Abstract
A wide variety of environmental factors including physical and biochemical signals are responsible for stem cell behavior and function. In particular, matrix elasticity and cell shape have been shown to determine stem cell function, yet little is known about the interplay between how these physical cues control cell differentiation. For the first time, by using ultraviolet (UV) lithography to pattern poly(ethylene) glycol (PEG) hydrogels we are able to manufacture microenvironments capable of parsing the effects of matrix elasticity, cell shape, and cell size in order to explore the relationship between matrix elasticity and cell shape in mesenchymal stem cell (MSC) lineage commitment. Our data shows that cells cultured on 1,000 μm2 circles, squares, and rectangles were primarily adipogenic lineage regardless of matrix elasticity, while cells cultured on 2,500 and 5,000 μm2 shapes more heavily depended on shape and elasticity for lineage specification. We further went on to characterize how modifying the cell cytoskeleton through pharmacological inhibitors can modify cell behavior. By showing MSC lineage commitment relationships due to physical signals, this study highlights the importance of cell shape and matrix elasticity in further understanding stem cell behavior for future tissue engineering strategies.
Keywords: Micropatterning, UV Lithography, Mesenchymal Stem Cells, Matrix Elasticity, Cell Shape
Introduction
The study of regenerative medicine, stem cells in particular, has become increasingly important in scientific and medical fields due to their potential to restore or replace injured tissue and organs.[1-5] The use of MSCs as a therapy option has been progressively more promising in scientific fields by possessing the ability to differentiate into bone cells (osteoblasts), cartilage cells (chondrocytes), and fat cells (adipocytes) among other potential lineages.[6, 7] MSCs may potentially demonstrate to be vital to tissue engineering bone replacements as the need for bone tissue repair in patients suffering from critical bone defects continues to rise.[3, 8-11] Complex combinations of physical, chemical, and biological signals are used to direct stem cell fate and control the natural healing of bone and other tissues in vivo.[5, 12-14] In order to fully elucidate these healing and regeneration principles, we must first understand the complexity of the underlying cellular and biomolecular factors that promote each tissue. To be fully realized as a potential treatment option, numerous cellular responses to microenvironmental cues as well as directed differentiation capacity of these stem cells need to be addressed. A significant challenge facing researchers is the ability to differentiate a stem cell into a certain programmed lineage. In particular, physical and geometric cues have emerged as significant factors in directing stem cell behavior.[15-19]
Physical signals derived from the stem cell microenvironment have been established as increasingly important to the lineage commitment of stem cells.[15, 18, 20-22] These signals were previously recognized as far back as the 1940s with tensile stresses leading to bone formation and compressive stresses leading to cartilage formation in cultured chick rudiments.[23] Further work has been aided with the implementation of microscale technologies to mimic the stem cell microenvironment in vitro.[24, 25] These microscale experiments have shown the critical importance of the cell microenvironment to cell behaviors such as apoptosis, migration, and differentiation.[15, 17, 26] In a recent key study, the importance of matrix elasticity has been presented by culturing MSCs on gels of differing elasticity with soft gels (<1 kPa) promoting neurogenic differentiation, intermediate gels (~12 kPa) promoting myocytes, and stiff gels (>25 kPa) promoting osteoblasts.[18] Other studies also showcase the importance of cell geometry on the lineage specification of stem cells with differing densities of cells promoting differing cell lineages as well as micropatterned shapes confirming these findings.[17, 21] These studies both found increasing levels of GTPase RhoA and downstream effectors promoting osteogenesis with lower levels of RhoA signaling being a signal for adipogenesis and neurogenesis. Within these studies it is clear that RhoA and the corresponding actomyosin contractions play a role in the lineage specifications of these stem cells, and thus a correlation between physical signals determining fate. Yet little is known about the cooperative interplay between these types of physical signaling. Therefore, there is a clear need for research determining the interplay between matrix elasticity and cell shape and how this ultimately effects cell lineage specification.
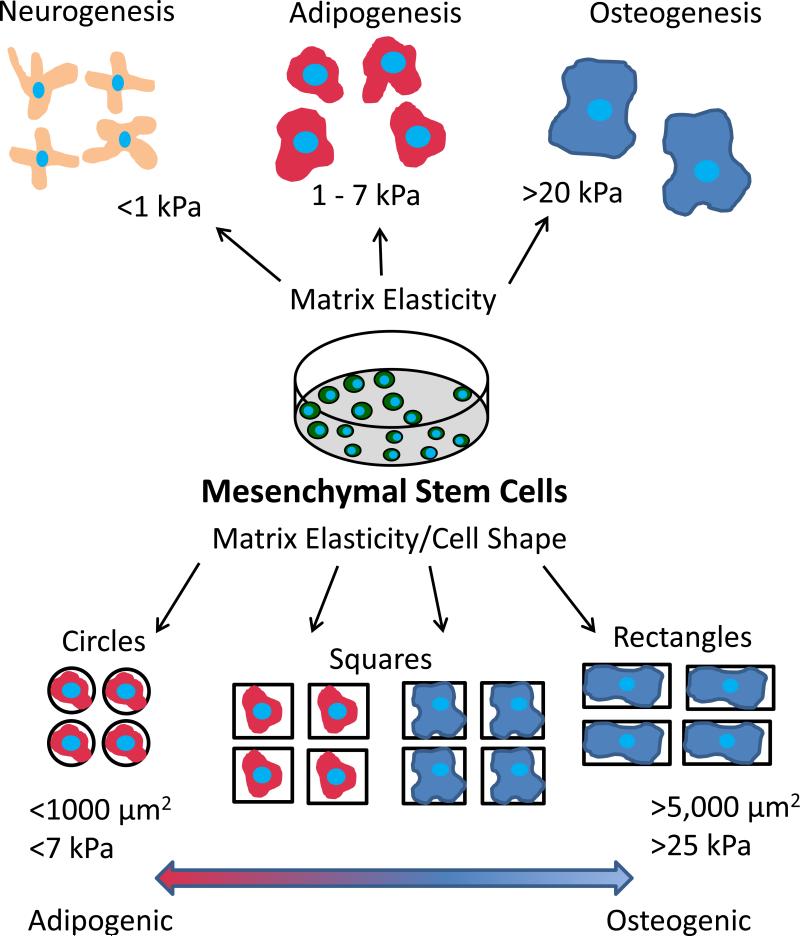
In this work, we present a novel method to decouple multiple physical signals including substrate elasticity, cell shape, and cell size in determining MSC lineage commitment as shown in Figure 1. This strategy uses micropatterned PEG hydrogels to vary the elasticity, size, and shape of adhesive area presented to cells cultured in a mixture of adipogenic and osteogenic differentiation medium to direct cell fate. By regulating the physical signals presented, we show that 1,000 μm2 areas promote adipogenic differentiation regardless of shape and elasticity while 2,500 and 5,000 μm2 areas are more heavily dependent on shape and elasticity in cell fate commitment. The importance of cytoskeletal tension on patterned areas in MSC differentiation was especially prevalent when cells were treated with Y-27632 and nacadazole and primarily committed to adipocyte and osteoblast lineage respectively. This work is able to further establish the cooperative roles presented through physical signaling due to elasticity and cell shape that are able to promote MSC fate commitment.
Figure 1.
Schematic of the methodology to determine the cooperative effects of cell shape, cell size, and matrix elasticity on the lineage commitment of mesenchymal stem cells.
Methods and Materials
Substrate Preparation
Glass coverslips (22×22 mm, Fisher Scientific) were washed with 70% ethanol and ozone treated (BioforceNano, Ames, IA) for 30 minutes to remove surface contaminants. Cover slips were then sputter coated with a 5 nm titanium adhesion layer (Denton Desk II Turbo, Moorestown, NJ) followed by approximately 40 nm of gold (Denton Desk II, Moorestown, NJ). Coverslips were then stored at room temperature until use.
Micropatterning Hydrogels
PEG precursor solution was assembled using 700 MW PEG diacrylate (PEG-DA) (Aldrich, Milwaukee, WI) mixed with 2000 MW 4-arm PEG thiol (PEG-SH)(CreativePEGWorks, Raleigh, NC) in H2O using 0.5% (v/v) 2-hydroxy-2-methylpropiophenone (Aldrich,Milwaukee, WI). Photomasks were produced using autocad software (AutoDesk, San Rafael, CA) and printed on transparencies (CAD/Art Services, Inc, Bandon, OR). PEG precursor was placed onto cover slip, covered with photomask, and placed under approximately 4 mW/cm2 Blak Ray UV light (UVP, Upland, CA) to polymerize. The patterned cover slip was then incubated in triethylene glycol mono-mercaptoundecyl ether (50 mM)(Aldrich, Allentown, PA) for 20 minutes to render unpatterned surfaces non-adhesive to proteins and rinsed with 70% ethanol and subsequently sterile PBS three times. Fibronectin (Sigma, St. Louis, MO) was treated with a heterogenous maleimide/N-hydroxysuccinimide bi-functional linker (ThermoFisher, Rockford, IL)[27] to allow functionalization of protein in order to attach to PEG patterns. Fibronectin was incubated at room temperature for 1 hour then separated from unreacted crosslinker using a Zeba Spin desalting column (Thermo Fisher, Rockford, IL). PEG patterns were then incubated in functionalized proteins at room temperature for 4 hours and 4°C overnight to allow covalent attachment of proteins to hydrogels. Hydrogel patterns were visualized and characterized by brightfield and fluorescent microscopy to confirm attachment.
Hydrogel Characterization
PEG hydrogel samples were created 5 mm in diameter and 3 mm height at desired ratio and let soak in deionized water for 48 hours at 37°C. Samples were tested in unconfined compression,[28-31] in short, the Young's modulus of each sample was determined using an ElectroForce 3200 (Bose, Eden Prairie, MN) in unconfined compression at 0.05 mm/sec between parallel nonporous plates while compressive force and displacement were recorded.
Cell Culture
Human bone marrow-derived mesenchymal stem cells were obtained from Lonza (Walkersville, MD). hMSCs were cultured in basal growth media (Lonza, Walkersville, NC) in culture flasks. The growth medium contained hMSC basal medium (440 mL), mesenchymal cell growth supplement (50 mL), L-glutamine (10 mL of 200 mM), and penicillin/streptomycin (0.5 mL). The cells were passaged after reaching 90% confluence and collected with 0.05% trypsin/EDTA solution. All cells were plated onto substrates under passage 6 at 5,000 cells/cm2. Cells were allowed 1 day for adhesion onto substrate before being placed in mixed medium which consisted of a 1:1 ratio of adipogenic to osteogenic medium. Adipogenic medium contained DMEM (444 mL)(Invitrogen), fetal bovine serum (50 mL)(Atlas), dexamethasone (0.5 mL of 1 μM), insulin (0.5 mL of 10 μM)(Sigma), indomethacin (200 μM)(Sigma), isobutyl-methylxanthine (0.5 mM)(Sigma), and penicillin/streptomyacin (5 mL). Osteogenic medium consisted of DMEM F/12 (444 mL)(Invitrogen), fetal bovine serum (50 mL), β-glycerophosphate (10 mM), ascorbic acid (50 μg/ml)(Sigma), dexamethasone (1 μM )(Sigma), and penicillin/streptomyacin (5 ml). For ROCK inhibited cells differentiation medium was changed daily and Y-27632 (2 μM)(Calbiochem, Rockaway, NJ) was added. For nocodazole treated cells differentiation medium was changed daily and nocodazole (1 μM)(Sigma) was added.
Immunocytochemistry and Histological Staining
Cells were fixed with 4% paraformaldehyde, permealized with 0.2% Triton X-100, and blocked with 1% BSA solution. The cytoskeleton, focal adhesions, and nuclei of cells were stained with a rhodamine-phalloidin conjugate (Invitrogen, Grand Island, NY), vinculin (Sigma, St. Louis, MO), and Floroshield with Dapi (Sigma, St. Louis, MO) respectively. Fluorescent photographs of the stained hMSCs were captured by a Nikkon Eclipse 80i microscope with CoolSnap HQ camera. Non-fluorescent cells were analyzed using phase contrast microscopy utilizing NIS-Elements-AR 3.2 64 bit software (NIS-Elements, Melville, NY). Fate specified cells were analyzed using dual alkaline phosphatase[21, 32] and Oil Red O staining[33, 34] for osteogenesis and adipogenesis using a Nikon Eclipse E600 microscope (Nikon, Melville, NY) with color camera. Cells containing lipid vacuoles stained red were counted as adipocyte specification while cells staining deep blue/purple were counted as osteoblast specification. Rare cells that exhibited both lipid vacuoles and osteoblast staining were not counted. Tiff images were taken of patterned areas and cells were counted individually.
Statistics
P-values were calculated using one way ANOVA function in Excel (Microsoft, Seattle, WA). Errors are standard error of the mean.
Results and Discussion
Effect of Soluble Factors, Cell Density, and Matrix Elasticity on MSC Differentiation
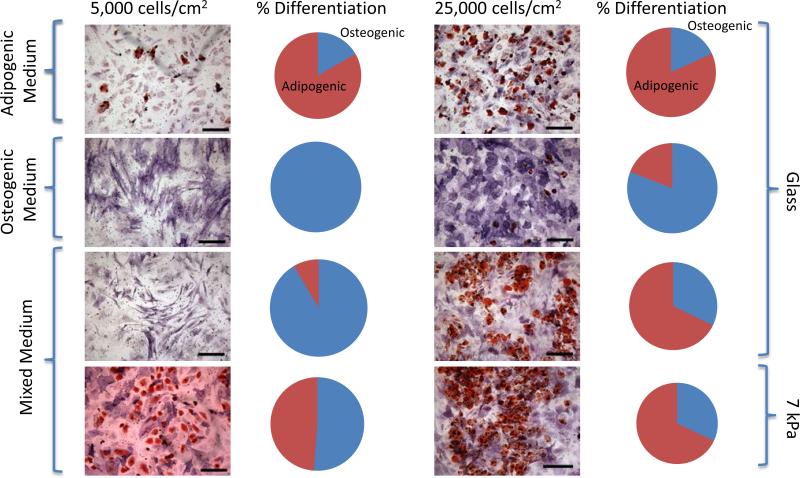
MSCs have previously been shown to be extremely sensitive to passage number in vitro when evaluating lineage commitment and differentiation of cells.[35-37] To address this concern of diminished differentiation capability, trials to assess the ability of MSCs to commit to adipocytes and osteoblasts under passage 6 were first run with lineage specific medium and soluble cues for 7 days. In strictly adipogenic medium, we observed 80.3% and 81.9% adipogenic lineage commitment at 5,000 cells/cm2 and 25,000 cells/cm2. Alternatively, in osteogenic medium we observed 100% and 80.9% osteogenic lineage commitment (Figure 2). Further evaluations were done using MSCs in a 1:1 mixture of adipogenic and osteogenic medium for 7 days on unpatterned substrates. As previously shown, [21, 38] we confirmed cell density contributed to lineage commitment when looking at the differentiation of MSCs at a density of 5,000 cells/cm2 and 25,000 cells/cm2. Our findings show that on glass coverslips, cells continued to show 100% osteogenic differentiation with 5,000 cm2 density while only 40.6% osteogenic differentiation with 25,000 cells/cm2. We then coated coverslips with 10% PEG (~7 kPa) and found the softer substrate contributed to 40.4% greater adipogenic differentiation in low plating densities and similar adipogenic differentiation in higher plating densities (Figure 2).
Figure 2.
MSCs showed multilineage capabilities when cultured in medium containing growth factors promoting osteogenesis and adipogenesis. Dual staining of MSCs after 1 week for osteogenesis (alkaline phosphatase-purple/blue) and adipogenesis (lipids-red). Each line of images and graphs represents a differing culture condition with both 5,000 cells/cm2 and 25,000 cells/cm2. Conditions tested were adipogenic medium alone on glass, osteogenic medium alone on glass, mixed medium on glass, and mixed medium on 7 kPa extracellular matrix. Pie charts show the percentage of differentiation to each lineage (red-adipocyte, blue-osteoblast).
These results compare similarly to previous studies using differing cell densities and show the effects of cell density and substrate stiffness on the differentiation potential of MSCs in mixed medium. As cell density increases, cell adhesion and spreading are decreased and cell-cell contact is increased which leads to enhanced signaling. This aspect has been confirmed by several studies to control cell behavior[21, 39] and we further show that substrate elasticity along with cell density can control lineage commitment of MSCs. To address the interplay between cell size, shape, and substrate elasticity remaining experiments were conducted using patterned cells cultured in mixed media conditions.
Micropatterning and Adhesion of Mesenchymal Cells
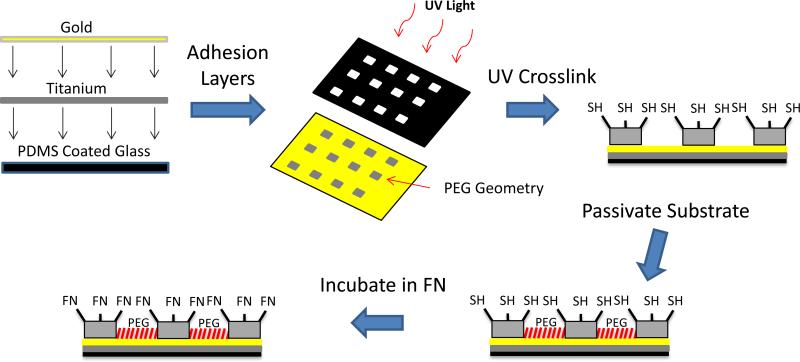
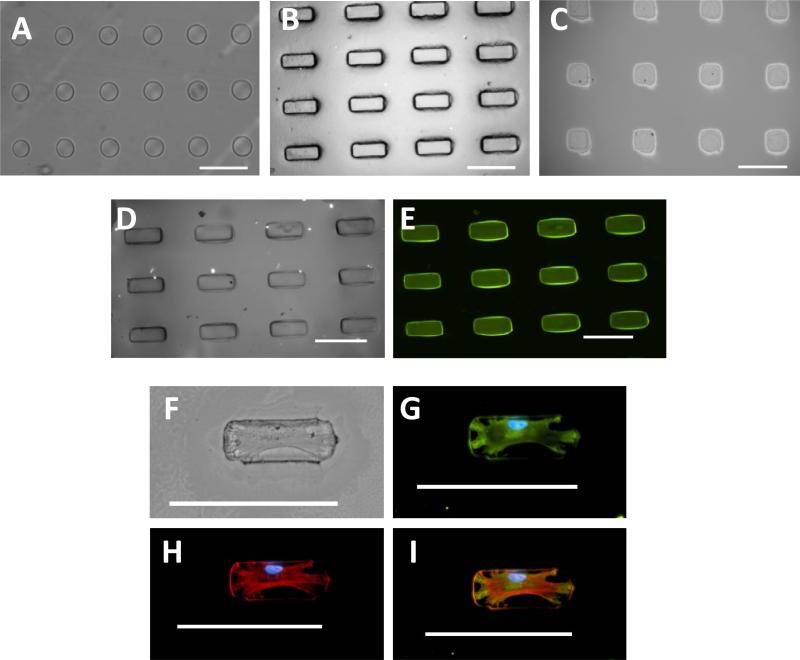
UV lithography techniques were used to restrict the shape of individual cells into circles, squares, and rectangles onto coverslips (Figure 3). A photomask was utilized to control size and shape of the islands with a mixture of PEG-SH and PEG-DA used as the precursor solution for the hydrogels. UV light was employed to selectively crosslink hydrogels into circles, squares, and rectangles on a gold coated glass coverslip through the photomask (Figure 4A-C). The remaining regions of the coverslip were then rendered non-adhesive with a tri(ethylene glycol)-terminated monolayer to prevent non-specific binding of protein or cells. Patterns were incubated in maleimide-modified fibronectin solution to absorb protein exclusively to hydrogel islands to allow cell attachment as seen in Figure 4D and 4E. MSCs were then able to attach to the hydrogel islands and spread to assume distinct shapes of the underlying islands (Figure 4F-I). Cells were able to attach and spread on patterns while remaining viable and constrained to hydrogel islands for one week in culture to determine the lineage commitment effects due to size, shape and elasticity of the microenvironment. MSCs were plated onto hydrogel islands using MSC growth medium initially, switched to a 50:50 mixture of adipogenic and osteogenic differentiation media, and cultured for 7 days. Cells were then analyzed by staining for lineage specific markers Oil Red O and alkaline phosphatase for adipogenic and osteogenic differentiation respectively.
Figure 3.
Schematic showing UV lithography process used to create hydrogel shapes of varying elasticity. Hydrogel shapes were functionalized with thiol to promote fibronectin binding exclusively to hydrogels.
Figure 4.
Hydrogel islands were fabricated with protein exclusively attached to islands facilitating cell adhesion. Presented are microscopy images with micropatterned shapes showing (A) 5,000 μm2 circles (B) 5,000 μm2 rectangles and (C) 5,000 μm2 squares. Fluorescent bovine serum albumin was used as a model protein to determine protein attachment to micropatterned areas with (D) brightfield microscopy image of 5,000 μm2 rectangles and (E) bovine serum albumin exclusively attached to hydrogel rectangles. MSC attachment shown with (F) brightfield microscopy and immunofluorescence stained for (G) vinculin to reveal focal adhesions, (H) F-actin, and (I) merged image. Scale bars are 100 μm.
MSC Differentiation Directed by Shape, Size, and Elasticity
MSCs were confined to 1,000, 2,500, and 5,000 μm2 area circle, square, and rectangular patterns with a substrate elasticity of 7, 47, and 105 kPa. This range of geometric features was considered to promote both adipogenic and osteogenic lineages with circles, squares, and rectangles previously shown capable of directing cell behavior and differentiation.[16, 17] Substrate elasticity was also considered and values were chosen to promote multiple lineages and cell behavior[18, 20] in order to parse differences in physical effects on cell differentiation.
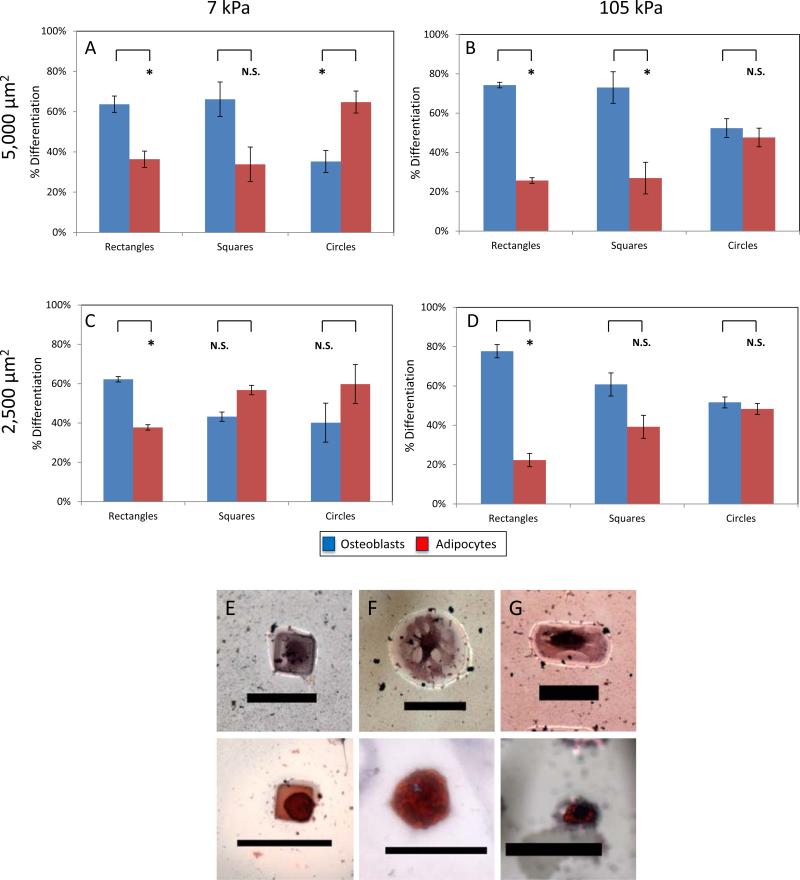
For 1,000 μm2 islands, we observed primarily adipogenic differentiation in all cases of elasticity and shape. This is consistent with previously reported micropatterning studies as well as matrix elasticity studies observing cell size to be a regulator of lineage commitment.[17, 21, 40] When looking at cells on 2,500 and 5,000 μm2 patterns with different shape and elasticity we found a more mixed population of adipocytes and osteoblasts. With 5,000 μm2 shapes we found at higher elasticity the cells behaved similar to glass with 74%, 73%, and 52% osteogenic differentiation on rectangles, squares, and circles respectively (Figure 5B). When switched to 7 kPa hydrogels, osteogenic differentiation decreased to 61%, 66%, and 35% on these identical shapes (Figure 5A). When switching to 2,500 μm2 shapes, we saw a much higher variation in lineage commitment with 78%, 61%, and 52% osteogenesis on 105 kPa substrates and 62%, 43%, and 40% osteogenesis on 7 kPa substrates (Figure 5C-D). We also found that our 47 kPa matrix elasticity had similar values to the 105 kPa experiments for each shape excluding the 53% and 52% osteogenic differentiation for rectangles and squares on 2,500 μm2 patterns (data not shown).
Figure 5.
By modifying the geometry and matrix elasticity of the underlying patterns, cells were able to choose lineage commitment based on the physical cues presented. (A)-(D) Shown in these graphs are the effect of shape, size, and matrix elasticity on MSC lineage commitment (E)-(F) Shown on these patterns are adipogenesis and osteogenesis on rectangles, squares, and circles (Scale bars 50 μm). Error bars are standard error from at minimum 2 separate experiments with over 75 cells per condition.
These results remain consistent when looking at patterning studies showing both cell shape and size to be a factor in osteogenic differentiation[17, 21, 33, 41-43] as well as other groups showing the role of matrix elasticity in osteogenic differentiation.[18, 22, 44-46] These studies have further shown that higher levels of RhoA lead to a higher degree of cell spreading and osteogenesis of MSCs[21, 47-49] on micropatterned surfaces along with similar RhoA pathways being responsible for enhanced cytoskeletal tension and osteogensis on stiffer extracellular matrices.[18] Our studies are able to highlight these cooperative signaling effects from both matrix elasticity and cell shape on the lineage commitment of MSCs. Our interpretation shows that cell size was responsible for lineage commitment choices at 1,000 μm2 in all cases regardless of matrix elasticity or shape. At larger cell sizes cell shape and matrix elasticity both played a role in the lineage commitment of MSCs with cell shape appearing to play the larger role. As to shape, in all cases rectangles were shown to have higher osteogenesis when compared with circles, showing the immense importance of curvature and cytoskeletal tension in lineage commitment. It is particularly interesting that cell shape seemed to be a more governing physical cue than matrix elasticity, but has been a theme of recent articles highlighting elasticity and shape as intertwined.[18, 22, 50] This study implies that by controlling cell shape initially and thus RhoA signaling, it is able to lessen the effects of matrix elasticity on lineage commitment.
MSC Differentiation altered by Cytoskeletal Modifications
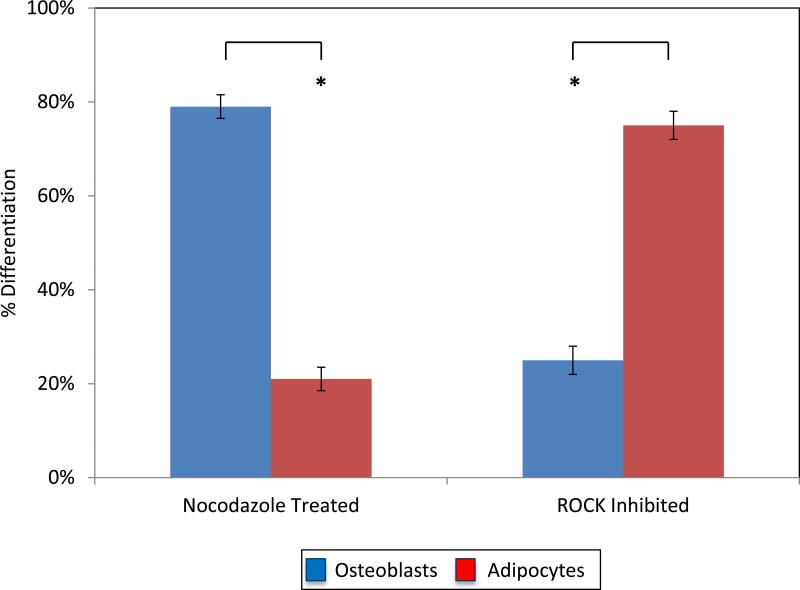
The following experiments further proceeded to characterize the differentiation of MSCs on patterns under cytoskeletal manipulation to observe how a contractile cytoskeleton directs cell behavior. The cytoskeleton has previously been shown to strongly guide cell adhesion and behavior on micropatterned geometric shapes.[15-17, 21, 51, 52] To further confirm our findings that cell spreading and cytoskeletal tension are primarily responsible for osteogenic differentiation in combination with substrate elasticity, we evaluated patterned cells in mixed medium with Y-27632 and nocodazole added, which are pharmacological agents designed to modify the cytoskeleton.[53-56] Cells were plated onto 2,500 μm2 square patterned surfaces with growth medium and inhibitors and mixed medium was added the following day to ensure cells complete spreading over patterns. Cells patterned on these 2,500 μm2 squares with 47 kPa matrix elasticity without inhibitors were shown to have 52% osteogenic differentiation. In the presence of nocodazole, a microtubule depolymerizing agent shown to increase cell contractility,[57] cells were shown to have 84% osteogenic differentiation. Y-27632, an agent that inhibits ROCK causing a decrease in cell contractility,[58] was shown to have 69% adipogenesis on the same patterns (Figure 6).
Figure 6.
Graph showing the percentage of cells committing to adipogenic or osteogenic lineage in the presence of pharmacological agents nocodazole and Y-27632 (ROCK Inhibitor) on 2,500 μm2 squares with a 47 kPa matrix elasticity.
These results further confirm that actomyosin contractility is a key regulator in the lineage commitment of MSCs. It is generally accepted that higher degrees of cell spreading promote increased myosin-generated cytoskeletal tension leading to increased levels of RhoA and ROCK.[17, 21] It has also been well noted that as matrix elasticity increases, RhoA and ROCK levels increase as well.[18] Therefore, by inhibiting or promoting ROCK, we observed that with constant matrix stiffness, shape, and size we could promote either osteogenic or adipogenic lineages confirming that ROCK signaling remains vital to lineage commitment when presenting cells with differing physical cues. This work further supports the immense importance of the cytoskeleton in looking at osteogenic differentiation in the presence of physical microenvironmental characteristics, and in our work, the presence of multiple conflicting physical characteristics.
Conclusion
Through the development of micropatterned hydrogels, we were able to ascertain the relationship between size, shape, and matrix elasticity for the first time in single MSC lineage commitment. UV lithography of PEG hydrogels was employed to provide a platform to study single MSCs in a manner capable of decoupling these physical signaling cues. This work has combined the ability to control cell size and spreading with the ability to adjust matrix elasticity to regulate stem cell lineage commitment and demonstrated that the size, shape, and matrix elasticity possess the ability to use physical characteristics to tune differentiation. The physical signals were critical to lineage commitment with cell size proving to be most significant to lineage commitment at lower adhesive areas and shape being most significant at larger adhesive areas. The use of single cells to determine lineage commitment parameters of stem cells has become paramount to engineering homogenous populations of stem cells for use in tissue engineering. Our study is one of the first to be able to present tools and insight into combining these physical characteristics directing stem cell lineage commitment for possible use in designing materials and scaffolds for future regenerative medicine.
Acknowledgements
We gratefully acknowledge support from the South Carolina Space Grant Consortium, National Institute of Health (Grant NIH P20 GM103641), and the National Science Foundation (Grant EPS-0903795). The authors would also like to acknowledge the University of South Carolina Electron Microscopy Center for instrument use, scientific and technical assistance.
References
- 1.Hutmacher DW. Biomaterials. 2000;21(24):2529. doi: 10.1016/s0142-9612(00)00121-6. [DOI] [PubMed] [Google Scholar]
- 2.Langer R, Vacanti JP. Science. 1993;260(5110):920. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 3.Lutolf MP, Hubbell JA. Nature Biotechnology. 2005;23(1):47. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 4.Griffith LG, Naughton G. Science. 2002;295(5557):1009. doi: 10.1126/science.1069210. [DOI] [PubMed] [Google Scholar]
- 5.Reya T, Morrison SJ, Clarke MF, Weissman IL. Nature. 2001;414(6859):105. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 6.Bosnakovski D, Mizuno M, Kim G, Takagi S, Okumura M, Fujinaga T. Cell and Tissue Research. 2005;319(2):243. doi: 10.1007/s00441-004-1012-5. [DOI] [PubMed] [Google Scholar]
- 7.Jiang Y, Jahagirdar BN, Reinhardt RL, Schwartz RE, Keene CD, Ortiz-Gonzalez XR, Reyes M, Lenvik T, Lund T, Blackstad M, Du J, Aldrich S, Lisberg A, Low WC, Largaespada DA, Verfaillie CM. Nature. 2002;418(6893):41. doi: 10.1038/nature00870. [DOI] [PubMed] [Google Scholar]
- 8.Park J, Ries J, Gelse K, Kloss F, von der Mark K, Wiltfang J, Neukam FW, Schneider H. Gene Therapy. 2003;10(13):1089. doi: 10.1038/sj.gt.3301960. [DOI] [PubMed] [Google Scholar]
- 9.Dupont KM, Sharma K, Stevens HY, Boerckel JD, Garcia AJ, Guldberg RE. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(8):3305. doi: 10.1073/pnas.0905444107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winnier G, Blessing M, Labosky PA, Hogan BLM. Genes & Development. 1995;9(17):2105. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- 11.Charge SBP, Rudnicki MA. Physiological Reviews. 2004;84(1):209. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 12.Ohlstein B, Kai T, Decotto E, Spradling A. Current Opinion in Cell Biology. 2004;16(6):693. doi: 10.1016/j.ceb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 13.Watt FM, Hogan BLM. Science. 2000;287(5457):1427. doi: 10.1126/science.287.5457.1427. [DOI] [PubMed] [Google Scholar]
- 14.Fuchs E, Tumbar T, Guasch G. Cell. 2004;116(6):769. doi: 10.1016/s0092-8674(04)00255-7. [DOI] [PubMed] [Google Scholar]
- 15.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Science. 1997;276(5317):1425. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 16.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Biotechnology Progress. 1998;14(3):356. doi: 10.1021/bp980031m. [DOI] [PubMed] [Google Scholar]
- 17.Kilian KA, Bugarija B, Lahn BT, Mrksich M. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(11):4872. doi: 10.1073/pnas.0903269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126(4):677. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 19.Guilak F, Cohen DM, Estes BT, Gimble JM, Liedtke W, Chen CS. Cell Stem Cell. 2009;5(1):17. doi: 10.1016/j.stem.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pelham RJ, Jr., Wang Y. Proc Natl Acad Sci U S A. 1997;94(25):13661. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Dev Cell. 2004;6(4):483. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 22.Engler AJ, Griffin MA, Sen S, Bonnetnann CG, Sweeney HL, Discher DE. Journal of Cell Biology. 2004;166(6):877. doi: 10.1083/jcb.200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glucksmann A. Journal of Anatomy. 1942;76(1):231. [PMC free article] [PubMed] [Google Scholar]
- 24.Khademhosseini A, Langer R, Borenstein J, Vacanti JP. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(8):2480. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikkhah M, Edalat F, Manoucheri S, Khademhosseini A. Biomaterials. 2012;33(21):5230. doi: 10.1016/j.biomaterials.2012.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gunawan RC, Silvestre J, Gaskins HR, Kenis PJA, Leckband DE. Langmuir. 2006;22(9):4250. doi: 10.1021/la0531493. [DOI] [PubMed] [Google Scholar]
- 27.Yoshitake S, Imagawa M, Ishikawa E, Niitsu Y, Urushizaki I, Nishiura M, Kanazawa R, Kurosaki H, Tachibana S, Nakazawa N, Ogawa H. Journal of Biochemistry. 1982;92(5):1413. doi: 10.1093/oxfordjournals.jbchem.a134065. [DOI] [PubMed] [Google Scholar]
- 28.Bryant SJ, Chowdhury TT, Lee DA, Bader DL, Anseth KS. Annals of Biomedical Engineering. 2004;32(3):407. doi: 10.1023/b:abme.0000017535.00602.ca. [DOI] [PubMed] [Google Scholar]
- 29.Huang T, Xu HG, Jiao KX, Zhu LP, Brown HR, Wang HL. Advanced Materials. 2007;19(12):1622. [Google Scholar]
- 30.Stammen JA, Williams S, Ku DN, Guldberg RE. Biomaterials. 2001;22(8):799. doi: 10.1016/s0142-9612(00)00242-8. [DOI] [PubMed] [Google Scholar]
- 31.Weinand C, Pomerantseva I, Neville CM, Gupta R, Weinberg E, Madisch I, Shapiro F, Abukawa H, Troulis MJ, Vacanti JP. Bone. 2006;38(4):555. doi: 10.1016/j.bone.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 32.Sila-Asna M, Bunyaratvej A, Maeda S, Kitaguchi H, Bunyaratavej N. The Kobe journal of medical sciences. 2007;53(1-2):25. [PubMed] [Google Scholar]
- 33.Song W, Lu HX, Kawazoe N, Chen GP. Langmuir. 2011;27(10):6155. doi: 10.1021/la200487w. [DOI] [PubMed] [Google Scholar]
- 34.Koopman R, Schaart G, Hesselink MK. Histochemistry and cell biology. 2001;116(1):63. doi: 10.1007/s004180100297. [DOI] [PubMed] [Google Scholar]
- 35.Sarugaser R, Hanoun L, Keating A, Stanford WL, Davies JE. Plos One. 2009;4(8) doi: 10.1371/journal.pone.0006498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crisostomo PR, Wang MJ, Wairiuko GM, Morrell ED, Terrell AM, Seshadri P, Nam UH, Meldrum DR. Shock. 2006;26(6):575. doi: 10.1097/01.shk.0000235087.45798.93. [DOI] [PubMed] [Google Scholar]
- 37.Wagner W, Horn P, Castoldi M, Diehlmann A, Bork S, Saffrich R, Benes V, Blake J, Pfister S, Eckstein V, Ho AD. Plos One. 2008;3(5) doi: 10.1371/journal.pone.0002213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Science. 1999;284(5411):143. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 39.Nelson CM, Chen CS. Febs Letters. 2002;514(2-3):238. doi: 10.1016/s0014-5793(02)02370-0. [DOI] [PubMed] [Google Scholar]
- 40.Fu JP, Wang YK, Yang MT, Desai RA, Yu XA, Liu ZJ, Chen CS. Nature Methods. 2010;7(9):733. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng SL, Yang JW, Rifas L, Zhang SF, Avioli LV. Endocrinology. 1994;134(1):277. doi: 10.1210/endo.134.1.8275945. [DOI] [PubMed] [Google Scholar]
- 42.Ruiz SA, Chen CS. Stem Cells. 2008;26(11):2921. doi: 10.1634/stemcells.2008-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oh S, Brammer KS, Li YS, Teng D, Engler AJ, Chien S, Jin S. Proc Natl Acad Sci U S A. 2009;106(7):2130. doi: 10.1073/pnas.0813200106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rowlands AS, George PA, Cooper-White JJ. American Journal of Physiology-Cell Physiology. 2008;295(4):C1037. doi: 10.1152/ajpcell.67.2008. [DOI] [PubMed] [Google Scholar]
- 45.Evans ND, Minelli C, Gentleman E, LaPointe V, Patankar SN, Kallivretaki M, Chen XY, Roberts CJ, Stevens MM. Eur Cell Mater. 2009;18:1. doi: 10.22203/ecm.v018a01. [DOI] [PubMed] [Google Scholar]
- 46.Shih YRV, Tseng KF, Lai HY, Lin CH, Lee OK. Journal of Bone and Mineral Research. 2011;26(4):730. doi: 10.1002/jbmr.278. [DOI] [PubMed] [Google Scholar]
- 47.Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(4):1484. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arthur WT, Burridge K. Molecular Biology of the Cell. 2001;12(9):2711. doi: 10.1091/mbc.12.9.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang YK, Yu X, Cohen DM, Wozniak MA, Yang MT, Gao L, Eyckmans J, Chen CS. Stem Cells and Development. 2012;21(7):1176. doi: 10.1089/scd.2011.0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Docheva D, Padula D, Popov C, Mutschler W, Clausen-Schaumann H, Schieker M. Journal of Cellular and Molecular Medicine. 2008;12(2):537. doi: 10.1111/j.1582-4934.2007.00138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thery M, Pepin A, Dressaire E, Chen Y, Bornens M. Cell Motility and the Cytoskeleton. 2006;63(6):341. doi: 10.1002/cm.20126. [DOI] [PubMed] [Google Scholar]
- 52.Chen CS, Alonso JL, Ostuni E, Whitesides GM, Ingber DE. Biochemical and Biophysical Research Communications. 2003;307(2):355. doi: 10.1016/s0006-291x(03)01165-3. [DOI] [PubMed] [Google Scholar]
- 53.Maekawa M, Ishizaki T, Boku S, Watanabe N, Fujita A, Iwamatsu A, Obinata T, Ohashi K, Mizuno K, Narumiya S. Science. 1999;285(5429):895. doi: 10.1126/science.285.5429.895. [DOI] [PubMed] [Google Scholar]
- 54.Wojciak-Stothard B, Potempa S, Eichholtz T, Ridley AJ. Journal of Cell Science. 2001;114(7):1343. doi: 10.1242/jcs.114.7.1343. [DOI] [PubMed] [Google Scholar]
- 55.Apodaca G. Traffic. 2001;2(3):149. doi: 10.1034/j.1600-0854.2001.020301.x. [DOI] [PubMed] [Google Scholar]
- 56.Malek AM, Izumo S. Journal of Cell Science. 1996;109(4):713. doi: 10.1242/jcs.109.4.713. [DOI] [PubMed] [Google Scholar]
- 57.Chang YC, Nalbant P, Birkenfeld J, Chang ZF, Bokoch GM. Molecular Biology of the Cell. 2008;19(5):2147. doi: 10.1091/mbc.E07-12-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bourgier C, Haydont V, Milliat F, Francois A, Holler V, Lasser P, Bourhis J, Mathe D, Vozenin-Brotons MC. Gut. 2005;54(3):336. doi: 10.1136/gut.2004.051169. [DOI] [PMC free article] [PubMed] [Google Scholar]