantes-Oliveira N, Apfeld J, Dillin A, Kenyon C. Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science. 2002;295:502–505. doi: 10.1126/science.1065768. [DOI] [PubMed] [Google Scholar]
- Baker M. Metabolomics: From small molecules to big ideas. Nature Methods. 2011;8:117–121. [Google Scholar]
- Baumeister R, Schaffitzel E, Hertweck M. Endocrine signaling in Caenorhabditis elegans controls stress response and longevity. Journal of Endocrinology. 2006;190:191–202. doi: 10.1677/joe.1.06856. [DOI] [PubMed] [Google Scholar]
- Broughton SJ, et al. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:3105–3110. doi: 10.1073/pnas.0405775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews B, et al. Variability analysis of human plasma and cerebral spinal fluid reveals statistical significance of changes in mass spectrometry-based metabolomics data. Analytical Chemistry. 2009;81:8538–8544. doi: 10.1021/ac9014947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong MQ, et al. Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science. 2007;317:660–663. doi: 10.1126/science.1139952. [DOI] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Bussiere F, Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Developmental Cell. 2001;1:633–644. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- Gerisch B, et al. A bile acid-like steroid modulates Caenorhabditis elegans lifespan through nuclear receptor signaling. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:5014–5019. doi: 10.1073/pnas.0700847104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden TR, Melov S. Gene expression changes associated with aging in C. elegans. Worm Book. 2007;12:1–12. doi: 10.1895/wormbook.1.127.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herndon LA, et al. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature. 2002;419:808–814. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- Hwangbo DS, Gershman B, Tu MP, Palmer M, Tatar M. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 2004;429:562–566. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- Imagita H, Yamano S, Tobimatsu Y, Miyata H. Age-related changes in contraction and relaxation of rat diaphragm. Biomedical Research. 2009;30:337–342. doi: 10.2220/biomedres.30.337. [DOI] [PubMed] [Google Scholar]
- Kashyap L, Perera S, Fisher AL. Identification of Novel Genes Involved in Sarcopenia Through RNAi Screening in Caenorhabditis elegans. Journals of Gerontology. Series A. 2011 doi: 10.1093/gerona/glr072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. The plasticity of aging: Insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Kleemann GA, Murphy CT. The endocrine regulation of aging in Caenorhabditis elegans. Molecular and Cellular Endocrinology. 2009;299:51–57. doi: 10.1016/j.mce.2008.10.048. [DOI] [PubMed] [Google Scholar]
- Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- Motola DL, et al. Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell. 2006;124:1209–1223. doi: 10.1016/j.cell.2006.01.037. [DOI] [PubMed] [Google Scholar]
- Northen TR, et al. Clathrate nanostructures for mass spectrometry. Nature. 2007;449:1033–1036. doi: 10.1038/nature06195. [DOI] [PubMed] [Google Scholar]
- Panowski SH, Dillin A. Signals of youth: Endocrine regulation of aging in Caenorhabditis elegans. Trends in Endocrinology and Metabolism. 2009;20:259–264. doi: 10.1016/j.tem.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Patti GJ, Tautenhahn R, Siuzdak G. Meta-analysis of untargeted metabolomic data from multiple profiling experiments. Nature Protocols. 2012;7:508–516. doi: 10.1038/nprot.2011.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti GJ, et al. Nanostructure-initiator mass spectrometry (NIMS) imaging of brain cholesterol metabolites in Smith–Lemli–Opitz syndrome. Neuroscience. 2010;170:858–864. doi: 10.1016/j.neuroscience.2010.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychogios N, et al. The human serum metabolome. PLoS ONE. 2011;6:e16957. doi: 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottiers V, et al. Hormonal control of C. elegans dauer formation and life span by a Rieske-like oxygenase. Developmental Cell. 2006;10:473–482. doi: 10.1016/j.devcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Russell SJ, Kahn CR. Endocrine regulation of ageing. Nature Reviews Molecular Cell Biology. 2007;8:681–691. doi: 10.1038/nrm2234. [DOI] [PubMed] [Google Scholar]
- Shim YH, Paik YK. Caenorhabditis elegans proteomics comes of age. Proteomics. 2010;10:846–857. doi: 10.1002/pmic.200900542. [DOI] [PubMed] [Google Scholar]
- Sluder AE, Maina CV. Nuclear receptors in nematodes: Themes and variations. Trends in Genetics. 2001;17:206–213. doi: 10.1016/s0168-9525(01)02242-9. [DOI] [PubMed] [Google Scholar]
- Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Analytical Chemistry. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- Smith CA, et al. METLIN: A metabolite mass spectral database. Therapeutic Drug Monitoring. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- Tatar M, Bartke A, Antebi A. The endocrine regulation of aging by insulin-like signals. Science. 2003;299:1346–1351. doi: 10.1126/science.1081447. [DOI] [PubMed] [Google Scholar]
- Tautenhahn R, et al. metaXCMS: Second-order analysis of untargeted metabolomics data. Analytical Chemistry. 2011;83:696–700. doi: 10.1021/ac102980g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, et al. HMDB: A knowledgebase for the human metabolome. Nucleic Acids Research. 2009;37:D603–D610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolkow CA, Kimura KD, Lee MS, Ruvkun G. Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science. 2000;290:147–150. doi: 10.1126/science.290.5489.147. [DOI] [PubMed] [Google Scholar]
- Woo HK, Northen TR, Yanes O, Siuzdak G. Nanostructure-initiator mass spectrometry: A protocol for preparing and applying NIMS surfaces for high-sensitivity mass analysis. Nature Protocols. 2008;3:1341–1349. doi: 10.1038/nprot.2008.110. [DOI] [PubMed] [Google Scholar]
- Yanes O, Tautenhahn R, Patti GJ, Siuzdak G. Expanding coverage of the metabolome for global metabolite profiling. Analytical Chemistry. 2011;83:2152–2161. doi: 10.1021/ac102981k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanes O, et al. Metabolic oxidation regulates embryonic stem cell differentiation. Nature Chemical Biology. 2010a;6:411–417. doi: 10.1038/nchembio.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanes O, et al. Metabolic oxidation regulates embryonic stem cell differentiation. Nature Chemical Biology. 2010b;6:411–417. doi: 10.1038/nchembio.364. [DOI] [PMC free article] [PubMed] [Google Scholar]